Abstract
Actin is a ubiquitous eukaryotic protein that is responsible for cellular scaffolding, motility and division. The ability of actin to form a helical filament is the driving force behind these cellular activities. Formation of a filament is dependent the successful exchange of actin’s ADP for ATP. Mammalian profilin is a small actin binding protein that catalyzes the exchange of nucleotide and facilitates the addition of an actin monomer to a growing filament. Here, crystal structures of profilin:actin have been determined showing an actively exchanging ATP. The structural analysis shows how the binding of profilin to the barbed end of actin causes a rotation of the small domain relative to the large domain. This conformational change is propagated to the ATP site and causes a shift in the nucleotide loops which in turn causes a repositioning of Ca2+ to its canonical position as the cleft closes around ATP. Reversing the solvent exposure of Trp-356 is also involved in cleft closure. In addition, secondary calcium binding sites were identified.
Keywords: protein-protein interaction, ATP, calcium, conformational change, domain motion
Introduction
Actin is a biologically important molecule that is highly conserved and has many different functions1. The dynamic nature of actin is central to all forms of cellular motility, including actin-based motility of bacteria2 and eukaryotic cells where actin filament growth at the leading edge drives cellular motion3. Due to the high demand for actin to participate in a large number of processes, it is no surprise that actin has been shown to adopt many structures4,5. Actin has two main forms: a monomeric globular G-actin bound to an actin binding protein and filamentous F-actin composed of an actin polymer. There are hundreds of actin binding proteins that regulate the polymerization, organization and function of actin filaments7. F-actin hydrolyzes ATP to ADP, with the aid of a bound metal (e.g. Mg2+)6. This event occurs shortly after G-actin is added to the barbed end of F-actin. Actin-based motility is achieved by a treadmilling mechanism where F-actin moves by growing at one end (barbed end) while disassociating at the other end (pointed end). Actin monomers that have been released from the pointed end can eventually be reused, providing that the ADP is exchanged for ATP.
In mammals, profilin facilitates this nucleotide exchange and then helps deliver actin to the barbed end of a new filament8,9, although this function of profilin is not completely evolutionarily conserved10–12. In mammals, profilin binds to actin and accelerates the exchange of ADP for ATP8. Profilin binds to the barbed end of actin and enhances barbed end elongation13. It is also thought that the free energy of ATP hydrolysis for filament growth is used by profilin, though this topic is still debatable14. Formin dimers nucleate unbranched actin filaments and require profilin15. Nucleation is achieved by binding actin to the FH2 domain of formin, this stabilizes an actin dimer16. The FH1 domain of formin contains a proline-rich region that binds profilin. Profilin binding to FH1 has been shown to stimulate elongation by increasing the concentration of profilin bound G-actin at the filament barbed end17. Once actin binds to formin a stable nucleus is formed, formin remains bound to the barbed end and acts as a processive elongation factor18. Profilin affinity for actin is also greatly lowered by binding of phosphatidylinositol-4,5-bisphosphate (PIP2) at the inner leaflet of the plasma membrane11. Evidence suggests that this process of PIP2-mediated actin release is coordinated by a series of cell signals, which leads to actin incorporation into a growing filament19,20. Clearly the profilin:actin complex is important in actin filament formation.
The first crystal structures of profilin:actin showed that profilin binds to W- and C-terminal-loops on the barbed end of actin (Fig. 1(a)). The profilin:actin structures had two forms: one that was wide-open to solvent, and another with a closed nucleotide cleft4,21,22. The domain motions between these two structures have been described as two rigid cores whose relative motion occurs at two shear regions23. Several exterior loops were also found to move in a hinge-like fashion (see Fig 4 in reference 23). Also, a structure of profilin:actin with a VASP202-244 fragment bound to profilin was solved that has a partially-open nucleotide-binding cleft24. This provides an excellent intermediary to study actin cleft closure and a rotation of 4.7° was observed between the two major actin domains24. Also, a computer simulation showed that wide-open actin closes when profilin disassociates25. The closure of the active site around ATP completes nucleotide exchange.
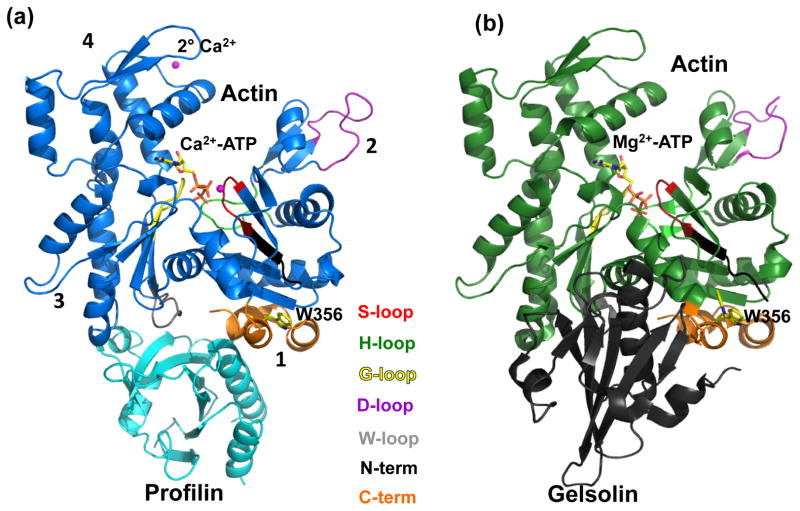
Figure 1.
Actin structure. (a) Wide-open state of bovine profilin-β:actin (PDB 3UB5). (b) Nonexchanging-closed state of gelsolin:pY53-actin. Subdomains 3 and 4 make up the large domain where Subdomans 1 and 2 make up the small domain. ATP is shown as sticks bound to Ca2+ (magenta sphere). Profilin (cyan) is bound to actin at subdomains 1 and 3. Gelsolin (grey) is bound to actin (green) at Subdomains 1 and 3. Actin loops are colored as follows: Sloop (residues 11 to 16) is red; H-loop (residues 70 to 78) is green; G-loop (residues 154-161) is yellow; D-loop (residues 38-52) is purple, W-loop (residues 165-172) is grey; N-terminus (residues 1-10) is black; and C-terminus (residues 349-375) is orange.
Gelsolin is an actin binding protein that has filament severing and capping functions, which lead to the disintegration of F-actin (Fig. 1(b))26. Gelsolin first binds to the side of F-actin and causes a kink. Once a kink is made, gelsolin severs F-actin and remains bound to cap the barbed end. The capping of the barbed end ensures that this smaller and newly-formed filament cannot further polymerize. In effect, the severing of F-actin into smaller fragments by gelsolin decreases the viscosity of the cytoplasm, creating the “gel” to “sol” transformation that gave gelsolin its name27.
