Introduction to zebrafish as a cancer model
In recent years, the zebrafish has emerged as an important model in cancer biology. The fish was originally developed in the 1950s as a model for toxicity testing [2] but during the 1960s–1990s it emerged as a powerhouse of developmental genetics [4]. The realization that the zebrafish was amenable to ENU-based, forward genetic screens led to the eventual large-scale effort to create large pools of mutant zebrafish each with a specific phenotype linked to an individual genetic mutation [5,6]. Initially, many of these phenotypes centered around specific cell types or tissues [7] but it was recognized early on that the zebrafish was especially sensitive to neoplasia [8]. Many of these tumors developed spontaneously or in p53 deficient backgrounds [9] but could be rapidly accelerated by mutagens such as DMBA [10].
The emergence of rapid and efficient transgenic technologies revolutionized the use of zebrafish in cancer research [11,12]. Because each pair of fish mates rapidly and produce hundreds of embryos per day, it was clear that it was a model amenable to large scale, unbiased approaches to cancer phenotypes. In its most straightforward iteration, dominant acting oncogenes under cell-type specific promoters can be used to produce a wide variety of tumors such as melanoma, as shown in Figure 1 [1,3,13]. More recently, increasingly complex models of cancer have been developed using a variety of overexpression and knockout technologies. A range of the available cancer models in zebrafish is shown in Table 1.
Figure 1.
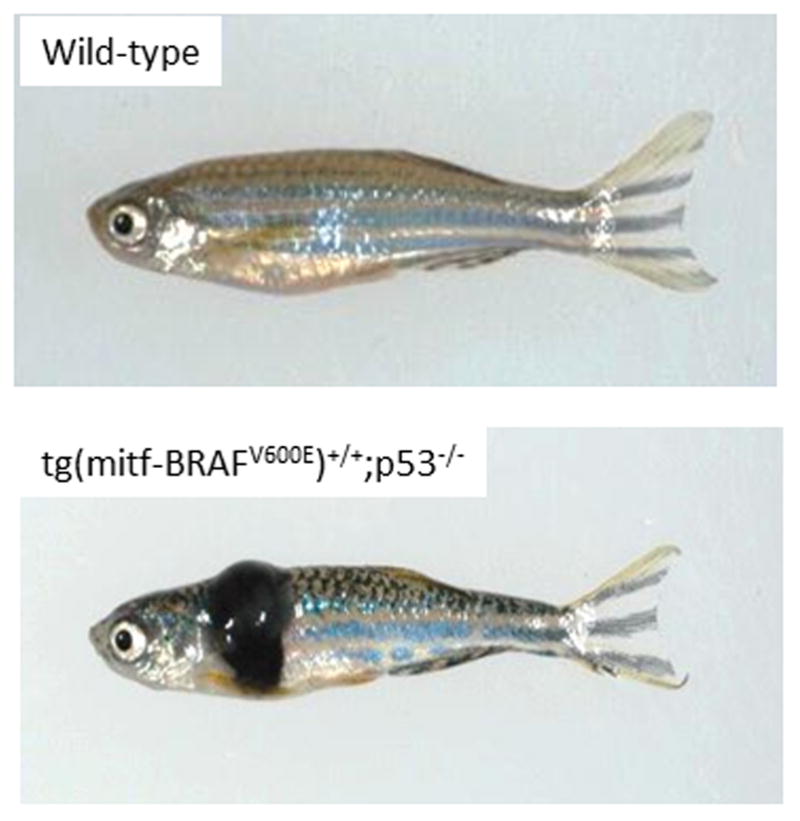
A transgenic model of melanoma in the zebrafish. On the top is a wild-type fish, and on the bottom an engineered fish expressing the human BRAFV600E gene under the melanocyte specific mitf promoter. Adapted from [1]
Table 1.
