Abstract
Understanding how non-dividing cells remain viable over long periods of time, which may be decades in humans, is of central importance in understanding mechanisms of aging and longevity. The long-term viability of non-dividing cells, known as chronological longevity, relies on cellular processes that degrade old components and replace them with new ones. Key among these processes is amino acid homeostasis. Amino acid homeostasis requires three principal functions: amino acid uptake, de novo synthesis, and recycling. Autophagy plays a key role in recycling amino acids and other metabolic building blocks, while at the same time removing damaged cellular components such as mitochondria and other organelles. Regulation of amino acid homeostasis and autophagy is accomplished by a complex web of pathways that interact because of the functional overlap at the level of recycling. It is becoming increasingly clear that amino acid homeostasis and autophagy play important roles in chronological longevity in yeast and higher organisms. Our goal in this chapter is to focus on mechanisms and pathways that link amino acid homeostasis, autophagy, and chronological longevity in yeast, and explore their relevance to aging and longevity in higher eukaryotes.
Keywords: Amino acid, Homeostasis, Chronological longevity, Caloric restriction, Autophagy
Replicative and Chronological Aging
The budding yeast Saccharomyces cerevisiae is a free-living unicellular eukaryote that has proven to be a valuable genetic model system to investigate mechanisms of aging that operate at the cellular level. Budding yeast undergo two types of aging: replicative and chronological. When nutrients are readily available in the environment, yeast reproduce by an asymmetric cell division process known as budding, during which a bud emerges, grows, and separates to yield a smaller daughter cell and a larger mother cell. Mother cells do not divide indefinitely and the number of daughter cells produced by a mother cell is known as the replicative life span (Mortimer and Johnson 1959). When nutrients in the environment become limiting, cell division ceases and yeast remain viable in a non-dividing state for a period of time known as the chronological life span (CLS) (Müller et al. 1980). Viability in this context is defined as the ability to resume the budding process when nutrients become available in the environment, which is a biologically meaningful measure of how well yeast cells survive in a non-dividing state. Exciting progress has been made in uncovering the molecular events that underlie aging in yeast, which has contributed to our understanding of mechanisms of aging and longevity that are evolutionarily conserved from lower to higher organisms (reviewed in Bishop and Guarente 2007; Fontana et al. 2010; Kaeberlein 2010; Kenyon 2005; Laun et al. 2008; Parrella and Longo 2010; Sinclair 2005; Smith et al. 2007; Srinivasan et al. 2010; Vijg and Campisi 2008).
Chronological Aging and Stationary Phase
Chronological aging in yeast is the process by which non-dividing cells lose viability due to a series of time- and metabolism-dependent changes and is distinguished from loss of viability due to the effects of deleterious mutations or adverse environmental factors (e.g., temperature extremes, toxic substances). Chronological aging in yeast takes place during a relatively inactive non-dividing state known as stationary phase. After preferred nutrients, such as glucose, are exhausted during logarithmic growth, alternative nutrients, such as ethanol or amino acids, support growth at a reduced rate during the diauxic shift growth phase. Following the diauxic shift is a period of slow growth termed the post-diauxic phase, during which yeast continue to adapt to nutrient scarcity and prepare for stationary phase. Depending on the nutrient limitation(s) faced during stationary phase, most yeast cells typically arrest as unbudded cells early in the cell cycle prior to the G1/S transition (Hartwell 1974). During stationary phase, yeast cells derive energy from respiration, exhibit reductions in transcription and translation, accumulate storage carbohydrates, reinforce the cell wall, and acquire resistance to a variety of environmental stresses, yet remain responsive to environmental stimuli (reviewed in Gray et al. 2004; Herman 2002). In addition to exhibiting resistance to environmental stress, stationary phase yeast also up-regulate pathways that reduce or remove molecular damage due to metabolic stress. For example, stationary phase cells employ multiple mechanisms to combat reactive oxygen species (ROS) damage and up-regulate pathways that degrade oxidatively damaged macromolecules (Chen et al. 2004, 2005; Longo et al. 1996).
Thus, stationary phase is not simply a winding down of cellular processes that drive growth and cell division in the presence of nutrients. Rather, yeast in stationary phase actively respond to reductions in nutrient availability by undertaking programmatic changes in gene expression and cell physiology that favor long-term survival in the non-dividing state. Because they retain the capacity to resume mitosis, viable stationary phase yeast are quiescent but not senescent, which is defined by the inability to resume or continue cell division. Stationary phase yeast are similar in many respects to non-dividing differentiated cells in the G0 state that age chronologically in human tissues. This similarity combined with the evolutionary conservation of regulatory pathways that affect aging highlights the relevance of the budding yeast model system for the investigation of fundamental processes of cellular aging in post-mitotic human cells.
Interestingly, stationary phase yeast derived from glucose-containing rich medium undergo differentiation into two cell types, quiescent and non-quiescent (Allen et al. 2006; Aragon et al. 2008; Davidson et al. 2011). Quiescent cells resemble G0 cells insofar as they are unbudded, resistant to stress, responsive to the environment, long-lived, and able to re-enter the cell cycle. Non-quiescent cells are more heterogeneous, contain budded and unbudded cells, lose the ability to re-enter the cell cycle relatively quickly, respire at a relatively low rate, give rise to an elevated number of respiration deficient (petite) colonies, and are more prone to apoptosis. The formation of these two cell types in stationary cultures may represent a survival strategy in which the population of replicatively older non-quiescent cells contribute nutrients (i.e., following cell death) and genetic diversity (i.e., due to mutation) to the population of replicatively young quiescent cells that persist the longest in the non-dividing state.
Adaptations that Circumvent Chronological Aging
It is important to note that yeast have evolved strategies to respond to nutrient scarcity other than stationary phase, such as filamentous growth and sporulation. Filamentous growth is a nutrient-seeking growth habit that depends on formation of elongate pseudohyphae. Filamentous growth is characterized by a number of cell biological changes, such as cell shape change, polar budding, suppression of asymmetric mother cell division, which lead to formation of chains of elongate cells arranged end-to-end (Gimeno et al. 1992). Diploid yeast strains respond to nitrogen limitation by forming pseudohyphae; haploid cells form filaments in response to glucose limitation (Zaman et al. 2008). Under conditions of nitrogen starvation in the presence of non-fermentable carbon sources, diploid yeast undergo meiosis and sporulation to generate ascospores that are resistant to desiccation and numerous environmental insults (Neiman 2005). Ascospores are capable of persisting in the environment for long periods of time. In addition to filamentous growth and sporulation, yeast are capable of adhesion, flocculation, and biofilm formation, typically in response to nutrient limitation or environmental stress (Verstrepen and Klis 2006). Adhesion allows yeast to remain attached to surfaces where nutrients are available, and flocculation is an adaptive response that protects cells at the center of flocs from environmental stress. Laboratory yeast strains typically harbor mutations in FLO genes (e.g., FLO8) that render them deficient in filamentous growth and flocculation (Liu et al. 1996), which is valuable for technical reasons, such as the isolation of clonal cell lines (i.e., pure colonies).
Nutrient Limitation Leads to Chronological Aging
Stationary phase and chronological aging in yeast are brought about by limitation of one or more nutrient(s) required for growth. In the natural environment, prototrophic yeast strains require simple carbon and nitrogen sources, salts, and trace elements for growth, and limitations of any required substance can lead to cessation of growth and chronological aging. Laboratory yeast strains typically contain auxotrophic marker alleles that are useful for genetic manipulation precisely because of the specific nature of the nutrient-limited growth these alleles confer. It has become clear that limitation of “natural” nutrients, such as ammonium sulfate, leads to changes in gene expression and cell physiology that are different from changes brought about by limitations in “supplemental” nutrients that complement auxotrophic deficiencies (Saldanha et al. 2004). For example, nitrogen or sulfur limitation leads to “glucose sparing” growth whereas leucine or uracil limitation (in leu2 or ura3 auxotrophs) leads to “glucose wasting” growth (Brauer et al. 2008). Differences in limitation of “natural” versus “supplemental” nutrients also lead to differences in stationary phase survival of yeast (Boer et al. 2008), and increased availability of essential “supplemental” nutrients extends the CLS of auxotrophic yeast strains (Gomes et al. 2007). These findings conform to our expectation that chronological aging in a free-living organism such as yeast is triggered by cessation of growth due to nutrient limitation. However, it is clear that not all nutrient limitations are equivalent in terms of their impact on cellular physiology, and that limitations in different required nutrients can elicit dramatically different physiological responses and states in stationary phase cells that undergo chronological aging.
