Abstract
BAF250a (ARID1A) loss is a frequent event in high-grade endometrial cancers. It has been proposed that ARID1A is a driver gene, with ARID1A mutations occurring secondary to deregulated mismatch repair mechanism in gastric cancers, representing an alternative oncogenic pathway to p53 alteration. The prognostic significance of ARID1A loss is controversial. In this study, we investigated the frequency of BAF250a immunohistochemical loss in a cohort of high-grade endometrial cancers (n = 190) and correlated it with mismatch repair (hMLH1, hMSH2, hMSH6, and hPMS2) and p53 protein expression. The 190 cases consisted of 82 high-grade endometrioid, 88 serous, 10 clear cell, and 10 mixed (carcinosarcomas and mixed histology). There was BAF250a loss in 55/190 (29%) cancers, most commonly in high-grade endometrioid carcinomas (46 vs 9% in serous carcinomas, P<0.0001). Loss of any mismatch repair proteins was observed in 63/190 (33%) cancers, most commonly in high-grade endometrioid carcinomas (57 vs 10% in serous carcinomas, P<0.0001). Aberrant p53 expression was found in 86/190 (45%) cancers, more commonly in serous carcinomas (77 vs 18% in high-grade endometrioid carcinomas, P<0.0001). BAF250a loss was associated with mismatch repair loss (P<0.0001) and normal p53 expression (P<0.0001). These associations were maintained in the subset analysis within the high-grade endometrioid (P = 0.026 and P = 0.0083, respectively) and serous carcinoma cases (P = 0.0031 and P<0.0001, respectively). Survival analysis revealed a superior progression-free survival (P = 0.017) for patients with BAF250a loss within the entire cohort but not within the high-grade endometrioid and serous subtypes. Additionally, data from The Cancer Genome Atlas were extracted to correlate mutations in ARID1A, TP53, and MMR genes; we found that ARID1A mutations were negatively associated with TP53 mutations but were unrelated to mismatch repair gene mutations. In conclusion, BAF250a loss is more common in high-grade endometrioid carcinomas than in other high-grade endometrial cancers and is associated with mismatch repair deficiency and normal p53 expression.
Keywords: ARID1A, BAF250a, endometrioid carcinoma, endometrium, mismatch repair, p53, serous carcinoma
The AT-rich interactive domain 1A (ARID1A) gene encodes the protein BAF250a, which is ubiquitous in eukaryotes and is a key component of the multi-protein SWI/SNF chromatin remodeling complex.1 BAF250a (ARID1A) alterations are frequent in high-grade endometrial cancers, with loss of expression reported in 39% of grade 3 endometrioid carcinomas, 18% of serous cancers and 26% of clear cell carcinomas,2 and mutations in 60 and 10% of high-grade endometrioid and serous carcinomas, respectively.3
ARID1A alterations have been associated with microsatellite instability (MSI) status. In gastric carcinomas, Wang et al4 demonstrated higher frequency of ARID1A alterations (inactivating mutations and/or protein deficiency) in carcinomas with MSI than microsatellite stable (MSS) EBV-negative gastric carcinomas (83 vs 11%, respectively, P = 0.001). The type of ARID1A mutations in MSI-associated carcinomas was recurrent, being mostly indels involving short mononucleotide tracts, occurring at a higher rate than the global background rate of indels in similar mononucleotide tracts.4 A similar ARID1A mutation spectrum was noted by Jones et al5 in 12 MSI-high tumors (6 colon, 5 gastric and 1 prostate). Wang et al4 cited the significantly higher ARID1A mutation rate in MSI-associated gastric cancers as being a result of clonal selection of a mutant driver gene that had been mutated as a result of disrupted DNA mismatch repair. Recognizing the similar spectrum of ARID1A mutations in gastric cancer and ovarian clear cell carcinoma, which may also be associated with MMR deficiency, they postulated that ARID1A mutations may be prevalent in MSI-associated tumors of diverse sites. In addition, it has been shown that ARID1A mutations in gastric and ovarian carcinomas are negatively associated with mutations in TP53 (P = 0.002 and 0.031, respectively), suggesting an alternate pathway of cell-cycle deregulation, independent of TP53.4,6
The prognostic significance of ARID1A loss is unclear. In gastric cancers, it is controversial; certain authors showed a trend to prolonged recurrence-free survival in gastric cancers with ARID1A alterations (P = 0.058),4 while other studies demonstrated poor prognosis with ARID1A loss.7,8 However, in the subset analysis, among the EBV-negative and MSS group, ARID1A loss was an independent poor prognostic marker.7 Other studies confirmed the association between BAF250A/ARID1A expression loss and poor prognosis in cervical (P = 0.047)9 and renal cell carcinomas (P = 0.003).10 Among endometrial malignancies, a recent report shows that ARID1A alterations did not affect overall survival or progression-free survival.11
As ARID1A has been implicated in high-grade endometrial cancers, and MSI occurs in about 30% of endometrial cancers, we hypothesized that BAF250A/ARID1A loss was associated with mismatch protein deficiency in high-grade endometrial cancers and explored its prognostic significance.