Gelsolin and profilin have opposite effects on actin nucleotide exchange. By measuring the fluorescent signal as ethano-ATP replaced bound ATP, Baek and coworkers showed that a complex of actin and gelsolin segment G1 inhibited nucleotide exchange24. They also showed that the profilin:actin complex increased nucleotide exchange. If the actin in either complex is phosphorylated at Tyr-53, the initial rate of nucleotide exchange decreases and the gelsolin complex does not exchange. Thus the structure of pY53-actin and gelsolin represents a “nonexchanging-closed” form of actin (Fig. 1(b)) and profilin:actin structures represents open conformations that can exchange nucleotide (Fig. 1(b)). A conformational change of actin’s D-loop was also implicated in nucleotide exchange28. Although there has been a great deal of research on the topic, the complete structural basis for the nucleotide-exchange mechanism has not been described. Here we describe what changes are induced to the bound ATP as profilin is exchanging actin’s nucleotide, and how do these structural changes propagate from the W- and C-terminal loops at the barbed end to close the S-, H- and D-loops in the nucleotide binding site and cleft.
Results and Discussion
Structure
In this study, the wide-open and closed profilin:actin crystal structures that were originally described by Schutt21,22 were completely redetermined using cryocooling techniques in order to obtain higher resolution structures. The best reagents for cryocooling the closed and wide-open PA crystals were 7M sodium formate and 35% glycerol, respectively. Also, to prevent spontaneous formation of higher-order actin structures in the crystal, excess Ca2+ was removed from the bathing solution. Overall, these actin crystal structures show an similarity with other reported structures4,22,24. Actin is composed of two major domains: a small domain, further divided into subdomains 1 and 2, and a large domain, consisting of subdomains 3 and 4 (Fig. 1(a))29. A fold consisting of a central five-stranded β-sheet and three α-helicies defines the general actin structure30. Wide-open profilin:actin was refined to 2.2 Å resolution with a final Rcryst/Rfree of 21.4%/29.1%. The actin of the refined wide-open state profilin:actin coordinates superpose with the starting structure (1HLU) with an r.m.s.d of 1.3 Å. Ramachandran analysis in Molprobity31 indicates that 93.1% lie in the most favored region, with 1.6% existing as outliers. Closed state profilin:actin was refined to 2.6 Å resolution with a final Rcryst/Rfree of 25.1%/32.7%. Superposition of actin molecules between our closed state and the starting structure (2BTF) overlay with an r.m.s.d of 0.8 Å. Ramachandran analysis in Molprobity indicates that 90.4% of the residues lie within the most favored regions, with 1.8% as outliers. Overall, the structures refined well, with reasonable stereochemistry (Table 1).
Table 1.
Data processing and refinement statistics.
| A. Data | Wide-open | Closed |
|---|---|---|
| PDB ID | 3UB5 | 3U4L |
| Spwace Group | P212121 | P212121 |
| Unit cell dimensions (Å) | a=38, b=72, c=186.8 | a=38.3, b=71.1, 172.0 |
| Resolution range (Å) | 85-2.2 | 85-2.6 |
| No. unique reflections | 26,998 | 14,947 |
| Average redundancy | 3.2 | 5.8 |
| Completeness (%)a | 95.4 (71.8) | 98.3 (97.4) |
| I/σa | 6.8 (2.2) | 5.4 (2.0) |
| Rsym (%)a,b | 5.9 (23.3) | 9.1 (37) |
|
| ||
| B. Refinement
| ||
| No. of protein atoms | 3914 | 3789 |
| No. of ATP atoms | 31 | 31 |
| No. of calcium atoms | 2 | 2 |
| No. of solvent atoms | 143 | 27 |
| Rcryst (%) | 21.4 | 25.1 |
| Rfree (%) | 29.1 | 32.7 |
| Estimated coordinate error (Å) | 0.35 | 0.54 |
| Geometry | ||
| r.m.s.d. bonds (Å) | 0.014 | 0.012 |
| r.m.s.d. angles (°) | 1.86 | 1.67 |
| Average B-factors (Å2) | ||
| Protein | 36.2 | 44.5 |
| Calcium | 51.7 | 34.8 |
| ATP | 40.8 | 51 |
| Water | 34.3 | 33.2 |
|
| ||
| C. Ramachandran analysis
| ||
| Bad rotamers (%) | 6.4 | 6.7 |
| Ramachandran outliers (%) | 1.4 | 1.8 |
| Ramachandran favored (%) | 92.9 | 90.4 |
All values in parentheses are for the highest resolution shell.
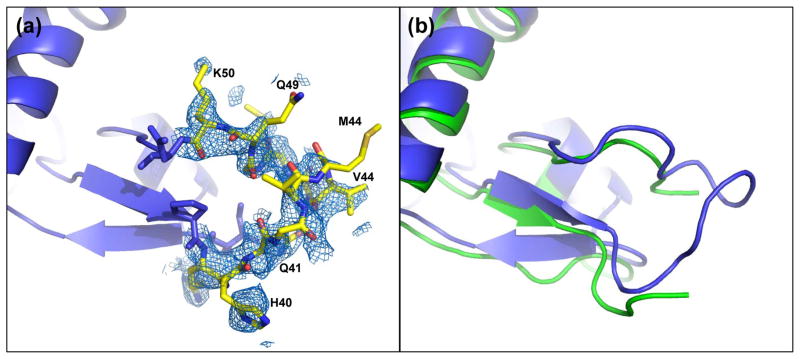
In order to determine the structure of the D-loop, residues 41-49 were omitted from the map calculation. Following refinement, omit FO-FC electron density was still weak, so omit-maps were recalculated at 3 and 4 Å. At lower resolution more clearly interpretable electron density was seen and each residue was manually fit and refined and the resolution was extended to 2.2 Å. The D-loop was partially ordered in the wide-open profilin:actin (Fig. (2a)). When compared to the nonexchanging-closed state structure (PDB 3CI5), of which a partial D-loop was determined, a superposition of the large domains show an overall similar secondary structure for the D-loop (Fig. 2(b)).
Figure 2.
D-loop conformation. (a) 2FO-FC electron density (0.8 σ) of the D-loop of wide-open state profilin:actin covering residues 41-49. (b) Overlay of the wide-open state profilin:actin (blue) with the partially modeled D-loop of nonexchanging-closed actin (green). The modeled loops are shifted, but show a similar overall structure.
