Significance
Sterol regulatory element-binding protein (SREBP) signaling is responsible for transcriptional regulation of cellular lipid homeostasis. Aberrant regulation of the feedback loop that switches on transcription of related genes upon depletion of fatty acids and cholesterol abrogates cellular integrity and is a hallmark of many cancers and other diseases. Regulated intramembrane proteolysis is a conserved mechanism also involved in other signaling processes like Wnt and Notch and defines cellular regulation from homeostasis to proliferation, compartmentalization, and differentiation. An understanding of basic features, however, such as substrate selectivity and specificity, has remained elusive to date. Understanding the proteolysis of the SREBP precursor is an important goal both for understanding and medically intervening in SREBP signaling and related diseases as well as for understanding other regulated intramembrane proteolysis-dependent processes.
Keywords: SREBP, regulated intramembrane proteolysis, membrane proteins, cellular lipid homeostasis, cancer
Abstract
Regulated intramembrane proteolysis (RIP) is a conserved mechanism crucial for numerous cellular processes, including signaling, transcriptional regulation, axon guidance, cell adhesion, cellular stress responses, and transmembrane protein fragment degradation. Importantly, it is relevant in various diseases including Alzheimer’s disease, cardiovascular diseases, and cancers. Even though a number of structures of different intramembrane proteases have been solved recently, fundamental questions concerning mechanistic underpinnings of RIP and therapeutic interventions remain. In particular, this includes substrate recognition, what properties render a given substrate amenable for RIP, and how the lipid environment affects the substrate cleavage. Members of the sterol regulatory element-binding protein (SREBP) family of transcription factors are critical regulators of genes involved in cholesterol/lipid homeostasis. After site-1 protease cleavage of the inactive SREBP transmembrane precursor protein, RIP of the anchor intermediate by site-2 protease generates the mature transcription factor. In this work, we have investigated the labile anchor intermediate of SREBP-1 using NMR spectroscopy. Surprisingly, NMR chemical shifts, site-resolved solvent exposure, and relaxation studies show that the cleavage site of the lipid-signaling protein intermediate bears rigid α-helical topology. An evolutionary conserved motif, by contrast, interrupts the secondary structure ∼9–10 residues C-terminal of the scissile bond and acts as an inducer of conformational flexibility within the carboxyl-terminal transmembrane region. These results are consistent with molecular dynamics simulations. Topology, stability, and site-resolved dynamics data suggest that the cleavage of the α-helical substrate in the case of RIP may be associated with a hinge motion triggered by the molecular environment.
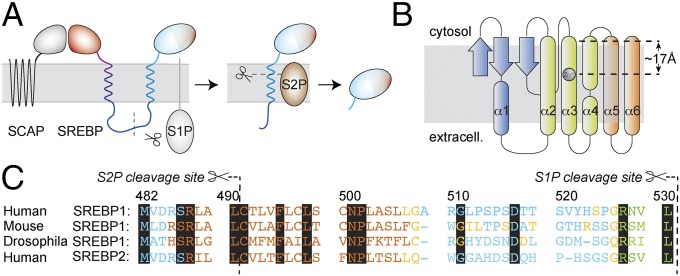
The expression of the cellular machinery responsible for cholesterol and fatty acid biosynthesis is tightly controlled by transcriptional regulation (1, 2). The negative-feedback loop, switching on transcription of target genes upon low fatty acid and cholesterol levels, is achieved via binding of transcriptional activators to the sterol regulatory element. This element is present in promoter regions near encoding genes (3, 4). Key players regulated include HMG-CoA reductase, the rate-limiting enzyme in cholesterol biosynthesis and target of statins, the low-density lipoprotein receptor, fatty acid synthase, and other enzymes vital for cellular lipid homeostasis. Consequently, aberrant regulation of these processes is linked to various conditions such as cardiometabolic disorders, inflammatory conditions, and aberrant development. Increased de novo fatty acid and cholesterol production has also been shown to be a hallmark of many human epithelial cancers (5, 6). Sterol regulatory element-binding proteins (SREBPs) are members of the bHLH-Zip transcription factor family using a basic helix–loop–helix–leucine zipper motif but are unique in that they are produced as inactive precursors anchored to the endoplasmic reticular (ER) membrane (3, 7). The SREBP precursor proteins consist of a two-helix membrane anchor, to which the amino (N)-terminal DNA-binding domain, including the transactivation domain (8), and a carboxyl (C)-terminal regulatory domain (CTD) are attached (Fig. 1A) (9). The extensive 31-residue hydrophilic loop linking the two transmembrane (TM) domains projects into the lumen of the ER. SREBP-2 has been shown to control the expression of genes governing the uptake and biosynthesis of cholesterol. Upon starvation of cholesterol, the SREBP-2 precursor is relocated from the ER to the Golgi by COPII vesicle transport. This involves dissociation of Insig1 from SCAP, which is bound to the CTD of SREBP. Site-1 protease (S1P) and S2P, located in the Golgi, in turn exert release of the active transcription factor (2). SREBP-1, which is linked to regulation of fatty acid, phospholipid, and triacylglycerol synthesis, can be activated by retrograde transport of the respective proteases from the Golgi into the ER (10). Cleavage of the SREBP precursor molecule arises in two steps. S1P separates the C-terminal TM helix, linked to the regulatory domain, from the N-terminal TM helix, tethered to the DNA-binding domain, at a cleavage site located within the long connecting loop (11). Subsequent cleavage by S2P occurs via regulated intramembrane proteolysis (RIP) (12) within the TM space to liberate the active transcription factor (see Fig. 1A).