Materials and methods
Patients and Tissue Samples
After approval of the University Health Network Research Ethics Board, consecutive cases of high-grade endometrial cancers (high-grade endometrioid FIGO III/III, serous, clear cell, and carcinomsarcoma) from 2001–2009 were retrospectively reviewed by a gynecologic pathologist (BC), and classified, according to the standard morphological criteria as defined by the World Health Organization classification of tumors,12 into serous, high-grade endometrioid, clear cell, or mixed carcinoma or carcinosarcoma. Relevant clinical data were obtained. Duplicate core tissue microarrays were constructed as previously described.13
Immunohistochemistry
Immunohistochemical staining was performed on 4-μm-thick paraffin sections of the tissue microarrays using anti-ARID1A (Sigma, clone: BAF250a, dilution: 1/75), hMLH1 (1:150, ES05, Novocastra, Vector Labs, Burlington, ON), hMSH2 (1:100, 25D12, Novocastra, Vector Labs), hMSH6 (1:600, PU29, Novocastra, Vector Labs), hPMS2 (1:150, MOR4G, Novocastra, Vector Labs), and p53 (Leica, clone: D07, dilution: 1/1000) antibodies, with appropriate positive and negative controls, according to the manufacturers’ standardized staining protocols.
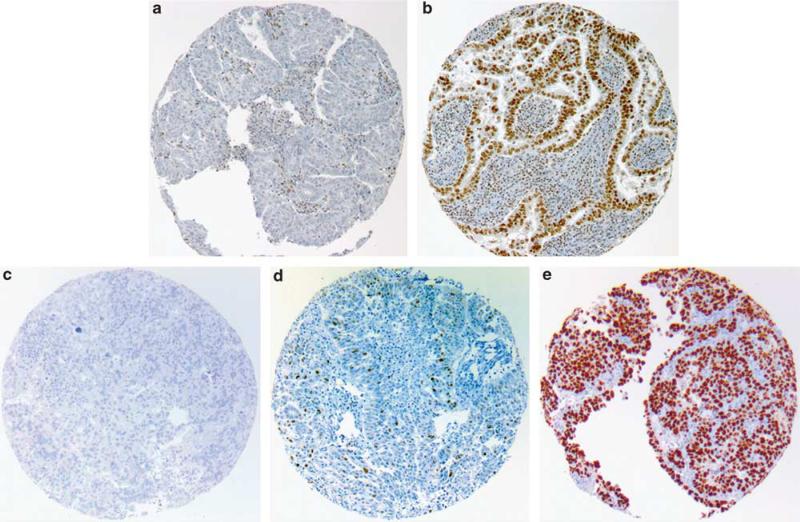
For BAF250a and MMR (hMLH1, hMSH2, hMSH6, hPMS2) immunohistochemistry, tumors were considered aberrant if tumor cells showed complete absence of nuclear staining with positive non-neoplastic internal control (Figure 1a), and intact if tumor cells showed nuclear positivity (Figure 1b), as previously described.14–16 The status of cases, in which tumor cores were negative or equivocal for any of the MMR proteins, was confirmed by full-section staining. Tumors were considered MMR deficient if there was loss of expression of at least one MMR protein. For p53, tumors were scored as follows: zero for complete loss of expression (Figure 1c), one for focal expression (1–50% of cells) (Figure 1d), or two for overexpression (defined as >50% of tumor cells showing strongly positive nuclear staining) (Figure 1e), and then binarized to aberrant (score of zero and two) and normal (score of one) as previously reported.17
Figure 1.
