Abstract
Objective
To identify a neuroimaging signature predictive of brain amyloidosis as a screening tool to identify individuals with mild cognitive impairment (MCI) that are most likely to have high levels of brain amyloidosis or to be amyloid-free.
Methods
The prediction model cohort included 62 MCI subjects with structural magnetic resonance imaging (MRI) and carbon-11-labeled Pittsburgh Compound B (11C-PiB) positron emission tomography (PET). We identified an anatomical shape variation based neuroimaging predictor of brain amyloidosis and defined a structural MRI-based brain amyloidosis score (sMRI-BAS). Amyloid beta positivity (Aβ+) predictive power of sMRI-BAS was validated on an independent cohort of 153 MCIs with cerebrospinal fluid (CSF) Aβ1-42 biomarker data but no amyloid PET scans. We compared the Aβ+ predictive power of sMRI-BAS to those of Apolipoprotein E (ApoE) genotype and hippocampal volume, two most relevant candidate biomarkers for the prediction of brain amyloidosis.
Results
Anatomical shape variations predictive of brain amyloidosis in MCI embraced a characteristic spatial pattern known for high vulnerability to Alzheimer’s disease (AD) pathology, including the medial temporal lobe, temporal-parietal association cortices, posterior cingulate, precuneus, hippocampus, amygdala, caudate, and fornix/stria terminals. Aβ+ prediction performance of sMRI-BAS and ApoE genotype jointly was significantly better than the performance of each predictor separately (AUC=0.88 versus AUC=0.70 and AUC=0.81, respectively) with greater than 90% sensitivity and specificity at 20% FPR and FNR thresholds. Performance of hippocampal volume as an independent predictor of brain amyloidosis in MCI was only marginally better than random chance (AUC=0.56).
Interpretation
As one of the first attempts to use an imaging technique that does not require amyloid-specific radioligands for identification of individuals with brain amyloidosis, our findings could lead to development of multidisciplinary/multimodality brain amyloidosis biomarkers that are reliable, minimally invasive, and widely available.
INTRODUCTION
Increasing evidence from in vivo imaging and post mortem studies indicates that the formation of amyloid plaques in the brain, mainly consisting of insoluble amyloid beta (Aβ) protein fragments, is a major factor that leads to a sequence of pathophysiological events occurring over two to three decades before the manifestation of Alzheimer’s Disease (AD) related dementia1. Brain amyloidosis can be detected by molecular imaging techniques such as positron emission tomography (PET) using an amyloid-specific radioligand (e.g., 11C-labelled Pittsburgh compound-B (11C-PiB)) or through measurement of cerebrospinal fluid (CSF) Aβ1-42 concentration2–4, both showing high correlations with postmortem measures of fibrillar Aβ5–7. As amyloid PET and CSF biomarker collection rapidly become an integral part of research studies on normal aging and AD8, 9, the research findings have raised the possibility that biomarkers of brain amyloidosis may detect and quantify disease associated changes in the latent (i.e., normal cognitive function with evidence of AD pathology) and prodromal (i.e., mild cognitive impairment with stronger evidence of AD pathology) AD stages when disease-modifying therapeutic intervention may be most effective. However, many subjects are resistant to lumbar puncture required for CSF sample collection, especially if they are asymptomatic. Furthermore, limited availability, exposure to radiation, and financial burden of amyloid PET techniques hinder their widespread acceptance in clinical practice. Recently, three independent studies have shown that a combination of AD-biomarkers, cognitive measures, ApoE genotype, and plasma protein measures, could be a predictor of amyloid-positivity (Aβ+) with accuracy high as 85%10–12. Different yet complementary to these approaches, our goal is to identify a low-cost and non-invasive neuroimaging signature predictive of brain amyloidosis as a screening tool to identify subjects that are most likely to have high levels of brain amyloid or to be amyloid-free.
Structural neuroimaging using magnetic resonance imaging (MRI), has been shown to be useful in AD13 diagnosis and is often used for assessment of subjects with memory problems, yet there is no protocol for identifying brain amyloidosis via MRI since no MR-based molecular imaging with a contrast specific to amyloid plaques has been demonstrated. Widespread availability, no radioligand requirement, no exposure to radiation, and lower cost are potential advantages of structural neuroimaging as a screening tool to identify brain amyloidosis. Various imaging studies related the brain amyloidosis to the downstream structural brain alterations across a wide range of cognitive impairments to better understand the sequences of AD-related pathophysiological events, to identify biomarkers of early AD-related brain changes, and to assess the effectiveness of these biomarkers on predicting individuals’ progression in the AD continuum14–20. Though neuroimaging research came a long way in its ability to link brain amyloidosis and AD-related neurodegeneration, to what extend structural imaging predicts brain amyloidosis has not been explored thoroughly yet. In this study, we aimed (1) to identify an anatomical shape variation based neuroimaging predictor of brain amyloidosis in mild cognitive impairment (MCI), a transitional stage between normal aging and dementia with an increased risk of developing AD, and (2) to define a structural MRI-based brain amyloidosis score (sMRI-BAS), whose Aβ+ predictive power was validated on an independent MCI cohort. Furthermore, we compared the Aβ+ predictive power of the sMRI-BAS to those of Apolipoprotein E (ApoE) genotype and hippocampal volume, two most relevant candidate biomarkers for the prediction of brain amyloidosis. Specifically, hippocampal atrophy, one of the most established and reliable AD imaging biomarker, progresses through the disease course, and is present even in healthy elders and MCIs who are most like to develop AD21. The relevance of ApoE-ε4 genotype for AD and its associations with AD-related biomarkers such as amyloid PET binding, CSF biomarker concentrations, structural, especially hippocampal atrophy, are well documented22–24.
METHODS
Participants
Participants were recruited through the Alzheimer’s Disease Neuroimaging Initiative (ADNI) from 56 centers in the U.S. and Canada25. Written consent was obtained from all subjects participating in the ADNI study. The ADNI study was approved by the institutional review board at each participating site. The study complies with the Health Insurance Portability and Accountability Act (HIPAA) guidelines. Details regarding subject inclusion and exclusion criteria are provided elsewhere25 (see Supplementary Material).
