Significance
Cell membranes are thought to partition into small (10–100 nm) and transient (<100 ms) lipid platforms or “rafts” to control signaling and trafficking across the membrane. The difficulty in observing such species has made “lipid rafts” a contentious topic. Indirect evidence for their existence has been available for decades, but it has not been possible to reveal their dynamics directly. By exploiting the differences in light scattering from different lipid phases, we achieve dynamic imaging of lipid nanodomains. We observe nanodomain formation, destruction, and coalescence—behaviors previously hypothesized but never observed, yet are critical to their proposed function.
Keywords: droplet interface bilayer, iSCAT, lipid nanodomains, label-free imaging, light scattering
Abstract
Lipid rafts are submicron proteolipid domains thought to be responsible for membrane trafficking and signaling. Their small size and transient nature put an understanding of their dynamics beyond the reach of existing techniques, leading to much contention as to their exact role. Here, we exploit the differences in light scattering from lipid bilayer phases to achieve dynamic imaging of nanoscopic lipid domains without any labels. Using phase-separated droplet interface bilayers we resolve the diffusion of domains as small as 50 nm in radius and observe nanodomain formation, destruction, and dynamic coalescence with a domain lifetime of 220 ± 60 ms. Domain dynamics on this timescale suggests an important role in modulating membrane protein function.
Cell membranes compartmentalize into lipid domains that enable the selective recruitment of specific proteins (1). These “lipid rafts” have been proposed to control many membrane processes including apical sorting, protein trafficking, and the clustering of proteins during signaling. The dynamic formation and destruction of lipid nanodomains are thought to provide the central mechanism to regulate this wide range of essential processes (2–4). Although many methods now provide strong evidence to support their existence in vivo (5), the combination of nanoscopic size and dynamics on millisecond timescales has placed the direct observation of their behavior beyond the scope of existing techniques. Consequently, although we know they exist, frustratingly little is known regarding their function and dynamics (6).
Recent advances in fluorescence nanoscopy provide the only time-dependent information on the behavior of lipid nanodomains (7–9). Stimulated emission depletion–fluorescence correlation spectroscopy has shown cholesterol-mediated hindered nanoscale diffusion of single labeled sphingomyelin lipids that is consistent with the lipid raft hypothesis and transient binding of lipids (9). Superresolution fluorescence microscopy has also revealed protein clusters in cell membranes with 0.5-s temporal resolution (7). All of these experiments, however, are limited in temporal resolution by fluorescence, and must infer lipid nanodomains from the addition of fluorescent labels.
Macroscopic phase separation in artificial lipid bilayers has been widely used to help understand the biological implications of domain formation. Different lipid phases can be visualized using fluorescence microscopy with labels that preferentially partition into a specific phase (10–12). This approach is successful for micrometer-sized domains but inevitably fails on the tens to few hundreds of nanometers scale due to limitations in phase specificity, the limited residence time of a label within a specific nanoscopic domain, and the achievable optical resolution (13). The fluorescent probe is itself an additional component that can perturb phase behavior, either directly or through photooxidation (14, 15). As a result, lipid nanodomain dynamics have not been observed directly even in artificial systems, although recent ensemble-based techniques report lipid heterogeneity on the appropriate length scales (13). In addition to fluorescence-based approaches, ellipsometry and reflection interference contrast microscopy have been used to characterize phase separation in lipid bilayers (16, 17), taking advantage of different bilayer thicknesses and refractive indices caused by varying degrees of cholesterol content and lipid packing. Given sufficient sensitivity and resolution, this approach should hold for arbitrarily small domains.
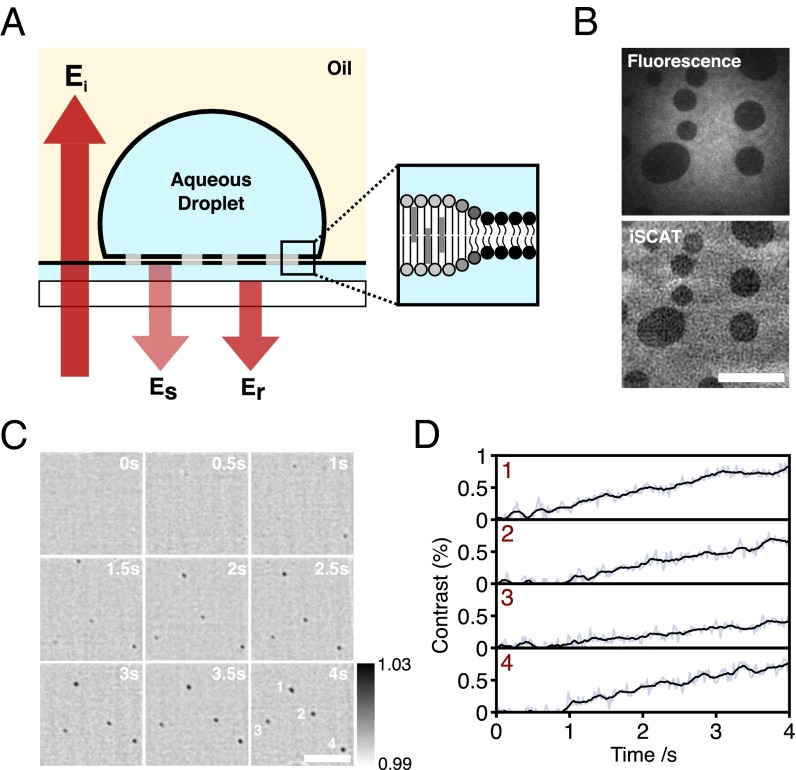
We recently developed interferometric scattering microscopy (iSCAT) (18–20) and achieved sensitivity to refractive index perturbations down to the level of a single unlabeled protein molecule in solution with millisecond time resolution (21, 22). Here, we exploit the unique sensitivity of iSCAT to overcome the limitations in temporal resolution and sensitivity to image, track, and characterize lipid nanodomains without requiring any labels. We use droplet interface bilayers (DIBs) as an artificial membrane model (23, 24) with phase-separated lipid mixtures (Fig. 1A). DIBs are formed by the contact of two lipid monolayers; in this case, a monolayer formed at the interface between an aqueous droplet and a solution of phospholipids in oil, and another between a thin hydrogel film and the oil. DIBs are robust, long-lived, and defect-free, show unrestricted diffusion, form gigaohm resistance seals, and are compatible with high-resolution optical microscopy (24).
