Abstract
Hepatitis C virus (HCV) infection is a major health problem recognized globally. HCV is a common cause of liver fibrosis that may lead to liver cirrhosis or hepatocellular carcinoma. The aim of this study was to estimate the prevalence of HCV infection and genotyping among Egyptian and Saudi Arabian chronic patients using different molecular techniques. HCV RNA viral load was assessed by real-time polymerase chain reaction (RT-PCR) technology. For HCV genotyping, RT-PCR hybridization fluorescence-based method and reverse hybridization line probe assay (INNO-LiPA) were used. A total of 40 anti-HCV-positive patients with chronic hepatitis C were examined for HCV RNA, genotyping, and different laboratory investigations. In the present study, HCV genotypes 4, mixed 4.1b, and 1 were detected in patients of both countries, while genotype 2 was only detected in Saudi Arabian patients. Genotyping methods for HCV showed no difference in the classification at the genotype level. With regard to HCV subtypes, INNO-LiPA assay was a reliable test in HCV genotyping for the detection of major genotypes and subtypes, while RT-PCR-based assay was a good test at the genotype level only. HCV genotype 4 was found to be the predominant genotype among Egyptian and Saudi Arabian chronic patients. In conclusion, data analysis for detecting and genotyping HCV was an important factor for understanding the epidemiology and treatment strategies of HCV among Egyptian and Saudi Arabian chronic patients.
Keywords: HCV RNA, detection, prevalence, genotyping
Introduction
Hepatitis C virus (HCV) is an enveloped positive single-stranded RNA virus that causes chronic hepatitis and liver disease worldwide.1,2 It is estimated that around 200 million people are infected with HCV worldwide, with a prevalence of 3.3% of the world’s population.3 Individuals infected with HCV are at risk of developing liver cirrhosis and hepatocellular carcinoma (HCC).4 The nucleotide sequence of the HCV genome is considered an important predictor of response to antiviral therapy.5 HCV demonstrates a high degree of sequence variability, resulting in the formation of quasi-species. However, the levels of heterogeneity differ considerably among the various regions of the virus, ranging from 19% in the 5′ untranslated region (5′ UTR) to 50% or more in the E1 region.6,7
HCV’s genome sequence is highly variable worldwide; six major types and approximately 80 subtypes have been recognized.8 The nucleotide diversity is about 31%–33% among genotypes and 20%–25% among subtypes.9 The region encoding envelope glycoproteins was the most variable when compared to the highly conserved 5′ UTR.10 Genotype 1 is the predominant genotype in many geographic regions, including Europe and North America, where it accounts for 50%–90% of cases. Genotypes 2 and 3 are also widely distributed. Genotype 2 is relatively common in Europe, North America, and Japan. Genotype 3 is common in Southeast Asia, Australia, South America, and northern Europe, in particular in intravenous drug users. Genotype 4 is found mainly in Egypt, the Middle East, and central Africa, genotype 5 in southern Africa, and genotype 6 in Southeast Asia. Despite the fact that they differ from each other by more than 30% of the RNA sequence, pathogenic differences between genotypes have not been demonstrated, except for steatosis associated with genotype 3a.11 However, because the genotype dictates the chance of therapeutic response and duration of treatment, genotyping has become fundamental in the clinical analysis of patients with hepatitis C.11 Genotypes 2 and 3 respond favorably, reaching 75% sustained response (SR) to 24 weeks of treatment with PEGylated interferon and ribavirin, while genotype 1 responds poorly, at 50% SR even after 48 weeks of treatment. Suitable treatment regimen for genotypes 4–6 is not yet established.11,12
The methods developed to investigate HCV genotypes are based on molecular biology or serological techniques. The reference methods for genotyping are sequencing and phylogenetic analysis.13 Although sequencing may be rationalized,14–16 simplified methods are preferable in the clinical routine and for epidemiology studies. Such methods include genotype-specific polymerase chain reaction (PCR),17 restriction fragment length polymorphism,18 serotyping,19,20 and INNO-LiPA.21,22 Overall, there is an acceptable concordance between these methods.23–25 Recently, real-time PCR (RT-PCR) on the Light Cycler instrument using hybridization probes has been applied for HCV genotyping.26,27 This report describes an alternative RT-PCR technique for simple and fast genotyping of HCV.
The prevalent genotype in Egypt is type 4, but other genotypes are also present. Genotype 4 was the commonest (73%) followed by genotype 1 (26%), and mixed infection was found in 15.7% of the studied samples.28 Genotype 4 followed by genotypes 1a and 1b are predominant in Saudi patients, but genotypes 2, 3, 5, and 6 are rare.29,30 Concurrent infection with more than one genotype, referred to as mixed genotype, can be found in the circulation of some HCV-infected patients, particularly in drug abusers and individuals who have received multiple transfusions.31 The rate of HCV mixed-genotype infections is variable in some groups of patients tested by different assays.23 It is difficult to assess the true prevalence of mixed-genotype infections by currently available assays, including direct DNA sequencing, since they are designed to identify only the HCV genotype dominant in the population.24,32 As a result, genotypes present at lower proportional levels in a mix could be missed or mistyped.33 Mixed infections with more than one genotype were observed in some studies. Two drugs have been approved for the treatment of chronic HCV infections. Interferon (IFN)-α has been approved for the treatment of compensated chronic hepatitis C. In addition, ribavirin, a guanosine analog, has been approved for use in combination with IFN-α. Recently, successful anti-HCV therapeutics such as sofosbuvir and ledipasvir have become available.