Modeling cohort consisted of 46 cognitively normal elderly individuals (CN) and 62 MCI subjects. Baseline MRIs of CN subjects were used to model the normal confounding effects of age, sex, and education on anatomical shape variation measure. Given that cumulative and regional Aβ burden in CNs correlate with regional brain atrophy, particularly in hippocampus, parietal cortex, and posterior cingulate regions, in a pattern similar to AD-related atrophy pattern16, 26, modeling cohort included only the CNs identified as Aβ− either by PiB-PET or CSF biomarker data. Furthermore, ApoE ε2-allele or ApoE ε4-allele carrier CNs were excluded due to sample size limitations. 62 MCI subjects in the prediction model cohort had an ADNI imaging session when both structural MRI and 11C-PiB PET scans were acquired. A brain amyloidosis prediction model and an sMRI-BAS model were derived from the modeling MCI cohort data. Validation cohort to assess the Aβ+ predictive power of the sMRI-BAS consisted of an additional 153 MCIs, who had baseline CSF Aβ1-42 biomarker data but no amyloid PET at any time points during ADNI project period. Individuals with ApoE ε2-allele were excluded from the modeling and the validation cohorts due to sample size limitations. Study group demographics are summarized in Table 1. We downloaded all related 11C-PiB PET, CSF biomarker, and structural MRI data from http://adni.loni.ucla.edu/.
Table 1.
Study group demographics
| Prediction model cohort | Validation cohort | ||
|---|---|---|---|
|
| |||
| CN (@baseline) | MCI (@ 1st 11C-PiB PET imaging) | MCI (@baseline) | |
| N | 46 | 62 | 153 |
| Age, years | 75.87 ± 5.38 | 74.24 ± 7.77 | 74.43 ± 7.52 |
| Sex, % female | 51% | 32% | 35% |
| Education, years | 15.91 ± 2.22 | 16.26 ± 2.77 | 15.47 ± 3.00 |
| ApoE- ε3/ε4 carriers, % | 0% | 42% | 40% |
| ApoE- ε4/ε4 carriers, % | 0% | 11% | 11% |
| MMSE | 29.20 ± 0.92 | 26.98 ± 2.331 | 26.72 ± 1.861 |
| CDR-SB | 0.02 ± 0.10 | 2.24 ± 1.601 | 1.61 ± 0.921,2 |
| ADAS-Cog | 6.00 ± 3.08 | 11.11 ± 5.411 | 11.94 ± 4.531 |
|
| |||
| Composite PiB-SUVR | - | 1.63 ± 0.29 | - |
| % Aβ+ (i.e., Composite PiB-SUVR > 1.478) | - | 66% | - |
|
| |||
| CSF Aβ1-42 | 243.00 ± 22.99 | 162.23 ± 54.071, 3 | 162.29 ± 53.831 |
| % Aβ+ (i.e., CSF Aβ1-42 < 1929) | 0% | 76%3 | 73% |
Two sample t-test group difference in comparison to CN at p<10−9 level
Two sample t-test group difference in comparison to prediction model MCI cohort at p=0.017 level
Based on 34 MCI subjects who had baseline CSF protein assay
Measures of brain amyloidosis
The PET-Core of ADNI developed standardized protocols to compute uniform resolution 11C-PiB PET images normalized to the mean 11C-PiB retention value of the cerebellar cortex, i.e., 11C-PiB standardized uptake value ratio (SUVR), and further provided average PiB-SUVR measures of automatically identified cortical and subcortical regions3. We used a composite PiB-SUVR measure as the average PiB-SUVR of frontal, lateral/mesial temporal, parietal, anterior cingulate, and precuneus regions. The Biomarker-Core of ADNI who conducts studies on ADNI-derived lumbar puncture CSF samples provided measures of CSF Aβ1-42 in standardized assays2. Further details are provided in Supplementary Material.
Structural MRI
The participants at each site underwent a standardized 1.5 Tesla MRI protocol. Two T1-weighted MRIs, using a sagittal volumetric magnetization prepared rapid gradient echo (MP-RAGE) sequence, with an echo time (TE) of 4ms, repetition time (TR) of 9ms, flip angle of 8°, and a nominal voxel size of 0.94×0.94×1.2mm3. A designated center selected the MP-RAGE image with higher quality and corrected for system-specific image artifacts such as geometry distortion, B1 non-uniformity, and intensity inhomogeneities25, 27.
Anatomical shape variations
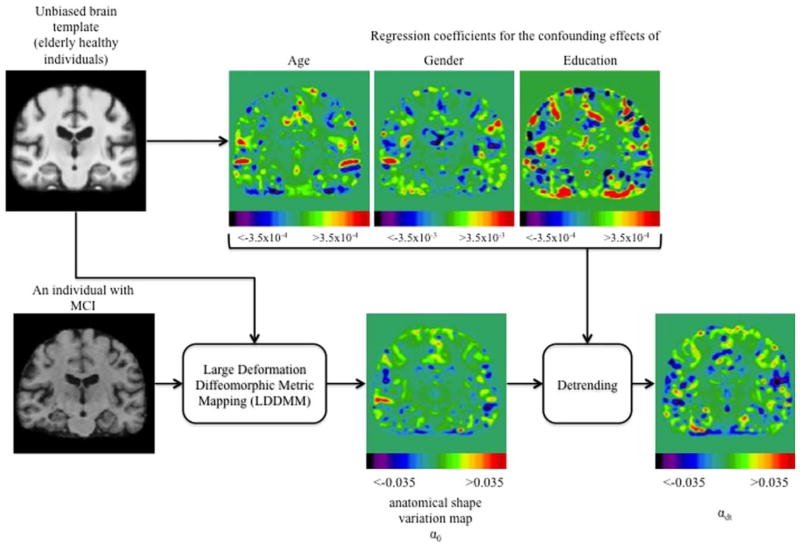
Skull, scalp, and extra-cranial tissue were removed from each T1-weighted MRI using the automated Brain Surface Extraction (BSE) software28, followed by manual refinement if required. To avoid bias towards a particular subject’s geometry in analysis of anatomical shape variations, we used the MRI data from CNs to create a study-specific unbiased large deformation brain image template (ULD-template) by applying a framework of large deformation diffeomorphic metric mapping (LDDMM) as described in full elsewhere29. ULD-template generation incorporated an unbiased approach where all brain images were first simultaneously affine transformed to adjust for global variations in brain positioning and scale, and then simultaneously deformed. MCI brain images were first affine aligned and then nonlinearly warped to this ULD-template using the LDDMM framework. The LDDMM was modeled as an evolution in time, with an associated smooth velocity vector field controlling this evolution. A scalar initial momentum map, α0 parameterized the entire geodesic with which the optimal trajectory emanated from the ULD-template to reach a subject brain image on a Riemannian manifold of diffeomorphism29. These momentum maps uniquely encoded the anatomical shape variations of individual brains relative to the ULD-template.