It is tempting to conclude that studies of chronological aging and longevity in yeast should focus on limitations of “natural” nutrients to avoid potentially artificial or artifactual effects of limitations of “supplemental” nutrients needed by auxotrophic strains. However, this conclusion may be premature at this point because it is not yet clear what conditions for nutrient limitation are relevant to human cells. Importantly, humans are not prototrophic organisms. In normal healthy humans, nine amino acids are essential and must be obtained from the diet, and another six amino acids are conditionally essential depending on growth rate (Pencharz and Ball 2003). Amino acid requirements for human cells are important in the context of aging for at least four reasons. First, the level of essential amino acids in toto is an important factor in maintaining cell and tissue function during aging. For example, elevated levels of essential amino acids offset the debilitating effects of sarcopenia in older women (Dillon et al. 2009). Second, certain essential amino acids appear to be more important than others for optimal function in certain cell and tissue types (Millward et al. 2008). For example, branched side chain amino acids (BCAA; isoleucine, leucine, and valine), especially leucine, play important roles in maintaining function in skeletal muscle and nervous tissue (Drummond and Rasmussen 2008; Layman 2003; Murin and Hamprecht 2008). Third, the requirements for individual essential amino acids in the diet change with age (McLarney et al. 1996; Young and Borgonha 2000), which likely reflects age-dependent changes in requirements at the cell and tissue level. Fourth, amino acid requirements vary with pathophysiological state in diseased cells and tissues (Soeters et al. 2004). Such altered requirements in aging-related diseases could contribute to the aging process, as well as provide a potential opportunity to be exploited as part of a therapeutic regimen. The absolute amount of essential amino acids, the ratio of individual essential amino acids, and age-dependent changes in these two parameters contribute to the optimum dietary intake of essential amino acids that support human health and longevity. Model organisms such as yeast may be valuable in this context. Although it is difficult to study how amino acids affect longevity in humans, it should be relatively easy to genetically engineer experimental organisms (such as yeast) to recapitulate specific amino acid requirements in human cells (at the level of specific enzymatic deficiencies) to test relationships between levels of essential amino acids, chronological aging, and longevity.
Calorie Restriction Promotes Chronological Longevity
Whereas nutrient limitation induces chronological aging in yeast, calorie restriction (CR) induces chronological longevity. CR is arguably the most established and promising intervention for life span extension based on the wide variety of species studied so far. Because reduced levels of nutrients, rather than reduced levels of calories per se, extend life span in some species (see below), this intervention is also known as dietary restriction (DR), a term that allows more latitude in terms of underlying mechanism(s). In many animals, the maximum life span is obtained when intake during CR is restricted to approximately two-thirds of the amount of food consumed ad libitum. This does not mean that CR is without consequences to the organism; the fecundity of many species is reduced by CR. Depending on the species, CR can be achieved with different feeding schedules, including continuous (i.e., daily) CR, intermittent CR, modest CR, or mid- to late-life CR (Piper and Bartke 2008). Even modulation of the perception of food availability appears to mimic the life span extending effects of CR in Drosophila (Libert et al. 2007). In general, life span extension by CR can be interpreted as a universal response to nutrient scarcity that has evolved to maximize survival until nutrient availability is sufficient for optimal reproduction.
In terms of chronological longevity in yeast, CR is achieved by either growth in low (e.g., 0.4%) glucose medium or growth in normal (e.g., 2%) glucose medium followed by transfer to, and washing with, water (Fabrizio and Longo 2003; Piper 2006). In low glucose medium, “adaptation and regrowth” may be observed, depending on the yeast strain, and is due to resumption of growth of surviving cells that utilize metabolites released from dead and apoptotic cells (Fabrizio et al. 2004; Herker et al. 2004). This phenomenon has been termed “altruistic aging” (Longo et al. 2005) and makes sense as a survival strategy for a free living single-celled microorganism that has evolved to grow quickly in the presence of food to maximize population size rather than individual longevity. Similarly, prolonged life span in water is likely an adaptation to adverse situations that arise naturally in the wild (e.g., when rain washes yeast cells from fruit into soil). Life span extension by CR in yeast appears to depend on the specific conditions experienced by non-dividing cells. Extracellular levels of organic acids, such as acetic acid, and pH play critical roles in determining chronological longevity (Burtner et al. 2009). Thus, although there is considerable interest in understanding intracellular signaling pathways that regulate the effect of CR on life span (see below), the question of what external factors are integrated to yield the CR response has only been answered in part.
It is worth noting that the difference between chronological longevity brought about by CR and chronological aging brought about by nutrient limitation is more than a matter of “less” nutrients versus “no” nutrients, respectively. In other words, there is a qualitative component as well as a quantitative component to this difference. Two important considerations are the nature of the conditions that constitute “less” versus “no” nutrients and the nature of the transition from “less” to “no” nutrients. For example, in higher organisms, life span extension is only attained if the calorie restricted diet is high quality (i.e., it contains sufficient essential vitamins, trace elements, and so forth, to support growth, albeit at reduced rates or with reductions in adult body mass). In yeast, a series of metabolic decisions are made during the transition from “less” to “no” nutrients that affect energy storage and resource utilization that have a significant impact on CLS. The availability of amino acids, which serve both as building blocks for protein synthesis as well as a non-fermentable carbon source, is an important factor. Thus, one of the key decisions made by yeast in response to amino acid depletion during this transition is the up-regulation of autophagy.
Autophagy Supports Chronological Longevity
Autophagy is a lysosome- or vacuole-mediated degradation system that responds to reductions in available nutrients, especially amino acids. Autophagy is constitutively active at low levels in virtually all eukaryotic cells, and is induced by nutrient limitation or CR as well as different forms of environmental and cellular stress. Autophagy is generally considered to be a non-specific process, but specialized autophagic pathways exist to handle specific targets (Yang and Klionsky 2009). Autophagy accomplishes two main functions – recycling and removal. Recycling maintains intracellular pools of building blocks, such as amino acids, that are needed for de novo synthesis. Removal targets cellular components ranging in size from small molecules to large organelles that are functionally impaired due to molecular damage, misfolding, and/or aggregation. Recycling and removal are particularly important during chronological aging because nutrient-limited non-dividing cells accumulate damage for two principal reasons: reduced rates of de novo synthesis and the absence of “dilution” of cellular damage during cell growth and division.
Studies in many organisms have revealed important roles for autophagy in forestalling chronological aging and promoting chronological longevity (Cuervo 2008a; Hubbard et al. 2011; Madeo et al. 2010; Vellai et al. 2009; Yen and Klionsky 2008). In C. elegans, autophagy is required for normal longevity (Melendez et al. 2003). In mammalian cells, autophagic protein turnover undergoes age-related decline (Cuervo and Dice 2000; Del Roso et al. 2003; Ward 2002). The decline in turnover of mitochondria by autophagy has been proposed to exacerbate ROS production with age (Kundu and Thompson 2005; Lemasters 2005). In skeletal muscle, this age-related decline in autophagy is attenuated by CR (Wohlgemuth et al. 2010).
Our studies have shown that autophagy is required for chronological longevity of yeast grown to stationary phase in synthetic media (Alvers et al. 2009). We found that autophagy-deficient mutant strains exhibited reduced chronological longevity compared to autophagy-proficient control strains in three different standard synthetic minimal and complete media. Genome-wide screens for mutations that reduce CLS in synthetic complete media likewise uncovered an important role for autophagy in maintaining chronological longevity (Fabrizio et al. 2010; Matecic et al. 2010). We also found that autophagy-deficient strains were as long lived as control strains in rich undefined medium, in agreement with the findings of Powers et al. (2006). This indicates that autophagy is required for longevity when amino acid availability is limited, but not when amino acid levels are high prior to or during stationary phase.