Fig. 1.
The proteolytic cascade for maturation of SREBPs. (A) After COPII vesicle transport to the Golgi, the C-terminal part of the SREBP precursor, attached to SCAP, is shed by S1P. The active transcription factor (light blue) is then liberated by RIP of the single-TM S2P substrate. (B) S2P topology derived from Protein Data Bank ID code 3B4R (13). (C) Alignment of full-length SREBP anchor sequences. Key residues, which are conserved among different kingdoms and in between SREBP-1 and -2, are shown on black background. JPRED prediction (40) of protein TM regions (brown, helix and less than 25% accessible to solvent; blue, random coil and more than 25% solvent accessible; yellow, coil and less than 25% accessible; green, β-strand and less than 25% accessible). The region predicted as a β-strand obtains random-coil propensity when only the strip until the S1P cleavage site is subjected to the prediction (SI Appendix).
Although the core domain of S2P has successfully been crystallized (13), details of the cleavage mechanism, such as domain mobility, substrate entry, and recognition, remain elusive to date. Despite a number of structures solved recently (14–17), these mechanisms are poorly understood for the entire family of RIP proteases, including other important members such as presenilins (16, 18–20) and signal peptide peptidases (15, 21). Apart from regulating SREBP transcription factors, RIP has also been associated with several processes determining cell homeostasis and proliferation, involving Wnt and Notch signaling, β-catenin signaling, and cleavage of the amyloid precursor protein (APP) (22). Accordingly, dysfunction of these proteases plays a role in manifold diseases. All substrates of intramembrane proteases are assumed to be TM α-helices, even though protein helices are considered poor substrates for proteolysis (23). Protease substrates typically exhibit extended conformation (24, 25), and side chains bind to the active site for substrate recognition. Currently, it is largely unclear what makes a substrate suitable or unsuitable for RIP and how the lipid environment within the membrane space impinges on these properties (26, 27). For example, γ-secretase, involved in cleavage of the APP and the Notch receptor, has a large number of different substrates, for which only a loose definition of common features can be given (28, 29). In rhomboids, the membrane-integral serine protease relatives of S2Ps, which regulate epidermal growth factor receptor (EGFR) signaling in Drosophila (17, 30), common features for substrate recognition (31, 32) have long been elusive. They are now thought to involve a recognition motif and a helix-destabilizing motif (33), dependent on whether this and the cleavage site are inside the predicted TM helix (34, 35). A bound substrate-derived inhibitor cocrystalized with rhomboid (36) has indeed been found to be in an elongated β-strand form.
In the case of the S2P metalloproteases, a deeper understanding of the factors that make the SREBP membrane anchor a suitable target for recognition and enzymatic processing is still missing. Peptide bonds within regular α-helices are sterically hindered due to helix-stabilizing hydrogen bonds (37). Given its enthalpic stability, unfolding of a prototypical α-helix seems energetically unfavorable. Three SREBP sequence motives that are conserved and imperative for regulatory functionality have been described by Brown and Goldstein and coworkers (38): (i) The recognition site is constituted by a DRSR sequence just preceding the first predicted TM helix, (ii) the cleavage site is located at a Leu–Cys bond three residues into the hydrophobic segment (39), and (iii) an Asn–Pro motif is conserved in all SREBPs from worms to flies and humans at a position 11 amino acids distal to the cleavage site. Interestingly, replacement of any amino acid in the TM helix primary sequence with alanine does not alter cleavage of the helix in vivo (39). However, replacement of both N and P in the conserved NP motif has been shown to abrogate S2P activity; on the other hand, this motif can be shifted up to five positions with little effect on cleavage activity. In work by Ye et al. (38), the region around this Pro has been suspected to induce a straightening of the helix and as such push the cleavage site out of the intramembrane space for proteolytic cleavage. After communication of the S2P X-ray structure (13), which bears an active center within the membrane space (Fig. 1B), this mechanism seems unlikely. Here we describe the topology and stability of the SREBP-1 membrane anchor to obtain information about RIP substrate characteristics as related to lipid regulation by S2P.
Results
We cloned the SREBP-1 membrane anchor in accordance with the conserved sequence and in line with the topology and membrane-integration predictions from JPRED (40) (depicted in Fig. 1C). Bacterial expression of SREBP-1 with or without fusion to different solubility-enhancing proteins in different Escherichia coli cell lines failed consistently, probably due to protease degradation of the partly unfolded peptides (41). Therefore, after extensive testing and optimization, SREBP-1 was eventually produced using cell-free expression and reconstituted in 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (LMPG) micelles (SI Appendix, Details). The SREBP-1 anchor topology is expected to differ from helix bundle membrane proteins, in which stable packing is required for physiological function. The SREBP-1 anchor contains no apparent packing motifs and is not predicted to pack by prediction routines. Instead the membrane anchor appears to consist of a combination of two individual membrane-embedded stretches, which are separated by a 31-amino-acid linker. This long luminal linker enables an independent function of the two TM helices after S1P action. Consequently, detergent screens pursued here (SI Appendix, Fig. S3) focused on the S2P substrate as truncated after S1P cleavage and were limited to folding, solubilization, and suitability for NMR characterization of this single-TM intermediate.