Scoring of immunohistochemistry antibodies. BAF250a is scored either as immunohistochemistry aberrant (a) or intact (b). Mismatch repair markers are scored similarly. p53 is given scores of zero—negative (c), one—focally positive (d), and two—diffusely positive (e).
Mutation Analysis
Somatic mutation data of 186 uterine corpus endometrioid carcinomas from The Cancer Genome Atlas (TCGA) were downloaded via cBioPortal (http://www.cbioportal.org/public-portal/, case_list_id = ucec_tcga_endo_core), including 103 non-hypermutated samples (case_list_id = ucec_tcga_endo_nonhypermut) and 83 hypermutated (case_list_id = ucec_tcga_endo_hypermut & ucec_tcga_endo_ultramut). These cases were selected from those prepared for TCGA publication as of 9 April 2013.
Statistical Analysis
The association of categorical clinico-pathological characteristics and differential biomarker expression and the association between ARID1A mutation and TP53 mutation, MMR genes mutation, and hypermutated phenotype were evaluated by Fisher's exact test for two-by-two comparisons and by Pearson's χ2 test for comparisons that exceeded the two-by-two criterion. Associations with continuous clinico-pathological characteristics and differential biomarker expression were quantified using the Welch's ANOVA. Univariable overall and progression-free survival were assessed by the generation of Kaplan–Meier curves and quantified with the log-rank statistic. Multivariable survival analysis was carried out using the Cox proportional hazards model. Analyses were not corrected for multiple comparisons, and p-values <0.05 were considered statistically significant. All analyses were computed with JMP v10.0, Cary, NC, USA.
Results
Study Cases
The age, stage, and the histological subtypes of this cohort of high-grade EC cases are summarized in Table 1. The median duration of follow-up was 1.32 (0.12–9.32) years. There was no significant association between the histological subtype and the age of diagnosis (P = 0.25). Serous carcinomas presented at a higher stage than endometrioid (P = 0.0014), and were associated with worse progression-free survival (P = 0.0061).
Table 1.
Demographics of study cases
|
Stage
|
|||||||
|---|---|---|---|---|---|---|---|
| n | % | Age mean (Years) | P | Low (I-II) | High (III-IV) | P | |
| Histological type | |||||||
| Endometrioid | 82 | 43% | 66 | 0.25 | 62 (76%) | 20 (24%) | 0.004 |
| Serous | 88 | 47% | 68 | 45 (51%) | 43 (49%) | ||
| Clear cell | 10 | 5% | 71 | 5 (50%) | 5 (50%) | ||
| Mixed | 10 | 5% | 65 | 8 (80%) | 2 (20%) | ||
| Total | 190 | 100% | 68 | 120 (63%) | 70 (37%) | ||
Association of BAF250a, MMR, and p53 Expression with Clinico-Pathological Characteristics
BAF250a loss was observed in 29% of the study cases, was more common in younger patients (P = 0.0008), and was significantly associated with non-serous histology (P<0.0001) (Table 2).
Table 2.