Detrending α0 maps
We used a linear detrending method in terms of general linear model to control for normal confounding effects of age, sex, and education on the brain anatomy based on the anatomical shape variations in the CN cohort. Robustness of the fit was achieved by weighting the least squares residual at each iterative step using a bisquare weighting function. For each MCI subject, a detrended (i.e., corrected for confounding effects) deformation momenta map, αdt, was computed voxel-wise by removing each subject’s individual age, sex, and education variations from the original estimate of deformation momenta α0 (see Supplementary Material).
High dimensionality neuroimaging predictors of brain amyloidosis
To find the anatomical shape variations that best predict brain Aβ burden, we performed a partial least squares (PLS)30 regression of the composite PiB-SUVR measure against the αdt of MCIs from the prediction model cohort. PLS regression determines the principal mode within a set of multiple explanatory variables (e.g., αdt values from every ULD-template imaging voxel) to predict a second set of response variable (e.g., composite PiB-SUVR)30. Once the mode, also known as latent variable (LV), have been determined, a set of weights can be calculated to predict an instance of the response variable from an explanatory variable (e.g., imaging data from a new subject). Specifically, by projecting αdt onto the LV, an sMRI-BAS was estimated for each MCI. We used non-parametric permutation tests (n=1000) to assess the significance of the regression of projected composite PiB-SUVR on projected αdt map and used the R2 as the test statistics. The distribution of the R2 statistic under the null hypothesis was calculated by randomly reordering the αdt map and composite PiB-SUVR pairings and then recalculating the new projection and the associated R2 each time. The significance at the 5% level of the LV was measured by the p-value from the empirical distribution.
Assessing the Aβ+ predictive power of sMRI-BAS in the validation MCI cohort
Only a small fraction of the ADNI MCI cohort received PiB-PET, limiting our statistical power for building and validating a brain amyloidosis prediction model using a single MCI cohort. However, the two measures of brain amyloidosis obtained by either PET or CSF sample are substantially related to one another regardless of whether evaluated as continuous or dichotomous variables. Specifically, dichotomous categorization (i.e., Aβ+ versus Aβ−) showed 91% agreement (κ=0.74) between PiB-PET and CSF Aβ1-4231. Majority of the disagreements between PiB-PET and CSF Aβ1-42 involved subjects whose biomarker values were close to the Aβ+/− dichotomization cut-offs. In this study, CSF Aβ1-42 levels of the MCI subjects in the validation cohort were outside the ±4 range of CSF Aβ1-42 cut-off. Taking advantage of the strong agreement between dichotomized PiB-PET and CSF Aβ1-42 measures, we assessed the Aβ+ predictive power of sMRI-BAS in a validation MCI cohort with CSF Aβ1-42 biomarker data but no PiB-PET scans. We performed an Aβ+ versus Aβ− classification analysis on the validation cohort of 153 MCIs using a linear logistic regression model with sMRI-BAS as the independent predictor. The validation cohort was dichotomized into Aβ+ and Aβ− subgroups based on a CSF Aβ1-42 biomarker cutoff (i.e., Aβ+ if CSF Aβ1-42 < 192) derived and validated in an autopsy-confirmed AD cohort2. Receiver Operation Characteristic (ROC) curve metrics including the area under the curve (AUC), sensitivity at 20% false positive rate (FPR), and specificity at 20% false negative rate (FNR) were used to assess the performance of the sMRI-BAS as a classifier. Furthermore, we compared the performance of sMRI-BAS to the performances of ApoE genotype (ApoE-ε3/ε3, ApoE-ε3/ε4, or ApoE-ε4/ε4) and intra-cranial vault volume (ICV)-adjusted hippocampal volume as independent predictors of Aβ+ in MCI. FreeSurfer (version 4.5) estimates of hippocampal volume and ICV were downloaded from the ADNI database. We also considered joint predictive power of sMRI-BAS and ApoE genotype in a multimodality Aβ+ classification model.
RESULTS
Demographic characteristics of the subjects are shown in Table 1. There were no significant age, sex, and education group differences among the three cohorts. As expected, MCIs had significantly (p<10−4) lower MMSE and higher CDR-SB and ADAS-Cog scores relative to the CNs. In terms of the cognitive performance, the two MCI groups differed only in their CDR-SB scores (p=0.017).
Neuroimaging predictors of brain amyloidosis
Regression coefficients for controlling normal confounding effects of age, sex, and education as well as the raw and detrended (i.e., α0 and αdt, respectively) anatomical shape variation maps are shown in Fig. 1.
FIGURE 1.
Flowchart of the creation of maps of anatomical shape variation, controlling for the confounding effects including age, sex, and education. MCI = mild cognitive impairment. [Color figure can be viewed in the online issue, which is available at www.annalsofneurology.org.]
