Abstract
It is widely accepted that cancer results from an array of epigenetic and genetic alterations, particularly aberrant epigenetic patterns that are a hallmark of every cancer type studied. Another well-known feature of cancer cells is the array of abnormalities in their nuclear structure. Although it is known that nuclear structure has an important role in the regulation of gene expression, we know little about the direct relationship between nuclear structural alterations and aberrant epigenetic patterns in cancer. Here, we discuss some of the recent studies from our lab and others to understand the relationship between alterations of nuclear architecture and aberrant epigenetic patterns in cancer cells. Although the precise relationship remains elusive, we suggest that changes in nuclear structure and composition could alter long-range genomic interactions and cause global epigenetic changes during tumorigenesis. We emphasize the need for further studies to elucidate the direct relationship between nuclear structure alterations and aberrant epigenetic patterns in cancers.
Multiple chapters in this volume emphasize the key role of nuclear structure in “packaging” DNA, via its organization by histone and nonhistone proteins, to subserve its gene expression and structural function. Epigenetic mechanisms are obviously intimately tied to this context of nuclear structure. The purpose of this chapter is to explore how nuclear structure may relate to epigenetically controlled abnormalities of gene expression in cancer.
EPIGENETIC DEREGULATION IN CANCER
Cancer cells undergo global changes in gene expression compared with their normal counterparts. An important observation regarding tumors is that key regulatory genes have been shown to undergo silencing by epigenetic processes at various stages of tumorigenesis, including very early stages (Jones and Baylin 2007). Hundreds of genes have been observed to undergo silencing by de novo promoter DNA methylation in various cancer types. DNA methylation is the covalent modification of cytosines to 5-methylcytosine (5mC) at cytosine-phospho-guanine (CpG) dyads that are enriched, in so-called “CpG islands,” at more than half of the gene promoters in the human genome. Methylated CpGs are bound by specific proteins, such as MeCP2, that contain a methyl-CpG binding domain (MBD) that in turn recruits various chromatin remodelers and histone modifiers that mediate gene silencing. It has been established that the silencing is ultimately mediated by promoter CpG methylation, histone modifications, and nucleosome remodeling (Jones and Baylin 2007; Cedar and Bergman 2009). Aberrantly hypermethyated genes show a drastic decrease of the activating H3K4Me2 mark and variable retention of the inactivating H3K27Me3 mark at their promoters (McGarvey et al. 2008).
One of the key unanswered questions in cancer epigenetics is how patterns of de novo methylation arise during tumorigenesis. One model postulates that abnormal de novo methylation in cancer arises stochastically due to abnormal overexpression of DNA methyltransferase 1 (DNMT1) (Vertino et al. 1996; De Marzo et al. 1999) and events involving loss of gene function that are advantageous to tumor formation are, therefore, naturally selected. However, a series of more recent observations has fostered the hypothesis that for many of the abnormally DNA-methylated genes in cancer, the de novo DNA methylation reflects a “program(s)” that renders hundreds of genes vulnerable to undergoing this change. Thus, studies from three different labs show that hypermethylated gene promoters in cancer tend to be significantly enriched for genes marked by the long-term silencing protein complex, Polycomb (PcG) in embryonic stem (ES) and progenitor cells (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). Furthermore, global analysis of DNA methylation patterns have revealed that de novo methylation in cancer can occur in clusters of genes and is targeted to promoters that are already repressed in normal tissue (Keshet et al. 2006). Developmentally, the above PcG regulation serves to maintain the genes at a low level of transcription in ES or embryonic progenitor cells without contribution from promoter DNA methylation. This helps to keep these cells in a totipotent or multipotent state until the genes are activated, or further repressed, via signals for lineage commitment (Bernstein et al. 2006; Mikkelsen et al. 2007; Meissner et al. 2008). These above relationships support the possibility of an instructive mechanism wherein many cancer-specific, hypermethylated genes are targeted at promoters that are already repressed and marked by PcG in normal tissues or cancer progenitor cells (Ohm and Baylin 2007). Interestingly, tumor cells are especially competent in initiating X-chromosome inactivation, a property unique only to cells of the early embryo and hematopoietic stem cells during a narrow window in their differentiation (Agrelo and Wutz 2009). A direct interpretation of these latter observations is that tumor cells seem to have some parallel mechanisms to those of stem cells in their ability to cause global epigenetic reprogramming. However, tumor cells couple this with the DNA methylation machinery to induce silencing of various target genes to a more stable silencing state. Especially for true tumor-suppressor genes, this can programmatically give cancer cells a selective advantage.