Prevalence of BAF250a (ARID1A), MMR and p53 immunohistochemical expression and in relation to age and histological subtype
| BAF250a+ | BAF250a – | P | MMR retained | MMR lost | P | p53 normal | p53 aberrant | P | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years), mean | 68.9 | 62.8 | 0.0003 | 67.6 | 66.4 | 0.471 | 66.2 | 68.3 | 0.201 | ||||||
| Histological type | <0.0001 | <0.0001 | <0.0001 | ||||||||||||
| Endometrioid | 44 | 54% | 38 | 46% | 35 | 43% | 47 | 57% | 67 | 82% | 15 | 18% | |||
| Serous | 80 | 91% | 8 | 9% | 79 | 90% | 9 | 10% | 20 | 22% | 68 | 78% | |||
| Clear cell | 6 | 60% | 4 | 40% | 9 | 90% | 1 | 10% | 9 | 90% | 1 | 10% | |||
| Mixed | 5 | 50% | 5 | 50% | 4 | 40% | 6 | 60% | 8 | 80% | 2 | 20% | |||
| Total | 135 | 71% | 55 | 29% | 127 | 67% | 63 | 33% | 104 | 55% | 86 | 45% | |||
Aberrations in MMR protein expression were identified in 33% of all high-grade endometrial cancers, the majority affecting MLH1 and PMS2 (Table 3). Across the entire cohort, loss of MMR proteins was significantly associated with endometrioid and mixed histologies rather than the serous or clear cell subtypes (P<0.0001). Aberrant p53 expression was discovered in 45% of cases and was significantly associated with serous subtype (P = 0.0001; Table 2).
Table 3.
Immunohistochemical loss of the four mismatch repair proteins among various histological subtypes
| Endometrioid | Serous | Clear cell | Mixed | P | |
|---|---|---|---|---|---|
| hMLH1 loss | 34 (59%) | 6 (7%) | 0 (0%) | 6 (60%) | <0.0001 |
| hMSH2 loss | 6 (7%) | 2 (2%) | 1 (10%) | 1 (10%) | 0.8 |
| hMSH6 loss | 11 (13%) | 3 (3%) | 0 (0%) | 1 (10%) | 0.13 |
| hPMS2 loss | 36 (44%) | 6 (7%) | 0 (0%) | 4 (40%) | <0.0001 |
| Total, n | 82 | 88 | 10 | 10 |
BAF250a Expression and Status of MMR Protein Immunohistochemistry
In the entire cohort, loss of BAF250a was significantly associated with MMR loss (65% of BAF250a negative are MMR deficient, and 57% of MMR-deficient endometrial cancers are BAF250a negative, P<0.0001; Table 4). This association was maintained within the endometrioid and serous carcinoma subtype (P = 0.026 and P = 0.0031, respectively; Table 4). In addition, of the 10 clear cell carcinomas, the single case with abnormal MMR status had BAF250a loss and 4 of the 6 mixed carcinomas (67%) with abnormal MMR had BAF250a loss.
Table 4.
Correlation between immunohistochemical expression of BAF250a (ARID1A), mismatch repair proteins, and p53 within the entire cohort and within the endometrioid and serous subtypes
|
BAF250a+
|
BAF250a
–
|
||||
|---|---|---|---|---|---|
| n | % | n | % | P | |
| Entire cohort | |||||
| MMR intact | 108 | 85% | 19 | 15% | <0.0001 |
| MMR lost | 27 | 43% | 36 | 57% | |
| p53 normal | 52 | 50% | 52 | 50% | <0.0001 |
| p53 aberrant | 83 | 96% | 3 | 4% | |
| Endometrioid carcinomas | |||||
| MMR intact | 24 | 69% | 11 | 31% | 0.026 |
| MMR lost | 20 | 42% | 27 | 58% | |
| p53 normal | 31 | 46% | 36 | 54% | 0.0083 |
| p53 aberrant | 13 | 87% | 2 | 13% | |
| Serous carcinomas | |||||
| MMR intact | 75 | 95% | 4 | 5% | 0.0031 |
| MMR lost | 5 | 56% | 4 | 44% | |
| p53 normal | 13 | 65% | 7 | 35% | <0.0001 |
| p53 aberrant | 67 | 98% | 1 | 2% | |
BAF250a and p53 Expression Status
BAF250a expression loss was negatively associated with aberrant p53 expression (95% of BAF250a-negative endometrial cancers showed normal p53 expression, and 97% of carcinomas with aberrant p53 expression were positive for BAF250a, P<0.0001; Table 4). This association was also observed among the endometrioid and serous histotypes (P≤0.0083, and P<0.0001, respectively; Table 4). In addition, only a single case of clear cell carcinoma (10%) had aberrant p53 expression, and this case had retained expression of BAF250a. Among the remaining nine clear cell carcinomas with normal p53, four (44%) were BAF250a negative. Moreover, 2 of the 10 (20%) mixed endometrial cancer subtype had aberrant p53, both of which were BAF250a positive, while 5 of the 8 (62%) mixed cases with normal p53 were BAF250a negative.