Fig. 2 shows the spatial signature of the LV inferred by PLS regression. Dark red/blue and white colors indicate greater contribution of the local anatomical shape variations to the LV, therefore to the brain amyloidosis prediction model. The anatomical shape variations predictive of brain amyloidosis were concentrated on the inferior parietal, precuneus, entorhinal, supramarginal, middle temporal cortices, hippocampus, amygdala, caudate nucleus, and fornix/stria terminals.
FIGURE 2.
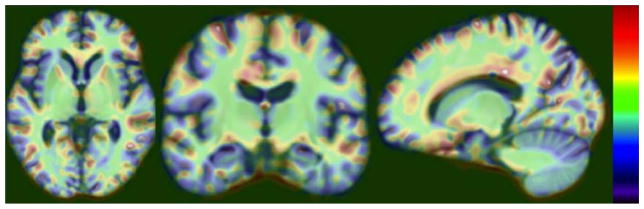
Neuroimaging predictors of brain amyloidosis in mild cognitive impairment. Spatial signature of the latent variable inferred by partial least squares regression for the prediction of brain amyloidosis is shown. The dependent outcome measure is the composite Pittsburgh compound B standardized uptake value ratio. The independent predictors are the detrended anatomical shape variation measures from every imaging voxel in the unbiased brain template space. [Color figure can be viewed in the online issue, which is available at www.annalsofneurology.org.]
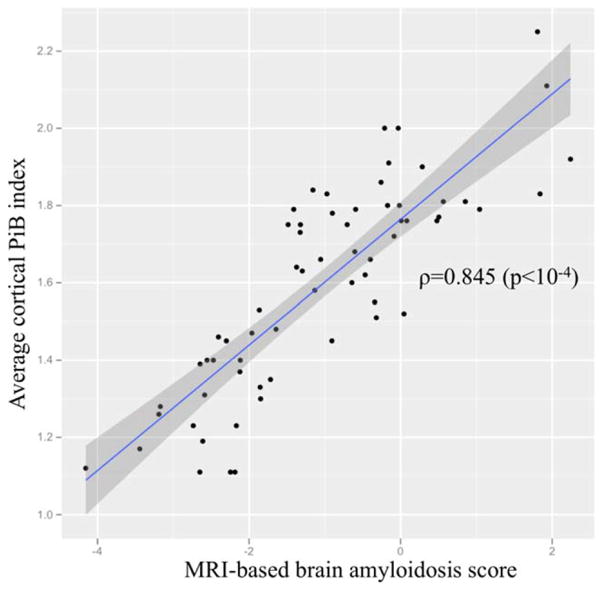
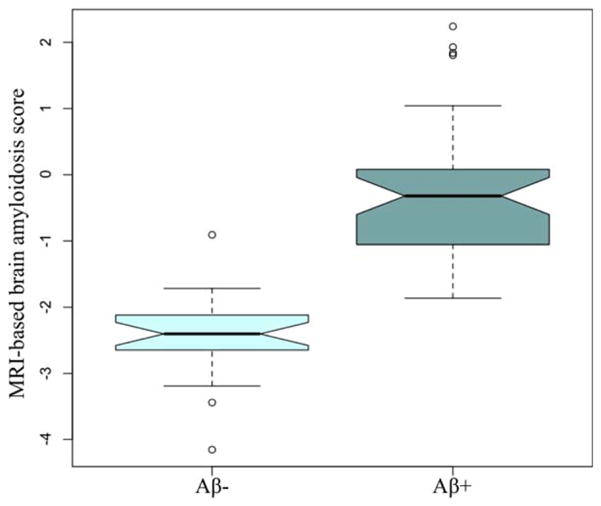
Fig. 3 shows the relation of sMRI-BAS against the actual composite PiB-SUVR for MCIs in the prediction model cohort, with an estimated correlation coefficient of 0.845 (p<10−4). Distributions of the sMRI-BAS for Aβ+ and Aβ− MCIs in the prediction model cohort are box-plotted in Fig. 4 to illustrate the excellent group separation. The established composite PiB-SUVR threshold of 1.47 based on a PiB-PET versus CSF biomarker study3 was used to dichotomize the prediction model MCIs as Aβ+ and Aβ−.
FIGURE 3.
Structural magnetic resonance imaging (MRI)-based brain amyloidosis score against actual composite Pittsburgh compound B (PiB) standardized uptake value ratio measures for mild cognitive impairment subjects in the prediction model cohort. [Color figure can be viewed in the online issue, which is available at www.annalsofneurology.org.]
FIGURE 4.
Distributions of the structural magnetic resonance imaging (MRI)-based brain amyloidosis score for amyloid beta-positive (Aβ+; ie, composite Pittsburgh compound B standardized uptake value ratio > 1.47) and Aβ− MCI subjects in the prediction model cohort. [Color figure can be viewed in the online issue, which is available at www.annalsofneurology.org.]
Aβ+ predictive power of the sMRI-BAS in the validation MCI cohort
The estimated performances of the proposed sMRI-BAS, ApoE genotype, and hippocampal volume as independent predictors of brain amyloidosis in MCI are summarized in Table 2. As an independent predictor of brain amyloidosis in MCI, ApoE genotype outperformed both sMRI-BAS and hippocampal volume neuroimaging measures (AUC=0.81 versus AUC=0.70 and AUC=0.56, respectively). sMRI-BAS as an independent predictor of brain amyloidosis in MCI performed with greater sensitivity (84.81% at 20% FPR threshold) and specificity (81.25% at 20% FNR threshold) compared to ApoE genotype and hippocampal volume. Performance of hippocampal volume as a predictor of brain amyloidosis in MCI was only marginally better than random chance (AUC=0.56). Finally, the joint performance of sMRI-BAS and ApoE genotype as predictors of brain amyloidosis in MCI was significantly better than the performance of each predictor separately (AUC=0.88) with greater than 90% sensitivity and specificity at 20% FPR and FNR thresholds.