Solvent Accessibility in LMPG Micelles.
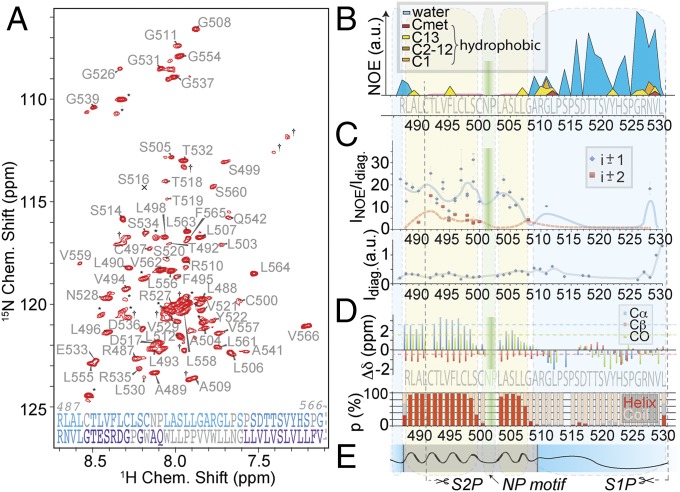
Fig. 2B depicts the accessibility of the anchor residues to solvent as measured with amide-to-water nuclear Overhauser effect (NOE). The first assigned residue, R487, which is the last residue of the DRSR sequence, still shows clear solvent accessibility. The following stretch until six residues after the conserved NP motif shows no significant NOEs to water, whereas strong amide–amide NOEs are observed instead (see NOEs). Obvious water accessibility is again observed for the C-terminal linker residues. NOE cross-peaks to lipid hydrogens are found only at the interface between putative lipid-embedded and solvent-exposed residues. Interestingly, the C terminus also harbors some contacts to fatty acid hydrogens in addition to (more intense) water–NOE cross-peaks. This may be induced by contacts to the micelle or transient binding of single detergent molecules to these residues under in vitro conditions. The contacts point to some hydrophobicity of the five C-terminal residues and could impose an amphipathic effect of the truncated protein. The observation is in line with a temporary interaction of the C-terminal linker residues with the membrane in molecular dynamics (MD) simulations (see Molecular Dynamics). See SI Appendix, Fig. S7 for water/lipid NOE data on a construct including the C-terminal TM helix.
Fig. 2.
Topology and putative membrane insertion of the SREBP-1 membrane anchor after S1P cleavage. (A) Two-dimensional proton–nitrogen correlation (TROSY) spectrum with assigned residues at 800 MHz 1H Larmor frequency of the complete anchor (M482 to V566). Asterisks mark residues assigned but belonging to the uncleaved N-terminal tag. Crosses mark unassigned residues. The S516 peak is below the contours shown in the plot. (Bottom) Gray, light blue, and purple denote unassigned amino acids of the construct, those present, and those absent after S1P cleavage, respectively. (B) NOE contacts of amide protons to water (blue) and aliphatic (fatty acid) protons of the S2P substrate (M482 to L530). Hydrophobic contacts (yellow, orange, and red) are broken up into such methyl and methylene protons with approximate assignments within the chain. (C) Numbers, kind, and peak heights of NOE contacts observed for sequential (i ± 1, blue) and i ± 2 correlations (red) as a qualitative measure of perseverance of secondary structure. Blue (solid) and red (dashed) lines represent the trends (floating average) along the primary sequence for abundance of NOE transfer from direct and longer range contacts, respectively. NOE intensities were corrected for relaxation differences using diagonal-peak intensities shown below. The figure displays resolved peaks only. (D) Experimental SCS values represent differences to random coil values for Cα (blue), Cβ (red), and CO shifts (green) averaged over a triple of adjacent amino acids. For the deuterated cell-free expression, deuterated amino acids Gln, Asn, Cys, and Trp were not available and were used in their protonated form, which often prohibits sufficient signal intensity for sequential assignments of these amino acids. The secondary structure in terms of TALOS+ prediction (42) is depicted below. (E) Topology model of the substrate (black line) concluded from the above data, representing helical (sinosoidal) and unstructured regions (stretched) as well as lipid (gray) and aqueous embedding (blue).
Secondary Chemical Shifts.
Fig. 2D shows the secondary chemical shifts (SCSs) of Cα, Cβ, and CO as obtained from sequential backbone assignment experiments as well as a TALOS-derived (42) secondary structure. For respective plots including the second helix, see SI Appendix, Figs. S7 and S8. From the first assigned residue, the second Arg of the DRSR motif, we observe a clear α-helical pattern. This continues through the S2P cleavage site until two residues before the conserved NP motif, where reduced helicity is found. The NP motif itself is unassignable due to the absence of the Pro HN and the reduced viability of coherences from (undeuterated) Q, N, W, and C under slow-motional dynamics (SI Appendix). The three-residue stretch just after the NP motif shows increasing helicity propensities, which then decrease to very low values for another three amino acids. The remaining residues until the C terminus of the construct, as defined by the S1P cleavage site, possess a random-coil conformation.