Other features of cancer-specific epigenetic deregulation remain confounding. For instance, in addition to the local gain in CpG methylation and changes in chromatin modifications at CpG-island promoters in cancer cells, there is a global reduction in 5mC content (Cadieux et al. 2006; Ehrlich 2009). Global hypomethylation can potentially lead to changes in genomic instability that might induce tumor formation (Gaudet et al. 2003). Reduction in DNA methylation at the centromeric regions of chromosomes 1, 9, and 16 in ICF (immunodeficiency, centromeric region instability, and facial anomalies) syndrome is caused by mutations in DNMT3B, resulting in alterations of heterochromatin organization (Jefferson et al. 2010). Furthermore, of great interest are observations that large stretches of contiguous genes can undergo silencing, in a phenomenon termed long-range epigenetic silencing (LRES), which can be reactivated by 5-azacytidine, a drug that causes DNA demethylation (Frigola et al. 2006; Hitchins et al. 2007). Interestingly, not all genes in the loci that undergo such LRES harbor promoter hypermethylation. Explorations of the mechanisms underlying alterations of DNA methylation should take into account these features of epigenetic deregulation in cancer.
NUCLEAR ORGANIZATION OF GENE REGULATION
As covered by multiple chapters in this volume, the nucleus is a nonrandom organization of the genomic content and protein machineries in distinct nuclear domains. In addition to the compartmentalization of the nucleus into nucleoli, one of the earliest-discovered concepts in nuclear organization is that the genome is distributed as tightly packed heterochromatin near the nuclear envelope, and in the perinucleolar region, and as loosely packed euchromatin in the interior of the nucleus (Fedorova and Zink 2008). This organization is further manifest in the presence of more distinct spatiotemporal functional domains such as DNA replication and DNA-damage-repair foci, gene transcription foci, RNA splicing speckles, etc. (Spector 2003, 2006). Another key aspect of the above functional domains is that genes move and associate with specific nuclear domains depending on whether they are in an active or silent transcription state (Osborne et al. 2004; Ragoczy et al. 2006; Brown and Silver 2007). For example, it has been demonstrated that genes that are active cluster together when the gene is in the “on” (active) state. These observations are suggestive of a model wherein nuclear compartmentalization into functional domains serves to provide critical concentrations of regulatory molecules for optimal regulation of gene expression (Fraser and Bickmore 2007; Zhao et al. 2009).
Furthermore, chromosomes are nonrandomly distributed in the nucleus, with each occupying distinct regions termed chromosome territories (CTs) (Cremer et al. 2006). CTs are organized in such a manner that gene-dense chromosomes tend to be more internal, whereas gene-poor chromosomes tend to be found near the periphery. Such a distribution is most likely a consequence of the distribution of proteins that differentially occupies genetic elements and where in the nucleus that these proteins tend to crowd (Misteli 2007). Early on, it was shown that active genes tend to be more internally located, whereas inactive genes tend to be at the nuclear periphery. The causal relationship between gene positioning and gene expression has been of great interest (Takizawa et al. 2008). However, such generalized relationships between gene expression and radial position are challenged in recent studies that have demonstrated that genes may or may not undergo changes in radial position in response to expression changes (Meaburn and Misteli 2008; Takizawa et al. 2008). Thus, it rather seems that nuclear position with respect to functional domains per se is more instructive in regulating gene expression (Zhao et al. 2009; Kress et al. 2010). This is supported by an array of studies that showed that artificial relocation of gene loci to the perinuclear heterochromatin compartment induced silencing of some of the genes in the locus (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008). A well-studied model system to understand the relationship between gene expression and gene position is the changes in nuclear position of hematopoietic loci during differentiation of hematopoietic progenitors. Earlier studies had shown that these loci get positioned close to the pericentromeric heterochromatin when committed to lineage-specific silencing. A recent study showed that changes in gene expression and chromatin modification are followed by changes in gene position based on whether the gene will be permanently silenced or activated in later stages of differentiation (Guillemin et al. 2009). Thus, the general model that can be observed from these studies is that although genes can interact with regulatory factors anywhere in the nucleus and be expressed, genes subjected to permanent silencing might in some instances be positioned close to heterochromatic compartments.
STRUCTURAL AND COMPOSITIONAL CHANGES OF CANCER NUCLEI
At the cellular level, tumor cells show a variety of changes in their nuclear morphology, including changes in the nuclear envelope, laminar invaginations, chromatin structure, and number of nucleoli (Zink et al. 2004; Zaidi et al. 2007). Morphological changes in nuclear structure are so well documented that they form the primary basis in diagnosing and staging different tumor types (Coffey 2002). Changes in nucleolar activity, size, numbers, and location within the nucleus can be indicators to differentiate tumor from normal cells. In prostate cancer, nucleolar composition can be used to distinguish benign from metastatic cancer (Fischer et al. 2004). Various features of nuclear structure, such as perinuclear and perinucleolar heterochromatin organization, nuclear matrix proteins, nuclear lamina, and splicing speckles have been observed to be affected in in vitro models of breast cancer cells (Chandramouly et al. 2007). From the point of view of the spatial organization of the genome, accruing evidence suggests that cancer cells harbor many abnormalities in distribution patterns of centromeres, telomeres, genes, and chromosome territories (Cremer et al. 2003; Chuang et al. 2004; Sarkar et al. 2007; Meaburn and Misteli 2008). Although the majority of these studies have been done in cultured cells, analysis of breast cancer tissue has shown cancer-specific alterations in gene position (Meaburn et al. 2009). These changes in the nuclear distribution of genes have been proposed to have potential in enhancing diagnosis of breast cancer.