Available transgenic zebrafish models of cancer. Adapted from [3].
| Cancer | Oncogene | Tumor suppressor | Use in cancer biology | PMID |
|---|---|---|---|---|
| Melanoma | mitfa-BRAFV600E | p53−/− | Genetic and chemical modifier screens | 15694309 [14] |
| mitfa:EGFP:NRASQ61K | p53−/− | 19954345 [33] | ||
| kita-Gal4 x UAS-HRAS | 21170325 [45] | |||
| Pancreatic | ptf1a-KRASG12V-GFP | Genetic modifier screens | 18549880 [46] | |
| ptf1a:Gal4-VP16 x UAS-KRASG12V-GFP | 21951538 [47] | |||
| T-cell lymphoma/leukemia | rag2-myc | Cancer modeling, in vivo imaging | 12574629 [13]; 15827121 [48] | |
| rag2-s lox-dsRED2-lox-EGFP-mMyc x hsp70-cre | Inducible cancer models | 17593023 [49] | ||
| rag2-NOTCH1 | Notch1 interaction with bcl2 | 17252014 [50]; 22538478 [51] | ||
| rag2-myc x rag2-bcl2 | Mechanisms of leukemia dissemination | 20951945 [52] | ||
| B-cell leukemia | Xenopus EF1a or zebrafish B actin – TEL-AML1 (ETV6-RUNX1) | Initiating events in B-cell leukema | 17015828 [53] | |
| Numerous | b-actin-lox-GFP-lox-KRASG12D x hsp70-cre | Inducible cancer models | 17517602 [54] | |
| krt4:Gal4VP16;14 x UAS:smoa1-EGFP x UAS:myrhAKT1 | Cooperation of hedgehog and Akt pathways | 19555497 [55] | ||
| Rhabdomyosarcoma | rag2-KRASG12D | Identification of tumor initiating cell populations | 17510286 [15] | |
| Neuroblastoma | dβh:EGFP-MYCN | Cooperation of MYCN and ALK | 22439933 [56] | |
| dβh:EGFP and dβh:ALKF1174L | Cooperation of MYCN and ALK | 22439933 [56] | ||
| AML | spi1(pu.1)-MYST3/NCOA2-EGFP | First model of AML in zebrafish | 18729850 [57] | |
| MPNST | p53−/− | Conservation of tumor suppressor pathways in fish Major tumor type found in p53-deficient zebrafish |
15630097 [9] | |
| Lipoma | krt4Hsa.myrAkt1 | Platform for the study of drugs to treat lipoma and/or obesity | 22623957 [58] | |
| Ewing’s sarcoma | hsp70 or beta actin-EWS/FLI1 | p53−/− | Conserved function of EWS-FLI1 fusion protein from human to fish | 21979944 [59] |
| Liver | fabp10:LexPR; LexA:EGFP x cryB:mCherry; LexA:EGFP-krasV12 | Inducible KRAS hepatocellular cancer model | 21903676 [60] | |
| fabp10:TA; TRE:xmrk; krt4:GFP | Inducible EGFR-homolog hepatocellular cancer model | 21888874 [61] | ||
| Pancreatic neuroendocrine | zmyod–MYCN | Pancreatic neuroendocrine model as a platform for downstream MYCN targets | 15492244 [62] | |
| Myeloproliferative neoplams | sp1(pu.1)-NUP98-HOXA9 | NUP98-HOXA9-induced oncogenesis from defects in haematopoiesis and aberrant DNA damage response | 21810091 [63] | |
| Corticotroph adenoma/neoplasm | POMC-PTTG (securin) | Identification of CDK inhibitors as possible treatment of corticotrph tumors | 21536883 [64] | |
| Testicular germ cell tumor | fugu flck-SV40 large T | Platform for modifier screens of testicular tumors | 21158563 [65] |
Cancer genomics in zebrafish models
All animal models, not just zebrafish, have a variety of uses in cancer genomics. These can be categorized into two main strategies. The first is to use the fish to functionally test candidate genes that emerge from human cancer genomic studies such as The Cancer Genome Atlas (TCGA). An example of this would be the overexpression of BRAFV600E in melanocytes, which produces melanoma [14]. A second approach is comparative oncogenomics, which is the method of comparing a zebrafish tumor (however it is generated) to the human counterpart in order to find the most functionally important, and perhaps “driver” events in that given cancer type. An example of this is using RNA microarrays in zebrafish and human rhabdomyosarcoma to find a “common” RAS-driven signature [15]. More recent work has established the ability of the fish to be used for chromatin immunprecipitation/ChIP [15] and promoter/enhancer elements [16,17]. Each of the given approaches (DNA, RNA, chromatin) requires specific technological and analytic approaches. The purpose of this review is to discuss selected examples of cancer genomics in the fish, with an emphasis on the methodologies used for these studies.