The atg1Δ and atg7Δ mutants used in our studies are deficient in multiple selective and non-selective autophagic pathways, including macroautophagy, microautophagy, the cytoplasm to vacuole targeting (CVT) pathway, piecemeal microautophagy of the nucleus (PMN), ribophagy, ER-phagy, turnover of fructose-1,6-bisphosphatase by vacuole import and degradation (VID), and selective degradation of mitochondria (mitophagy) and peroxisomes (pexophagy) (Nair and Klionsky 2005; Yang and Klionsky 2009). Chaperone mediated autophagy is not present in budding yeast. In our experiments, an atg11Δ strain exhibited a normal CLS, indicating that the CVT, pexophagy, and mitophagy pathways are not required for chronological longevity in synthetic media (Alvers et al. 2009; Kanki et al. 2009; Okamoto et al. 2009). Thus, selective mitophagy per se does not appear to play a significant role in maintaining chronological longevity. Taken together, these findings support a role for macroautophagic turnover and amino acid recycling in chronological longevity.
Amino Acid Homeostasis Promotes Chronological Longevity
Amino acid homeostasis consists of five primary processes: uptake, synthesis, utilization, recycling, and catabolism. In yeast, these processes are regulated by multiple, overlapping, global, and amino acid-specific pathways. Amino acids are among the most important nutrients recycled by autophagy, and maintenance of amino acid levels by autophagy is required for cell survival during nitrogen starvation (Onodera and Ohsumi 2005). Amino acid limitation is a potent inducer of autophagy in a wide range of eukaryotes from yeast to mammalian cells, and amino acid starvation in autophagy-deficient cells leads to rapid loss of viability. Thus, it is not surprising that amino acid availability has been found to play an important role in modulating yeast CLS. We and others have found that levels of essential amino acids affect chronological longevity (Alvers et al. 2009; Gomes et al. 2007). Traditionally, minimal and complete synthetic glucose media are prepared according to formulations that contain specific amounts of essential and nonessential amino acids (e.g., Amberg et al. 2005; Sherman 2002; Styles 2002). Inclusion of five-fold elevated concentrations of essential amino acids in minimal synthetic glucose medium extended CLS and increased resistance to oxidative, pH, and thermal stress during chronological aging (Gomes et al. 2007). Furthermore, Gomes et al. found that standard levels of essential amino acids resulted in aberrant cell cycle arrest and cellular DNA content consistent with programmed cell death. Thus, it is clear that limiting amounts of essential amino acids can have an adverse impact on chronological longevity that is distinct from the life span extending effects of CR. For this reason, yeast CLS measurements are routinely done using synthetic media that contain elevated levels of essential amino acids (Fabrizio and Longo 2003; Powers et al. 2006).
Interestingly, different essential amino acids are not equally effective in promoting chronological longevity. We analyzed the requirements for individual essential amino acids and learned that not all essential amino acids extend CLS when present at elevated levels (Alvers et al. 2009). This argues against the notion that elevated levels of essential amino acids extend CLS purely by a nitrogen supplementation mechanism. We tested three essential amino acids and found that elevated levels of leucine, but not histidine or lysine, resulted in a pronounced extension of CLS. Elevated levels of uracil, the only other essential nutrient, did not affect CLS. Furthermore, elevated leucine levels extended CLS in both autophagy-deficient and autophagy-proficient strains. In agreement with this, genetically complementing the leu2Δ auxotrophy in strain BY4742 with LEU2 extended CLS whereas complementation of his3Δ or ura3Δ had only minimal effects on CLS (Alvers et al. 2009). Complementation of the lys2Δ marker with LYS2 extended longevity in autophagy-proficient, but not autophagy-deficient strains. Thus, leucine promoted chronological longevity to the greatest extent among the essential nutrients we studied.
Moreover, nonessential amino acids promote chronological longevity in minimal media in both autophagy proficient and autophagy deficient strains. But, there is no strict correlation between number and amounts of nonessential amino acid supplements in synthetic media and chronological longevity (Alvers et al. 2009). Thus, the effect of nonessential amino acids is not simply a matter of providing an additional source of nitrogen. Furthermore, we found that the non-essential BCAA isoleucine and valine, and their precursor threonine, had the most pronounced effect on chronological longevity. To our knowledge, our studies were the first to demonstrate roles for specific individual nonessential amino acids in promoting chronological longevity in S. cerevisiae. This indicates that during chronological aging in synthetic media the availability of specific nonessential amino acids is more important to amino acid homeostasis than availability of all nonessential amino acids. This may also apply to the role of autophagy during chronological aging: recycling of specific amino acids by autophagy may be more important than recycling of bulk amino acids or nitrogen.
Down-Regulation of General Amino Acid Control Extends Chronological Life Span
The observation that nonessential amino acids extend CLS suggested a regulatory role rather than nutritional effect. Cellular responses to amino acid availability are regulated in large part by the target of rapamycin (TOR) and general amino acid control (GAAC) pathways, both of which are highly conserved from yeast to human. GAAC is an elaborate regulatory network that coordinates synthesis of amino acids, purines, and other metabolites when they are in short supply and need to be synthesized by the cell (Hinnebusch 2005). A hallmark of GAAC is that reduced levels of one amino acid can exert “general” or global control on anabolic pathways for many amino acids as well as other nutrients. At the heart of GAAC is the transcription factor Gcn4p that participates in regulation of transcription of hundreds of target genes. GCN4 expression is regulated at the translational level by Gcn2p. Gcn2p indirectly senses amino acid levels by responding to levels of uncharged tRNAs and regulating translation initiation accordingly, including translation of GCN4 mRNA (Hinnebusch 2005).
Given the prominent role of GAAC in responding to nutrient limitation, we tested if the non-essential amino acids isoleucine, valine, and threonine extended CLS by down-regulating the GAAC pathway. We showed that Gcn4p levels were reduced in synthetic minimal medium containing isoleucine, valine, and threonine compared to medium without these amino acids (Alvers et al. 2009). Furthermore, we showed that constitutive expression of GCN4 suppressed extension of CLS by isoleucine, valine, and threonine. Thus, at least some nonessential amino acids can bring about extension of CLS by down-regulating GAAC. However, deletion of GCN4 does not extend CLS (Fig. 8.1). Similar results have been obtained in the context of replicative life span (RLS). Steffen et al. reported that GCN4 is required for extension of RLS by 60S ribosomal protein mutants and CR, but deletion of GCN4 does not similarly extend RLS (Steffen et al. 2008). To our knowledge, our studies were the first to demonstrate a link between down-regulation of GAAC and chronological longevity in S. cerevisiae.
Fig. 8.1.
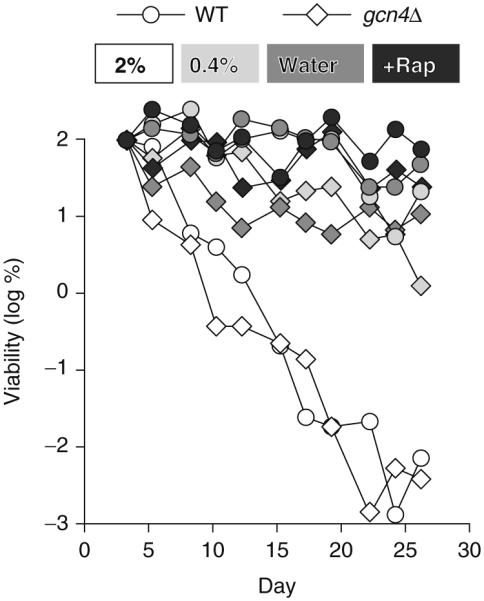
General amino acid control is not required for extension of CLS by CR or rapamycin. CLS measurements were done with yeast strains in the BY4742 genetic background (MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) grown in synthetic dextrose (SD) minimal medium as described (Alvers et al. 2009). Viability is expressed in terms of colony forming units (CFU) per mL and is plotted as the log of the percent viability on day three of the experiment. Wild type (WT, hoΔ) and gcn4Δ strains were grown in: normal (2%) glucose SD; low (0.4%) glucose SD; normal glucose SD followed by washing with water beginning on day three (Water); or normal glucose SD plus 10 nM rapamycin (+Rap)
To extend these studies, we wished to determine if GAAC played a role in the extension of CLS by rapamycin-dependent inhibition of the TOR pathway. Generally speaking, GAAC and TOR signaling act in a “yin-yang” fashion to manage cellular growth in relation to nutrient availability. GAAC up-regulates autophagy and synthesis of amino acids, purines, and other metabolites at a global level in response to amino acid limitation whereas the TOR pathway up-regulates protein synthesis and cell growth, and down-regulates autophagy and cellular stress response when nutrients are plentiful (De Virgilio and Loewith 2006a). S. cerevisiae has two partially functionally redundant TOR complexes. TOR complex 1 (TORC1) up-regulates anabolic processes, including amino acid utilization, and down-regulates catabolic processes, including autophagy, as well as expression of a number of stress response transcription factors. The functions of TORC2 are less well understood, but regulation of the actin cytoskeleton and cell polarity are important regulatory targets. In humans, aberrant regulation of TOR signaling is associated with aging and pathophysiological changes in multiple diseases (De Virgilio and Loewith 2006b).