Fig. 1.
Detection of lipid nanodomains using iSCAT. (A) Schematic of a DIB showing ordered (light gray) and disordered (black) phases. The interference between scattered and reflected fields (Es and Er) is detected in the far field using a digital camera. (B) The 100-ms TIRF (Top) and iSCAT (Bottom) images of a DIB containing So domains within a bulk Ld phase (1:1 DOPC: bSM plus 1 mol% Atto488-DPPE). The static background due to scattering from the agarose substrate can been seen in this raw iSCAT image. This background is subtracted in subsequent images. (C) Time-lapse sequence of iSCAT images of Lo nanodomains appearing from a uniform Ld phase upon cooling of a DIB below the phase transition temperature. The droplet was heated to 45 °C for 10 min. Nanodomains appeared 2–5 min after heating was stopped. Composition, 1:1:1 DPhPC:bSM:Chol. Greyscale values are of the normalized reflected intensity. (D) Trajectories corresponding to average pixel contrast within a 900 × 900-nm window centered on each nanodomain shown in C. Values before the appearance of the domain are representative of the background fluctuations at the position where the domain first becomes visible. (Scale bars: 5 μm.)
Results
Fig. 1B shows a DIB formed from a binary mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and (brain) sphingomyelin (bSM) at a molar ratio of 1:1. 1 mol% Atto488-labeled 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (Atto488-DPPE) is also present to permit fluorescence visualization of lipid domains. Macroscopic solid-ordered (So)/liquid-disordered (Ld) phase coexistence is visible both in total internal reflection fluorescence (TIRF) (Fig. 1B, Top) and iSCAT images (Fig. 1B, Bottom). Additional experimental details are described in Figs. S1 and S2. For the So phase, the signal-to-noise ratio for a single 10-ms frame is 13:1 with a Weber contrast [the ratio of the time-dependent intensity (the signal) to the time-independent intensity (the background)] of 2%, limited by fluctuations in the substrate roughness (20).
Fig. S1.
Diagram of experimental setup. AOD, acousto-optic deflector; DM, dichroic mirror; O, objective; PBS, polarizing beam splitter; QWP, quarter wave plate. In essence, iSCAT simply requires the interference between light scattered from the sample and nearby substrate. A high-speed CMOS camera then detects this interferometric signal as a perturbation on the large reflected background from the substrate (20). A 473 nm laser is used for TIRF microscopy. The returning TIRF beam is imaged on a CCD camera to provide feedback-control of pizeo-mounted sample focus.
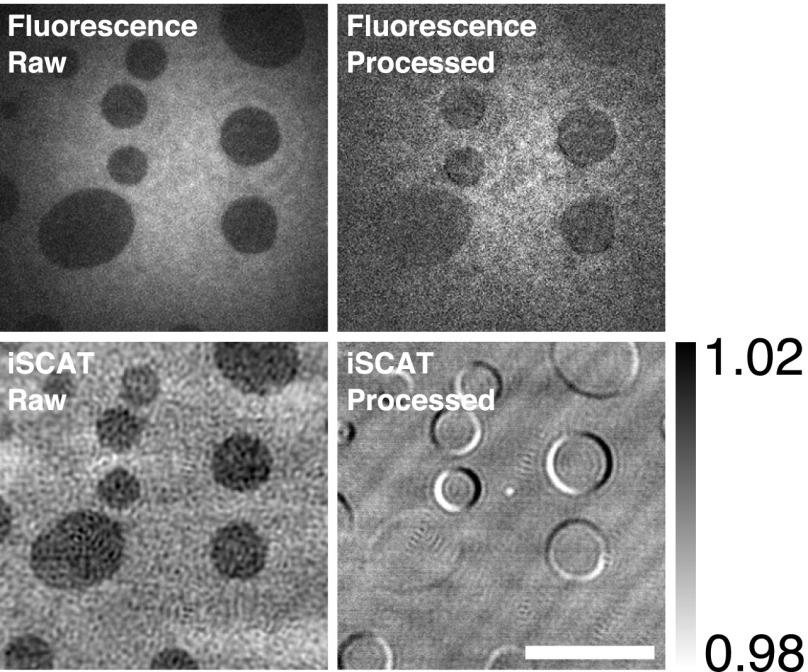
Fig. S2.