DNA-based approaches
a. Array CGH
Array CGH (comparative genomic hybridization) has been used for over a decade to assess large and small scale copy number changes in cancer genomes [17]. The technology relies upon hybridization of fragmented, fluorescently labelled sample DNA to complementary sequence probes embedded on a solid chip or glass substrate. This technology is essentially a high resolution view of the entire genome of a given sample, which had traditionally been done at the chromosomal level using karyotyping and metaphase chromosome spreads. The level of resolution for array CGH depends on two factors: how many individual “spots” are on the chip, and what size is the probe (i.e. a 25bp oligonucleotide, longer PCR products, cDNA clones or BAC fragments). The changes in copy number – amplification or deletion – then depends on the fluorescence intensity of the two samples. In most cases, these samples represent cancer vs. normal tissue, but could also represent cancer vs. cancer samples.
A zebrafish array CGH platform was developed in 2008 using BAC (bacterial artificial chromosome) technology [18]. Previous work had cloned nearly the entire zebrafish genome into BAC libraries (CHORI-211, CHORI-73 and Danio key). Each BAC clone had between 100–200 kb of DNA, and after confirming BAC fragment identity, 207 clones were ultimately spotted onto glass slides. This design also included 109 previously identified BAC clones which served as chromosomal location markers. These and similar arrays were then used to analyze the genomes of three transgenic zebrafish models of cancer: KRASG12D-driven rhabdomyosarcoma (RMS), MYC-driven T-cell leukemia, and BRAFV600E-driven melanoma. These experiments yielded multiple genomic abnormalities in each tumor type. For the RMS, each sample had an average of 6–14 significant genome alterations, including some aberrancies on fish chromosome 17 that was observed in multiple tumor samples. The recurrent changes were mostly skewed towards amplifications, with most of the deletions being unique to each sample, an observation we have seen as well in other tumor types. In the T-ALL model, each sample had between 1–17 alterations, and in this case common gains and losses were seen across samples on multiple chromosomes, suggesting selection for these events. Finally, in the BRAF-induced melanoma model, an average of 6–28 genomic lesions were seen, including regions of abnormalities on contiguous BAC clones. A recurrent gain in half the melanoma samples was seen for 5 BAC clones, suggesting possible functional importance of this region. Interestingly, when comparing the copy number changes across all 3 tumor types, several recurrent regions of amplifications and losses were seen across multiple tumor types, suggesting that some of the genes in these regions may serve as general tumor promoting alterations. Although the resolution of this BAC array was not high enough to definitively assess the contribution of individual genes within the region, several candidates that are thought to play important roles in human cancer were suggested by the authors: EP300, PIM3, COL4A2, KIT, MITF and BRAF itself. This early report was an important proof-of-principle that copy number changes could be assessed in zebrafish tumors, and point towards genes with potential cancer-specific functions.
Since then, several other zebrafish cancer models have been subject to similar analyses. A series of malignant peripheral nerve sheet tumors (MPNSTs) were examined for copy number changes and karyotypic abnormalities [19,20]. These were found to have several genetic changes reminiscent of this relatively rare human tumor, including changes in MET, CYCLIND2, SLC45A3 and CDK6. One tumor was found to have a focal amplification of FGF8, and overexpression of FGF8 in the p53−/− background was found to accelerate tumorigenesis, suggesting that zebrafish aCGH could be used to identify a specific functional oncogene. Other tumor types analyzed in this manner include KRAS-driven rhabdomyosarcoma [21] and T-cell ALL [22], as followups to the above broad aCGH study.
b. Exome sequencing
Moving beyond large-scale copy number changes, our group has extensively utilized exome sequencing to examine both point mutations and copy number changes in a diverse set of zebrafish melanomas [23]. The rationale here was that melanoma is a notoriously heterogeneous tumor at the genetic level, which has been assumed to be related to the effects of chronic UV-exposure and a high background mutation rate. For this reason, identifying the “drivers” in a sea of very noisy passenger mutations has been exceptionally difficult. We reasoned that finding the overlap between fish and human melanomas would be one way of identifying “true” driver events conserved across species.