In addition to structural changes of the nuclei in cancer cells, proteomic analysis comparing the nuclear matrix of cancer and normal cells has uncovered important compositional differences in nuclear matrix proteins (Coffey 2002; Leman and Getzenberg 2008). Many nuclear matrix proteins are involved in organizing global chromatin loops/domains and can induce tumorigenesis in various model systems. Table 1 summarizes some well-studied matrix proteins in cancer cells.
Table 1.
Nuclear matrix proteins involved in organizing chromatin domain in nucleus that have been shown to be deregulated in various tumors
| Molecule | Properties | Association in cancer | References |
|---|---|---|---|
| SATB1 | Matrix protein that binds to matrix attachment region (MAR) and scaffold attachment region (SAR) sequences; organizes chromatin and chromatin loops | Expressed in aggressive and metastatic breast tumors; proposed to impart stem-cell-line epigenetic plasticity to cancer cells | Han et al. (2008); Agrelo and Wutz (2009); Patani et al. (2009) |
| SATB2 | Matrix protein | Expression correlated with tumor grade in breast and colon cancers | Patani et al. (2009); Wang et al. (2009) |
| CTCF | Zn-finger DNA-binding protein that binds to specific unmethylated DNA target sites; functions as transcription factor or chromatin insulator protein | Tumor-specific mutations in the Zn-finger domain; dysregulated posttranslational modification; CTCF-haploinsufficiency correlated with LOI of IGF2 in Wilms tumor | Mummert et al. (2005); Filippova (2008); Docquier et al. (2009); Witcher and Emerson (2009) |
| CTCFL (BORIS) | Zn-finger DNA-binding protein that binds to CTCF target sites independent of DNA methylation; embryonic male germ-cell-specific expression | Up-regulated in various cancer types; can potentially compete with CTCF for unmethylated CTCF target sites and influence gene expression | Klenova et al. (2002); Hong et al. (2005); Woloszynska-Read et al. (2007); D’Arcy et al. (2008) |
| SAFB | Binds to MARs/SARs | Expression correlated with disease outcome in breast cancers | Garee and Oesterreich (2009) |
| HMG proteins | Nonhistone chromatin-binding proteins | Genomic rearrangements and expression changes in tumors; causally related to unbalanced chromosomal rearrangements | Takaha et al. (2002); Fusco and Fedele (2007); Di Cello et al. (2008) |
Given that the cancer nucleus is disorganized, it is not known whether the changes in nuclear structure have a role in the global changes in DNA methylation in tumors or vice versa. The majority of current studies aimed at understanding epigenetic deregulation in cancer focus on promoter-specific changes in DNA methylation and histone modifications. Considering that large-scale changes in nuclear organization have been observed in cancer cells, it is imperative to understand how these organizational changes impinge on epigenetic deregulation. Changes in nuclear organization could potentially lead to altered long-range genomic interactions that could cause epigenetic deregulation in cancer cells. Thus, our view of the changes in gene expression and epigenetic deregulation in cancer-related genes has to accommodate the global changes in nuclear organization observed in cancer cells. Our knowledge of specific examples of this, however, is in its infancy. Below, we explore some aspects of changes in nuclear organization of the genome that might accompany specific abnormal epigenetic patterns in cancer cells.
ROLE OF NUCLEAR STRUCTURE ALTERATIONS IN EPIGENETIC DEREGULATION IN CANCER
The first systematic analysis of altered gene position, measured as a change in radial position of the gene as a function of gene-expression changes in an in vitro breast cancer model system, showed definite cancer-specific repositioning of various gene loci (Meaburn and Misteli 2008). However, no correlation between radial repositioning and altered gene expression was observed. This study clearly showed that radial gene position is not correlated with gene activity, unlike that projected from previous studies. Because the study did not analyze repositioning with respect to nuclear subcompartments, such as heterochromatic or euchromatic domains, it remains unclear whether there is any correlation between changes in gene position accompanying transformation and nuclear subcompartments. In a recent study, we analyzed the link between aberrantly hypermethylated genes and functional organization of the nucleus into euchromatic (H4K4Me3-labeled) and heterochromatic (H4K27Me3-labeled) domains in the nucleus (Easwaran et al. 2010). Genes that undergo frequent hypermethylation in colon cancer were compared in colon cancer cell lines where the genes are methylated and silenced versus those in which they are unmethylated and expressed. Interestingly, it was observed that the genes were positioned either at euchromatic or heterochromatic domains independent of methylation status. This suggests that gene position relative to euchromatic or heterochromatic domains might not be essential for maintaining, in cancer cells, the DNA hypermethylation status of CpG-island-containing promoters. Furthermore, it was observed that genomic loci that undergo long-range silencing of neighboring genes did not show a requirement to be in heterochromatic domains and that such silencing could also occur in euchromatic domains. Thus, silencing of genes individually, or in clusters, does not require positioning close to heterochromatic domains. However, it is possible that during initial stages of tumor development, large-scale changes in nuclear structure might have a role in the initial establishment of methylation states. A limitation of such studies has been the feasibility of doing the analyses in normal tissues. With advances in microscopy and image analysis of normal tissue sections, future studies should focus on directly comparing normal and tumor cells for nuclear structural alterations and changes in gene positions relative to a full repertoire of functional nuclear domains.