Comparison of raw and processed TIRF and iSCAT images. Images of domains in a DOPC:bSM (1:1) bilayer with 1 mol% Atto 488. Images were taken at 100 Hz (iSCAT) and 20 Hz (fluorescence). (Scale bar: 10 μm.)
The area fraction and hence domain size in lipid bilayers varies with the lipid composition and temperature. Above the phase transition temperature, lipid components are mixed. We formed a DIB from a 1:1:1 ternary lipid mixture of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC), bSM and cholesterol (Chol), which partitions into liquid-ordered (Lo) and Ld phases at room temperature. We heated this bilayer above the transition temperature and watched it cool. We observed the appearance of nanoscopic domains (Fig. 1 C and D, and Movie S1) that grew in size as the sample cooled back to room temperature. The sensitivity limits of these measurements (0.3% contrast) were much better than the images in Fig. 1B because the static background could be subtracted more efficiently for diffusing domains.
To further quantify these observations, we tracked the diffusion of So nanodomains. Many theoretical models of domain diffusion (25–27) are based on rigid, cylindrical inclusions, for which thermodynamically stable and essentially noninteracting So domains are a good approximation. Most importantly, we chose to examine So domains as they can be kinetically trapped at a range of different sizes, and thus could be used to examine closely how diffusion and contrast vary with domain size.
We prepared DIBs with So domains (DOPC:bSM, 1:1) and tracked domains using 2D Gaussian fitting. Rather than cooling, here we heated existing domains by exposing them to laser illumination 30 times greater than our regular imaging conditions. This enabled more rapid control of local temperature than by heating the entire apparatus. We observed changes in contrast and diffusion coefficient from individual domains consistent with heating-induced reduction in domain size to below the diffraction limit (Fig. 2A). The measured half-width half-maximum (HWHM) decreased until we reached the Abbe diffraction limit. Below this limit, domains could still be detected, but they are nanoscopic in size (Fig. 2B).
Fig. 2.
Quantification of solid-ordered (So) nanodomains. (A) A 15-s, time-lapse sequence of iSCAT images showing shrinking So nanodomains in a DIB (1:1 DOPC:bSM, plus 1 mol% Atto488-DPPE). (Scale bar: 0.5 μm.) (B) HWHM vs. contrast for the same dataset as shown in A. HWHM tends to the resolution limit of our microscope as the contrast decreases. Our limiting detectable contrast is 0.5% at 0.5 kHz. Error bars show the average error in the HWHM, based on the covariance of the Gaussian fitting parameters. (C) Diffusion coefficient (red) vs. radius calculated by calibration to microdomain diffusion for the same bilayer composition. Contrast (blue) vs. radius calculated from the measured diffusion coefficient. Contrast increases with increasing radius and can be approximated by a sigmoid (dotted line). Error bars show the values of the radius corresponding to ±1 SD in the measured diffusion coefficient. Calibration details are given in Figs. S3 and S5.
Although these nanodomains are subdiffraction in size, we were able to determine their position with 10-nm precision, and thus use their diffusive motion to estimate their size. We tracked domains both larger and smaller than the diffraction limit to determine the relation between the diffusion coefficient and domain radius. Notably, the Saffman–Delbrück model of 2D diffusion in a viscous fluid (28) does not adequately describe our data (Fig. S3). We compared a number of alternative models (Fig. S3), and our data were best fit by the recent model of Guigas and Weiss, which takes into account internal domain motions (29). We note that the Evans–Sackmann model of a rigid inclusion with a velocity-dependent drag (26) also fits our data (Fig. S3), albeit less well than the Guigas–Weiss model. The Evans–Sackmann model requires viscous drag between the bilayer and the agarose substrate. We do not detect any deviation from pure Brownian diffusion (Fig. S4); for the Evans–Sackmann model to be correct, this would imply homogenous drag, even at the nanoscale.
Fig. S3.
Tracking of small So microdomains. Data were recorded at 500 Hz, using 662-nm illumination at a power density of 0.7 kW⋅cm2. The radii of domains smaller than the diffraction limit (338.6 nm) were obtained by deconvolution of the PSF. Diffusion coefficient vs. domain radus was fit to the large limit of the Saffman–Delbrück (SD) (yellow), Guigas–Weiss (GW) (green), Petrov–Schwille (38) (PS) (blue), and Evans–Sackmann (ES) models (red). The GW model best describes the data (GW: χ2 = 2.957, ηc= 0.00505 N⋅s⋅m−2, c = 1.377 × 10−6; ES: χ2 = 3.002, ηm/h = 0.20596 N⋅s⋅m−2, bs = 13,561.8 N⋅s⋅m−3; PS: χ2 = 6.110, ηc = 0.0083 N⋅s⋅m−2, ηm = 2.5 × 10−12 N⋅s⋅m−1; SD: χ2 = 9.02, ηc = 0.00306 N⋅s⋅m−2, ηm/h = 1.51 N⋅s⋅m−2) where h is the bilayer height, ηc is the solvent viscosity and ηm is the membrane viscosity. This scaling enables the estimation of So and Lo nanodomain radii.