Table 2.
Estimated performances of the proposed sMRI-BAS, ApoE genotype, and hippocampal volume in predicting Aβ+ in the validation MCI cohort.
| Brain amyloidosis predictor | Area under the ROC curve | Sensitivity at 20% FPR | Specificity at 20% FNR |
|---|---|---|---|
| sMRI-BAS | 0.70 ± 0.05 | 84.81 ± 10.73 | 81.25 ± 8.53 |
| ApoE genotype | 0.81 ± 0.021 | 80.80 ± 2.641 | 50.90 ± 3.461 |
| Hippocampal volume (ICV-adjusted) | 0.56 ± 0.041,2,3 | 58.44 ± 8.461,2 | 57.89 ± 9.311,2 |
| sMRI-BAS + ApoE genotype | 0.88 ± 0.031,2 | 90.97 ± 4.491,2 | 92.82 ± 7.171,2 |
Statistical significance (p<10−6) in two-sample t-test comparison to the performance of sMRI-BAS as an independent predictor of brain amyloidosis in MCI.
Statistical significance (p<10−6) in two-sample t-test comparison to the performance of ApoE genotype as an independent predictor of brain amyloidosis in MCI.
Statistically no different than random chance.
ROC: Receiver Operating Characteristic
ICV: Intra-Cranial Vault volume
FPR: False Positive Rate
FNR: False Negative Rate
Subgroup analysis of the validation MCI cohort by median age split
To assess the performance of amyloid prediction models in different age groups of the validation MCI cohort, age was dichotomized using a median split (median age = 74.9 years), yielding a younger MCI validation cohort (MCI-Y) and an older MCI validation cohort (MCI-O). The group demographics and the performance of brain amyloidosis predictors for each age group are summarized in Table 3. The MCI-Y and MCI-O groups did not differ on any cognitive test scores, suggesting comparable MCI symptom severity between the two age groups. MCI-Y included a higher percent of female subjects compared to MCI-O. As an independent predictor of brain amyloidosis, sMRI-BAS performed similarly in both age groups, i.e., AUC = 0.69 in discriminating Aβ+ versus Aβ− MCI-Y and AUC = 0.71 in discriminating Aβ+ versus Aβ− MCI-O. The performance of ApoE genotype as an independent predictor of brain amyloidosis was better in MCI-O group than in MCI-Y group (AUC = 0.77 versus AUC = 0.70), but slightly worse in both age groups than in full validation cohort. Joint performance of sMRI-BAS and ApoE genotype in predicting the brain amyloidosis was better in MCI-O than in MCI-Y (AUC = 0.88 versus AUC = 0.81). In MCI-Y, ICV-adjusted hippocampal volume was a predictor of the brain amyloidosis with an AUC = 0.75. ICV-adjusted hippocampal volume neuroimaging measure was not able to discriminate Aβ+ versus Aβ− MCI-O at a rate any better than chance.
Table 3.
Subgroup analyses of the validation MCI cohort
| Median age split | Median CDR-SB split | |||
|---|---|---|---|---|
|
| ||||
| MCI-Y | MCI-O | MCI-E | MCI-L | |
| N | 71 | 78 | 61 | 53 |
| Age, years | 68.35 ± 4.97 | 80.23 ± 3.911 | 76.03 ± 6.91 | 72.59 ± 7.354 |
| Sex, % female | 46% | 24%1 | 30% | 42% |
| Education, years | 15.52 ± 3.02 | 15.42 ± 3.06 | 15.33 ± 3.21 | 15.2 ± 2.75 |
| ApoE-ε3/ε4 carriers, % | 48% | 40% | 34% | 51%4 |
| ApoE-ε4/ε4 carriers, % | 17% | 6% | 16% | 9% |
| MMSE | 26.87 ± 1.80 | 26.55 ± 1.91 | 27.00 ± 1.99 | 26.62 ± 1.78 |
| CDR-SB | 1.59 ± 0.85 | 1.63 ± 1.00 | 0.80 ± 0.25 | 2.61 ± 0.774 |
| ADAS-Cog | 11.89 ± 4.99 | 12.04 ± 4.14 | 11.05 ± 4.37 | 13.11 ± 4.394 |
|
| ||||
| CSF Aβ1-42 | 155.78 ± 50.02 | 168.18 ± 56.66 | 177.41 ± 63.86 | 145.12 ± 34.384 |
| % Aβ+ (i.e., CSF Aβ1-42 < 1929) | 76% | 70% | 61% | 88%4 |
|
| ||||
| Brain amyloidosis predictor | Area under the ROC curve | |||
|
| ||||
| sMRI-BAS | 0.69 ± 0.06 | 0.71 ± 0.05 | 0.68 ± 0.05 | 0.87 ± 0.054 |
| ApoE genotype | 0.70 ± 0.042 | 0.77 ± 0.041,2 | 0.75 ± 0.052 | 0.67 ± 0.082,4 |
| Hippocampal volume (ICV-adjusted) | 0.75 ± 0.062,3 | 0.59 ± 0.051, 2,3 | 0.61 ± 0.062,3 | 0.75 ± 0.092,3, 4 |
| sMRI-BAS + ApoE-ε4 genotype | 0.81 ± 0.052,3 | 0.88 ± 0.041, 2,3 | 0.84 ± 0.052,3 | 0.92 ± 0.042,3, 4 |
Two sample t-test or Fisher’s exact test group difference in comparison to MCI-Y at p<0.001 level
Statistical significance (p<10−6) in two-sample t-test comparison to the performance of sMRI-BAS as an independent predictor of brain amyloidosis in MCI.
Statistical significance (p<10−6) in two-sample t-test comparison to the performance of ApoE genotype as an independent predictor of brain amyloidosis in MCI.