Comparison of these structures with those determined by Baek and coworkers24 allowed for the construction of a continuum of states, which actively show the conformational changes required for actin cleft closure involved in nucleotide exchange. In this report we examine the major differences in actin structure and function between the wide-open, partially-open and nonexchanging-closed states. Comparison of these structures has allowed us to analyze the major conformational changes that occur when actin is opened by profilin and binds ATP, and the nucleotide binding cleft is closed by gelsolin binding. Specifically, the contributions of the individual actin loops (W-, S-, D-, H-, G- and C-terminal) were analyzed. The closure of the actin cleft involves a rotation of the large actin domain, relative to the small domain as ATP lands into the binding pocket. Also, we discuss implication of profilin binding the actin nucleotide-sensing W-loop and how the conformational changes induced by profilin propagate to the nucleotide binding loops and cleft.
Nucleotide binding
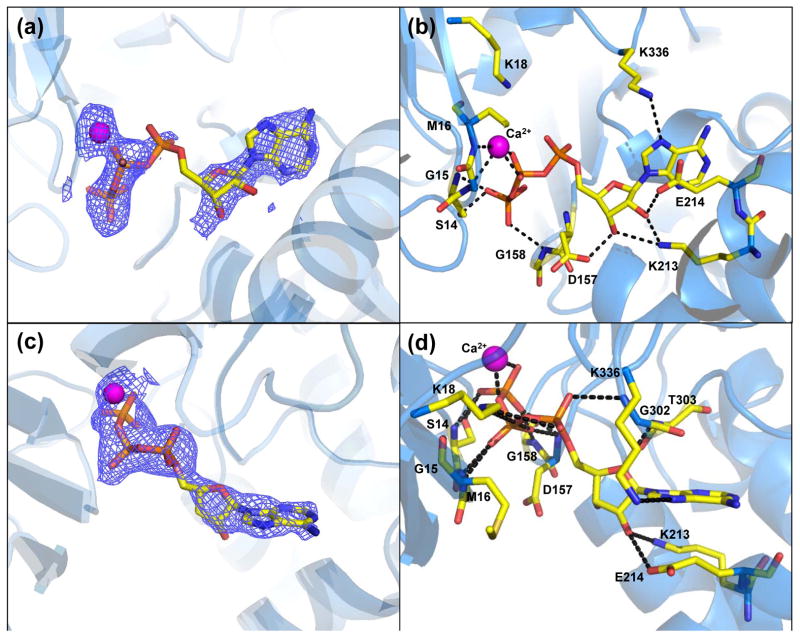
In both the wide-open and closed states there was clear interpretable density for modeling ATP binding (Fig. 3(a,c)). The hydrogen bonding between ATP and the surrounding residues is less extensive for the wide-open structure (Fig. 3(b,d)). The position and orientation of the wide-open state ATP was modeled differently than in the original search model (PDB 1HLU and is now in agreement with other actin structures. One major feature is the upward kinking of the α-phosphate in the wide-open state. In fact, this phosphate position appears to have more than one position as indicated by a lack of electron density for α-phosphate compared to β and γ-phosphates (Fig. 3(a)). Therefore, the ATP seems to be flexible and partially exchanging. The position of the Ca2+ is also different than in the 1HLU structure and is positioned on the other side of the β-phosphate, where it makes hydrogen bonds with the backbone residues Gly-15 and Met-16.
Figure 3.
Profilin:actin nucleotide binding sites (a) Wide-open state omit FO-FC electron density map (2.5 σ). Most of the nucleotide has ordered density, but the α-phosphate is in a bent conformation, and lacks clear density. (b) Wide-open state ATP binding site. (c) Closed state omit FO-FC map (2.5 σ) showing continuous, well ordered density for the nucleotide and bound Ca2+. (d) Closed state ATP binding site. For parts (b) and (d) dashed lines represent hydrogen bonds.
The closed state ATP can be seen in the standard conformation along with bound Ca2+. The electron density is continuous, indicating that the ATP is not moving, when compared to the wide-open state. When the large domains of our closed state actin are superposed with the nonexchanging-closed state actin (PDB 3CI5) the ATP molecules overlay in the same conformation (Fig. S1). The Ca2+ ion makes two coordinating bonds to oxygens on the β and γphosphates of ATP. From the FO-FC omit density it can be seen that there is one clear position for the nucleotide and metal. The position of the Ca2+ also agrees well with the nonexchanging-closed state. Whereas the nucleotide in the wide-open state is slightly shifted out of the binding cavity, the closed state nucleotide assumes the standard position.
Interesting differences exist in ATP binding between the wide-open and nonexchanging-closed actin. A list of close contacts between ATP and actin up to 3.5 Å for these two structures are given in Table 2 and are shown in supplemental Figure S2. As the nucleotide binding cleft closes, the S-loop (residues 14-16) moves towards the ATP by 2.9 Å (Fig. 4) and creates hydrogen bonds with ATP (Table 2). Also, Lys18 moves in and provides charge compensation for the buried phosphates. Asp-157 and Gly-302 also form hydrogen bonds with ATP in the nonexchanging-closed state. The conformation of ATP is similar, but the liganding metal positions are very different (Fig. 4 and Fig. S2), with the closed state metal facing the surface and coordinating 4 water molecules (not shown). The interaction between Gly-158 and the ATP γ-phosphate is broken, however an additional contact is created between Asp-157 and the same ATP γ-phosphate oxygen (Table 2). Note, Asp-157 was modeled with two conformations in the nonexchanging-closed state. With these structural changes actin is able to close with ATP in the active site.
Table 2.
Interactions between ATP and coordinating residuesa.