NOEs.
We characterized the amide-to-amide NOE patterns that the anchor residues show in 15N-edited NOESY experiments. In line with secondary structural characterization by SCSs, we find that significant interresidue NOE intensities are present only in the stretch between R487 and G508. In addition, an i – 1 cross-peak is also observed for G511. We exclusively observe i ± 1 and i ± 2 contacts, as is expected for α-helical domains. Interestingly, also in combination with hydrophobicity and solvent accessibility of the C-terminal residues in the linker, N528 bears a sequential NOE cross-peak to R527 of significant intensity. This confirmed NOE is the only amide–amide contact observed for the solvent-exposed C-terminal part of the anchor. Fig. 2C shows the intensity and kind of NOE cross-peaks to each anchor residue as a function of residue. Intensity data were corrected by the height of the diagonal peak to account for differences in transfer efficiencies and line width. The intensity and number of contacts throughout the helical stretch differ, however a qualitative trend can be derived by fitting the residue-specific NOE abundance over the sequence, using an interpolation of the running average. This approximate trend line, which is depicted in red/blue in the same plot as a qualitative measure of conformational stability, is in agreement with the SCSs described above. A lower degree of order is found at the C-terminal end of the lipid-embedded motif, commensurate to the picture obtained from SCSs. Lower structural stability is found directly next to the NP motif (with three comparably weak cross-peak totals in the two adjacent amino acids), which is in line with the helix-breaking tendency of Pro residues generally, and interestingly also around position F495. A lower helicity of this site is not suggested by the SCSs. However, low structural stability here is also apparent in relaxation experiments (see Backbone Dynamics). Diagonal-peak intensities used for computing the relative NOE cross-peak intensities are plotted in Fig. 2 C, Bottom. The high intensity at the C-terminal residues is due to little longitudinal and transverse relaxation (see also Backbone Dynamics) and is an indication of high mobility. The concluded anchor topology is represented in Fig. 2E.
SCSs as interpreted by TALOS (Fig. 2D) and NOE data were used to generate a topology model from CNS (43) (SI Appendix, Fig. S10). In contrast to the local structural features, however, no concise relative orientation of the upper and lower part of the helical elements could be obtained from paramagnetic relaxation enhancement (PRE) distance restraints (SI Appendix, Fig. S9) in micelles.
Backbone Dynamics.
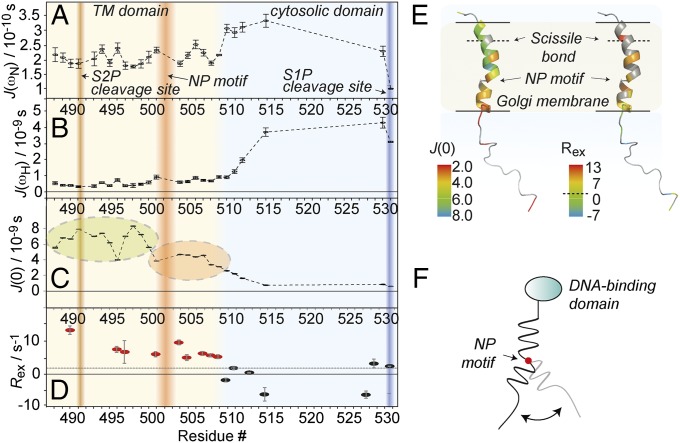
We pursued characterization of backbone dynamics using longitudinal (T1) and transverse relaxation times (T2) of 15NH as well as heteronuclear NOE (1H, 15N hetNOE). In addition, we acquired longitudinal and transverse CSA/dipole–dipole 1H/15N cross-correlated cross-relaxation rates ηz and ηxy. These experiments were recorded on the SREBP-1 membrane anchor as processed by S1P, reflecting the anchor dynamics relevant at the stage of S2P integral-membrane protease cleavage. hetNOE ratios (SI Appendix, Fig. S11A) follow a consistent global trend reflecting the decrease of effective correlation times toward the C-terminal end of the protein. Only a weak decrease in hetNOE values around the NP motif and toward the membrane/solvent interface is observed. The hydrophilic linker increases in flexibility at the C terminus reaching negative hetNOE effects, as is similar for small molecules and intrinsically disordered proteins (IDPs). Transverse and longitudinal 15NH relaxation (SI Appendix, Fig. S11 B and C) reflects the same overall trend, however with a higher sensitivity to local differences, particularly for T2. Notably, steadily increasing T2 times at the C terminus are opposed by initially decreasing T1 times, which then revert to the same values found for the membrane-integral residues. This behavior resembles characteristics of unfolded polypeptides. Within the TM segment, transverse relaxation rates surpass low rates at F495, which is in line with low amide–amide NOE magnetization transfer (see NOEs). T1 times for this residue are only marginally longer than for the remainder of the membrane-imbedded residues but still represent a local maximum also. We pursued reduced spectral density mapping to obtain the values of the spectral density function at zero, 1H, and 15N frequency [J(0), J(ωH), and J(ωN)] (Fig. 3 A–C). J(0) thereby represents the effective correlation time at each protein site, if we assume the spectral density function to be a Lorentzian (44). Its average value amounts to ∼8 ns, which is significantly lower than what would be expected for a globular membrane protein of this size in micelles (45). In addition to the highly flexible C terminus, the data confirm the presence of motion in the C-terminal part of the membrane-embedded anchor region. The J(0) data as a function of residue point to increasing flexibility at the membrane:solvent interfaces and lower amplitude dynamics in the center. The region between the NP motif and the solvent-exposed region deviates from this bell and provides evidence for increased motility also within the membrane-embedded part. Here J(0) values are halfway in between those in the more rigid N-terminal helical element and those in the neighboring flexible C-terminal loop region. Slightly enhanced motility is also found for the region around the conserved F495, which is six residues upstream of the NP motif. In addition to J(0), also J(ωH) and J(ωN) values show that the C-terminal loop region is much more flexible than the helical element, but without providing any detail about the motional heterogeneity in the helix. The comprehensive relaxation dataset enabled us to determine the slow motion-derived (chemical exchange) contribution (μs to ms motions) to the 15N transverse relaxation using the Kroenke approach (Fig. 3D) (46). These data provide evidence of slow-motional contributions to 15N transverse relaxation within the putative membrane-embedded space, as opposed to the linker. Together with the J(0) data, this may indicate that fast and slow motion coexist for part of the lipid-embedded stretch. Fig. 3E represents local relaxation properties mapped onto a topology model of the S1P-processed SREBP-1 membrane anchor as obtained after equilibrating the CNS model in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid environment by molecular dynamics (see Molecular Dynamics).