In our same above study, it was observed that MLH1, SFRP4, and HBB loci are consistently positioned close to the heterochromatic domain independent of its radial position. Even aneuploid SFRP4 and HBB loci were positioned close to perinuclear or perinucleolar regions, indicating that the alleles tend to associate with heterochromatic compartments independent of the radial position in the nucleus. Many studies have focused on the relationship between the radial position of genes and their expression status. Our observation, similar to that of Meaburn et al. (2009), indicates that the reference point in such gene position studies should be a functional domain, such as the heterochromatic domain analyzed in our study rather than the radial position.
The role of nuclear structural alterations in tumorigenesis is unclear. The nuclear self-organization model (Misteli 2007) would predict that the absence of gross changes in nuclear position of hypermethylated genes in our study could reflect the fact that differences in regional gene expression in the analyzed loci are not great enough to change the position of loci with respect to nuclear functional domains. This is supported by the observation that genes that showed association with heterochromatic foci or euchromatic foci tended to have low or high gene density, respectively, as observed in previous studies (Zinner et al. 2007).
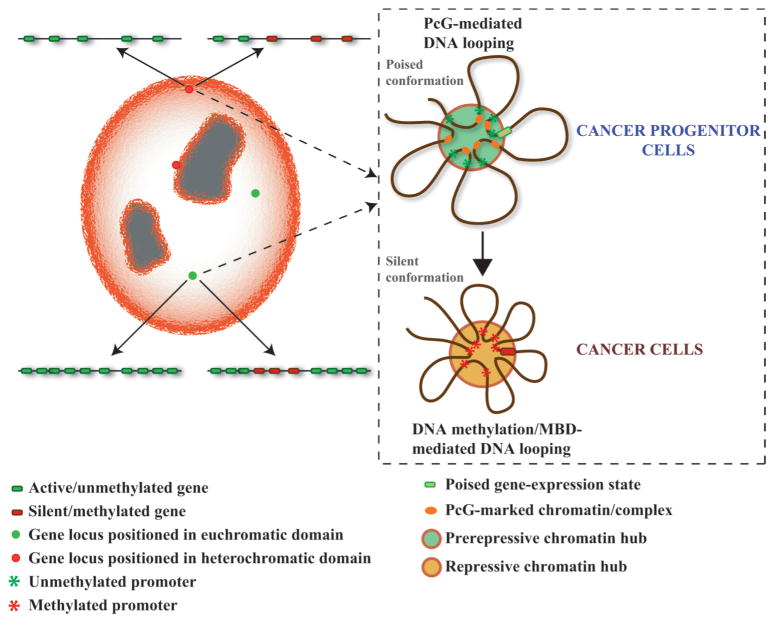
On the basis of some of these studies, it seems to be plausible that hypermethylated genes can exist in euchromatic or heterochromatic domains. However, it remains possible that finer, local changes that could contribute to epigenetic deregulation of individual genes or gene clusters, such as the local DNA looping and genomic interactions, could be altered in cancer cells even in the absence of drastic changes in gene position (Fig. 1). A few recent studies, described below, indicate that such increasingly gene-centric interactions may have a role in aberrant silencing in cancer.
Figure 1.
Genes positioned in euchromatic (green dots) and or heterochromatic (red dots) domains may undergo DNA- methylation-mediated gene silencing in cancer. (Left) Nucleus with regions of the nucleus visible as heterochromatic or euchromatic. Heterochromatin (orange) is shown to be present in the perinuclear and perinucleolar region (nucleoli are gray). Euchromatic or heterochromatic position of a locus is correlated with gene density with low or high gene density localized to heterochromatic or euchromatic regions, respectively. DNA-methylation-dependent silencing, including in the context of LRES (clusters of red bars), may occur at both euchromatic and heterochromatic domains. (Right) Role of DNA looping around genes with CpG-island-containing promoters. Repressed genes in the context of PcG-mediated loops have a low poised transcription, characteristic of many genes in ES cells that become DNA hypermethylated in adult cancers. This looping, mediated by PcG proteins and H3K27Me3 marks (orange ovals), maintains the gene in a prerepressive chromatin hub (pre-RCH). In cancer cells, DNA methylation of the gene promoter and other CpG islands (red asterisks) in the vicinity mediates formation of tighter DNA looping via methylcytosine-binding protein complexes to form the RCH. Frequency of long-range interactions with the RCH increases and interactions are stabilized, resulting in stable gene silencing. Poised or silent chromatin conformation could occur in regions of the nucleus that are euchromatic or heterochromatic.