| Res 1 | Res # | Atom | Res 2 | Atom | Distance (Å) Wide-open state | Distance (Å) Nonexchanging-closed state |
|---|---|---|---|---|---|---|
| LYS | 336 | NZ | ATP | N7 | 3.3 | 3.2 |
| SER | 14 | OG | ATP | O3G | 2.4 | 2.6 |
| SER | 14 | N | ATP | O3G | 2.9 | 2.8 |
| LYS | 213 | NZ | ATP | O3′ | 3.1 | 3.2 |
| LYS | 213 | NZ | ATP | O2′ | 2.6 | 2.9 |
| GLU | 214 | OE2 | ATP | O2′ | 2.8 | 2.7 |
| ASP | 157 | OD2 | ATP | O3′ | 2.5 | |
| GLY | 158 | N | ATP | O1G | 3.1 | |
| ASP | 157 | N | ATP | O1G | 2.9 | |
| MET | 16 | N | ATP | O1B | 2.8 | |
| GLY | 15 | N | ATP | O1B | 2.9 | |
| LYS | 18 | NZ | ATP | O2B | 2.8 | |
| LYS | 18 | NZ | ATP | O1A | 2.8 | |
| GLY | 302 | N | ATP | O2A | 2.8 | |
| Ca2+ | 377 | Ca | ATP | O1B | 2.6 | |
| Ca2+ | 377 | Ca | ATP | O2B | 2.3 | |
| MET | 16 | N | Ca2+ | Ca | 2.4 | |
| GLY | 15 | N | Ca2+ | Ca | 3.1 | |
| Mg2+ | 402 | Mg | ATP | O2G | 1.9 | |
| Mg2+ | 402 | Mg | ATP | O2B | 2.3 |
S-loop residues 11-16; H-loop residues 70-78; G-loop residues 154-161
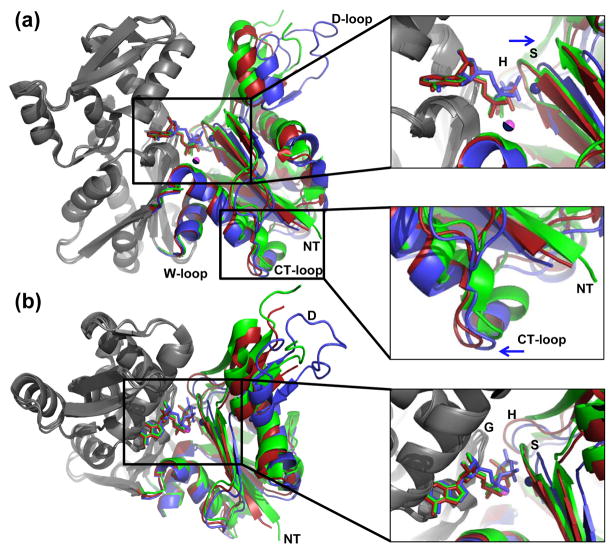
Figure 4.
The domain motions that close actin when ATP is loaded. (a) Wide-open state actin (blue) was superimposed with the partially-open state (red, PDB 3CHW) and nonexchanging-closed state (green, PDB 3CI5) actins. Their large domains (grey residues 150-331) were aligned using the program Superpose47(rmsd=0.45 Å and 0.65 Å, respectively). (b) Top view of the active site and entrance to the nucleotide binding cleft. Zoomed-in views are on the right. The Ca2+ and Mg2+ (pink) for the closed state and partially-open state have identical positions, and therefore only one atom is visible. The wide-open state Ca2+ is shown in the non-canonical position and is colored blue.
Concerted domain motions that close off the ATP binding cleft
The wide-open state represents the immediate binding of ATP. To visualize changes in nucleotide binding and the protein during cleft closure, a superposition of wide-open profilin:actin, partially-open profilin:actin and nonexchanging-closed actin was carried out. The actins were aligned by their large domains (residues 150-331; grey; Fig. 4). Analysis in Dyndom32 shows a 9° rotation between the two major domains as the wide-open conformation (Fig. 4, blue) completely closes to the nonexchanging-closed state (Fig. 4, green). A movie morphing these two structures is provided in the supplement. When the S-loop closes, ATP and Ca2+ shift down and are then stably bound by actin (Fig. 3(c)). A concerted motion moves the D-loop 6 Å from the His-40 mainchain, which closes off the solvent channel, protecting ATP from solvent. As was observed by Baek and coworkers, the partially-open conformation (Fig. 4, red) transitions to the nonexchanging-closed state (Fig. 4, green) which involves a smaller rotation of 5° between the large and small domains24. Also, to see if these features were unique to the nonexchanging-closed state, a superposition of our closed state actin with the nonexchanging-closed state actin was done by superposing the large domains (Fig. S1). There is a striking similarity between the two structures, especially in the ATP and bound metal positions.
From a close-up of the active-site (Fig. 4, right; Fig. S2) the overall conformation of the ATP molecules are the same. In Figure 4a, the wide-open state ATP can clearly be seen landing into the binding pocket and shifts into the active site of the more closed states (see the supplemental movie). In the nucleotide binding cleft (Fig. (4b)), all ATP atoms show close agreement at the adenine ring positions. The adenine rings are practically in the same position, and the nucleotide seems to form a hinge at ribose C1′, where the wide-open state ATP starts to rotate out of the cleft (Fig. 4(a)). Thus it appears that the incoming ATP from wide-open profilin:actin aligns first at the base and sugar, followed by the shifting of the phosphates into the canonical position as the small domain rotates and closes (Fig. 4(a)). At the one extreme, the phosphorus atoms of the γ-phosphate are shifted by 1.8 Å. This distance decreases from the γphosphate back to the sugar, where the O4′ atoms are shifted by 0.6 Å. As the phosphates begin to move into the canonical position, the S-loop (containing actin residues 11 to 16) shifts 2.9 Å into the cavity, which in turn moves the Ca2+ into its final closed state position. In the top views (Fig. 4(a,b); right panels) the wide-open state Ca2+ is closer to the S-loop. In fact, rearrangements of the H-bonding are pronounced here, as the oxygen atoms of the β-phosphate are now coordinated to the main chain via Ca2+, and not by direct H-bonding.
Motion of the well-ordered actin W-loop propagates to the active site
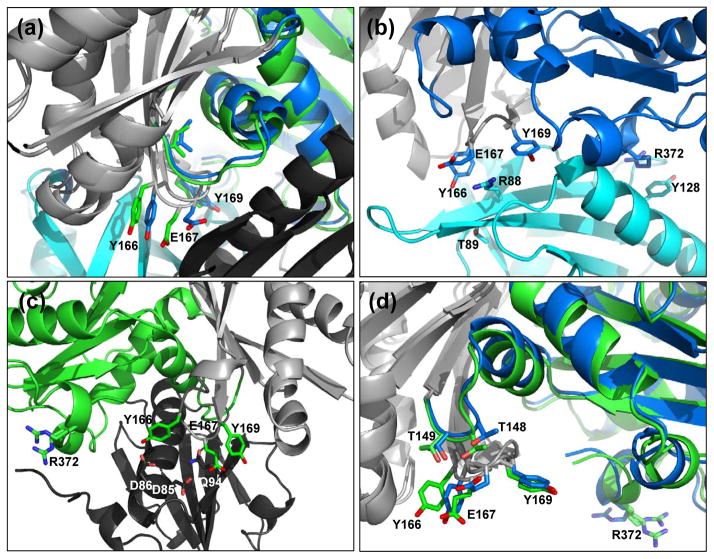
The actin W-loop (residues 165-172) is located at the actin barbed end, and makes contacts to stabilize profilin binding. The W-loop is interesting because it lies near a hydrophobic patch (called the “hot spot”) which makes up part of an important binding site for barbed-end actin-binding proteins12. A recent study has shown that the conformation of the W-loop is conformationally and functionally linked to the nucleotide-binding pocket33. Also, molecular dynamics simulations predicted that the W-loop transitions from an unstructured coil to an ordered β-turn upon ATP hydrolysis34. However, for both the wide-open and nonexchanging-closed states, the W-loop retains an ordered β-turn that is stabilized by backbone interactions between Tyr-166 and Tyr-169 (Fig. 5(a)). Although the conformation is the same, the W-loop is shifted 1.0 Å in the wide-open state compared to the nonexchanging-closed state. This movement of the nucleotide sensing W-loop propagates into a corresponding 1.0 Å movement of the G-loop and 2.9 Å movement of the S-loop.