Fig. 3.
Backbone dynamics of the membrane anchor as processed by S1P. (A–C) Spectral density mapping [J(0), J(ωH), and J(ωN)] derived from 1H/15N hetNOE, transverse and longitudinal 15N autocorrelation rates R2 and R1, and transverse and longitudinal 1H/15N cross-relaxation rates ηz and ηxy data. Also see SI Appendix, Fig. S11. The S1P and S2P cleavage sites are marked by blue and brown shading, respectively; the NP motif is depicted in red. The background color of the plots denotes the position of the amino acids as membrane-imbedded (brown) or solvent-exposed (blue), according to Fig. 2. (D) Relaxation contributions from slow motion (μs to ns timescale motion). The dashed black line represents the average value. Residues with significant contributions from chemical exchange are depicted in red. (E) Model for the conformational flexibility derived from dynamics data, with the NP motif acting like a hinge. (F) Spectral density values at zero frequency (Left) and chemical-exchange contributions to R2 (Right) depicted on a membrane anchor topology model (see Molecular Dynamics for details) as expected in a membrane environment. Residues with increased fast motional amplitudes would be marked by lower J(0) values (orange to red colors). The same colorization was used to mark significant exchange contributions (yellow to red colors). The membrane boundaries (as derived from water NOEs in micelles) are marked by the black lines; gray residues mark those with incomplete datasets for the determination for each parameter due to unclear assignments, overlapping H/N signals, or insufficient signal-to-noise ratio. Determination of relaxation parameters involves only dispersed peaks in all experiments. In addition, the Ser and Thr-rich solvent-exposed terminus is largely exchange-broadened below detection in the T1 and T2 relaxation experiments. The dashed line in the color legend represents the average Rex throughout the protein.
Molecular Dynamics.
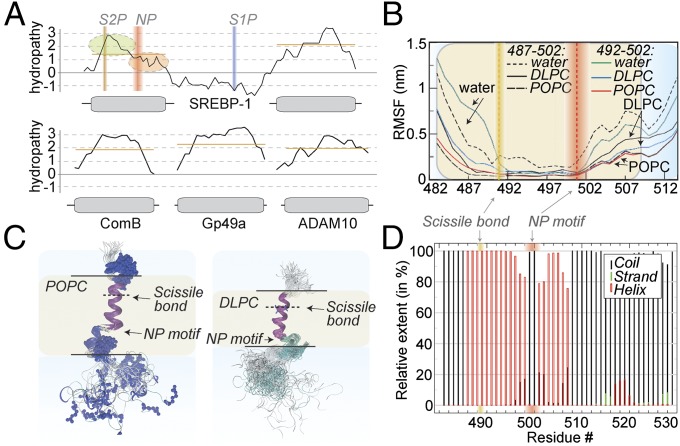
Using MD simulations, we equilibrated the SREBP-1 anchor structural model obtained from NMR data in micelles (SI Appendix, Fig. S10) into different lipid bilayers and monitored the secondary-structure stability over a period of up to 1 μs as well as the backbone root-mean-square fluctuations (RMSFs) as a function of residue number (Fig. 4 B and D and SI Appendix, Fig. S12). In accordance with the NMR data, the stretch around the scissile bond remains α-helical and consistently rigid. The C-terminal helical element of the membrane-embedded part also retains its α-helical topology, and a hinge motion is observed with respect to the N-terminal helical element, again in full agreement with the NMR data. Remarkably, when we use membranes of lower thickness [1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) instead of POPC lipids; Fig. 4C], we obtain an anchor with a steeper tilt angle (DLPC mean values of 48.3°, fluctuating between ∼30 and 80°, opposed to a mean of 24.3°, with fluctuations only between 15 and 35°, for POPC) between the two helical elements, with the NP motif functioning as a hinge. A consistent trend from tilted conformations toward the more linear conformation (for detergent micelles via thin membranes to thick membranes) hints to a stretching effect of the anchor in response to the environment. In addition, slightly different amplitudes of the hinge motion (∼17.7° and 8.7° for DLPC and POPC, respectively) are present in the two membranes (Fig. 4 B and C and SI Appendix, Fig. S13). Upon replacement of the lipids by water, fluctuations are significantly increased throughout the sequence, however complete unfolding is not observed within the timescale of the simulation (SI Appendix, Fig. S14). SI Appendix, Fig. S15 shows the structural model docked into the S2P structure (Protein Data Bank ID code 3B4R) (13). The open conformation is sufficiently wide to accommodate the substrate in its α-helical conformation. This protease–substrate combination is, however, artificial and has to be treated with extreme care.