We engineered a series of 53 transgenic melanomas, driven primarily by either BRAFV600E or NRASQ61K expressed under the melanocyte specific mitfa promoter. These were driven in the context of a p53−/− animal, with in many cases additional “initiator” genes added on using the miniCoopR transgenic system [24,25]. There was no UV-exposure in our fish. Tumor and normal DNA were enriched for exonic/5′UTR/3′UTR sequences using Agilent SureSelect technology, followed by Illumina sequencing. Single nucleotide variants and copy number changes were analyzed by multiple algorithms. Surprisingly, for highly engineered transgenic tumors, there was tremendous mutational heterogeneity across the 53 tumors. In total, 403 mutations were seen in the 53 tumors, but about half of those mutations were found in just 8 of the tumors, suggesting that some tumors have very high mutation rates and others barely any additional mutations. Indeed, a close examination of the mutation frequencies (Figure 2) shows that the number of exonic mutations did not strongly depend upon the initiating oncogene (i.e. BRAF or NRAS), although there was a trend towards more mutations in those tumor with two rather than three initiating events. The overall number of mutations seen in these tumors is consistent with a non-UV induced tumor type in humans: whereas human cutaneous (sun and UV-exposed) melanomas have a median of 171 exonic mutations [26,27], mucosal (non-sun and UV-exposed) melanomas have a median on 9 exonic mutations. As is the case with most human cancers, C→T substitutions were the most common mutation type, with no strand bias or evidence of transcription-coupled mutational processes. Very few recurrent mutations were found across the 53 tumors.
Figure 2.
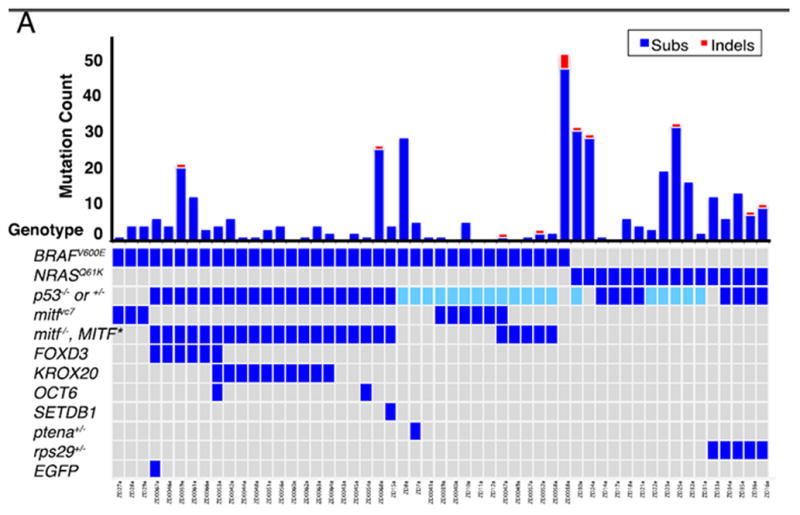
The spectrum of mutation pattern seen in zebrafish melanomas. Individual fish are along the x-axis, with mutation count along the top y-axis. The initiating driver events are shown along the y-axis on the bottom. Adapted from [23]
Other structural changes could also be analyzed from this dataset, including insertion-deletions (indels) and copy number changes. Overall, indels were relatively rare (only 13 across all the 53 samples), but recurrent copy number changes, especially amplifications, were common. Overall, 991 copy number changes were seen across the samples, but again with marked variation across the samples. Despite this heterogeneity, at least one recurrent event on zebrafish chromosome 3 was found in 10 tumors, which encompasses a region containing several potential tumor promoting genes: prkacaa, samd1, asf1ba, wu:fj41e11 and tecra. Whether these genes represent functional “drivers” in human melanoma awaits further analysis using shRNA or CRISPR approaches in appropriate human cell lines.