Figure 5.
Actin W-loop conformations in the wide-open and nonexchanging-closed states. (a) W-loop residues for the wide-open state (blue) and the nonexchanging-closed state (green) rotated +45° from the standard view. Light gray denotes the superimposed regions (same as in Fig. 4) and profilin and gelsolin are colored cyan and black, respectively. (b) The W-loop of wide-open state actin interacts with profilin to stabilize the interaction. Note the stacking interactions between actin residue Arg-372 and profilin residue Tyr-128. Here, actin is in the standard view. (c) Gelsolin binds the opposite face of actin. Nonexchanging-closed state actin W-loop residues interact with gelsolin to stabilize the interaction. Here, the structure is rotated 180° from the standard view. (d) Intramolecular interactions between the actin W-loop actin residues Thr-148 and Thr-149. Here, actin is in the standard view.
Examination of the side-chain conformations and interactions explains how this movement is propagated from the W-loop to the nucleotide binding site. For the side-chains of the W-loop residues, the major difference is the conformation of Glu-167. In the profilin:actin interface, Glu-167 stacks against profilin residue Arg-88 in a strong ionic interaction. Arg-88 holds Glu-167 in this conformation (Fig. 5(b)). In the nonexchanging actin:gelsolin structure, Glu-167 is oriented in a different way that allows for stacking with gelsolin residue Gln-94 (Fig. 5(c)). The conformation of Glu-167 when profilin is bound to actin causes a steric conflict with Thr-148 of actin (Fig. 5(d)) that pushes the structural elements away in a rigid manner that propagates all the way to the nucleotide binding site and opens it. Profilin binds the actin barbed end in a bidentate fashion that include interactions between actin Arg-372 of the C-terminal domain and profilin Tyr-128 (Fig. 5(b); right side). Gelsolin binds actin differently, so in the nonexchanging-closed state the C-terminal domain is free to move. The net effect is the opposite motions of the C-terminal domain and S-loop that opens and closes the cleft (Fig. 4(a) compare green and blue structure; see also the movie in the supplement). Finally, these structures show that movements in the W-loop upon actin cleft opening by profilin are linked to conformational changes in the nucleotide-binding loop. These structural observations of the reciprocal communication between the W-loop and the nucleotide binding cleft are in agreement with the experimental measurements of Kudryashov and coworkers33.
The solvent accessibility of the ATP binding site
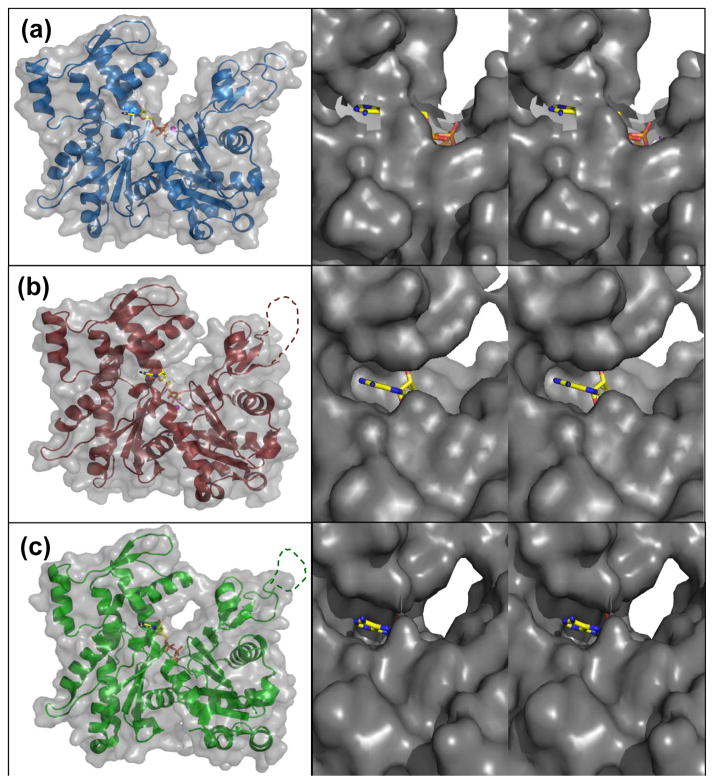
For the ADP to be replaced with ATP, it must have a clear path to and from the surrounding solvent. Solvent accessible surface (SAS) for the wide-open state shows a clear path for ATP to enter the active site, or alternatively for the ADP to be released to solvent (Fig. 6(a)). From the positions of the phosphates, a large cavity opens up into a channel that leads directly to the solvent. The nucleotide base is buried and tucked into its binding pocket. When partially-open profilin:actin is viewed in the same way, the channel begins to close off to the solvent (Fig. 6(b)). The large cavity that existed for the wide-open state has now closed off around the ATP phosphates. It can also be seen that the channel that leads to the surrounding solvent is now starting to pinch off, further preventing nucleotide exchange. The SAS for nonexchanging-closed actin shows the solvent channel is completely closed so it is not possible for nucleotide to exchange (Fig. 6(c)). The SAS areas for ATP in the wide-open, partially-open and nonexchanging-closed state are 95.1, 56.1 and 38.5 Å2, respectively.
Figure 6.
The wide-open state ATP of profilin:actin is more solvent exposed. (a) Wide-open state profilin:actin solvent accessible surface (SAS). (b) SAS of partially-open state profilin:actin. (c) SAS of nonexchanging-closed state actin. Zoomed-in crosseye stereo pairs (right panels) shows the solvent accessibility of ATP looking down the nucleotide cleft. For the zoomed views, the molecule was rotated 90° about the z-axis.