First, the potential role of DNA looping across a broad region has been studied for GATA-4, a gene frequently DNA hypermethylated in colon cancer (Tiwari et al. 2008). In embryonic carcinoma (EC) cells, this gene is held in a state of low, but poised, expression in a multiloop configuration associated with the Polycomb mark H3K27Me3 and EZH2 that spans the whole gene. In this state, phosphorylated polymerase II (Pol II) is associated with the transcription start site. This configuration dissolves completely following activation of GATA-4 expression, with no need for changes in levels of associated Pol II, during retinoic-acid-induced differentiation of the cells. In a colon cancer cell line, where GATA-4 is silenced in association with DNA hypermethylation, the frequency and intensity of long-range looping increases. The gene is completely silenced and no Pol II is present at the start site. Along the loops are multiple abnormally DNA-hypermethylated CpG islands that cosegregate with the methylated-CpG-binding protein MBD2. Interestingly, in an isogenic line of these colon cancer cells in which two DNA methyltransferases have been genetically disrupted, DNA methylation has been erased, and MBD2 association is lost, the looping intensity and frequency have been reduced. However, looping still remains in a conformation that is between that observed in EC and colon cancer cells, and a poised state of GATA-4 expression similar to that in EC cells has been restored. Thus, both Polycomb and DNA-methylation-mediated silencing bring about a similar three-dimensional chromatin configuration, with the important difference being that DNA methylation increases the stability of this configuration by multiple long-range interactions that effectively cause complete silencing of GATA-4.
Second, in another recent study, estrogen administration to breast epithelial cells growing in a mammosphere induced silencing of gene clusters associated with occupancy by estrogen receptor α (ESR1) in these and in cultured breast cancer cells sensitive to estrogen (Hsu et al. 2010). This silencing was abrogated by DNMT and histone deacetylase (HDAC) inhibitors. One such cluster that was analyzed in normal epithelial cells showed dynamic interaction of the promoters of silenced genes with ESR1-binding sites associated with DNA looping. Estrogen-responsive cancer cells, which already maintain this locus in a repressed state, displayed basal, low-level looping interactions. However, no increase in looping interactions was reported after further estrogen-mediated silencing. The study indicates that, unlike normal epithelial cells, cancer cells maintain less plastic and stable looping interactions that are probably part of the epigenetic program keeping this cluster repressed. Direct knockdown of components of DNA methylation and Polycomb machinery will be required to unravel the causative relationship between DNA looping and aberrant silencing.
In a third study, long-range DNA looping was mapped for the imprinted IGF2 locus in normal cells and compared to cancer cells that either maintain or display loss of imprinting (LOI) of this locus (Vu et al. 2010). In normal genomic imprinting, the IGF2 locus is regulated by differentially methylated regions (DMRs). Normal and cancer cells that have imprinted IGF2 display multiple long-range interactions. However, in cancer cells that have lost genomic imprinting of the IGF2 locus, these long-range interactions are grossly disrupted. Because parental alleles were not experimentally analyzed, the mapped interactions were an average of the steady-state interactions of the paternal and maternal alleles. Thus, the drastic deviation of long-range interactions most probably occurs at both parental alleles and not just the allele that has lost imprinting, indicating that disruption of long-range interactions itself might be a general feature in certain tumors. Altered long-range interactions could possibly dysregulate other genes that gain or lose interacting genomic regions of the IGF2 locus and possibly aid in oncogenic transformation.
Whether changes in long-range interactions is a cause or consequence of epigenetic changes and whether long-range interactions are required for maintenance of DNA methylation remain to be analyzed. In line with the above outlined studies, it is important to note that various nuclear matrix proteins can be misregulated in cancers, thereby possibly contributing to aberrant gene silencing or activation. In this regard, the nuclear matrix protein CTCF, with roles in transcription, chromatin insulation, and long-range genomic interactions is interesting (Ling et al. 2006). Witcher and Emerson (2009) showed that CTCF prevents hypermethylation of p16, RASSF1A, and CDH1 promoters by insulating the promoter from repressive marks. In addition, this study showed that cancer cells with aberrantly methylated and silenced p16 lose CTCF binding at an upstream promoter site because of defective posttranslational modification of CTCF by poly(ADP-ribosyl)ation (PARylation). Such protein PARylation is a modification also important in maintaining imprinting of the IGF2 locus (Yu et al. 2004). Consistent with this, loss of CTCF-PARylation frequently occurs in breast tumors and correlates with stage (Docquier et al. 2009). Interestingly, a paralog of CTCF, called BORIS (brother of the regulator of imprinted sites), with the same DNA-binding specificity as CTCF, is abnormally expressed in many tumors. It is proposed that BORIS can compete with CTCF and thus lead to abnormal gene regulation (Klenova et al. 2002; Hong et al. 2005). To what extent CTCF and BORIS cause global epigenetic deregulation remains to be explored. Because the binding of CTCF to its DNA targets is sensitive to CpG methylation of the target DNA, it is possible that abnormal demethylation can alter CTCF-binding properties, which in turn might influence DNA methylation.