Fig. S4.
Tracking of nanodomain (blue) and microdomain (red) diffusion shows invariance with respect to time lag. This suggests domain diffusion is purely Brownian and unrestricted with no anomalous component. These results are in agreement with our previous work where the presence of the hydrated agarose substrate did not affect protein diffusion in DIBs (39).
Given these observations, and the better fit by Guigas–Weiss, we chose to use this scaling of the diffusion coefficient with domain radius to estimate the size of domains much smaller than the diffraction limit and whose radius could not be measured directly. Using this calibration, we measured So domains down to 50 nm in radius with 0.6% contrast at 0.5-kHz frame rate. The contrast increases with radius (dotted line, Fig. 2C; and Fig. S5) until domains exceed the diffraction limit (radius, ∼200 nm), where the contrast plateaus. These results demonstrate the capability of iSCAT in imaging and tracking nanoscopic domains in bilayer membranes without addition of any labels.
Fig. S5.
Logarithmic (A) and linear (B) plots of the variation of the contrast (c) of So domains with radius (r). Dashed lines are power [green, ln(c) = 1.462 ln[r] −6.377], and sigmoid (blue, c = 0.61081 + 3.3037/{1 + exp[(131.72 − r)/25.146]}) fits to the data.
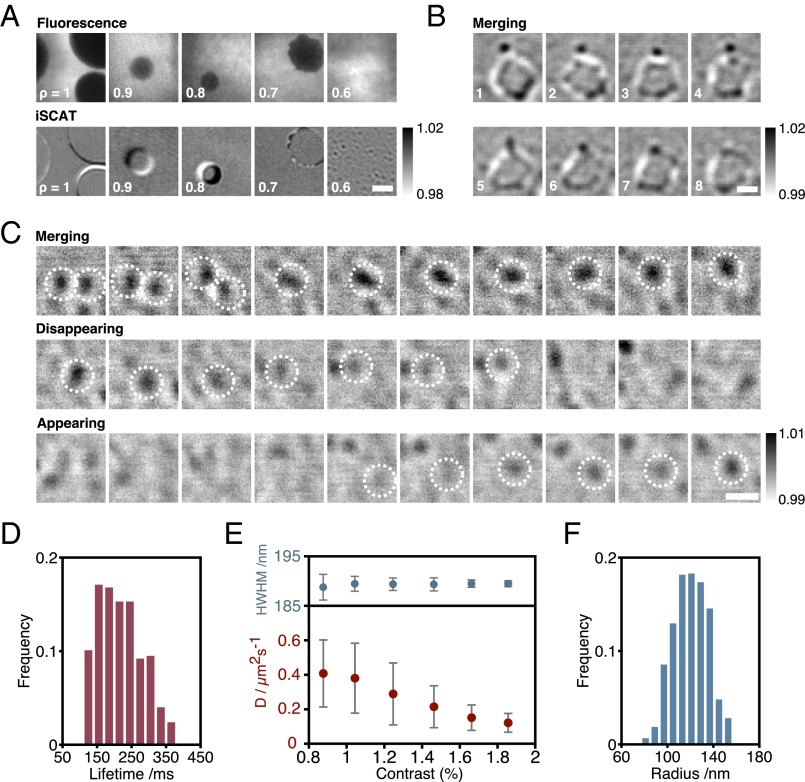
Although So/Ld phase coexistence does play a limited role in vivo (30, 31), it is nanoscopic Lo/Ld phase coexistence that is the hallmark of lipid rafts. Ternary lipid mixtures of sphingomyelin, phosphatidylcholine, and cholesterol result in coexisting liquid phases, but nanoscopic fluctuations are expected only close to miscibility critical points (32). We observed nanodomains by heating ternary mixtures (Fig. 1 C and D); however, quaternary lipid mixtures, pioneered by Feigenson and coworkers (33), provided us with a model system with highly controllable nanoscopic phase separation. We exploited quaternary mixtures of DOPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), bSM, and cholesterol to create Lo nanodomains with compositions ρ:(1 − ρ):1:1, where ρ is the relative fraction of DOPC and POPC in the mixture [ is the mole fraction]. We observed a shift from microscopic to nanoscopic domain size similar to that reported previously (33). Only microdomains can be distinguished by TIRF, whereas both nanodomains and microdomains are visible in iSCAT as ρ is varied (Fig. 3A). For ρ = 0.7, both nanodomains and microdomains with fluctuating boundaries were present and we could reveal fusion events between nanodomains and microdomains (Fig. 3B and Movie S2). For ρ = 0.6, we observed nanodomains appearing, disappearing, merging, and splitting (Fig. 3C and Movie S3). Coalescence between domains was common, and by tracking individual domains we were able to determine a nanodomain lifetime of 0.22 ± 0.06 s (Fig. 3D). As the lifetime is related to the domain size, the lower limit of the distribution of lifetimes is determined not by the nanodomains themselves, but by the noise of our measurement.