Wide-open profilin:actin forces Trp-356 to become solvent exposed
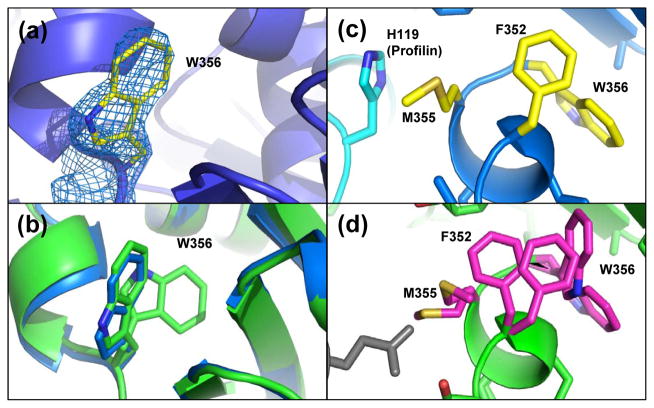
It was observed that when profilin binds to actin, a noticeable decrease in the intensity of tryptophan fluorescence occurs, which was thought to be caused by shielding of actin Trp-356 by profilin24. The wide-open state profilin:actin structure showed one clear position for Trp-356 (Fig. 7(a)). A comparison of Trp-356 side-chain orientations for the wide-open state with the partially-open state structure shows differing rotamer positions (Supplemental Fig. S3(a)). In the nonexchanging-closed actin, there are two positions for Trp-356 (Fig. 7(b) green); one of which overlays the wide-open Trp-356 position (Fig. 7(b) blue). In our closed state actin, there was evidence for two positions for Trp-356 and in the partially-open state Trp-356 is in the buried position (Supplemental Fig. S3(b)). In the structure of the nonexchanging profilin:actin, two conformations for Trp-356 and Phe-352 were determined and it can be seen how a second, and more buried conformer of Phe-352 compensates for the buried Trp-356 position (Fig. 7(d)). It can be seen in this structure how Trp-356 can be buried in the actin surface, due to a compensatory Phe-352 dual conformer that rotates to accommodate the indole ring (Figure 7(d)). Therefore, it appears that the interior position of Trp-356 occurs as the small domain rotates relative to the large domain and actin closes upon the ATP. Clearly, the binding of profilin in the wide-open state can cause Trp-356 to alter its position as was indicated by a change in tryptophan fluorescence24. Structurally, this is explained by the position of His-119 on profilin that contacts Met-355 across the interface. There are no contacts here in the nonexchanging-closed structure (Fig. 7(d)). The contact with profilin His-119 holds Met-355 in place, which consequently holds Phe-352 in place and squeezes Trp-356 out of the hydrophobic pocket and into the solvent (Fig. 7(c)). Interestingly, the unfavorable surface exposed position of the hydrophobic Trp-356 in the wide-open state probably helps make it a thermodynamically less stable state as was observed computationally25.
Figure 7.
Profilin prevents dual conformations for Trp-356 in the wide-open state. (a) 2FO-FC density (1.5 σ) for wide-open state Trp-356. (b) Trp-356 single conformation in the wide-open state (blue) and dual conformations in the nonexchanging-closed state (green). (c) In the wide-open state His-119 on profilin blocks the concerted motions of Met-355, Phe-352 and therefore, Trp-356 is extended to the surface. (d) Corresponding view for the nonexchaning-closed state showing the dual conformation for Met-355, Phe-352 and Trp-365 that are not blocked by gelsolin.
Secondary sites of Ca2+ binding
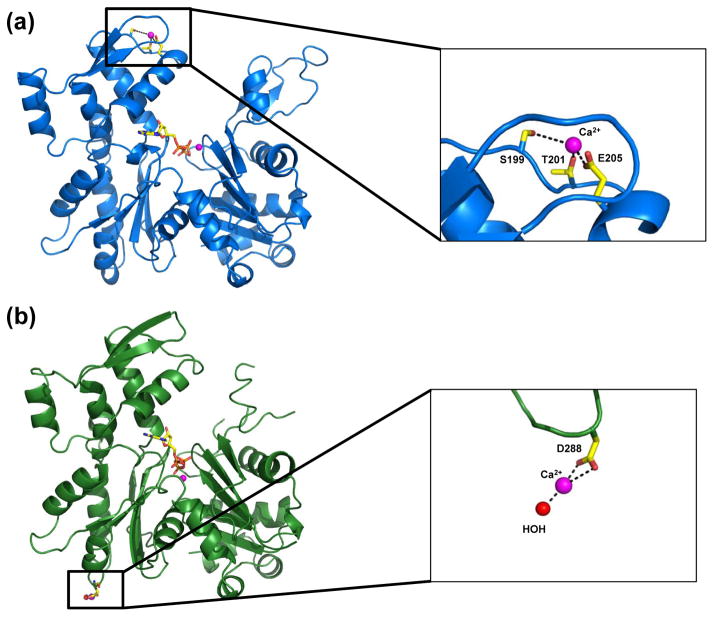
It is well documented that actin contains several low affinity binding sites for metals (i.e. Ca2+, Mg2+, K+)35,36. These sites also have known regulatory roles. For example, the transformation of Ca-G-actin to a polymerizable species is mediated by the binding of Mg2+ to a low affinity site, and not by replacing Ca2+ at the high affinity site35. In this study, a couple of secondary Ca2+ sites were found for profilin:actin. In the wide-open state, a Ca2+ binding site was found coordinating to Glu-205, Ser-199 and Thr-201 in subdomain 4 (Fig. 8(a)). This corresponds with a site found in un-complexed ADP actin (PDB 1J6Z)37. For the closed state profilin:actin, a secondary Ca2+ binding site was located off Asp-288. Ca2+ makes two bonds with the Asp sidechain, and is coordinated by a water molecule (Fig. 8(b)). There was no contribution from Asp-286. Similar binding of Ca2+ can been seen with structure (PDB 1NWK) of an uncomplexed actin in the ATP state (PDB 1NWK)38.
Figure 8.
Secondary Ca2+ binding sites on actin. (a) Secondary Ca2+ binding site on wide-open profilin:actin subdomain 4. (b) Secondary Ca2+ binding site on closed state profilin:actin subdomain 3.