Prognostic Significance of BAF250a, MMR, and p53 Expression
Examination of the entire cohort using univariable survival analysis revealed that loss of BAF250a did not affect overall survival but was associated with improved progression-free survival (log-rank P = 0.27, and 0.0107, respectively; Table 5). Although MMR loss and normal p53 expression were associated with improved overall survival (log rank P = 0.0315, and 0.0418, respectively) and progression-free survival (log rank P = 0.023, and 0.055, respectively) within each of the histological subtypes, BAF250a did not carry a prognostic significance (Table 5).
Table 5.
Univariable analysis of the prognostic significance of BAF250a, MMR, and p53
|
Overall survival
|
Progression-free survival
|
|||
|---|---|---|---|---|
| Hazard ratio | P | Hazard ratio | P | |
| Entire cohort (n = 190) | ||||
| BAF250a loss | 0.67 | 0.27 | 0.25 | 0.011 |
| MMR deficiency | 0.5 | 0.0315 | 0.43 | 0.023 |
| p53 aberrant | 1.67 | 0.0418 | 1.75 | 0.055 |
| Endometrioid (n = 82) | ||||
| BAF250a loss | 0.94 | 0.917 | 0.42 | 0.28 |
| MMR deficiency | 0.7 | 0.483 | 2.5 | 0.231 |
| p53 aberrant | 1.32 | 0.612 | 0.61 | 0.554 |
| Serous (n = 89) | ||||
| BAF250a loss | 0.48 | 0.234 | 0.34 | 0.268 |
| MMR deficiency | 0.54 | 0.251 | 0.23 | 0.117 |
| p53 aberrant | 2.34 | 0.0528 | 1.64 | 0.305 |
On multivariable analysis (Table 6), age at diagnosis, stage, and histological subtype were considered, revealing that BAF250a immunonegativity was associated with longer progression-free survival (P = 0.0062) but was not associated with overall survival (P = 0.098). Loss of any MMR proteins (P = 0.033) and normal p53 immunohistochemistry (P = 0.039) were associated with a better overall survival. There was no significant correlation between MMR and p53 immunohistochemistry with progression-free survival (P = 0.34 and 0.48, respectively).
Table 6.
Multivariable analysis of the prognostic significance of BAF250a, MMR, and p53
|
Overall survival
|
Progression-free survival
|
|||
|---|---|---|---|---|
| Hazard ratio | P | Hazard ratio | P | |
| Entire cohort (n = 190) | ||||
| BAF250a loss | 1.09 | 0.098 | 3.63 | 0.0062 |
| MMR deficiency | 2.12 | 0.033 | 1.48 | 0.34 |
| p53 aberrant | 0.52 | 0.039 | 0.76 | 0.48 |
Association Between ARID1A Mutation and Mutation in TP53 and MMR Genes
There is a significant negative correlation between ARID1A mutation and TP53 mutation in endometrioid carcinoma according to pathology reports, (odds ratio = 0.2284, P = 0.0051, Fisher's exact test, two tailed). No correlation is detected between ARID1A mutation and hypermutated phenotype (P = 0.2269, Fisher's exact test, 2 tailed). No correlation is observed between ARID1A mutation and mutations in any of the MMR genes, including PMS1, PMS2, MSH2, MSH3, MSH6, MLH1, and MLH3 (P = 0.1032, Fisher's exact test, two tailed).