Fig. 3.
Quantification of liquid-ordered (Lo) nanodomains. (A) TIRF and iSCAT images for quaternary mixtures of DOPC, POPC, bSM, and cholesterol [ρ:(1 − ρ):1:1 mol%], with the fraction, ρ, varying from 1 to 0.6. DiI was also added (1 mol%) for fluorescence imaging. iSCAT images were binned to 100 Hz. TIRF images were binned to 10 Hz. (Scale bar: 2.5 μm.) (B) Image sequences of nanodomain/microdomain merging. (Scale bar: 1 μm.) (C) Image sequence of nanodomains merging, disappearing, and appearing for ρ = 0.6. (Scale bar: 1.5 μm.) (D) Distribution of lifetimes of nanodomains for ρ = 0.6 (749 domains). (E) Measured diffusion vs. contrast and HWHM vs. contrast for ρ = 0.6. (F) Distribution of measured radii for ρ = 0.6.
In contrast to So domains, we observed no change in HWHM with contrast (Fig. 3E), consistent with scattering from domains significantly smaller than the diffraction limit. Although it is straightforward to detect these Lo nanodomains, we cannot use the diffusion of small Lo microdomains to calibrate our experiments as with the So domains (Fig. 2C), because all of the Lo domains are nanoscopic. An estimate of Lo nanodomain size can be made if we assume the scaling of contrast with radius observed for So domains (Fig. S5) can be applied to Lo domains (Fig. 2C). We multiply the function describing contrast vs. radius for So nanodomains (Fig. S5) by the ratio of the maximum detected contrast for Lo to So microdomains, and use the measured Lo nanodomain contrast to estimate Lo domain size. Our results exhibit a broad range of domain sizes (Fig. 3F), with a mean of 120 nm. The lower limit of this distribution is not determined by the nanodomains themselves, but by the noise level of our experiment.
Discussion
The nature of the relation between object size and diffusion coefficient in a bilayer is important for predicting the motions of both lipid nanodomains and proteins (29, 34). In this work, we have shown a failure of the hydrodynamic model of Saffmann and Delbrück, and importantly we have the temporal and spatial resolution to test the scaling of diffusion coefficient with object radius and thus distinguish between different theoretical models of these processes (Fig. S3).
iSCAT does not suffer from the uncertainties in studying lipid nanodomains by fluorescence as it is an inherently label-free technique and avoids the potential problems of fluorescent probes perturbing the phase behavior one would seek to study (14, 15). Even when taking advantage of FRET, current methods are restricted by the affinity of lipid probes for specific lipid phases (13) and cannot image lipid nanodomains. Here, we are able to overcome these limitations and image the motion of lipid nanodomains without any additional bilayer components, while retaining the nanometer precision inherent in other superresolved microscopies.
We remark that the sensitivity levels achieved with iSCAT in recent single protein landing assays (10, 11) are 50 times higher than this work, providing scope for studies of even smaller nanoscopic domains or faster dynamics. The contrast for our method simply relies on a difference in light scattering between phases, and although this is a primary advantage of this label-free method, it also presents a significant future challenge for applying this technique to real cell membranes, as all cell components will contribute to our signal. These measurements set out the potential for this technique to study fast, nanoscale fluctuations of phase boundaries in lipid bilayers and a wide range of other systems. Nevertheless, the strongest contributors to iSCAT signals even in a cellular setting will be from structures close to the interface. In addition, domain structures may be identified by using methods that are expected to change cellular phase behavior, such as changes in temperature or extraction of cholesterol. Furthermore, the facile combination of scattering and fluorescence imaging provides additional opportunities for identifying domains or domain-associated species.
The nanodomain lifetimes we measure are on a similar order to the kinetics reported for many membrane-signaling processes (2–4). This suggests that the dynamic creation and destruction of lipid nanodomains might indeed be a key modulator of protein signaling. One example is the dimerization of GPI-anchored receptors taking place on the timescale of several hundred milliseconds, where dimerization is altered by raft–lipid interactions (35). To our knowledge, our measurements represent the first step in exploiting iSCAT to provide a quantitative understanding of the role of nanoscopic lipid phase separation in membrane signaling.
Materials and Methods
Materials.
DPhPC, DOPC, POPC, bSM, and Chol were obtained from Avanti Polar Lipids. DiI-C18 was obtained from Invitrogen. ATTO-488 DPPE was obtained from ATTO-TEC Lipids purchased were used without further purification, dissolved in chloroform at 50 mg⋅mL−1 and stored at −20 °C until use. All other reagents were obtained from Sigma-Aldrich.
DIB Formation.