Two sample t-test or Fisher’s exact test group difference in comparison to MCI-E at p<0.001 level
Subgroup analysis of the validation MCI cohort by median CDR-SB split
The validation MCI cohort was dichotomized to early versus late MCI cases (MCI-E and MCI-L, respectively) using a median CDR-SB split (median CDR-SB=1.5) to indirectly assess the influence of MCI symptom severity on Aβ+ predictive power of the sMRI-BAS, ApoE genotype, and ICV-adjusted hippocampal volume (Table 3). MCI-E was significantly older than MCI-L, suggesting later age of onset in MCI-E group. Percentage of ApoE-ε3/ε4 carriers in MCI-L was significantly greater than the percentage in MCI-E. CSF Aβ1-42 levels in MCI-L were significantly lower than the levels in MCI-E with a greater percentage of Aβ+ cases. Although both imaging measures, sMRI-BAS and ICV-corrected hippocampal volume, independently and jointly with ApoE genotype yielded better discrimination of Aβ+ versus Aβ− subjects in MCI-L compared to their performances in MCI-E, the relatively higher level of Aβ+ versus Aβ− imbalance in MCI-L makes generalization of these results difficult.
DISCUSSION
In this study, we used a novel measure of regional anatomical shape variation and PLS regression to determine the neuroimaging predictors of brain amyloidosis and to derive an sMRI-BAS, whose predictive power in identifying Aβ+ and Aβ− MCIs were measured. Anatomical shape variations predictive of brain amyloidosis (detected by 11C-PiB PET) in MCI embraced a characteristic pattern of brain structures known for high vulnerability to AD pathology. The sMRI-BAS when combined with ApoE genotype, the most relevant genotype for AD, yielded a brain amyloidosis prediction with AUC=0.88 and greater than 90% sensitivity and specificity at 20% FPR and FNR thresholds in identifying Aβ+ and Aβ− MCIs in an independent validation cohort. Although as an independent predictor of brain amyloidosis ApoE genotype outperformed neuroimaging measures, we observed a significant 9% improvement in Aβ+ versus Aβ− classification accuracy and a drastic increase in sensitivity and specificity of the prediction model when sMRI-BAS and ApoE genotype jointly act as the predictors of brain amyloidosis. These results implicate independent contribution of each predictor in identifying MCIs with brain amyloidosis and the added value of the MRI measures beyond the effects of ApoE genotype. This study represents one of the first attempts to use an imaging technique that do not require amyloid-specific radioligands for identification of individuals with brain amyloidosis. Although, the dosimetry of amyloid PET is suitable for clinical and research applications, the effective doses range from 1.6 to 3.7 mSv32, 33, on the average greater than the natural radiation exposure of 2.0 mSv per year. Thus, amyloid PET contributes to a patient’s overall long-term cumulative radiation exposure, which is associated with an increased risk of cancer34.
The spatial extent of neuroimaging predictors of brain amyloidosis observed in the prediction model MCI cohort is consistent with previous studies on biomarkers for early diagnosis of AD25. Multiple groups have now reported associations between Aβ levels and brain tissue atrophy rates, especially the hippocampal and medial temporal cortical atrophy rates and ventricular expansion, even at mild stages of cognitive deficits15, 35, 36. Furthermore, brain atrophy in a characteristic pattern involving the medial temporal lobes (i.e., hippocampus and entorhinal cortex), paralimbic and temporoparietal cortices is a biomarker of AD-related neurodegeneration25. A surprising finding in this study is though the poor performance of hippocampal volume in classification of Aβ+ and Aβ− MCIs. One explanation could be the fact that both prediction model and validation cohorts included primarily amnestic MCIs to whom atrophy within the hippocampal complex25 is common but might not be differentiating. Several research groups have independently investigated the relationship between brain amyloidosis and hippocampal volume in MCIs. It has been consistently reported that the global, regional, and also voxel-based correlations between 11C-PiB PET brain amyloidosis measures and hippocampal volume are not significant15, 16, 37. Similarly, correlation between CSF Aβ1-42 and hippocampal volume failed to reach statistical significance in various MCI studies38–40.
Our finding of a distributed spatial pattern of anatomical shape variations as the neuroimaging predictor of brain amyloidosis suggests a greater involvement of a memory network including hippocampal complex in amyloidosis-related pathology in MCI. The transneuronal-spread hypothesis41 proposes that an injury in a vulnerable site triggers a process, which then spreads across interconnected components. In a similar manner, the anatomical shape variations at the primary AD-pathology brain sites (e.g., middle temporal regions) may have driven the contribution from the brain regions traditionally not considered as AD-pathology-specific via the brain’s intrinsic connectivity.
Strengths of this study include the use of an advanced anatomical shape variation measure, unique detrending of cofounding effects, and use of a modern statistical analysis method. Most of the earlier studies on characterization of neuroanatomical changes have focused on the study of deformation maps, either using the associated Jacobian determinate of the transformations, as in the now ubiquitous deformation-based morphometry42, or have done the analysis directly on the displacement maps. Rather than using the associated Jacobian determinate of transformations or the vector-valued velocity or deformation fields, we formulated the prediction problem in terms of scalar initial momenta maps that completely encoded the geodesics on the manifold of diffeomorphisms. Use of a modern statistical method, PLS, allowed us to identify co-varying brain networks, providing maximum predictive value for brain amyloidosis in MCI. Conventional voxel-based methods, such as statistical parametric mapping, are not suitable for prediction models where the data from all imaging voxels are the high-dimensional and partly collinear predictors. When the space of the explanatory variables is of full rank, ordinary multilinear regression can be used but in practice the explanatory variable space is likely to be singular due to collinearity in the predictors. PLS is designed to deal with small sample size and multicollinearity issues in high-dimensional prediction modeling and searching for modes that explain maximal covariance between the explanatory and response spaces30.