In yeast, unlike in higher eukaryotes, the GAAC and TOR pathways communicate at the level of Gcn2p, which functions immediately upstream of Gcn4p. In the presence of ample amino acids, Gcn2p activity is down-regulated by phosphorylation under control of the TORC1 complex (De Virgilio and Loewith 2006a). Thus, in yeast, it is conceivable that Gcn4p may contribute to extension of CLS due to inhibition of TORC1 activity by rapamycin. However, we find that GCN4 is not required for extension of CLS by rapamycin treatment (Fig. 8.1). Furthermore, we showed that GCN4 is not required for extension of CLS by two CR interventions: growth in low glucose and washing with water (Fig. 8.1). Thus, although down-regulation of GAAC can extend CLS in yeast, as discussed above, extension of CLS by rapamycin and CR apparently involves branches of the TORC1 signaling pathway that do not include GAAC.
Deletion of the GAAC Target Gene LEU3 Extends Chronological Life Span
Because GCN4 regulates diverse cellular functions, there are many potential mechanisms by which down-regulation of GCN4 may extend CLS. Down-regulation of GCN4 down-regulates autophagy, but it is unlikely that this extends CLS given that autophagy is up-regulated during chronological aging and is required for chronological longevity, as discussed above. GCN4 is required for cellular responses to oxidative stress and DNA damage (Mascarenhas et al. 2008), and thus down-regulation of GCN4 should shorten CLS. Given the large number of potential avenues by which GCN4 might influence CLS, we focused on the link between chronological longevity and BCAA that emerged in our studies. The best known GCN4 target in this context is LEU3. Leu3p is a key transcriptional regulator of BCAA synthesis and is known to regulate transcription of LEU1, LEU2, LEU4, ILV2, and ILV5 as well as genes involved in BCAA uptake (Kohlhaw 2003). We found that deletion of LEU3 resulted in a profound extension of CLS, and that elevated levels of leucine, which extend CLS in a LEU3 strain, did not further extend CLS in the leu3Δ strain (Alvers et al. 2009). This points to down-regulation of LEU3 expression as an important mechanism by which down-regulation of GCN4 expression extends CLS.
These results raise an interesting question: how does deletion of LEU3 extend CLS? We originally suggested that deletion of LEU3 extends CLS by increasing cellular leucine availability and relieving an imbalance in synthesis of BCAA (Alvers et al. 2009). This suggestion was based on the observation that three conditions extended CLS in our studies: supplemental BCAA, conversion of leu2Δ to LEU2, or deletion of LEU3 (Alvers et al. 2009). The notion that balanced synthesis of BCAA contributes to chronological longevity is consistent with the relative abundance of BCAA in the yeast proteome. Leucine, isoleucine, and valine codons account for 9.6, 6.5, and 5.6%, respectively, of codons present in annotated verified open reading frames (ORFs) in the Saccharomyces Genome Database (Alvers et al. 2009). Because BCAA account for 21.7% of the amino acids in the yeast proteome, reduced amino acid availability may exert a direct effect on protein synthesis. Alternatively, or in addition, levels of the nonessential amino acids isoleucine and valine may exert an indirect effect on the levels of the essential amino acid leucine. Isoleucine and valine not only regulate enzymes required for their synthesis (i.e., threonine deaminase Ilv1p and acetolactate synthase Ilv2p) by feedback inhibition, but also regulate Leu1p by cross-pathway (non-end product) control (Jones and Fink 1982). Thus, for these reasons, we proposed that leu3Δ extended CLS by balancing BCAA levels (Alvers et al. 2009).
However, regulation of CLS in a leu3Δ strain may be more complicated and other factors may also contribute to extension of CLS. We have found that autophagy is up-regulated in a leu3Δ strain compared to a WT control (Fig. 8.2a, b). However, the regulation of autophagy does not appear to be altered in a leu3Δ strain insofar as autophagy is down-regulated by elevated levels of supplemental amino acids (Fig. 8.2c, d). This suggests that enhanced autophagy may contribute to extension of CLS in a leu3Δ strain, which agrees with our previous observations that autophagy promotes chronological longevity.
Fig. 8.2.
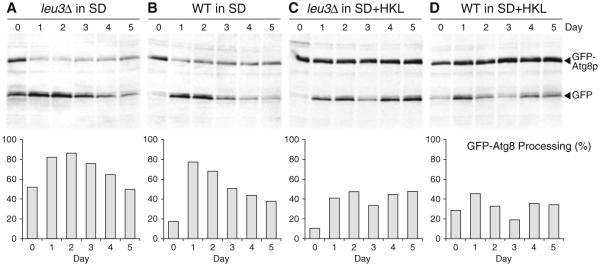
Macroautophagy is up-regulated in a leu3Δ strain. Activation of autophagy in leu3Δ and wild type (WT) strains was measured over 5 days of a CLS experiment performed as described (Alvers et al. 2009). Strains in the BY4742 background were transformed with plasmid pCuGF-PAUT7(416) (URA3), grown in synthetic dextrose medium (Sherman 2002) with standard (SD) or three-fold elevated levels (SD+HKL) of histidine, lysine, and leucine, and a western blotting assay was used to measure cellular levels of GFP and the GFP-Atg8p fusion protein (Klionsky et al. 2007). The percent conversion of GFP-Atg8p to GFP on each day reflects the extent of autophagic activation and is plotted in graphs below western blots. Equivalent amounts of cells, based on cell density measurements, were collected, processed, and analyzed on blots. Day zero samples were collected during mid-log growth. Quantification was done using ImageJ software [% processing = 100 × GFP ÷ (GFP + GFP−Atg8p)]
The fact that autophagy is upregulated in a leu3Δ strain is not unexpected given the number of cellular processes affected by Leu3p transcriptional regulation. At first glance, the Leu3p regulon appears relatively simple in terms of the number of genes that bind Leu3p and the “condition invariant” nature of regulation by Leu3p (Harbison et al. 2004). However, Leu3p binds at detectable levels to only about 3% of genes whose expression is affected by LEU3 deletion (Boer et al. 2005; Tang et al. 2006). Because of this, it has been estimated that Leu3p may contribute indirectly to the regulation of up to 10% of all yeast genes (Tang et al. 2006). Importantly, deletion of LEU3 does not simply result in reduced transcriptional activation. Leu3p also functions as a transcriptional repressor when levels of its obligate coactivator 2-isopropyl malate are low (Kohlhaw 2003). Thus, deletion of LEU3 results in increased expression of many genes involved in amino acid biosynthesis, including five genes involved in BCAA synthesis, when glucose is in excess and nitrogen is limiting (Boer et al. 2005). In addition, deletion of LEU3 may influence CLS via its effect on nitrogen assimilation (see below).
We also tested the hypothesis that CLS is influenced by 3-isopropyl malate methyl ester, a secreted signaling molecule implicated in stimulation of filamentous growth during amino acid limitation (Dumlao et al. 2008). We reasoned that accumulation of 3-isopropyl malate in leu2Δ mutants may favor Tmt1p-dependent methyl esterification during chronological aging. However, we found that tmt1Δ and TMT1 strains (in a leu2 background) yield indistinguishable CLS (Fishwick and Aris, unpublished results), indicating that signaling by 3-isopropyl malate methyl ester does not influence CLS under the conditions we examined.