DIBs were prepared following our previously reported protocol (24). Briefly, 400-nL droplets of 90 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) were incubated for 40 min in a solution of the required lipids in hexadecane plus 10% (vol/vol) silicone oil (AR 20) at a concentration of 8.7 mg⋅mL−1 to form a lipid monolayer at the oil–water interface. This concentration is significantly below the solubility limit for our lipid mixtures (Fig. S6). Meanwhile, 140 μL of 0.75% (wt/vol) ultralow-melt agarose was spin-cast onto a plasma-cleaned coverslip at 1,800 × g for 30 s. The agarose-coated coverslip was sealed with a microfabricated poly(methyl methacrylate) (PMMA) device (24). One hundred eighty microliters of 2.8% (wt/vol) low-melt agarose was used to seal the gap between the coverslip and device. The device was filled with the aforementioned lipid/oil solution. Incubated droplets were transferred to the device after 20 min to form a bilayer upon contact with the agarose substrate. Based on our previous work (36), we estimate a maximum bilayer hydrocarbon content of 9.2%. Finally, the device was heated (46 °C) above the transition temperature of bSM, cooled to room temperature, and imaged using iSCAT and/or TIRF microscopy.
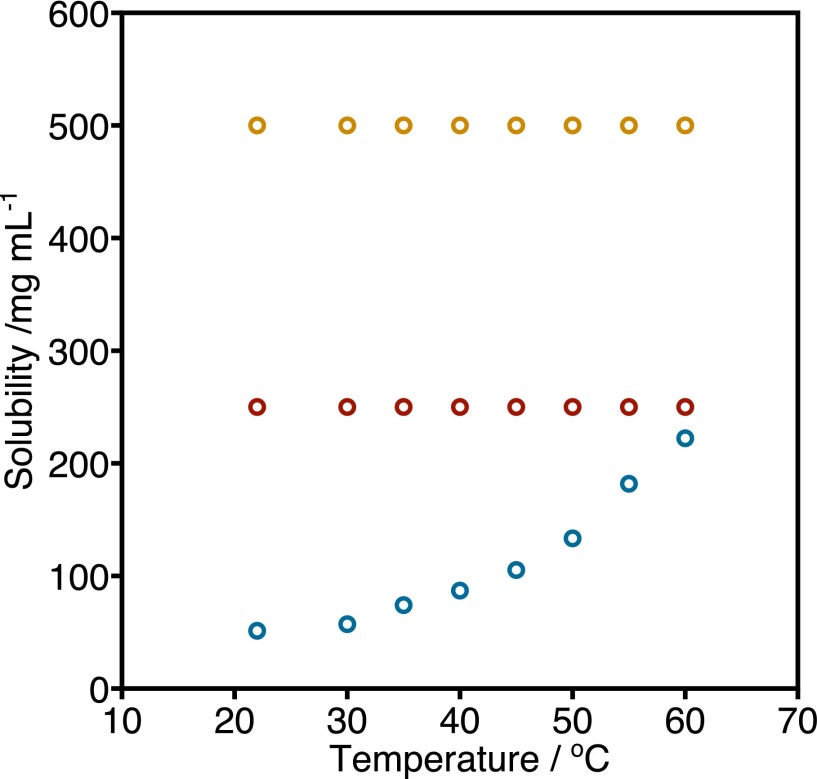
Fig. S6.
Solubility of DOPC:bSM, 1:1 (blue), DOPC:bSM:Chol, 1:1:1 (red), and DOPC:POPC:bSM:Chol, 1:1:1:1 (orange), in hexadecane. The solubility of all lipid mixtures is larger than the concentration of lipid in oil used in this work (8.7 mg⋅mL−1). With the exception of the solubility of the binary mixture of DOPC:bSM, which increases with temperature, all other compositions do not show a pronounced variation in solubility with temperature.
Experimental Setup.
All optical components were obtained from Thorlabs unless mentioned otherwise.
For iSCAT, the output of a 662-nm diode laser (Coherent Obis) was adjusted to 2-mm beam diameter before passing through two acousto-optic deflectors (AODs) (Gooch and Housego). The two AOD channels are scanned in a sawtooth by separate, phase-locked function generators (Rigol DG1022) at 79 and 80 kHz, respectively. Both the absolute and relative frequencies are chosen to induce the smallest detectable fluctuations in the background light intensity on the timescale of the camera exposure time. The beam deflected by the AODs is imaged with telecentric lenses onto the back focal plane of an oil immersion 1.42 N.A., 60× objective (Olympus PLAPON) after passing through a polarizing beam splitter (PBS). The 662-nm beam under fills the back aperture of the objective to generate a focal spot of ∼1 μm. A quarter wave plate before the objective causes the p-polarized incident light to be converted to s-polarized light after reflection from the sample. The returning light is reflected by the PBS and imaged onto a complementary metal–oxide–semiconductor (CMOS) camera (Photonfocus MV-D1024-160-CL-8; 10-μm pixels) at 166× or 250× magnification. Schematic for setup is shown in Fig. S1. Data in Fig. 1 C and D were recorded with a 445-nm laser, without the dichroic mirrors and fluorescence channel. Data in Fig. 2 A and B were recorded with a 638-nm laser; the remainder of the setup was the same. Data in Fig. 2C were recorded with a 532-nm laser, without the dichroic mirrors and fluorescence channel and without AOD scanning.
For TIRF, a 473-nm diode laser (Crystal Laser) is translated across the back aperture of the objective to totally internally reflect off the sample. The beam is focused onto the back focal plane with a 400-mm lens, to generate a wide-field illumination spot with FWHM ∼15 μm. The beam is reflected off a 505-nm long-pass dichroic and a 605-nm long-pass dichroic mirror, which passes the 662-nm beam so that both beams are overlapped entering the objective. Fluorescence in the 505- to 605-nm window is separated from both the reflected 662-nm beam and 473-nm beams by the two dichroic mirrors and, after passing through a 550-nm long-pass filter, is imaged onto an emCCD camera (Andor iXon3 860).