One important take-away from this study is that mutational heterogeneity, so common in human tumors, is not merely due to time and UV-induced mutational processes. Even in these highly engineered fish melanomas, with defined initiating events, there was tremendous variation in both the number and genomic location of mutations. Why this occurs remains unclear, but may point to mutational processes that are unleashed by strong drivers such as BRAF or NRAS that yield subsequent genetic heterogeneity. The other important conclusion from this study is that a simple comparative oncogenic analysis of DNA, directly comparing human to fish mutations to discover drivers, will not be straightforward.
RNA-based approaches
In addition to comparing somatic DNA changes across species, several studies have now done similar analyses using RNA quantification technologies, either microarrays or RNA-seq. The underlying assumption of these studies, like the DNA studies, is that there will be a core set of transcriptional pathways altered in both species, and help to elucidate the driver transcriptional programs for that given tumor type. In many ways, these RNA studies have yielded a more straightforward commonality between the two species than DNA studies.
Several investigations of hepatocellular carcinoma (HCC) in fish and humans have been performed using a variety of induced models in the fish. Zebrafish HCC was induced by carcinogen treatment of adult fish, followed by RNA isolation and microarray analysis [28]. When compared to normal liver, the zebrafish HCC’s had 2315 abnormally expressed genes, which ultimately mapped to 1920 human orthologs. These genes were then compared to human cancer gene signatures using Gene Set Enrichment Analysis (GSEA) [29]. Overall, the fish tumors were found to be similar to cancer (compared to normal) in general, but were most similar to the subset of human liver tumors. In a direct comparison of the fish HCC to human HCC, gastric and prostate tumors, the authors were able to identify a set of 76 genes that were specifically enriched in both fish and human HCC. This signature was enriched for genes in the Wnt-beta catenin and MAP kinase pathways, both of which are known to be strongly associated with human HCC. Similar types of analyses have been performed for other induced models of HCC, including a KRASV12 [30], xmrk [31] and RAF [32].
Using a KRASG12D model of rhabdomyosarcoma (RMS), Langenau employed microarray technology to identify a signature of these tumors compared to normal muscle [15], and then used GSEA to compare these signatures to signatures of human Alevolar RMS (ARMS) and Embryonal RMS (ERMS). This revealed a significant enrichment of the fish tumors in ERMS, but not ARMS. To determine whether this enrichment was solely due to the muscle origin of these tumors, the authors also compared their fish signature to other human cancers, which surprisingly yielded a similarity between the fish RMS and human pancreatic adenocarcinoma (PDAC). Because PDAC is well known to be a KRAS driven tumor, this suggested a common “RAS-signature” embedded within the fish RMS dataset, including known RAS target genes such as mcl1, pim1 and g3bp.
Our own group has similarly used GSEA to compare BRAF and NRAS driven melanomas to human melanoma [1,33]. This revealed not only the expected core signature related to RAS and MAP kinase activation, but also a strong enrichment for lineage-specific genes in the neural crest. This includes genes such as sox10, mitf and ednrb. This cross-species enrichment of neural crest and melanocyte genes has led to the concept of “lineage addiction” in melanoma [34], a hypothesis that was tested in the zebrafish melanoma model using a chemical genetic suppressor screen [1]. This yielded inhibitors of the enzyme dihydroorotate dehydrogenase (DHODH) as potent suppressors of the neural crest signature in fish and human melanomas, which may have clinical utility in the treatment of the disease.
These data suggest that cross-species RNA analysis can be used to very clearly define signature associated with that disease state, and point directly to potentially targetable pathways in a given cancer. With the advent of increasingly sophisticated RNA-seq methodologies in the fish [35], and ever-increasing databases of human cancer transcriptomes, this approach is likely to have significant and ongoing utility in the near future.
Future directions
The ability to model cancer in the zebrafish offers unique capabilities in terms of high-throughput and high-content screening approaches. In terms of how these models contribute to human cancer biology, it is clear that cross-species oncogenomics represents one major approach, because it allows for fine-tuning of DNA or RNA signatures that likely act as drivers of the disease. As newer zebrafish models come on board, both DNA and RNA methods will continue to contribute to our understanding of cancer.