Nitrogen Assimilation and Chronological Longevity
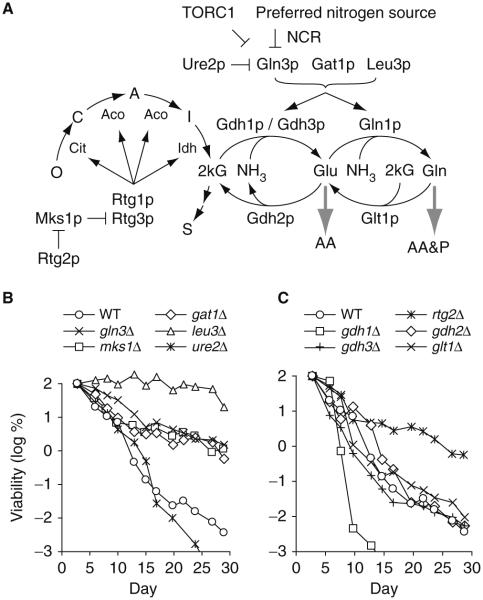
Given the influence of autophagy and amino acid homeostasis on CLS, we wished to explore potential relationships between nitrogen utilization and chronological longevity. S. cerevisiae can utilize a wide range of nitrogen sources, but exhibits preferences based on the metabolic investment required to catabolize them (best = glutamine > serine, ammonia > glutamate > proline = poor). In the presence of preferred nitrogen sources, nitrogen catabolite repression (NCR) down-regulates a wide variety of genes needed for utilization of nonpreferred nitrogen sources (Hofman-Bang 1999). In the absence of preferred nitrogen sources and NCR, a number of genes required for nitrogen assimilation are activated, including GAT1, GDH1, GLN1, and GLN3 (Fig. 8.3a). The transcription factors Gat1p and Gln3p are arguably the most important regulators of nitrogen assimilation, although nitrogen assimilation is known to be regulated at multiple levels (Zaman et al. 2008). Gln3p is the major regulator of NCR-sensitive gene expression, although many gene products are also controlled at the post-transcriptional level (Kolkman et al. 2006). Gln3p target genes are also up-regulated during release from NCR brought about by rapamycin mediated inhibition of TORC1, although rapamycin treatment does not faithfully mimic the pattern of Gln3p dephosphorylation observed during down-regulation of NCR (Tate et al. 2009). Gln3p stimulates expression of Gat1p, and both of these transcription factors stimulate expression of multiple downstream targets, including Gdh1p and Gln1p (Fig. 8.3a). GDH1 expression is also regulated by Leu3p as well as the HAP transcriptional complex, which regulates many aspects of mitochondrial function (Buschlen et al. 2003). GDH1 encodes the major NADP+-linked glutamate dehydrogenase isozyme that catalyzes the first step in nitrogen assimilation: formation of glutamate from ammonia and 2-ketoglutarate (2kG) (Fig. 8.3a). Nitrogen is also assimilated during conversion of glutamate to glutamine by glutamine synthetase encoded by GLN1. Because glutamate and glutamine serve as primary amine donors for synthesis of amino acids and other amine containing molecules, Gdh1p and Gln1p are central regulators of nitrogen assimilation (Hu et al. 1995). GDH3 encodes an isozyme that consumes 2kG at a lower rate and is glucose repressed. The third enzyme capable of synthesizing glutamate, NAD+-dependent glutamate synthase, is encoded by GLT1. GDH2 encodes an enzyme that catalyzes an anaplerotic reaction that regenerates 2kG.
Fig. 8.3.
Role for nitrogen assimilation in chronological longevity. a Nitrogen / ammonia assimilation pathway in S. cerevisiae. The TCA cycle intermediates are oxaloacetate (O), citrate (C), cis-aconitate (A), isocitrate (I), 2-ketoglutarate (2-kG), and succinate (S). Gene products are discussed in the text. AA, amino acids. P, purines and pyrimidines. Consumption of ATP, NAD+, and NADP+ are omitted for simplicity. Inhibition by TORC1 and Ure2p are diagrammed separately but participate in nitrogen catabolite repression (NCR). Interactions between NCR and the retrograde signaling pathway as well as negative regulation by Dal80p and Deh1p are not shown. b and c CLS measurements were done using deletion mutants in the BY4742 genetic background grown in SD minimal medium as described in Fig. 8.1. Viability is expressed in terms of colony forming units (CFU) per mL of culture and is plotted as the log of the percent viability on day three of the experiment. The results shown are representative of at least two independent experiments
To investigate the relationship between nitrogen assimilation and chronological longevity, we measured the CLS of a number of mutant strains impaired in different aspects of nitrogen utilization. Deletion of GLN3 or GAT1, as well as LEU3, extends CLS, although deleting GLN3 or GAT1 does not prolong CLS to the same extent as deleting LEU3 (Fig. 8.3b). Deletion of GLN3 has been previously shown to extend CLS in synthetic complete media (Matecic et al. 2010; Powers et al. 2006). Deletion of URE2, which encodes an inhibitor of Gat1p and Gln3p, yields a CLS equivalent to the wild type control, suggesting that increased Gat1p and Gln3p activities do not reduce CLS. Extension of CLS in gln3Δ, gat1Δ, and leu3Δ strains suggests that reductions in assimilation of ammonia may contribute to chronological longevity. This raises the question: does elimination of the enzymatic machinery required for assimilation of ammonia extend CLS? The answer to this question appears to be “no” because a gdh1Δ strain is relatively short-lived, and gdh2Δ, gdh3Δ, or glt1Δ deletion strains are not more long-lived than the wild type control strain (Fig. 8.3c). The reduced CLS in the gdh1Δ mutant may be due to the pleiotropic effects of a greater than ten-fold decline in cytoplasmic glutamate concentration that is characteristic of gdh1Δ strains (Hofman-Bang 1999). We were not able to test deletion of GLN1 because it is an essential gene. Deletion of GAP1, which encodes a general amino acid permease controlled by Gln3p and Gat1p, has no effect on CLS (data not shown).
Also important in this context is the retrograde signaling (mitochondrion to nucleus) pathway, which coordinates cellular and mitochondrial function (Liu and Butow 2006). Retrograde signaling has been implicated in coordinating metabolism, stress resistance, and genome stability during yeast replicative aging (Borghouts et al. 2004; Jazwinski 2005), but relatively little is known about the role of the retrograde response in chronological aging in yeast. One of the primary functions of retrograde signaling is to regulate carbon flow through the tricarboxylic acid (TCA) cycle and production of 2kG. The retrograde transcription factors Rtg1p and Rtg3p achieve this by regulating expression of four TCA pathway enzymes upstream of 2kG (Fig. 8.3a). Nuclear localization of Rtg1p and Rtg3p is inhibited by the cytoplasmic sequestration factor Mks1p that is regulated in turn by Rtg2p. Puzzlingly, deletion of either MKS1 or RTG2 results in extension of CLS and to an extent similar to deletion of GLN3 or GAT1 (Fig. 8.3b, c). The result with rtg2Δ notwithstanding, the extension of CLS in the gln3Δ, gat1Δ, and mks1Δ strains is consistent with increased aerobic respiration due to increased flow of carbon through the TCA cycle. That is, reduced consumption of 2kG by ammonia assimilation and increased utilization of 2kG by the TCA cycle is consistent with increased aerobic energy production. Increased CLS in tor1Δ strains has been attributed to increased aerobic respiration due to increased mitochondrial gene expression (Bonawitz et al. 2007). Such an increase in aerobic respiration would require increased carbon flow through the TCA cycle. This is consistent with the general idea that under the nutrient limiting conditions experienced during chronological aging, the balance between production of energy and synthesis of amino acids plays an important role in post-mitotic cell survival.
In the paragraphs above, we have discussed how autophagy, amino acid availability, and nitrogen assimilation are associated with chronological longevity in yeast. In the paragraphs below, we address connections between amino acids and two important paradigms in the aging field: the mitochondrial theory of aging and life span extension by CR.
Amino Acid Homeostasis and the Mitochondrial Theory of Aging
A group of well-known and closely allied mitochondria-related theories of aging have provided a mechanistic basis for aging at the cellular level that underscores the importance of molecular damage due to mitochondrially produced ROS. The free radical theory of aging and the mitochondrial theory of aging constitute central paradigms in aging research (Attardi 2002; Beckman and Ames 1998; Harman 1956, 1972, 2003; Miquel et al. 1980; Wallace 2005). Mitochondrial dysfunction, ROS production, and oxidative stress have been linked to chronological aging in many species and life span extending interventions generally mitigate these problems (Merry 2004).