Conclusions
The role of the D-loop in actin nucleotide exchange has remained unclear. In an experiment that tested the proteolytic susceptibility of actin upon exchanging ATP for ADP (with Mg2+ as the tightly bound cation), the D-loop became more resistant to cleavage by subtilisin and a novel E. Coli protease28. It seems likely that this is the result of a conformational change in the D-loop, where it assumes a more compact structure. It is interesting that in our wide-open profilin:actin, where the nucleotide is exchanging, the D-loop is much more far away from the ATP site than in other structures. What has most frustrated our understanding of the structural basis of actin nucleotide exchange is a lack of actin structures in the open state with a modeled D-loop. Considering that the D-loop is highly mobile, this difficulty in obtaining D-loop structures likely stems from issues with crystal packing this flexible region into a crystal lattice. It was suggested that although the partially-open state profilin:actin could explain the stimulation of nucleotide exchange, the short-lived state by which the nucleotide is actually released may be described by a more wide-open cleft24. This finding is also supported by a molecular dynamics simulation of profilin:actin, where it was shown that the wide-open state of the original room-temperature structure was a short lived state, and that the cleft immediately closed upon profilin disassociation25. For the ATP to enter or exit the binding pocket, the cleft would have to be wide-open, and this tertiary state would likely be unstable so it closes and becomes protected from solvent.
For the bound nucleotide to be exchanged, it must be given proper space to enter or exit the nucleotide binding cleft. Before profilin binds, the actin nucleotide binding cleft is in the closed position. Profilin then binds to actin subdomains 1 and 3, which rotate relative to each other by 4.7° to pinch around the profilin24. An opening of the nucleotide binding cleft subdomains 2 and 4 complements this rotation. When profilin binds the actin W-loop is moved by 1 Å in the wide-open state, a movement that propagates to the G-loop in the nucleotide-binding pocket, causing another 1 Å shift that further stabilizes ATP binding. Therefore, it would seem that as profilin binds to the actin barbed end, movement of the W-loop is structurally linked to the nucleotide-binding site. From the SAS (Fig. 6) it was shown that a partially opened profilin:actin has a tightly packed ATP, and although the cleft is slightly open, is still insufficient for releasing a bound nucleotide. Therefore, actin needs a mechanism by which the cleft can fully open to exchange nucleotide. The wide-open profilin:actin is shown to have a clear solvent channel leading from the nucleotide binding cleft to the bulk solvent (Fig. 6(b)).
Analysis of the close contacts and hydrogen bonds between the wide-open and the nonexchanging-closed state actin shows a change in the hydrogen bonding pattern (Table 2). A 9° rotation of the small domain relative to the large domain causes propagates to the nucleotide binding loops, causing them to shift into the nucleotide binding cleft. The bound Ca2+, which is coordinated to residues G15 and M16 is pushed into the canonical position as it coordinates to the β and γ-phosphates. The results further show how it is the wide-open state of PA that is actively exchanging nucleotide. Since it is known that mammalian profilin rapidly increases the rate of nucleotide exchange when binding actin, it is now clearer how this mechanism occurs, through the opening of the cleft, and formation of an active channel by which the nucleotide can be exchanged. On the other hand, for actin to be opened and for ADP to leave it may follow a nearly identical, yet reverse pathway.
Materials and Methods
Bovine profilin-β-actin was purified in the presence of Ca2+-ATP in the open form following published protocols39,40. Profilin:β-actin was crystallized using batch crystallization and microseeding. The purified precipitated protein was resolubilized at 10–15 mg ml−1 in a 5 mM phosphate (pH 7.6) buffer containing 0.5 mM adenosine-5′ triphosphate (ATP), 0.2 mM CaCl2 and 1.0 mM dithiothreitol (DTT), and clarified by centrifugation. The resolubilized protein was then dialyzed against 1.3 M phosphate (pH 7.3) containing 0.5 mM ATP, 0.2 mM CaCl2 and 1.0 mM DTT. After 8 h, a microcrystalline precipitate of actin paracrystals formed and was removed by ultracentrifugation. The supernatant was filtered with 0.22 mm Millex-GV filter units from Millipore, and 5–30 ml hanging drops were suspended above the dialysis solution for crystallization. Microseeds were grown in unfiltered drops and were used to seed the filtered hanging drops. Crystals grew in 24–36 h, to an average size of 0.5 × 0.35 × 0.15 mm. Crystallization was performed in a cold room at 277 K. For the wide-open state data, a profilin:actin crystal of dimensions 0.5 × 0.3 × 0.1 was soaked in a solution of 1.8 M KPO4 pH 7.36 with 5 mM DTT and 1 mM ATP. The crystal was then transferred to a 10 μL drop of the same buffer, but with 35% glycerol to act as a cryoprotectant. All manipulations of profilin:actin crystals took place in a cold room at 4 °C. The crystal was mounted on a Mitegen micromount41, plunged in liquid N2 and transferred to a N2 gas stream. Data were collected using a Rigaku FR-E superbright Cu Kα rotating-anode generator operating at 45 kV and 45 mA and fitted with a quarter-χ goniometer. Beam focusing was carried out using VariMaxHR optics for crystals with large unit cells. Diffraction images were collected on an R-AXIS IV++ image-plate detector and were processed using Eval1542. For the partially closed data, a crystal was soaked in a solution of mother liquor with 7 M sodium formate to act as a cryoprotectant.
The wide-open cryocooled structure was solved by molecular replacement with Molrep software43 and the original room-temperature profilin:actin coordinates (1HLU) as a search model22 with Ca2+, ATP and solvent atoms removed from the initial model. The starting working R-value was 37% at 2.2 Å resolution and was reduced by cycles of rigid-body refinement, followed by restrained refinement using Refmac software44. The model was then improved by iterative cycles of crystallographic refinement and model building with Coot45 as follows. FO-FC maps showed clear density for bound ATP and Ca2+, as well as a site for a second Ca2+. Also present was clear density for an acetylated N-terminus for profilin. Due to a lack of interpretable electron density, actin N-terminal residues 1-5 and 41-49 of the D-loop were deleted. Low resolution omit maps calculated at 3 and 4 Å were used to retrace the D-loop. Omit phases with minimal model bias were calculated by deleting the region of interest and neighbors within 4 Å, by randomly displacing the remaining atoms 0.3 Å with Moleman246, followed by restrained refinement. During the last stage of refinement, the solvent waters were added. Final refinement statistics are listed in Table 1.
A cryocooled closed state profilin:β-actin structure was solved by molecular replacement in Molrep using PDB coordinates for room temperature profilin:actin (2BTF)21 as the search model, with ATP, Ca2+ and solvent atoms removed. The starting R-value was 44% at 2.6 Å resolution and was reduced by rigid body refinement, followed by restrained refinement using Refmac. FO-FC maps showed clear density for ATP, two Ca2+ and a Cl− ion that was fit using Coot. Due to a lack of interpretable electron density, the following regions were removed from the structure: actin N-terminal residues 1-5, subdomain 2 residues 37-50 and residues 65-67. The following residues were converted to Ala, due to a lack of side-chain density: Y53, I64, K84, K95, R210, E224, K291, K315 of actin, and M122 of profilin. Final refinement statistics are listed in Table 1.