That genes marked by PcG in ES cells tend to be hypermethylated suggests a direct progression of progenitor cells to cancer cells (Ohm and Baylin 2007). The model put forth by us and advanced by others (Feinberg et al. 2006; Ohm and Baylin 2007) posits that a key contribution to tumorigenesis may be epigenetic alterations in progenitor cells such that they adopt a neoplasia-ready state, which is epigenetically plastic and has the potential to gradually progress to a full-blown tumor. As discussed earlier, one mediator for creating such an epigenetic state may be the molecular progression from a state of promoter PcG marking to abnormal promoter DNA methylation (Ohm and Baylin 2007). According to the cancer epigenetic progenitor cell model, epigenetic plasticity is a crucial requirement during the various stages of tumorigenesis to constantly reprogram the epigenome during progression to tumor. In this context, alterations in nuclear structural proteins may provide the ability to reprogram epigenetic information. For example, the nuclear matrix protein SATB1 binds to AT-rich sequences and functions as a tether for chromatin loops (Cai et al. 2003). SATB1 coordinately regulates gene expression by recruiting chromatin-modifying and/or -remodeling enzymes to the tethered DNA (Yasui et al. 2002). SATB1 expression is associated with aggressive breast tumors and is important in metastasis (Han et al. 2008). A recent study showed that SATB1 mediates initiation of X inactivation in a lymphoma model (Agrelo et al. 2009). Normally, the ability to inactivate the X chromosome is confined to early embryonic and hematopoietic precursors. Thus, expression of SATB1 in tumor cells probably provides an epigenetic environment that mimics that of embryonic progenitor cells and aids in epigenetic reprogramming conducive with neoplastic evolution (Agrelo and Wutz 2009). By contributing to epigenetic reprogramming through altering genomic interactions, nuclear structural proteins such as SATB1 could be important factors in initiating and/or maintaining epigenetic abnormalities arising during tumorigenesis.
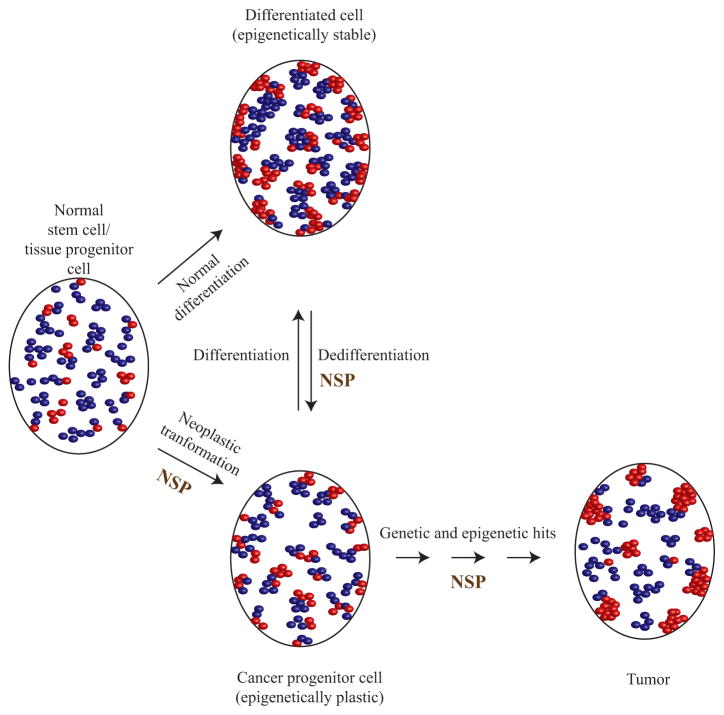
Furthermore, decondensed chromatin is a feature of the pluripotent status of ES cells, which may bestow an epigenetically plastic genome (Meshorer and Misteli 2006; Meshorer et al. 2006). Epigenetic plasticity helps to maintain developmental genes in a poised expression state responsive to specific developmental cues. Regions of decondensed chromatin that coexist with regions of abnormally condensed chromatin, however, are also a feature of cancer cell nuclei (Frost 1986). Several observations indicate that nuclear structural proteins might have a crucial role in redistributing chromatin condensation patterns in cancer cells. Ras-transformed mouse fibroblasts have been shown to result in less condensed chromatin structure due to increased phosphorylation of HMGN1, which allows histone H1 and H3 phosphorylation (Chadee et al. 1995; Lim et al. 2004; He et al. 2008). Chromatin decondensation induced by nuclear structural proteins seems to be an important initial player in oncogenic transformation (He et al. 2008). In a breast cancer model system, nuclear organization is lost in the malignant cells, which can be reversed by inducing partial differentiation. Interestingly, disruption of nuclear organization by introduction of an antibody against the nuclear structural protein NuMA (nuclear mitotic apparatus) stimulates the partially differentiated cells to enter the cell cycle, highlighting the fact that disruption of nuclear structure may be influential in tumorigenesis. The decondensed chromatin in cancer cells may have a role similar to that in ES cells, i.e., to confer epigenetic plasticity to the cancer genome. On the basis of the epigenetic progenitor model for tumor initiation and progression, epigenetic plasticity is required for accumulation of genetic and epigenetic changes favoring tumor proliferation and heterogeneity. The observation that cancer cells have decondensed chromatin is consistent with such a model. Given the link between decondensed chromatin and nuclear structural proteins, maintenance of epigenetic plasticity might rely on altered nuclear structural proteins, which may have an important role during various stages of tumor progression (Fig. 2).