During chronological aging in yeast, a main source of cellular damage is ROS produced by mitochondria (Chen et al. 2005). To combat oxidative damage, yeast possess multiple oxidant defense systems, including catalases, glutathione, glutathione peroxidase and oxidoreductase, and superoxide dismutases (SOD). These mechanisms minimize oxidative stress during stationary phase and are key factors in influencing CLS (Fabrizio and Longo 2003; Garrido and Grant 2002; Longo 2004; Piper 2006). Yeast lacking Sod1p (cytoplasmic Cu/ZnSOD) or Sod2p (mitochondrial MnSOD) have reduced viability in stationary phase (Longo et al. 1996, 1999) and over-expression of SOD1 or SOD2 increases CLS (Bonawitz et al. 2006; Harris et al. 2003, 2005). Yeast in stationary phase up-regulate transcription of SOD1, SOD2, and GLR1 (glutathione reductase) following exposure to menadione (Cyrne et al. 2003). Oxidative damage to mitochondrial DNA (mtDNA) is linked to instability of the mitochondrial genome and loss of respiration (Doudican et al. 2005).
Although the mitochondrial / ROS theory of aging is well established, there are exceptions to the existence of a strict correlation between aerobic respiration, ROS production, and chronological aging. In fact, there is a large and growing body of evidence that runs counter to mitochondrial theories of aging, which has lead to revision and reconsideration of some key elements of these theories (e.g., Lapointe and Hekimi 2010; Ristow and Zarse 2010). In yeast, deletion of TOR1, CR, and mild uncoupling increase respiration, but reduce ROS production and oxidative damage, which are linked to extended CLS (Barros et al. 2004; Bonawitz et al. 2007; Gomes et al. 2007). Similar salubrious effects of mild uncoupling on life span have been observed in higher organisms (e.g., Andrews 2010; Dietrich and Horvath 2010). Conversely, reductions in gene products required for oxidative phosphorylation do not necessarily lower ROS production and extend CLS. On the contrary, a mutation in mitochondrial RNA polymerase (Rpo41p) that causes a defect in mitochondrial translation of gene products required for oxidative phosphorylation results in a significant increase in ROS production and a severe reduction in CLS that can be rescued by over-expression of SOD1 or SOD2 (Bonawitz et al. 2006). These results indicate that increased longevity is associated with increased respiration, which may be due to increased electron transport chain efficiency and/or reductions in ROS production and concomitant molecular damage.
Amino acids are important in this context because they are able to serve as an aerobic carbon source that supports aerobic energy production during chronological aging. Rich media contain abundant nonessential amino acids that can serve this purpose. Certain amino acids, such as BCAA, phenylalanine, and methonine, cannot serve as carbon sources because they are metabolized via the Ehrlich pathway (Hazelwood et al. 2008). Nevertheless, other amino acids function as aerobic carbon sources following exhaustion of glucose during the diauxic shift, post-diauxic growth, and stationary phase. Aerobic growth prior to stationary phase has been shown to promote chronological longevity (Brauer et al. 2005; Piper et al. 2006). Furthermore, amino acids are also important because that are used as building blocks for protein synthesis that accompanies reprogramming of the cellular proteome during adaptation to the non-dividing state.
It is also important to consider the relevance of amino acid biosynthetic pathways to chronological longevity. De novo synthesis of many amino acids involves at least one step carried out in mitochondria. BCAA are synthesized by a super-pathway in which catalytically similar steps are carried out and regulated by shared gene products, namely Bat1p, Bat2p, Ilv2p, Ilv3p, Ilv5p, and Ilv6p. All of these enzymes, except Bat2p, function in the mitochondrial matrix and are potential targets of ROS damage. Perhaps the best understood case of oxidative damage to a mitochondrial matrix enzyme is aconitase. Aconitase is a bifunctional iron sulfur cluster (ISC) containing enzyme that catalyzes two steps in the TCA cycle and stabilizes the mitochondrial genome. Over-expression of mitochondrial Sod2p extends CLS by protecting aconitase from oxidative damage during chronological aging (Fabrizio et al. 2003; Harris et al. 2003). ISC synthesis is known to take place in the mitochondrial matrix due to the oxidizing environment present there. However, ISC enzymes are susceptible to oxidative damage, irrespective of their localization within the cell. Leu1p is a cytoplasmic ISC containing enzyme required for synthesis of leucine that is susceptible to inactivation by ROS, but is protected under aerobic conditions by SOD (Wallace et al. 2004). The same is true for Lys4p, a cytoplasmic ISC enzyme required for lysine synthesis. Thus, the extent to which amino acid biosynthetic enzymes are targets of oxidative inactivation may also play an important role in chronological longevity.
In addition to amino acid availability and de novo synthesis, amino acid recycling may play an important role in chronological longevity. The main cellular pathways for amino acid recycling – autophagy and the ubiquitin/proteasome system – function in both recycling and damage removal. Because of this, it is difficult to parse the relative significance of recycling and removal. Clearly, maintaining intracellular amino acid pools by recycling is important, especially when levels of environmentally available amino acids are low, as discussed above. Damage removal is important because it eliminates damage after exposure to ROS whereas cellular oxidant defense systems inactivate ROS before damage occurs. This is particularly valuable for non-dividing cells wherein damage cannot be “diluted out” by cellular growth and division, as discussed above. Autophagy is the only known mechanism for degrading large intracellular structures and organelles such as mitochondria, the importance of which has been codified in the lysosomal-mitochondrial axis theory of aging (Terman et al. 2006). Recent studies of ours indicate that when amino acids are limiting, autophagy is needed to turnover damaged mitochondria in order to promote chronological longevity, but under certain conditions of amino acid availability mitochondrial damage is reduced and autophagy is not required for chronological longevity (Aris et al. unpublished results). This suggests that interrelationship between amino acid availability, mitochondrial damage, and autophagy is important for chronological longevity.
Amino Acid Homeostasis and Extension of Life Span by Calorie Restriction
CR is the most widely recognized intervention for extension of life span and there has been considerable debate about the mechanism(s) by which CR extends life span in yeast and other species, including the way in which CR affects mitochondrial ROS production. Multiple signaling pathways have been implicated in life span extension by CR, including the insulin/insulin-like growth factor (I/IGF), Ras/protein kinase A, and TOR pathways (Stanfel et al. 2009). These pathways play crucial roles in integrating information from growth signals and levels of available nutrients, including levels of amino acids. In budding yeast, inhibition or abrogation of TOR signaling appears to mimic CR and promotes chronological longevity via a cell autogenous mechanism that involves modulation of mitochondrial translation and aerobic energy production (Bonawitz et al. 2007; Cheng et al. 2007; Powers et al. 2006). Similarly, inactivation of the TOR pathway extends CLS in Drosophila (Kapahi et al. 2004), and inhibition of TOR or translation extends life span in C. elegans (Hansen et al. 2007; Henderson et al. 2006; Pan et al. 2007). Multiple downstream targets have been the focus of studies seeking to elucidate underlying mechanisms, including up-regulation of multiple stress response pathways that impact production, repair, accumulation, and clearance of different types of molecular damage (Cuervo 2008b; Guarente 2008).
An exciting development in the field is the emerging importance of diet composition in life span extension by CR. It is becoming increasingly clear that CR-mediated life span extension is not simply a matter of reduced caloric intake and that amino acid sensing and utilization play key roles (Mair et al. 2005; Piper et al. 2005). Restriction of specific essential amino acids has been found to extend life span in animals and appears to do so by mimicking CR. Reducing the amount of the essential amino acid methionine can mimic many of the effects of CR / DR in rats, including increased life span (Orentreich et al. 1993). A diet deficient in methionine also extends life span in mice and yields many of the physiological benefits associated with CR (Miller et al. 2005). A careful analysis of the effects of specific essential amino acids in Drosophila reveals that methionine restriction confers extended life span without the decrease in fecundity observed under traditional CR conditions (Grandison et al. 2009). Moreover, the Drosophila I/IGF signaling pathway links amino acid sensing to reduced life span when amino acids are abundant (Grandison et al. 2009). The benefit of restricting nutrients, rather than calories, likely involves, at least in part, a reduction in oxidative stress and damage. Methionine restriction reduces oxidative damage and increases mitochondrial biogenesis as well as expression of an uncoupling protein in rat brain (Naudi et al. 2007). Reduced consumption of protein, but not fat or carbohydrates, leads to lower levels of ROS and oxidative stress in rat liver, which may be attributable to methionine restriction (Ayala et al. 2007). These results highlight the importance of amino acids as modulators of life span and point to amino acid sensing pathways and effects on translation and energy production / utilization as potential mediators.