Fig. 4.
Bioinformatics and molecular dynamics. (A) Hydropathy as a function of residue of SREBP-1 in comparison with other (non-protease substrate, single-TM helix) membrane proteins. The SREBP anchor is characterized by an asymmetric distribution of hydropathy values, implying very low hydropathy at the C-terminal end of the first TM helix. Orange bars depict the approximate average of hydropathy within each TM region. (B) RMSFs of the anchor in hydrophobic (DLPC and POPC membranes) and polar environment (exchange of lipids by water). Even in a polar environment, the C-terminal part of the helix shows a certain stability (also compare SI Appendix, Figs. S12 and S13). For alignment, residues 487–502 (black curves) or 492–502 (colored curves) were used. (C) Ensembles of equidistant snapshots in molecular dynamics trajectories over 1 μs. When accommodated in POPC (Left) and DLPC membranes (Right), the anchor displays significantly different tilt angles between the N-terminal and C-terminal part of the helix (see Molecular Dynamics). (D) Secondary-structural stability over a 1-μs molecular dynamics simulation in POPC. Random coil, β-strand, and α-helical structure is represented as black, green, and red bars, respectively.
Hydropathy.
We asked if the distinct motional features of the anchor are associated with characteristics on the primary sequence level. We used the European Molecular Biology Open Software Suite (EMBOSS) Pepwindow framework to obtain information about the hydropathy of the sequence. In fact, compared with a series of single-spanning membrane proteins (Fig. 4A), one obtains a strikingly asymmetric profile for the first helix of the SREBP-1 membrane anchor. In contrast to the N-terminal part of the helix, which accommodates the scissile bond, hydropathy values between the NP motif and the lipid:cytosol interface are very low and show a continuous, shallow slope. The shallow slope and low hydrophathy values in the C-terminal helical region of the S2P substrate are conserved among different kingdoms and between SREBP-1 and -2 (SI Appendix, Fig. S16).
Discussion
The characterization of the SREBP-1 membrane anchor both by NMR in micelles and by MD in lipid bilayers shows a topology consisting of two membrane-integral α-helical amino acid stretches separated by a flexible, solvent-accessible linker domain with no clear secondary structure. This linker was speculated in the work by Brown and Goldstein and coworkers (11) to be required for sufficient separability of the two helices for S1P action and to facilitate independent migration of the N-terminal part before S2P processing. In accordance with its considerably large expansion well beyond the common loop lengths in helix bundles, the C-terminal linker region indeed turns out to be highly mobile in our studies.
Peptide bonds within α-helices are poorly accessible to proteases because of the steric hindrance provided by the framework of helix-stabilizing hydrogen bonds (37). Accordingly, cytosolic proteases normally involve substrates in a β-strand extended conformation rather than relying on the poor cleavage properties of α-helices (24). Our data show that in the case of SREBP-1, similar to what is assumed for the other integral membrane proteases, the enzyme supposedly acts on a generally α-helical substrate. Interestingly, around the scissile peptide bond, the α-helical secondary structure even seems to be exceptionally stable, showing the lowest backbone flexibility throughout the primary sequence of the S2P substrate in terms of SCSs, NOEs, and relaxation data, and remains partly folded even upon switching the polarity of the environment in MD simulations. The length of the second helix (∼29 residues) derived from the different experimental data are in the range of known single spanning TM helices. The first helix with only on the order of 22 nonsolvent accessible residues, however, involves significantly fewer residues within the membrane under the same conditions. This feature is in line with the imperfect α-helical topology derived from secondary chemical-shift data and an according stretched conformation around the NP motif. The stretching was originally hypothesized to push the scissile bond out of the hydrophobic space to allow protease cleavage (38), which seemed unlikely after finding the S2P active site within the hydrophobic space (13). Instead, the low helical SCSs, low numbers and intensities of sequential amide NOEs, and the local minimum of H/N hetNOE intensity at C500 just before the NP motif in this study strongly suggest this site to act as a helix-breaking motif that reduces local conformational stability. A high level of mobility, starting from just before the NP motif toward the C-terminal solvent interface, also within the membrane-spanning region, goes hand in hand with the interrupted helicity. This mobility, which is obvious from transverse and longitudinal relaxation rates in this element, with a local maximum at C500, is represented by a conformational flexibility as observed in MD simulations. Even though SCSs of this element clearly state a reversion to a loose α-helical topology for the remaining residues after the NP motif, the C-terminal integral-membrane part of the S2P substrate represents a structured but still dynamic element. Its dynamics increasing C-terminally and already approaching the flexibility of the solvent-exposed residues near the lipid–cytosol interface as seen by NMR are in line with a flexure of the element using the helix breaker as a hinge.