Discussion
Our data show that BAF250a negativity, present in 29% of high-grade endometrial carcinomas, is associated with loss of MMR protein expression and inversely correlated with aberrant p53 expression. Although BAF250a negativity is associated with progression-free survival in the whole cohort, histotype-specific analyses failed to demonstrate an association with prognosis.
In this study, the rate of BAF250a loss is concordant with previous studies showing mutations in ARID1A gene in 29% (32/109) of cases3 and loss of the expression of its protein BAF250a in 24% (85/358).2 BAF250a negativity is more common in endometrioid than in serous carcinomas (46 and 9%, respectively, P<0.0001), similar to previous studies showing a higher rate of ARID1A mutations in the endometrioid subtype than serous endometrial carcinomas (60 vs 11%, P = 2.4E-53) and more frequent loss of BAF250a expression (39 vs 18%, P = 0.0012). The limited number of clear cell carcinomas in this study precludes making an inference with regards to the frequency of BAF250a negativity in clear cell carcinomas. The significant differential expression of BAF250a in our study suggests that, in the context of high-grade endometrioid and serous uterine carcinomas, loss of BAF250a expression is mostly but not exclusively an endometrioid phenomenon. The higher frequency of BAF250a loss is similar to a recent report demonstrating that the percentage of complete ARID1A loss increased from 0% in complex atypical hyperplasia to 25% in low-grade endometrioid carcinoma and to 44% in high-grade endometrioid carcinoma.18 As neither mutation of ARID1A or loss of BAF250a expression was observed in morphologically pure uterine serous carcinomas,19 it is interesting to note that a handful of serous carcinomas exhibited BAF250a negati vity. These cases demonstrated an unusual immunophenotype, with the majority (7/8, 88%) showing normal p53 expression and half showing MMR deficiency. Cognizant of the poor reproducibility of cell type in the high-grade endometrial spectrum,20 this suggests that this small subset of BAF250a-negative serous carcinomas may indeed be endometrioid carcinomas mimicking the serous histomorphology.
Wang et al4 demonstrated collusion between MSI and ARID1A mutations or protein deficiency in gastric cancer. They suggested that this was applicable across various tumor sites. In this study, we confirmed that BAF250a negativity is significantly associated with MMR protein defects in high-grade endometrial carcinomas (P<0.0001). However, the biological significance of this correlation is not clear; Wang et al4 reported that the rate of indels in ARID1A in gastric carcinomas with MSI, which is a manifestation of MMR defects, was higher than that in MSS carcinomas. Furthermore, with MSI gastric cancers, the indel rate is significantly higher in ARID1A than similar sized mononucleotide tracts. Based on this, they postulated that this frequent recurrent mutational mechanisms might represent targeting of a tumor-driver gene.4 In addition, indels and frameshift mutations, which can occur frequently in the context of MMR deficiency, were found to be the most common types of ARID1A mutations over a variety of cancer types, including gynecological malignancies (overall, 67%; gynecological tumors, 64%).21 These studies suggest a causal relationship between MMR deficiency and ARID1A mutations. It is worth noting that 34% (19/56) of BAF250a-negative high-grade endometrial carcinomas in this study displayed intact MMR profile and normal p53 expression, suggesting an independent pathway of MMR loss in a subset of cases. Further studies are needed to investigate whether BAF250a/ARID1A alterations directly result from defective MMR mechanisms in endometrial carcinomas and to discover other pathways that may result in inactivation of ARID1A.