Finally, the availability of sugars / carbon sources is integrated with the availability of amino acids / nitrogen sources to influence chronological longevity. Glucose is unique in terms of the metabolic preference exhibited for this sugar by many organisms including yeast. In yeast, sugar / carbon source availability is detected by the Ras/PKA pathway whereas amino acid / nitrogen source availability is detected by the TOR pathway. When glucose is readily available, S. cerevisiae grows with minimal respiration, even under aerobic conditions, a phenomenon known as the Crabtree effect. To achieve this, numerous genes involved in aerobic respiration and utilization of alternative carbon sources are subject to glucose repression (Carlson 1999; Verstrepen et al. 2004). In contrast, Kluyveromyces lactis is a budding yeast closely related to S. cerevisiae that is Crabtree negative, meaning that glucose fails to down-regulate respiration and mitochondrial function when oxygen is available for aerobic respiration. An interesting result in this regard is that K. lactis does not appear to enjoy extension of CLS due to CR in low glucose medium (Oliveira et al. 2008).
One possibility is that S. cerevisiae favors fermentation and rapid growth when glucose is plentiful but must shift to aerobic respiration to achieve chronological longevity when glucose is exhausted. This physiological strategy to ferment glucose and produce ethanol may have conferred a selective advantage to yeast during evolution because of the protection of environmental resources afforded by ethanol (Thomson et al. 2005), and its oxidation product acetic acid. But, when glucose is exhausted, S. cerevisiae must carry out extensive metabolic reprogramming to aerobic metabolism to support long-term survival in the non-dividing state. This metabolic and proteomic reprogramming takes place during the transition from rapid fermentative growth in log phase to slow aerobic growth during the diauxic and post-diauxic phases to low level aerobic respiration in stationary phase. If there is sufficient availability of amino acids and non-fermentable carbon sources (including certain amino acids), then yeast are able to complete this transition to aerobic metabolism, which supports chronological longevity.
Concluding Perspectives
As our knowledge of how diet influences longevity has increased, relatively simple ideas have given way to increasingly complex ones. The idea that longevity is simply a matter of reducing caloric intake and respiration-dependent energy production has given way to a more complex set of ideas that relate nutrient availability to cellular metabolic decision-making, damage control, and longevity. Dietary nutrient balance is emerging as an important longevity factor that affects a multitude of metabolic processes, including amino acid and protein homeostasis. But, many significant questions remain. For example, increased essential amino acid availability supports chronological longevity in yeast, yet limitation of a specific essential amino acid in mice and flies brings about extension of life span, apparently in a manner that mimics CR. Perhaps the most exciting aspect of the relationship between nutrient balance and longevity is the potential for human dietary intervention. Altering the composition of the diet offers fundamental advantages over adding a pharmacological compound to the diet. Perhaps the most important advantage is the potential for widespread acceptance and implementation in society. Put simply, a “nutrient balance” strategy for extending life span and/or health span has the potential to reach a much larger portion of our population than a “calorie restriction” strategy.
Acknowledgements
We are grateful for the support that we have received from the NIH (AG023719 to JPA; AG17994 to CL; CA95552 to WAD), including the Claude D. Pepper Older Americans Independence Center (AG028740), and the University of Florida Institute on Aging. We acknowledge the Honors Program at the University of Florida, which has facilitated the participation of undergraduate students in our research efforts.
Abbreviations
- BCAA
branched side-chain amino acids
- CLS
chronological life span
- CR
calorie restriction
- GAAC
general amino acid control
- ISC
iron sulfur cluster
- NCR
nitrogen catabolite repression
- ROS
reactive oxygen species
- TOR
target of rapamycin
References
- Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- Andrews ZB. Uncoupling protein-2 and the potential link between metabolism and longevity. Curr Aging Sci. 2010;3:102–112. doi: 10.2174/1874609811003020102. [DOI] [PubMed] [Google Scholar]
- Aragon AD, Rodriguez AL, Meirelles O, Roy S, Davidson GS, Tapia PH, Allen C, Joe R, Benn D, Werner-Washburne M. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol Biol Cell. 2008;19:1271–1280. doi: 10.1091/mbc.E07-07-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G. Role of mitochondrial DNA in human aging. Mitochondrion. 2002;2:27–37. doi: 10.1016/s1567-7249(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Ayala V, Naudi A, Sanz A, Caro P, Portero-Otin M, Barja G, Pamplona R. Dietary protein restriction decreases oxidative protein damage, peroxidizability index, and mitochondrial complex I content in rat liver. J Gerontol A Biol Sci Med Sci. 2007;62:352–360. doi: 10.1093/gerona/62.4.352. [DOI] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci USA. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Daran JM, Almering MJ, de Winde JH, Pronk JT. Contribution of the Saccharomyces cerevisiae transcriptional regulator Leu3p to physiology and gene expression in nitrogen- and carbon-limited chemostat cultures. FEMS Yeast Res. 2005;5:885–897. doi: 10.1016/j.femsyr.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol Cell Biol. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1093/genetics/166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschlen S, Amillet JM, Guiard B, Fournier A, Marcireau C, Bolotin-Fukuhara M. The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genomics. 2003;4:37–46. doi: 10.1002/cfg.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ding Q, Keller JN. The stationary phase model of aging in yeast for the study of oxidative stress and age-related neurodegeneration. Biogerontology. 2005;6:1–13. doi: 10.1007/s10522-004-7379-6. [DOI] [PubMed] [Google Scholar]
- Chen Q, Thorpe J, Ding Q, El-Amouri IS, Keller JN. Proteasome synthesis and assembly are required for survival during stationary phase. Free Radic Biol Med. 2004;37:859–868. doi: 10.1016/j.freeradbiomed.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Cheng C, Fabrizio P, Ge H, Wei M, Longo VD, Li LM. Significant and systematic expression differentiation in long-lived yeast strains. PLoS One. 2007;2:e1095. doi: 10.1371/journal.pone.0001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008a;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Calorie restriction and aging: the ultimate “cleansing diet”. J Gerontol A Biol Sci Med Sci. 2008b;63:547–549. doi: 10.1093/gerona/63.6.547. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- Cyrne L, Martins L, Fernandes L, Marinho HS. Regulation of antioxidant enzymes gene expression in the yeast Saccharomyces cerevisiae during stationary phase. Free Radic Biol Med. 2003;34:385–393. doi: 10.1016/s0891-5849(02)01300-x. [DOI] [PubMed] [Google Scholar]
- Davidson GS, Joe RM, Roy S, Meirelles O, Allen CP, Wilson MR, Tapia PH, Manzanilla EE, Dodson AE, Chakraborty S, Carter M, Young S, Edwards B, Sklar L, Werner-Washburne M. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol Biol Cell. 2011;22:988–998. doi: 10.1091/mbc.E10-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006a;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006b;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38:519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- Dietrich MO, Horvath TL. The role of mitochondrial uncoupling proteins in lifespan. Pflugers Arch. 2010;459:269–275. doi: 10.1007/s00424-009-0729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon EL, Sheffield-Moore M, Paddon-Jones D, Gilkison C, Sanford AP, Casperson SL, Jiang J, Chinkes DL, Urban RJ. Amino acid supplementation increases lean body mass, basal muscle protein synthesis, and insulin-like growth factor-I expression in older women. J Clin Endocrinol Metab. 2009;94:1630–1637. doi: 10.1210/jc.2008-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Rasmussen BB. Leucine-enriched nutrients and the regulation of mammalian target of rapamycin signalling and human skeletal muscle protein synthesis. Curr Opin Clin Nutr Metab Care. 2008;11:222–226. doi: 10.1097/MCO.0b013e3282fa17fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumlao DS, Hertz N, Clarke S. Secreted 3-isopropylmalate methyl ester signals invasive growth during amino acid starvation in Saccharomyces cerevisiae. Biochemistry. 2008;47:698–709. doi: 10.1021/bi7018157. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, SelverstoneValentine J, Gralla EB, Longo VD. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span – from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido EO, Grant CM. Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol Microbiol. 2002;43:993–1003. doi: 10.1046/j.1365-2958.2002.02795.x. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Gomes P, Sampaio-Marques B, Ludovico P, Rodrigues F, Leao C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech Ageing Dev. 2007;128:383–391. doi: 10.1016/j.mad.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Mitochondria – a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Harman D. The free radical theory of aging. Antioxid Redox Signal. 2003;5:557–561. doi: 10.1089/152308603770310202. [DOI] [PubMed] [Google Scholar]