Supplementary Material
Acknowledgments
This work was funded by NSF grant MCB-0718661 and the Nebraska Research Initiative and the Eppley Cancer Center Support Grant P30CA036727. We would like to thank Clarence E. Schutt (Princeton University) and Uno Lindberg (Stockholm University) for useful discussions. We would also like to thank Peter Simone for technical assistance.
Abbreviations used
- PIP2
phosphatidylinositol-4,5-bisphosphate
- SAS
solvent accessible surface
Footnotes
Accession Numbers
Coordinates and structure factors for both wide-open and closed structures have been deposited in the Protein Data Bank33 with accession numbers 3UB5 and 3U4L, respectively.
References
- 1.Cooper JA. The role of actin polymerization in cell motility. Annu Rev Physiol. 1991;53:585–605. doi: 10.1146/annurev.ph.53.030191.003101. [DOI] [PubMed] [Google Scholar]
- 2.Gouin E, Gantelet H, Egile C, Lasa I, Ohayon H, Villiers V, Gounon P, Sansonetti PJ, Cossart P. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci. 1999;112 ( Pt 11):1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 3.Hoglund AS, Karlsson R, Arro E, Fredriksson BA, Lindberg U. Visualization of the peripheral weave of microfilaments in glia cells. J Muscle Res Cell Motil. 1980;1:127–146. doi: 10.1007/BF00711795. [DOI] [PubMed] [Google Scholar]
- 4.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 5.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–9. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 6.Sheterline P, Clayton J, Sparrow JC. Actin: Protein profile. 2002;4:1–2. [Google Scholar]
- 7.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–73. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 8.Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt CE, Lindberg U, Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- 9.Nyman T, Page R, Schutt CE, Karlsson R, Lindberg U. A cross-linked profilin-actin heterodimer interferes with elongation at the fast-growing end of F-actin. J Biol Chem. 2002;277:15828–15833. doi: 10.1074/jbc.M112195200. [DOI] [PubMed] [Google Scholar]
- 10.Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–81. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- 11.Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- 12.Wen KK, McKane M, Stokasimov E, Rubenstein PA. Mutant profilin suppresses mutant actin-dependent mitochondrial phenotype in Saccharomyces cerevisiae. J Biol Chem. 2011;286:41745–57. doi: 10.1074/jbc.M110.217661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–70. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 14.Perelroizen I, Didry D, Christensen H, Chua NH, Carlier MF. Role of nucleotide exchange and hydrolysis in the function of profilin in action assembly. J Biol Chem. 1996;271:12302–9. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- 15.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 16.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–94. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 17.Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009;284:12533–40. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lassing I, Lindberg U. Specificity of the interaction between phosphatidylinositol 4,5-bisphosphate and the profilin:actin complex. J Cell Biochem. 1988;37:255–67. doi: 10.1002/jcb.240370302. [DOI] [PubMed] [Google Scholar]
- 20.Lassing I, Lindberg U. Evidence that the phosphatidylinositol cycle is linked to cell motility. Exp Cell Res. 1988;174:1–15. doi: 10.1016/0014-4827(88)90136-x. [DOI] [PubMed] [Google Scholar]
- 21.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–6. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 22.Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 23.Page R, Lindberg U, Schutt CE. Domain motions in actin. J Mol Biol. 1998;280:463–74. doi: 10.1006/jmbi.1998.1879. [DOI] [PubMed] [Google Scholar]
- 24.Baek K, Liu X, Ferron F, Shu S, Korn ED, Dominguez R. Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc Natl Acad Sci U S A. 2008;105:11748–11753. doi: 10.1073/pnas.0805852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minehardt TJ, Kollman PA, Cooke R, Pate E. The open nucleotide pocket of the profilin/actin x-ray structure is unstable and closes in the absence of profilin. Biophys J. 2006;90:2445–2449. doi: 10.1529/biophysj.105.072900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 27.De Bruyn PPH. Theories of amoeboid movement. Quart Rev Biol. 1947;22:1–24. doi: 10.1086/395577. [DOI] [PubMed] [Google Scholar]
- 28.Strzelecka-Golaszewska H, Moraczewska J, Khaitlina SY, Mossakowska M. Localization of the tightly bound divalent-cation-dependent and nucleotide-dependent conformation changes in G-actin using limited proteolytic digestion. Eur J Biochem. 1993;211:731–742. doi: 10.1111/j.1432-1033.1993.tb17603.x. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W, Holmes KC. The actin fold. FASEB J. 1995;9:167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 30.Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 31.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poornam GP, Matsumoto A, Ishida H, Hayward S. A method for the analysis of domain movements in large biomolecular complexes. Proteins. 2009;76:201–212. doi: 10.1002/prot.22339. [DOI] [PubMed] [Google Scholar]
- 33.Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E. A nucleotide state-sensing region on actin. J Biol Chem. 2010;285:25591–601. doi: 10.1074/jbc.M110.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X, Diraviyam K, Sept D. Nucleotide effects on the structure and dynamics of actin. Biophys J. 2007;93:1277–83. doi: 10.1529/biophysj.107.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlier MF, Pantaloni D, Korn ED. Fluorescence measurements of the binding of cations to high-affinity and low-affinity sites on ATP-G-actin. J Biol Chem. 1986;261:10778–10784. [PubMed] [Google Scholar]
- 36.Murakami K, Yasunaga T, Noguchi TQ, Gomibuchi Y, Ngo KX, Uyeda TQ, Wakabayashi T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 37.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 38.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 39.Lovelace JJ, Murphy CR, Daniels L, Narayan K, Schutt CE, Lindberg U, Svensson C, Borgstahl GEO. Protein crystals can be incommensurately modulated. J Appl Cryst. 2008;41:600–605. [Google Scholar]
- 40.Carlsson L. PhD Thesis. Uppsala University; 1979. Cell motility: the possible role of unpolymerized actin. [Google Scholar]
- 41.Mitegen. MiteGen Micromounts. 2011. [Google Scholar]
- 42.Duisenberg AJM, Kroon-Batenburg LMJ, Schreurs AMM. An intensity evaluation method: EVAL14. J Appl Cryst. 2003;36:220–229. [Google Scholar]
- 43.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleywegt GJ. Validation of protein models from Calpha coordinates alone. J Mol Biol. 1997;273:371–376. doi: 10.1006/jmbi.1997.1309. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.