A relative weakness of this study is related to the etiologic and pathologic uncertainty in the MCI diagnosis as at least 30% of the MCI subjects have been found to harbor a non-AD pathology43, 44. Specific to our prediction model cohort, 8% of the MCI subjects were diagnosed with additional non-memory features, 6% were identified as due to etiology other than AD, and about 4% presented with depressive symptoms. These heterogeneities in the prediction model MCI cohort may have contributed to the brain amyloidosis and atrophy covariations in unexpected brain regions including the mid-brain and basal ganglia regions45, 46. Validation cohort MCI subjects that are identified as due to etiology other than AD and/or with non-memory features presented with relatively lower MRI-BAS (i.e., mean MRI-BAS of −1.73 compared to mean MRI-BAS of −1.35 estimated for amnestic MCI due to AD), resulting in greater false positive rates in predicting brain amyloidosis in this subgroup. A proper study with a larger sample size is warranted to better model the etiological and pathological heterogeneities in prediction of brain amyloidosis in MCI. Another limitation of our study is that both CSF and amyloid-PET biomarkers are proxy measures for the retention of fibrillar form of Aβ and not necessarily provide a measure of oligomeric form of Aβ accumulation, which might be the most relevant to synaptic toxicity.
The pathophysiological process of AD is thought to begin many years before the diagnosis of AD dementia. An understanding of the neuroanatomy associated with brain amyloidosis is significant because it may aid to identify individuals in prodromal phase of AD providing a critical opportunity for therapeutic intervention. The extend to which biomarkers of AD pathology --- e.g., Aβ accumulation and structural alterations --- predict an MCI subject’s subsequent clinical course remains to be determined since some of these individuals with AD pathology will never manifest AD related dementia in their lifetime. Multi-center studies enriched with neuroimaging and biomarker data will ensure future studies focused on the prediction of brain amyloidosis in a wider AD cognitive continuum including the latent AD stage. Together with the work of others10, 11, our findings could result in development of multidisciplinary and multimodality biomarkers of brain amyloidosis that are reliable, minimally invasive, simple to perform, and widely available. These may include detailed demographic characteristics such as family history, cognitive performance, and high-throughput biomarker technologies such as genomics and proteomics.
Supplementary Material
Acknowledgments
This work was funded by a research resource grant from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (grant: P41 RR023953). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is Rev March 26, 2012 coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. The work was possible by using resources of the Veterans Affairs Medical Center, San Francisco, California.
References
- 1.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica. 1991;82(4):239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 2.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Annals of Neurology. 2009;65(4):403–13. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jagust WJ, Bandy D, Chen K, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s and Dementia. 2010;6(3):221–9. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister-James J, Pontecorvo MJ, Clark C, et al. Florbetapir F-18: A Histopathologically Validated Beta-Amyloid Positron Emission Tomography Imaging Agent. Seminars in Nuclear Medicine. 2011;41(4):300–4. doi: 10.1053/j.semnuclmed.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Clark CM, Pontecorvo MJ, Beach TG, et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-β plaques: a prospective cohort study. The Lancet Neurology. 2012;11(8):669–78. doi: 10.1016/S1474-4422(12)70142-4. [DOI] [PubMed] [Google Scholar]
- 6.Driscoll I, Troncoso J, Rudow G, et al. Correspondence between in vivo 11C-PiB-PET amyloid imaging and postmortem, region-matched assessment of plaques. Acta Neuropathologica. 2012:1–9. doi: 10.1007/s00401-012-1025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark CM, Xie S, Chittams J, et al. Cerebrospinal Fluid Tau and β-Amyloid: How Well Do These Biomarkers Reflect Autopsy-Confirmed Dementia Diagnoses? Arch Neurol. 2003 Dec 1;60(12):1696–702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- 8.Rabinovici GD, Jagust WJ. Amyloid imaging in aging and dementia: Testing the amyloid hypothesis in vivo. Behavioural Neurology. 2009;21(1):117–28. doi: 10.3233/BEN-2009-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordberg A. Amyloid imaging in Alzheimer’s disease. Current Opinion in Neurology. 2007;20(4):398–402. doi: 10.1097/WCO.0b013e3281a47744. [DOI] [PubMed] [Google Scholar]
- 10.Apostolova L, Hwang K, Kohannim O, editors. Predicting brain amyloidosis in MCI using clinical, cognitive, imaging and peripheral blood protein measures; Alzheimer’s Association International Conference; 2012 July 14–19; Vancouver, British Columbia, Canada. [Google Scholar]
- 11.Mielke MM, Wiste HJ, Weigand SD, et al. Indicators of amyloid burden in a population-based study of cognitively normal elderly. Neurology. 2012 Oct 9;79(15):1570–7. doi: 10.1212/WNL.0b013e31826e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahar-Fuchs A, Villemagne V, Ong K, et al. Prediction of amyloid-beta pathology in amnestic mild cognitive impairment with neuropsychological tests. J Alzheimers Dis. 2013;33(2):451–62. doi: 10.3233/JAD-2012-121315. [DOI] [PubMed] [Google Scholar]
- 13.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia. 2011;7(3):263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Jr, Lowe VJ, Senjem ML, et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain. 2008 Mar 1;131(3):665–80. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chetelat G, Villemagne VL, Bourgeat P, et al. Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Annals of Neurology. 2010;67(3):317–24. doi: 10.1002/ana.21955. [DOI] [PubMed] [Google Scholar]
- 16.Bourgeat P, Chetelat G, Villemagne VL, et al. β-Amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010 Jan 12;74(2):121–7. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- 17.Archer HA, Edison P, Brooks DJ, et al. Amyloid load and cerebral atrophy in Alzheimer’s disease: An 11C-PIB positron emission tomography study. Annals of Neurology. 2006;60(1):145–7. doi: 10.1002/ana.20889. [DOI] [PubMed] [Google Scholar]