- Harris N, Bachler M, Costa V, Mollapour M, Moradas-Ferreira P, Piper PW. Overexpressed Sod1p acts either to reduce or to increase the lifespans and stress resistance of yeast, depending on whether it is Cu(2+)-deficient or an active Cu,Zn-superoxide dismutase. Aging Cell. 2005;4:41–52. doi: 10.1111/j.1474-9726.2005.00142.x. [DOI] [PubMed] [Google Scholar]
- Harris N, Costa V, MacLean M, Mollapour M, Moradas-Ferreira P, Piper PW. Mnsod overexpression extends the yeast chronological (G0) life span but acts independently of Sir2p histone deacetylase to shorten the replicative life span of dividing cells. Free Radic Biol Med. 2003;34:1599–1606. doi: 10.1016/s0891-5849(03)00210-7. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK. Stationary phase in yeast. Curr Opin Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- Hu Y, Cooper TG, Kohlhaw GB. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol Cell Biol. 1995;15:52–57. doi: 10.1128/mcb.15.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard VM, Valdor R, Macian F, Cuervo AM. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology. 2011 doi: 10.1007/s10522-011-9331-x. doi:10.1007/s10522-011-9331-x. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Rtg2 protein: at the nexus of yeast longevity and aging. FEMS Yeast Res. 2005;5:1253–1259. doi: 10.1016/j.femsyr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Jones EW, Fink GR. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast saccharomyces metabolism and gene expression. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 1982. pp. 181–299. [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Kohlhaw GB. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67:1–15. doi: 10.1128/MMBR.67.1.1-15.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman A, Daran-Lapujade P, Fullaondo A, Olsthoorn MM, Pronk JT, Slijper M, Heck AJ. Proteome analysis of yeast response to various nutrient limitations. Mol Syst Biol. 2006;2:2006 0026. doi: 10.1038/msb4100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 2005;12(Suppl 2):1484–1489. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laun P, Heeren G, Rinnerthaler M, Rid R, Kossler S, Koller L, Breitenbach M. Senescence and apoptosis in yeast mother cell-specific aging and in higher cells: a short review. Biochim Biophys Acta. 2008;1783:1328–1334. doi: 10.1016/j.bbamcr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Layman DK. The role of leucine in weight loss diets and glucose homeostasis. J Nutr. 2003;133:261S–267S. doi: 10.1093/jn/133.1.261S. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Longo VD. Ras: the other pro-aging pathway. Sci Aging Knowledge Environ. 2004;2004:pe36. doi: 10.1126/sageke.2004.39.pe36. [DOI] [PubMed] [Google Scholar]
- Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- Longo VD, Liou LL, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- Longo VD, Mitteldorf J, Skulachev VP. Opinion: programmed and altruistic ageing. Nat Rev Genet. 2005;6:866–872. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas C, Edwards-Ingram LC, Zeef L, Shenton D, Ashe MP, Grant CM. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarney MJ, Pellett PL, Young VR. Pattern of amino acid requirements in humans: an interspecies comparison using published amino acid requirement recommendations. J Nutr. 1996;126:1871–1882. doi: 10.1093/jn/126.7.1871. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Merry BJ. Oxidative stress and mitochondrial function with aging – the effects of calorie restriction. Aging Cell. 2004;3:7–12. doi: 10.1046/j.1474-9728.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward DJ, Layman DK, Tome D, Schaafsma G. Protein quality assessment: impact of expanding understanding of protein and amino acid needs for optimal health. Am J Clin Nutr. 2008;87:1576S–1581S. doi: 10.1093/ajcn/87.5.1576S. [DOI] [PubMed] [Google Scholar]
- Miquel J, Economos AC, Fleming J, Johnson JE., Jr Mitochondrial role in cell aging. Exp Gerontol. 1980;15:575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnson JR. Life spans of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Müller I, Zimmermann M, Becker D, Flomer M. Calendar life span versus budding life span of Saccharomyces cerevisiae. Mech Ageing Dev. 1980;12:47–52. doi: 10.1016/0047-6374(80)90028-7. [DOI] [PubMed] [Google Scholar]
- Murin R, Hamprecht B. Metabolic and regulatory roles of leucine in neural cells. Neurochem Res. 2008;33:279–284. doi: 10.1007/s11064-007-9444-4. [DOI] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Naudi A, Caro P, Jove M, Gomez J, Boada J, Ayala V, Portero-Otin M, Barja G, Pamplona R. Methionine restriction decreases endogenous oxidative molecular damage and increases mitochondrial biogenesis and uncoupling protein 4 in rat brain. Rejuvenation Res. 2007;10:473–484. doi: 10.1089/rej.2007.0538. [DOI] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Oliveira GA, Tahara EB, Gombert AK, Barros MH, Kowaltowski AJ. Increased aerobic metabolism is essential for the beneficial effects of caloric restriction on yeast life span. J Bioenerg Biomembr. 2008;40:381–388. doi: 10.1007/s10863-008-9159-5. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrella E, Longo VD. Insulin/IGF-I and related signaling pathways regulate aging in nondividing cells: from yeast to the mammalian brain. Scient World J. 2010;10:161–177. doi: 10.1100/tsw.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencharz PB, Ball RO. Different approaches to define individual amino acid requirements. Annu Rev Nutr. 2003;23:101–116. doi: 10.1146/annurev.nutr.23.011702.073247. [DOI] [PubMed] [Google Scholar]
- Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Piper MD, Mair W, Partridge L. Counting the calories: the role of specific nutrients in extension of life span by food restriction. J Gerontol A Biol Sci Med Sci. 2005;60:549–555. doi: 10.1093/gerona/60.5.549. [DOI] [PubMed] [Google Scholar]
- Piper PW. Long-lived yeast as a model for ageing research. Yeast. 2006;23:215–226. doi: 10.1002/yea.1354. [DOI] [PubMed] [Google Scholar]
- Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev. 2006;127:733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kennedy BK, Kaeberlein M. Genome-wide identification of conserved longevity genes in yeast and worms. Mech Ageing Dev. 2007;128:106–111. doi: 10.1016/j.mad.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Soeters PB, van de Poll MC, van Gemert WG, Dejong CH. Amino acid adequacy in pathophysiological states. J Nutr. 2004;134:1575S–1582S. doi: 10.1093/jn/134.6.1575s. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Kriete A, Sacan A, Jazwinski SM. Comparing the yeast retrograde response and NF-kappaB stress responses: implications for aging. Aging Cell. 2010;9:933–941. doi: 10.1111/j.1474-9726.2010.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles C. How to set up a yeast laboratory. Methods Enzymol. 2002;350:42–71. doi: 10.1016/s0076-6879(02)50955-1. [DOI] [PubMed] [Google Scholar]
- Tang L, Liu X, Clarke ND. Inferring direct regulatory targets from expression and genome location analyses: a comparison of transcription factor deletion and overexpression. BMC Genomics. 2006;7:215. doi: 10.1186/1471-2164-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Georis I, Feller A, Dubois E, Cooper TG. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J Biol Chem. 2009;284:2522–2534. doi: 10.1074/jbc.M806162200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem-Biol Interact. 2006;163:29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Sass M, Klionsky DJ. The regulation of aging: does autophagy underlie longevity? Trends Cell Biol. 2009;19:487–494. doi: 10.1016/j.tcb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Iserentant D, Malcorps P, Derdelinckx G, Van Dijck P, Winderickx J, Pretorius IS, Thevelein JM, Delvaux FR. Glucose and sucrose: hazardous fast-food for industrial yeast? Trends Biotechnol. 2004;22:531–537. doi: 10.1016/j.tibtech.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MA, Liou LL, Martins J, Clement MH, Bailey S, Longo VD, Valentine JS, Gralla EB. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem. 2004;279:32055–32062. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- Ward WF. Protein degradation in the aging organism. Prog Mol Subcell Biol. 2002;29:35–42. doi: 10.1007/978-3-642-56373-7_3. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Klionsky DJ. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda) 2008;23:248–262. doi: 10.1152/physiol.00013.2008. [DOI] [PubMed] [Google Scholar]
- Young VR, Borgonha S. Nitrogen and amino acid requirements: the Massachusetts Institute of Technology amino acid requirement pattern. J Nutr. 2000;130:1841S–1849S. doi: 10.1093/jn/130.7.1841S. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]