Figure 2.
Alterations in nuclear structure caused by alterations in nuclear structural proteins (NSPs) may have an important role in imparting epigenetic plasticity to cancer-initiating cells. Stem cells have a decondensed chromatin (degree of condensation depicted by clustering of the red/blue ovals; red represents the heterochromatic regions and blue represents the euchromatic regions) that resolves into condensed chromatin during normal differentiation. During tumorigenesis, deregulation of structural proteins may perpetuate the decondensed chromatin state of expanding cancer progenitor cells. Through further epigenetic abnormalities, there may be serial epigenetic and genetic alterations, resulting in areas of increased condensation during tumor formation. As a result, tumor nuclei simultaneously maintain regions of decondensed and abnormally highly condensed, chromatin.
Nuclear structural and compositional changes have been known for a long time. As listed above, some of these changes might influence the epigenetic makeup of normal cells and drive them toward tumorigenesis. However, very few studies link nuclear structure to epigenetic changes in the context of cancer cells. Future studies should aim at understanding whether nuclear matrix proteins, some of which are important in genome organization and maintaining chromatin loops by binding to nuclear MARs/SARs (Table 1), influencing the global epigenetic changes that occur in tumor cells. Furthermore, a challenging task is to dissect whether epigenetic changes in tumor cells are the cause or effect of changes in chromatin configuration.
CONCLUSION
A building body of research is indicating that there may be no universal mechanism that leads to epigenetic dysregulation in cancer. The complexity of causes for such alterations may underlie the complexity of the disease itself. Careful dissection of the pathways that lead to epigenetic deregulation, ranging from misregulated protein complexes that have a role in epigenetic modulation to changes in genomic interactions as a result of changes in nuclear architecture, is not solely an intellectual endeavor in understanding regulation of gene expression. This forthcoming information will also increase our arsenal of diagnostic and possibly therapeutic tools for better characterization and management of cancer.
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences grant ES011858 and National Cancer Institute grant CA043318. H.P.E. was supported by a Hodson Foundation postdoctoral fellowship.