As it is increasingly recognized that cancer is as much a disease of the epigenome as it is of the genome [36], the zebrafish must now be used to address this important topic as well. The zebrafish genome undergoes all of the canonical epigenetic alterations described in mammalian cells, such as methylation, hydroxymethylation, and a wide-variety of chromatin modifications. Integrating methyl-seq, ChIP-seq and other chromatin analytic methods into studies of zebrafish cancer will be an important next step. Cross-species epigenomics has already been described [37], and several studies have now made landmark observations about conserved functions of chromatin modifiers such as SETDB1 in melanoma [25] and SUV39H1 in RMS [38]. Integrating the epigenomic and genomic landscapes of cancer in zebrafish models is one of the great strengths of the system, due to the ease of transgenic and CRISPR-based methodologies.
An emerging area of cancer biology relates to enhancer elements and other noncoding portions of the genome [39,40]. Although several studies have now identified mutations in these noncoding regions [41–43], it remains difficult to assign these changes functionality. Because of the remarkable advances in CRISPR technologies [44], it will now be relatively easy to specifically engineer such changes into zebrafish cancer models. Individually, many of these noncoding changes are likely to have subtle phenotypic effects, so it will be critical to identify how combinations of different changes across the noncoding genome collectively yield overt cancer phenotypes. The zebrafish is well poised to contribute to this specific area of cancer biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hisaoka KK. The effects of 4-acetylaminofluorene on the embryonic development of the zebrafish. I. Morphological studies. Cancer Res. 1958;18:527–535. [PubMed] [Google Scholar]
- **3.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13:624–636. doi: 10.1038/nrc3589. This paper provides the most comprehensive review of zebrafish cancer modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker C, Streisinger G. Induction of Mutations by gamma-Rays in Pregonial Germ Cells of Zebrafish Embryos. Genetics. 1983;103:125–136. doi: 10.1093/genetics/103.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grunwald DJ, Streisinger G. Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet Res. 1992;59:103–116. doi: 10.1017/s0016672300030317. [DOI] [PubMed] [Google Scholar]
- 6.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein BM, Schier AF, Abdelilah S, Malicki J, Solnica-Krezel L, Stemple DL, Stainier DY, Zwartkruis F, Driever W, Fishman MC. Hematopoietic mutations in the zebrafish. Development. 1996;123:303–309. doi: 10.1242/dev.123.1.303. [DOI] [PubMed] [Google Scholar]
- 8.Beckwith LG, Moore JL, Tsao-Wu GS, Harshbarger JC, Cheng KC. Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio) Lab Invest. 2000;80:379–385. doi: 10.1038/labinvest.3780042. [DOI] [PubMed] [Google Scholar]
- 9.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitsbergen JM, Tsai HW, Reddy A, Miller T, Arbogast D, Hendricks JD, Bailey GS. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol Pathol. 2000;28:705–715. doi: 10.1177/019262330002800511. [DOI] [PubMed] [Google Scholar]
- 11.Stuart GW, McMurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 12.Stuart GW, Vielkind JR, McMurray JV, Westerfield M. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development. 1990;109:577–584. doi: 10.1242/dev.109.3.577. [DOI] [PubMed] [Google Scholar]
- 13.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 14.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 2006;7:R71. doi: 10.1186/gb-2006-7-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **17.Lee HJ, Lowdon RF, Maricque B, Zhang B, Stevens M, Li D, Johnson SL, Wang T. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat Commun. 2015;6:6315. doi: 10.1038/ncomms7315. This paper is the first to comprehensively map methylation patterns during embryogenesis at multiple stages, providing a framework for enhancer utilization logic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman JL, Ceol C, Feng H, Langenau DM, Belair C, Stern HM, Song A, Paw BH, Look AT, Zhou Y, et al. Construction and application of a zebrafish array comparative genomic hybridization platform. Genes Chromosomes Cancer. 2009;48:155–170. doi: 10.1002/gcc.