The lower stability of the C-terminal helical element is paired with a conserved amphipathic primary sequence, which contains the Arg of the NP motif and a Ser, two hydrophilic residues, and may enable flexible accommodation in differentially hydrophobic environments. A tilt of this atypical element away from the remainder of the protein may be of relevance for a facilitated conformational adjustment for uptake by S2P and potentially differential unfolding upon contact with the protease. The bent conformation might also expose the polar Asn residue, which may be relevant for an initial interaction with S2P. Certainly, as derived from the different tilt angles in MD simulations of different membrane thickness, the flexure of the S2P substrate and all derived consequences will be directly influenced by the characteristics of the lipid membrane. Work by Walker et al. concluded from cell biological studies on Caenorhabditis elegans previously that membrane composition and curvature play a significant role for SREBP-1 cleavage in vivo, which could be related to the SREBP flexing (10).
In between the S2P cleavage site and the more mobile TM region toward the C terminus of the helix, F495 constitutes another site of enhanced mobility. For this region, we find significant contributions from slow-motional dynamics, as known from conformational preselection-type mechanisms. Whereas the functionally indispensable NP motif induces a required interruption of the α-helical conformation, the cleavage site seems completely buried before getting in contact with the protease and should require effective chaperone-like activity of the protease to unfold. This is in accordance with ample space in the S2P open conformation to accommodate the substrate in an α-helical (rather than extended) conformation. A similar picture has been obtained for APP, in which a flexible, bent TM domain might serve to most effectively interact with the protease (29). The TM region C-terminal to the hinge, however, is characterized by its more ambiguous hydropathy properties and the connection to the NP motif on one side and the highly mobile linker on the other. These destabilized residues in the C-terminal TM regions are thus likely to be unfoldable more easily in various conditions. Thus, apart from their tilt away from the NP motif, a partial unwinding of these residues might exist to further facilitate entry of the otherwise helical anchor into the protease. This could then exert any necessary unfolding of the internalized scissile bond by specific chaperone-like mechanisms.
For SREBP cleavage by S2P in the cell, quantitative details (dynamics time scales and amplitudes) and participants (constituting specific functions or crowding effects) will differ from the situation characterized under in vitro and in silico conditions. Still, the peculiar mechanical flexibility of the anchor is an intrinsic molecular property; thus, the observed flexure can well be expected to represent a significant feature of the substrate in vivo.
To summarize, the transcriptional activator SREBP regulates expression of machinery for cellular lipid and cholesterol homeostasis. A crucial step toward transcriptional activation, the liberation of the transcriptionally active regulatory domain, requires regulated intramembrane cleavage of the SREBP membrane anchor by S2P. Using solution NMR spectroscopy and molecular dynamics simulations, we find that the topology of this anchor is made up of a C-terminal regular TM helix, a long flexible linker, and an N-terminal TM helix. The substrate of the S2P cleavage, the anchor processed by S1P, bearing only the N-terminal TM domain and most of the linker, shows interrupted helicity at an evolutionarily conserved motif two-thirds down toward the linker. Whereas the clearly α-helical region bearing the scissile bond shows extraordinary rigidity, the lower elements of the TM domain, particularly the C-terminal helical element between the helix-breaking motif and the lipid-solvent interface, show reduced structural stability. The mobility of the residual linker outside the hydrophobic space increases to an extent comparable with IDPs C-terminally and is accompanied by fast-motional dynamics as well as chemical exchange within the destabilized TM region. Owing to the helix-breaking motif, which acts as a hinge in the S2P substrate, the substrate seems to be able to undergo conformational changes dependent on its molecular environment. In accordance with differential cleavage probabilities of SREBP-1 for different membranes reported previously, the conformational changes observed in vitro and in silico may be relevant for the initiation and general feasibility of RIP cleavage.