- 18.Mormino EC, Kluth JT, Madison CM, et al. Episodic memory loss is related to hippocampal-mediated β-amyloid deposition in elderly subjects. Brain. 2009 May 1;132(5):1310–23. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vemuri P, Wiste HJ, Weigand SD, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: Diagnostic discrimination and cognitive correlations. Neurology. 2009 Jul 28;73(4):287–93. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vemuri P, Wiste HJ, Weigand SD, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: Predicting future clinical change. Neurology. 2009 Jul 28;73(4):294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apostolova LG, Dutton RA, Dinov ID, et al. Conversion of Mild Cognitive Impairment to Alzheimer Disease Predicted by Hippocampal Atrophy Maps. Arch Neurol. 2006 May 1;63(5):693–9. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- 22.Vemuri P, Wiste HJ, Weigand SD, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Annals of Neurology. 2010;67(3):308–16. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pievani M, Rasser PE, Galluzzi S, et al. Mapping the effect of APOE ε4 on gray matter loss in Alzheimer’s disease in vivo. Neuro Image. 2009;45(4):1090–8. doi: 10.1016/j.neuroimage.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuff N, Woerner N, Boreta L, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009 Apr 1;132(4):1067–77. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiner MW, Veitch DP, Aisen PS, et al. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s and Dementia. 2012;8(1 Supplement):S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker JA, Hedden T, Carmasin J, et al. Amyloid-beta associated cortical thinning in clinically normal elderly. Ann Neurol. 2011 Jun;69(6):1032–42. doi: 10.1002/ana.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack CR, Bernstein MA, Fox NC, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002 Jun;6(2):129–42. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 29.Jiang T, Navab N, Pluim J, et al., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2010. Springer; Berlin/Heidelberg: 2010. Multivariate Statistical Analysis of Deformation Momenta Relating Anatomical Shape to Neuropsychological Measures. [DOI] [PubMed] [Google Scholar]
- 30.Wold S, Geladi P, Esbensen K, Öhman J. Multi-way principal components-and PLS-analysis. Journal of Chemometrics. 1987;1(1):41–56. [Google Scholar]
- 31.Jagust WJ, Landau SM, Shaw LM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009 Oct 13;73(15):1193–9. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheinin NM, Tolvanen TK, Wilson IA, Arponen EM, Nagren KA, Rinne JO. Biodistribution and radiation dosimetry of the amyloid imaging agent 11C-PIB in humans. J Nucl Med. 2007 Jan;48(1):128–33. [PubMed] [Google Scholar]
- 33.O’Keefe GJ, Saunder TH, Ng S, et al. Radiation dosimetry of beta-amyloid tracers 11C-PiB and 18F-BAY94-9172. J Nucl Med. 2009 Feb;50(2):309–15. doi: 10.2967/jnumed.108.055756. [DOI] [PubMed] [Google Scholar]
- 34.Griffey RT, Sodickson A. Cumulative radiation exposure and cancer risk estimates in emergency department patients undergoing repeat or multiple CT. AJR Am J Roentgenol. 2009 Apr;192(4):887–92. doi: 10.2214/AJR.08.1351. [DOI] [PubMed] [Google Scholar]
- 35.Tosun D, Schuff N, Mathis CA, Jagust W, Weiner MW Initiative AsDN. Spatial patterns of brain amyloid-β burden and atrophy rate associations in mild cognitive impairment. Brain. 2011 Apr 1;134(4):1077–88. doi: 10.1093/brain/awr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosun D, Schuff N, Truran-Sacrey D, et al. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiology of Aging. 2010;31(8):1340–54. doi: 10.1016/j.neurobiolaging.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chetelat G, Villemagne VL, Pike KE, et al. Independent contribution of temporal beta-amyloid deposition to memory decline in the pre-dementia phase of Alzheimer’s disease. Brain. 2011 Mar 1;134(3):798–807. doi: 10.1093/brain/awq383. [DOI] [PubMed] [Google Scholar]
- 38.Apostolova LG, Hwang KS, Andrawis JP, et al. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiology of Aging. 2010;31(8):1284–303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hampel H, Burger K, Pruessner JC, et al. Correlation of Cerebrospinal Fluid Levels of Tau Protein Phosphorylated at Threonine 231 With Rates of Hippocampal Atrophy in Alzheimer Disease. Arch Neurol. 2005 May 1;62(5):770–3. doi: 10.1001/archneur.62.5.770. [DOI] [PubMed] [Google Scholar]
- 40.Henneman WJP, Vrenken H, Barnes J, et al. Baseline CSF p-tau levels independently predict progression of hippocampal atrophy in Alzheimer disease. Neurology. 2009 Sep 22;73(12):935–40. doi: 10.1212/WNL.0b013e3181b879ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de LaCoste M-C, White CL. The role of cortical connectivity in Alzheimer’s disease pathogenesis: A review and model system. Neurobiology of Aging. 1993;14(1):1–16. doi: 10.1016/0197-4580(93)90015-4. [DOI] [PubMed] [Google Scholar]
- 42.Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: Deformation-based morphometry. Human Brain Mapping. 1998;6(5–6):348–57. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephan BCM, Hunter S, Harris D, et al. The neuropathological profile of mild cognitive impairment (MCI): a systematic review. Mol Psychiatry. 2012;17(11):1056–76. doi: 10.1038/mp.2011.147. [DOI] [PubMed] [Google Scholar]
- 44.Schneider JA, Aggarwal NT, Barnes L, Boyle P, Bennett DA. The neuropathology of older persons with and without dementia from community versus clinic cohorts. J Alzheimers Dis. 2009;18(3):691–701. doi: 10.3233/JAD-2009-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madsen SK, Ho AJ, Hua X, et al. 3D maps localize caudate nucleus atrophy in 400 Alzheimer’s disease, mild cognitive impairment, and healthy elderly subjects. Neurobiology of Aging. 2010;31(8):1312–25. doi: 10.1016/j.neurobiolaging.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Looi JC, Rajagopalan P, Walterfang M, et al. Differential putaminal morphology in Huntington’s disease, frontotemporal dementia and Alzheimer’s disease. Aust N Z J Psychiatry. 2012 Dec;46(12):1145–58. doi: 10.1177/0004867412457224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.