The data underpinning these results can be accessed at ora.ox.ac.uk.
Experimental Conditions for Fig. 1.
The appearing nanodomains (Fig. 1C) were recorded with 445-nm illumination with power density of 0.2 kW⋅cm−2. Images were collected at 50 Hz.
Experimental Conditions for Fig. 2.
The dataset for A and B was recorded with 662-nm illumination with power density of 13 kW/cm2. Images were collected at 2 kHz. Frames shown have been frame binned to 100 Hz. The data for A and B have been appended from three movies (each of a different nanodomain), rather than to attempt to follow the domain across a typical travel of several tens of micrometers. The dataset shown in C was recorded with 532-nm illumination with a power density of 20 kW⋅cm−2. Images were collected at 4 kHz, and binned to 400 Hz. Each nanodomain was tracked by fitting a 2D Gaussian in every frame. The trajectories were divided into 100 frame intervals, and the average contrast, average FWHM, and diffusion coefficient for each interval was calculated. The diffusion coefficient was calculated by fitting the line of best fit to the mean squared displacement (MSD) vs. Δt, for time intervals up to 20 frames (maximum time interval, 50 ms), where D = slope/4. In the final plot shown in Fig. 2C, the data have been averaged into 0.1% contrast bins.
Experimental Conditions for Fig. 3 A–E.
All compositions shown were recorded using 662-nm illumination at a power density of 13 kW⋅cm−2. Dynamic events have been frame binned to 50 fps for in the frame sequences shown. Fig. 3D includes data from 749 nanodomains. Data were recorded at 500 Hz and divided into 50 frame intervals, or 100 ms. For each interval, the average contrast, FWHM, and diffusion coefficient (best linear fit to MSD vs. time for Δt up to 10 frames or 20 ms, where D = slope/4) were calculated. The FWHM reported here is calculated from the maximum difference in mean contrast between consecutive concentric circles.
iSCAT Image Processing.
Image processing is achieved by removing any constant background caused by residual reflections and illumination inhomogeneity. To do so, 1,000 images are recorded while manually moving the sample stage, and each pixel is then replaced by the temporal median value of the frame sequence to generate an optimal flat-field image that is independent of the sample. Following division by the flat-field image, sample-specific images with shot noise-limited sensitivity are obtained. Finding the correct focal point is critical and is automatically achieved by locking the position of the reflected 473-nm beam by feeding back the positional difference to the sample stage Piezo controllers. The feedback loop operates at 20 Hz, keeping the z position within 5 nm of the target. To generate an image containing all of the static iSCAT features, we performed a stack average over the acquired frames. Domains were fitted to Gaussian point spread functions (PSFs) to obtain the center and contrast values.
So Nanodomains Tracking and Analysis.
Nanodomains were tracked using the ImageJ Trackmate plugin (37) (Q > 0.15; maximum link, 15 px; maximum gap = 15 px 2 frames). Domains were then fit to a Gaussian to determine their FWHM. The FWHM reaches a constant, minimum value for domains <2% contrast. We assume that the FWHM of the PSF is equal to this value (338.6 nm). Domain radii were determined by deconvolution of this PSF.
Lo Nanodomains Tracking and Analysis.
Nanodomain lifetime was determined as the track lengths returned by Trackmate (37). The nanodomains trajectories were divided into 50 frame subtrajectories for which the average HWHM, average contrast, and diffusion coefficients were calculated. The data from all subtrajectories was binned into 0.2% contrast intervals, to display the binned HWHM and diffusion coefficients as a function of the contrast (Fig. 3 E and F). Error bars are the SD of the data within each contrast bin.
Microdomains Tracking and Analysis.
Domains were delineated by thresholding a high-pass filtered (7 pixels) image to the half-maximal intensity. Domains in the resulting binary image were tracked by centroiding. The radius was found from the area of the domain, assuming a circular geometry.
Supplementary Material
Acknowledgments
This work was funded by the European Research Council [CoSMiC (to M.I.W.) and NanoScope (to P.K.)]. P.K. was supported by the John Fell Fund and a Career Acceleration Fellowship from the Engineering and Physical Sciences Research Council (EP/H003531). J.S.H.D. was funded by a Louis Dreyfus–Weidenfeld Scholarship, and G.d.W. was funded by a Clarendon Scholarship. Data are archived locally and will be provided on request to the corresponding authors.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508483112/-/DCSupplemental.