Materials and Methods
The deuterated, 13C/15N-labeled membrane anchor of the S2P substrate was expressed using cell-free expression. The purified protein was reconstituted into LMPG micelles. Samples were investigated by solution NMR at 800 MHz Larmor frequency. Established methodology was used for assignments and for characterization of topology and dynamics. MD simulations were performed in either POPC or DLPC lipid bilayers. Further details on the methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Anders Näär, Andras Böszörmenyi, Silvain Tourel, Thomas Raschle, Franz Hagn, Joshua Ziarek, and Haribabu Arthanari for helpful discussions and Anders Näär and Joshua Ziarek for critical reading of the manuscript. We acknowledge the Australian Research Council Discovery Early Career Research Award (to R.L.), a Liebig Fellowship of the Fonds der Chemischen Industrie (to R.L.), and the Emmy-Noether program (R.L.) and collaborative research project (SFB) 803 (project A04 to R.L. and project A03 to B.L.d.G. and R.B.) of the Deutsche Forschungsgemeinschaft for financial support. N.S. acknowledges a European Molecular Biology Organization Long-Term Fellowship (ALTF 612-2013) and an Early Postdoc Mobility Fellowship of the Swiss National Science Foundation. The project was supported by the Agilent Thought Leader Award and NIH Grants GM046476 and HL116391 (to G.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513782112/-/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. Sterol regulatory element binding proteins (SREBPs): Controllers of lipid synthesis and cellular uptake. Nutr Rev. 1998;56(2 Pt 2):S1–S3; S54–S75. doi: 10.1111/j.1753-4887.1998.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Cholesterol feedback: From Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50(Suppl):S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie. 2004;86(11):839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 6.Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: New players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9(4):358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- 7.Brown MS, Goldstein JL. Sterol regulatory element binding proteins (SREBPs): Controllers of lipid synthesis and cellular uptake. Nutr Rev. 1998;56(2 Pt 2):S1–S3; discussion S54–S75. doi: 10.1111/j.1753-4887.1998.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 8.Párraga A, Bellsolell L, Ferré-D’Amaré AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6(5):661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 9.Nohturfft A, Brown MS, Goldstein JL. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J Biol Chem. 1998;273(27):17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 10.Walker AK, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147(4):840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai J, et al. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85(7):1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 12.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 13.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318(5856):1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Xue Y, Lee S, Ha Y. The crystal structure of GXGD membrane protease FlaK. Nature. 2011;475(7357):528–531. doi: 10.1038/nature10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493(7430):56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 16.Lu P, et al. Three-dimensional structure of human γ-secretase. Nature. 2014;512(7513):166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107(2):173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe MS, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature. 1999;398(6727):513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 19.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398(6727):522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 20.De Strooper B, et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 21.Fluhrer R, Steiner H, Haass C. Intramembrane proteolysis by signal peptide peptidases: A comparative discussion of GXGD-type aspartyl proteases. J Biol Chem. 2009;284(21):13975–13979. doi: 10.1074/jbc.R800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal M, Caplan M. Regulated intramembrane proteolysis: Signaling pathways and biological functions. Physiology (Bethesda) 2011;26(1):34–44. doi: 10.1152/physiol.00028.2010. [DOI] [PubMed] [Google Scholar]
- 23.Loughlin WA, Tyndall JD, Glenn MP, Hill TA, Fairlie DP. Update 1 of: Beta-strand mimetics. Chem Rev. 2010;110(6):PR32–PR69. doi: 10.1021/cr900395y. [DOI] [PubMed] [Google Scholar]
- 24.Tyndall JD, Fairlie DP. Conformational homogeneity in molecular recognition by proteolytic enzymes. J Mol Recognit. 1999;12(6):363–370. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 25.Madala PK, Tyndall JD, Nall T, Fairlie DP. Update 1 of: Proteases universally recognize beta strands in their active sites. Chem Rev. 2010;110(6):PR1–PR31. doi: 10.1021/cr900368a. [DOI] [PubMed] [Google Scholar]
- 26.Lichtenthaler SF, Haass C, Steiner H. Regulated intramembrane proteolysis—Lessons from amyloid precursor protein processing. J Neurochem. 2011;117(5):779–796. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- 27.Langosch D, Scharnagl C, Steiner H, Lemberg MK. Understanding intramembrane proteolysis: From protein dynamics to reaction kinetics. Trends Biochem Sci. 2015;40(6):318–327. doi: 10.1016/j.tibs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Haapasalo A, Kovacs DM. The many substrates of presenilin/γ-secretase. J Alzheimers Dis. 2011;25(1):3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett PJ, et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336(6085):1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107(2):161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 31.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol. 2002;12(17):1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 32.Koide K, Ito K, Akiyama Y. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J Biol Chem. 2008;283(15):9562–9570. doi: 10.1074/jbc.M709984200. [DOI] [PubMed] [Google Scholar]
- 33.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11(6):1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 34.Strisovsky K, Sharpe HJ, Freeman M. Sequence-specific intramembrane proteolysis: Identification of a recognition motif in rhomboid substrates. Mol Cell. 2009;36(6):1048–1059. doi: 10.1016/j.molcel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strisovsky K. Structural and mechanistic principles of intramembrane proteolysis—Lessons from rhomboids. FEBS J. 2013;280(7):1579–1603. doi: 10.1111/febs.12199. [DOI] [PubMed] [Google Scholar]
- 36.Zoll S, et al. Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J. 2014;33(20):2408–2421. doi: 10.15252/embj.201489367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paetzel M, Dalbey RE, Strynadka NCJ. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396(6707):186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 38.Ye J, Davé UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc Natl Acad Sci USA. 2000;97(10):5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duncan EA, Davé UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273(28):17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 40.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linser R, et al. Selective methyl labeling of eukaryotic membrane proteins using cell-free expression. J Am Chem Soc. 2014;136(32):11308–11310. doi: 10.1021/ja504791j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Salvi N, Ulzega S, Ferrage F, Bodenhausen G. Time scales of slow motions in ubiquitin explored by heteronuclear double resonance. J Am Chem Soc. 2012;134(5):2481–2484. doi: 10.1021/ja210238g. [DOI] [PubMed] [Google Scholar]
- 45.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 46.Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AG. Longitudinal and transverse 1H-15N dipolar 15N chemical shift anisotropy relaxation interference: Unambiguous determination of rotational diffusion tensor and chemical exchange effect in biological macromolecules. J Am Chem Soc. 1998;120(31):7905–7915. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.