Furthermore, we demonstrate an inverse relationship between BAF250a expression loss and p53 aberrant expression in high-grade endometrial carcinomas (P<0.0001). This inverse correlation has been reported in gastric carcinomas (P = 0.002),15 esophageal adenocarcinomas (P = 0.028),22 and a cohort of ovarian clear cell and uterine endometrioid carcinomas (P = 0.031).6 Guan et al6 have studied the relationship and interaction between ARID1A and TP53 pathways and their tumor-suppressor function. They found that some downstream targets of ARID1A include CDKN1A/p21 and SMAD3, which are well-known p53 downstream target genes, and that ARID1A/BRG1 complex of SWI/SNF proteins interacts with the same promoter regions of CDKN1A and SMAD3 as p53. Knocking down ARID1A significantly reduced BRG1 binding to these regions, reducing the transcriptional activity of p21 and SMAD3, but it did not affect p53 binding to these regions.6 These findings suggest that inactivating mutations in either ARID1A or TP53 result in loss of transcriptional regulation of CDKN1A and SMAD36 and support the interpretation that there is some functional equivalence to loss of function of either protein, accounting for the uncommon occurrence of mutations in both the genes in the same tumor. This has been described previously for other genes impacting on the same signaling pathway, eg, HER2 amplification and KRAS mutation in mucinous carcinoma of the ovary.23
Different studies have reported inconsistent results on the prognostic significance of ARID1A alterations. In gastric carcinomas, these alterations have been reported to be associated with a trend towards better prognosis (P = 0.058).4 However, other authors described poor survival in gastric cancers with ARID1A/BAF250a expression loss.8 Furthermore, other reports showed no prognostic significance for ARID1A alterations;7 however, on subset analysis, in EBV-negative MLH1-preserved gastric carcinomas, BAF250A expression loss was significantly associated with worse overall survival (P = 0.027) and progression-free survival (P = 0.016).7 Similar variability have been reported in ovarian clear cell and endometrioid carcinomas, where Wiegand et al14 showed no association between survival and ARID1A mutations in ovarian carcinomas of either subtype, and Maeda et al24 also demonstrated no association between loss of BAF250a expression and clinical outcome of ovarian clear cell carcinoma, while Katagiri et al25 discovered worse progression-free survival (P<0.01) and resistance to platinum-based chemotherapy (P = 0.04) in patients with ovarian clear cell carcinoma. Moreover, ARID1A/BAF250a loss was significantly associated with worse overall survival in cervical adenocarcinoma9 and worse overall survival (P = 0.003) and progression-free survival (P = 0.01) in renal clear cell carcinomas. In this study, while there was no association between BAF250a expression status and age at presentation and tumor stage, BAF250a-negative high-grade endometrial carcinomas exhibited a better progression-free survival. The prognostic significance did not hold up within the endometrioid and the serous subtypes. This lack of significance within subtypes may be due to underpowered analysis because of lower number of subjects in the subset analysis, or it might represent a subtype bias with ARID1A/BAF250a loss identifying endometrioid subtypes, and the favorable association between progression-free survival and ARID1A/BAF250a loss may only reflect a preponderance of the less aggressive endometrioid cases.
The recent deposition of TCGA data on uterine corpus endometrial carcinoma allows us to validate the immunohistochemistry-based findings from this study in a large cohort of cases. Based on analyzing a total of 186 endometrioid carcinomas, we found negative correlation between ARID1A and TP53 mutations, consistent with the inverse association between BAF250a expression loss and p53 abnormal expression. However, the lack of correlation of ARID1A mutations with either hypermutation phenotype (characterized by hundreds of somatic mutations) or MMR gene mutation is in contrast to our immunostaining findings. The most likely explanation is that many cases with MMR dysfunction (lost expression) are caused by promoter methylation rather than mutation, supported by the fact that the majority of the losses in MMR proteins occurred in hMLH1 and hPMS2 (Table 3).26,27
In summary, BAF250a/ARID1A abnormalities occur in approximately 30% of high-grade endometrial carcinomas and are significantly associated with MMR protein deficiency and normal p53 expression. The constellation of BAF250a/ARID1A loss, MMR deficiency, and normal p53 expression is highly characteristic of the endometrioid subtype, and further studies of the rare cases showing this immunophenotype and serous morphological features are necessary to determine whether such cases are better classified as endometrioid, in light of their underlying molecular abnormalities.
Footnotes
Disclosure/conflict of interest
The authors declare no conflict of interest.