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Hoersch S, Amsterdam A, Whittaker CA, Lees JA, Hopkins N. Highly aneuploid zebrafish malignant peripheral nerve sheath tumors have genetic alterations similar to human cancers. Proc Natl Acad Sci U S A. 2010;107:16940–16945. doi: 10.1073/pnas.1011548107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Hoersch S, Amsterdam A, Whittaker CA, Beert E, Catchen JM, Farrington S, Postlethwait JH, Legius E, Hopkins N, et al. Comparative oncogenomic analysis of copy number alterations in human and zebrafish tumors enables cancer driver discovery. PLoS Genet. 2013;9:e1003734. doi: 10.1371/journal.pgen.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Chen EY, Dobrinski KP, Brown KH, Clagg R, Edelman E, Ignatius MS, Chen JY, Brockmann J, Nielsen GP, Ramaswamy S, et al. Cross-species array comparative genomic hybridization identifies novel oncogenic events in zebrafish and human embryonal rhabdomyosarcoma. PLoS Genet. 2013;9:e1003727. doi: 10.1371/journal.pgen.1003727. A good example of cross-species genomics, in this case using array CGH technology, to find common driver events in fish and human tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudner LA, Brown KH, Dobrinski KP, Bradley DF, Garcia MI, Smith AC, Downie JM, Meeker ND, Look AT, Downing JR, et al. Shared acquired genomic changes in zebrafish and human T-ALL. Oncogene. 2011;30:4289–4296. doi: 10.1038/onc.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Yen J, White RM, Wedge DC, Van Loo P, de Ridder J, Capper A, Richardson J, Jones D, Raine K, Watson IR, et al. The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biol. 2013;14:R113. doi: 10.1186/gb-2013-14-10-r113. The most comprehensive genomic characterization of any zebrafish cancer model, using exome sequencing of over 50 tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyengar S, Houvras Y, Ceol CJ. Screening for melanoma modifiers using a zebrafish autochthonous tumor model. J Vis Exp. 2012:e50086. doi: 10.3791/50086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. The definitive manuscript listing key genetic events in human melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, Zhan H, Govindarajan KR, Lee S, Mathavan S, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen AT, Emelyanov A, Koh CH, Spitsbergen JM, Lam SH, Mathavan S, Parinov S, Gong Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis Model Mech. 2011;4:801–813. doi: 10.1242/dmm.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Luo H, Li C, Huo X, Yan C, Huang X, Al-Haddawi M, Mathavan S, Gong Z. Transcriptomic analysis of a transgenic zebrafish hepatocellular carcinoma model reveals a prominent role of immune responses in tumour progression and regression. Int J Cancer. 2014;135:1564–1573. doi: 10.1002/ijc.28794. [DOI] [PubMed] [Google Scholar]
- 32.He S, Krens SG, Zhan H, Gong Z, Hogendoorn PC, Spaink HP, Snaar-Jagalska BE. A DeltaRaf1-ER-inducible oncogenic zebrafish liver cell model identifies hepatocellular carcinoma signatures. J Pathol. 2011;225:19–28. doi: 10.1002/path.2936. [DOI] [PubMed] [Google Scholar]
- 33.Dovey M, White RM, Zon LI. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish. 2009;6:397–404. doi: 10.1089/zeb.2009.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 35.Aanes H, Collas P, Alestrom P. Transcriptome dynamics and diversity in the early zebrafish embryo. Brief Funct Genomics. 2014;13:95–105. doi: 10.1093/bfgp/elt049. [DOI] [PubMed] [Google Scholar]
- **36.Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. A seminal review of epigenetics and its applicability to cancer biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tena JJ, Gonzalez-Aguilera C, Fernandez-Minan A, Vazquez-Marin J, Parra-Acero H, Cross JW, Rigby PW, Carvajal JJ, Wittbrodt J, Gomez-Skarmeta JL, et al. Comparative epigenomics in distantly related teleost species identifies conserved cis-regulatory nodes active during the vertebrate phylotypic period. Genome Res. 2014;24:1075–1085. doi: 10.1101/gr.163915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albacker CE, Storer NY, Langdon EM, Dibiase A, Zhou Y, Langenau DM, Zon LI. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS One. 2013;8:e64969. doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **39.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.02.014. A manuscript defining superenhancers, a functional regulatory element that is thought to control core elements of cell identify in normal tissues and in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. A discrete example of an oncogenic superenhancer that likely is the first of many to come. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 43.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. The most important enabling technology for genetic manipulation in fish and other species, now performed using tissue specific expression of Cas9. [DOI] [PMC free article] [PubMed] [Google Scholar]