References
- 1.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387(6633):569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14(1):111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 4.Truong-Quang B-A, Lenne P-F. Membrane microdomains: From seeing to understanding. Front Plant Sci. 2014;5:18. doi: 10.3389/fpls.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klotzsch E, Schütz GJ. A critical survey of methods to detect plasma membrane rafts. Philos Trans R Soc Lond B Biol Sci. 2013;368(1611):20120033. doi: 10.1098/rstb.2012.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simons K, Vaz WLC. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33(1):269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 7.Jones SA, Shim S-H, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8(6):499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hein B, et al. Stimulated emission depletion nanoscopy of living cells using SNAP-tag fusion proteins. Biophys J. 2010;98(1):158–163. doi: 10.1016/j.bpj.2009.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457(7233):1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 10.Veatch SL, Keller SL. Seeing spots: Complex phase behavior in simple membranes. Biochim Biophys Acta. 2005;1746(3):172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich C, et al. Lipid rafts reconstituted in model membranes. Biophys J. 2001;80(3):1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104(9):3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Šachl R, Humpolíčková J, Štefl M, Johansson LB-Å, Hof M. Limitations of electronic energy transfer in the determination of lipid nanodomain sizes. Biophys J. 2011;101(11):L60–L62. doi: 10.1016/j.bpj.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sezgin E, et al. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim Biophys Acta. 2012;1818(7):1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Skočaj M, et al. The sensing of membrane microdomains based on pore-forming toxins. Curr Med Chem. 2013;20(4):491–501. doi: 10.2174/0929867311320040002. [DOI] [PubMed] [Google Scholar]
- 16.Howland MC, Szmodis AW, Sanii B, Parikh AN. Characterization of physical properties of supported phospholipid membranes using imaging ellipsometry at optical wavelengths. Biophys J. 2007;92(4):1306–1317. doi: 10.1529/biophysj.106.097071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rädler J, Sackmann E. Imaging optical thicknesses and separation distances of phospholipid vesicles at solid surfaces. J Phys II Fr. 1993;3(5):727–748. [Google Scholar]
- 18.Jacobsen V, Stoller P, Brunner C, Vogel V, Sandoghdar V. Interferometric optical detection and tracking of very small gold nanoparticles at a water-glass interface. Opt Express. 2006;14(1):405–414. doi: 10.1364/opex.14.000405. [DOI] [PubMed] [Google Scholar]
- 19.Kukura P, et al. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat Methods. 2009;6(12):923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 20.Ortega-Arroyo J, Kukura P. Interferometric scattering microscopy (iSCAT): New frontiers in ultrafast and ultrasensitive optical microscopy. Phys Chem Chem Phys. 2012;14(45):15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 21.Ortega Arroyo J, et al. Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Lett. 2014;14(4):2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piliarik M, Sandoghdar V. Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nat Commun. 2014;5:4495. doi: 10.1038/ncomms5495. [DOI] [PubMed] [Google Scholar]
- 23.Bayley H, et al. Droplet interface bilayers. Mol Biosyst. 2008;4(12):1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leptihn S, et al. Constructing droplet interface bilayers from the contact of aqueous droplets in oil. Nat Protoc. 2013;8(6):1048–1057. doi: 10.1038/nprot.2013.061. [DOI] [PubMed] [Google Scholar]
- 25.Hughes B, Pailthorpe B, White L. The translational and rotational drag on a cylinder moving in a membrane. J Fluid Mech. 1981;110:349–372. [Google Scholar]
- 26.Evans E, Sackmann E. Translational and rotational drag coefficients for a disk moving in a liquid membrane associated with a rigid substrate. J Fluid Mech. 1988;194:553–561. [Google Scholar]
- 27.Petrov EP, Schwille P. Translational diffusion in lipid membranes beyond the Saffman-Delbruck approximation. Biophys J. 2008;94(5):L41–L43. doi: 10.1529/biophysj.107.126565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffman PG, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72(8):3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guigas G, Weiss M. Size-dependent diffusion of membrane inclusions. Biophys J. 2006;91(7):2393–2398. doi: 10.1529/biophysj.106.087031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plasencia I, Norlén L, Bagatolli LA. Direct visualization of lipid domains in human skin stratum corneum’s lipid membranes: Effect of pH and temperature. Biophys J. 2007;93(9):3142–3155. doi: 10.1529/biophysj.106.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruocco MJ, Shipley GG. Interaction of cholesterol with galactocerebroside and galactocerebroside-phosphatidylcholine bilayer membranes. Biophys J. 1984;46(6):695–707. doi: 10.1016/S0006-3495(84)84068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci USA. 2007;104(45):17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heberle FA, et al. Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc. 2013;135(18):6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 34.Gambin Y, et al. Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA. 2006;103(7):2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki KGN, et al. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat Chem Biol. 2012;8(9):774–783. doi: 10.1038/nchembio.1028. [DOI] [PubMed] [Google Scholar]
- 36.Gross LCM, Heron AJ, Baca SC, Wallace MI. Determining membrane capacitance by dynamic control of droplet interface bilayer area. Langmuir. 2011;27(23):14335–14342. doi: 10.1021/la203081v. [DOI] [PubMed] [Google Scholar]
- 37.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrov EP, Petrosyan R, Schwille P. Translational and rotational diffusion of micrometer-sized solid domains in lipid membranes. Soft Matter. 2012;8(29):7552–7555. [Google Scholar]
- 39.Thompson JR, Heron AJ, Santoso Y, Wallace MI. Enhanced stability and fluidity in droplet on hydrogel bilayers for measuring membrane protein diffusion. Nano Lett. 2007;7(12):3875–3878. doi: 10.1021/nl071943y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.