References
- 1.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 2.Wiegand KC, Lee AF, Al-Agha OM, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–333. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 3.McConechy MK, Ding J, Cheang MC, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abe H, Maeda D, Hino R, et al. ARID1A expression loss in gastric cancer: pathway-dependent roles with and without Epstein–Barr virus infection and microsatellite instability. Virchows Arch. 2012;461:367–377. doi: 10.1007/s00428-012-1303-2. [DOI] [PubMed] [Google Scholar]
- 8.Wang DD, Chen YB, Pan K, et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS ONE [Electronic Resource] 2012;7:e40364. doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Kim JS, Chung H, et al. Loss of ARID1A/ BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum Pathol. 2013;44:1365–1374. doi: 10.1016/j.humpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Lichner Z, Scorilas A, White NM, et al. The chromatin remodeling gene ARID1A is a new prognostic marker in clear cell renal cell carcinoma. Am J Pathol. 2013;182:1163–1170. doi: 10.1016/j.ajpath.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Rahman M, Nakayama K, Rahman MT, et al. Clinicopathologic analysis of loss of AT-rich interactive domain 1A expression in endometrial cancer. Hum Pathol. 2013;44:103–109. doi: 10.1016/j.humpath.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Tavassoli FA, Devilee P, editors. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. IARC Press; Lyon, France: 2003. World Health Organization Classification of Tumours. [Google Scholar]
- 13.Alkushi A, Clarke BA, Akbari M, et al. Identification of prognostically relevant and reproducible subsets of endometrial adenocarcinoma based on clustering analysis of immunostaining data. Mod Pathol. 2007;20:1156–1165. doi: 10.1038/modpathol.3800950. [DOI] [PubMed] [Google Scholar]
- 14.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu FI, Gilks CB, Mulligan AM, et al. Prevalence of loss of expression of DNA mismatch repair proteins in primary epithelial ovarian tumors. Int J Gynecol Pathol. 2012;31:524–531. doi: 10.1097/PGP.0b013e31824fe2aa. [DOI] [PubMed] [Google Scholar]
- 16.Alkushi A, Kobel M, Kalloger SE, et al. High-grade endometrial carcinoma: serous and grade 3 endometrioid carcinomas have different immunophenotypes and outcomes. Int J Gynecol Pathol. 2010;29:343–350. doi: 10.1097/PGP.0b013e3181cd6552. [DOI] [PubMed] [Google Scholar]
- 17.Kobel M, Reuss A, Bois A, et al. The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol. 2010;222:191–198. doi: 10.1002/path.2744. [DOI] [PubMed] [Google Scholar]
- 18.Mao TL, Ardighieri L, Ayhan A, et al. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013 doi: 10.1097/PAS.0b013e3182889dc3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan B, Mao TL, Panuganti PK, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Sug Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilks CB, Oliva E, Soslow RA. Poor inter-observer reproducibility in the diagnosisof high-grade endome-trial carcinoma. Am J Surg Pathol. 2013;37:874–881. doi: 10.1097/PAS.0b013e31827f576a. [DOI] [PubMed] [Google Scholar]
- 21.Guan B, Gao M, Wu CH, et al. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14:986–993. doi: 10.1593/neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streppel MM, Lata S, Delabastide M, et al. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus. Oncogene. 2013 doi: 10.1038/onc.2012.586. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anglesio MS, Kommoss S, Tolcher MC, et al. Molecular characterization of mucinous ovarian tumours supports a stratified treatment approach with HER2 targeting in 19% of carcinomas. J Pathol. 2013;229:111–120. doi: 10.1002/path.4088. [DOI] [PubMed] [Google Scholar]
- 24.Maeda D, Mao TL, Fukayama M, et al. Clinicopatho-logical significance of loss of ARID1A immunoreactivity in ovarian clear cell carcinoma. Int J Mol Sci. 2010;11:5120–5128. doi: 10.3390/ijms11125120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katagiri A, Nakayama K, Rahman MT, et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25:282–288. doi: 10.1038/modpathol.2011.161. [DOI] [PubMed] [Google Scholar]
- 26.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 27.Esteller M, Levine R, Baylin SB, et al. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]