References
- Agrelo R, Wutz A. Cancer progenitors and epigenetic contexts: An Xisting connection. Epigenetics. 2009;4:568–570. doi: 10.4161/epi.4.8.10186. [DOI] [PubMed] [Google Scholar]
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chadee DN, Taylor WR, Hurta RA, Allis CD, Wright JA, Davie JR. Increased phosphorylation of histone H1 in mouse fibroblasts transformed with oncogenes or constitutively active mitogen-activated protein kinase kinase. J Biol Chem. 1995;270:20098–20105. doi: 10.1074/jbc.270.34.20098. [DOI] [PubMed] [Google Scholar]
- Chandramouly G, Abad PC, Knowles DW, Lelievre SA. The control of tissue architecture over nuclear organization is crucial for epithelial cell fate. J Cell Sci. 2007;120:1596–1606. doi: 10.1242/jcs.03439. [DOI] [PubMed] [Google Scholar]
- Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey DS. Nuclear matrix proteins as proteomic markers of preneoplastic and cancer lesions: Commentary re: G. Brunagel et al., 2002, Nuclear matrix protein alterations associated with colon cancer metastasis to the liver Clinical Cancer Research 8: 3039–3045. Clin Cancer Res. 2002;8:3031–3033. [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories: A functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- D’Arcy V, Pore N, Docquier F, Abdullaev ZK, Chernukhin I, Kita GX, Rai S, Smart M, Farrar D, Pack S, et al. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br J Cancer. 2008;98:571–579. doi: 10.1038/sj.bjc.6604181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- Di Cello F, Hillion J, Hristov A, Wood LJ, Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R, Resar LM. HMGA2 participates in transformation in human lung cancer. Mol Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier F, Kita GX, Farrar D, Jat P, O’Hare M, Chernukhin I, Gretton S, Mandal A, Alldridge L, Klenova E. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin Cancer Res. 2009;15:5762–5771. doi: 10.1158/1078-0432.CCR-09-0329. [DOI] [PubMed] [Google Scholar]
- Easwaran HP, Van Neste L, Cope L, Sen S, Mohammad HP, Pageau GJ, Lawrence JB, Herman JG, Schuebel KE, Baylin SB. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Res. 2010;70:8015–8024. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova E, Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783:2174–2184. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AH, Bardarov S, Jr, Jiang Z. Molecular aspects of diagnostic nucleolar and nuclear envelope changes in prostate cancer. J Cell Biochem. 2004;91:170–184. doi: 10.1002/jcb.10735. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Frost JK. The cell in health and disease. An evaluation of cellular morphologic expression of biologic behavior (2nd ed, revised) Monogr Clin Cytol. 1986;2:1–304. [PubMed] [Google Scholar]
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Garee JP, Oesterreich S. SAFB1’s multiple functions in biological control-lots still to be done! J Cell Biochem. 2009;109:312–319. doi: 10.1002/jcb.22420. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Guillemin C, Maleszewska M, Guais A, Maes J, Rouyez MC, Yacia A, Fichelson S, Goodhardt M, Francastel C. Chromatin modifications in hematopoietic multipotent and committed progenitors are independent of gene subnuclear positioning relative to repressive compartments. Stem Cells. 2009;27:108–115. doi: 10.1634/stemcells.2008-0755. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- He S, Dunn KL, Espino PS, Drobic B, Li L, Yu J, Sun JM, Chen HY, Pritchard S, Davie JR. Chromatin organization and nuclear microenvironments in cancer cells. J Cell Biochem. 2008;104:2004–2015. doi: 10.1002/jcb.21485. [DOI] [PubMed] [Google Scholar]
- Hitchins MP, Lin VA, Buckle A, Cheong K, Halani N, Ku S, Kwok CT, Packham D, Suter CM, Meagher A, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, Fischette MR, Adnani MT, Loukinov DI, Vatolin S, Risinger JI, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–744. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson A, Colella S, Moralli D, Wilson N, Yusuf M, Gimelli G, Ragoussis J, Volpi EV. Altered intra-nuclear organisation of heterochromatin and genes in ICF syndrome. PLoS One. 2010;5:e11364. doi: 10.1371/journal.pone.0011364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Klenova EM, Morse HC, III, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- Kress C, Ballester M, Devinoy E, Rijnkels M. Epigenetic modifications in 3D: Nuclear organization of the differentiating mammary epithelial cell. J Mammary Gland Biol Neoplasia. 2010;15:73–83. doi: 10.1007/s10911-010-9169-x. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman ES, Getzenberg RH. Nuclear structure as a source of cancer specific biomarkers. J Cell Biochem. 2008;104:1988–1993. doi: 10.1002/jcb.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mummert SK, Lobanenkov VA, Feinberg AP. Association of chromosome arm 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer. 2005;43:155–161. doi: 10.1002/gcc.20176. [DOI] [PubMed] [Google Scholar]
- Ohm JE, Baylin SB. Stem cell chromatin patterns: An instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Patani N, Jiang W, Mansel R, Newbold R, Mokbel K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009;9:18. doi: 10.1186/1475-2867-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Sarkar R, Guffei A, Vermolen BJ, Garini Y, Mai S. Alterations of centromere positions in nuclei of immortalized and malignant mouse lymphocytes. Cytometry A. 2007;71:386–392. doi: 10.1002/cyto.a.20395. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Takaha N, Hawkins AL, Griffin CA, Isaacs WB, Coffey DS. High mobility group protein I(Y): A candidate architectural protein for chromosomal rearrangements in prostate cancer cells. Cancer Res. 2002;62:647–651. [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, Nguyen AH, Hoffman AR. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum Mol Genet. 2010;19:901–919. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM, Jin H, Liu L, Zhang YF, Liu J, et al. Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol. 2009;219:114–122. doi: 10.1002/path.2575. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- Zinner R, Teller K, Versteeg R, Cremer T, Cremer M. Biochemistry meets nuclear architecture: Multicolor immuno-FISH for co-localization analysis of chromosome segments and differentially expressed gene loci with various histone methylations. Adv Enzyme Regul. 2007;47:223–241. doi: 10.1016/j.advenzreg.2007.01.005. [DOI] [PubMed] [Google Scholar]