In the present study, we investigated the prevalence of HCV infection and genotyping among Egyptian and Saudi Arabian chronic patients using different molecular techniques. Our results showed no difference between genotyping molecular methods in classification at the genotype level. HCV genotype 4 was found to be the predominant genotype among Egyptian and Saudi Arabian chronic patients. The methods were accurate overall and provide an attractive alternative for HCV genotyping.
Patients and Methods
Study design and patient recruitment strategies
The study was conducted on 40 patients who were positive for anti-HCV by single enzyme-linked immunosorbent assay (ELISA). Recruitment of patients was random and in the absence of knowledge of HCV genotyping and subtypes. Our study was conducted on two different categories of patients; the first group comprised 20 patients (10 males, 10 females; mean age 50.35 years) from Alexandria Armed Forces Hospital in Egypt and 20 patients (14 males, 6 females; mean age 44.6 years) from King Fahd Specialist Hospital in Saudi Arabian. Informed written consent was obtained from each patient included in the study. The study protocol was approved by the ethics committee of the Alexandria Armed Forces Hospital, and the research was conducted in accordance with the principles of the Declaration of Helsinki. Overall samples were used for the establishment of the procedure for detection of HCV RNA, representing different HCV genotypes. All samples were genotyped by RT-PCR and line probe assay (INNO-LiPA), according to the manufacturer’s instructions. For the HCV RNA-positive patients, blood samples were collected for different biochemical analysis, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, platelets (PLTS), prothrombin time (PT), international normalized ratio (INR), and albumin. The whole blood sample (5 mL) was left to coagulate and then centrifuged at 3000 rpm/2100 × g for 10 minutes (Eppendorf centrifuge, model 5402). Serum was collected and stored in small aliquots at −80°C.
Serological detection of HCV
All serum samples were assayed for anti-HCV positivity by ELISA (third-generation ELISA, murex anti-HCV version III, VK 47), following the manufacturer’s instructions. Briefly, diluted samples or controls were loaded into a 96-well plate precoated with a recombinant HCV-specific antigen. The plate was then incubated for one hour at 37°C to allow for the formation of the Ag–Ab complex. The plate was washed, the conjugate was added, and the plate was incubated for 30 minutes at 37°C. After incubation, plate was washed and a TMB substrate solution (colorimetric microwell substrates HRP applications-based immunoassays) was added for detection. Finally, the reaction was stopped using H2SO4, and the colorimetric signal was measured by absorbance at 450 nm using a spectrophotometer (Multiscan “Plus” DASIT SPA).
Biochemical tests
To 500 μL of ALT or AST, reagent 1 was added 100 μL of serum or blank in the test tube. The tube was mixed and incubated for 30 minutes at 37°C. Then, 500 μL of ALT or AST reagent 2 was added to the tube, and the tube was mixed and incubated for 20 minutes at 37°C. After the incubation, 500 μL of sodium hydroxide was added to the tested tube. The reaction was measured for the absorbance of each at 546 nm after five minutes.
To measure bilirubin, 200 μL of reagent 1, one drop of reagent 2, 1000 μL of reagent 3, and 200 μL of serum samples or blank were added in the tested tube. The tube was mixed and incubated for 10 minutes at 20–25°C. After incubation, 1000 μL of reagent 4 was added. The reaction was measured for absorbance of each sample at 578 nm (560–600 nm), and the color intensity was stabled for 30 minutes.
Molecular detection of HCV
RNA extraction
Viral RNA was extracted using a viral RNA mini kit according to the manufacturer’s instructions by using spin column protocol (Applied Biosystems). Briefly, 560 μL of prepared viral lysis buffer (AVL) containing carrier RNA and 140 μL of serum were pipetted together in a 1.5-mL microcentrifuge tube and incubated at room temperature for 10 minutes. Then, 560 μL of ethanol (97%) was added to each sample and mixed by pulse-vortexing for 15 seconds. Next, 630 μL of the previous solution were carefully applied to the QIAamp spin column (in a 2-mL collection tube) and centrifuged at 8000 rpm/10.017 × g (Eppendorf centrifuge, model 5402) for one minute. The QIAamp spin column was placed into a clean 2 mL collection tube, and 500 μL of AW1 buffer was added and centrifuged at 8000 rpm/10.017 × g for one minute. The QIAamp spin column was placed again in a clean 2 mL collection tube, and 500 μL of buffer AW2 was added and centrifuged at full speed, 14000 rpm/20.913 × g for three minutes. Finally, 60 μL of AVE buffer was added, equilibrated to room temperature for one minute, then centrifuged at 8000 rpm/10.017 × g for one minute. The total HCV RNA was extracted and collected in sterile vials for amplification.
RT-PCR of HCV
A RT-PCR test was done using RT-PCR reagents that constitute a ready-to-use system for the detection of HCV RNA by PCR in a Stratagene’ Mx3000P quantitative RT-PCR system. The HCV RT-PCR kit included reagents and enzymes for the reverse transcription and specific amplification of a specific region of the HCV genome in a fluorescence detector FAM (reporter dye). The kit has a second heterologous amplification system to identify possible PCR inhibition. TaqMan Universal PCR Master Mix 2-fold (Applied Biosystems) was added including an optimized RT-PCR buffer, MgCl2, Taq DNA polymerase, and stabilizers. HCV-RNA was amplified by RT-PCR using primers KY80 (5′GCAGAAAGCGTCTAGCCATGGCGT) and KY78 (5′CTCGCAAGCACCCTATCAGGCAGT) targeting the 244-base region located within the highly conserved 5′ noncoding region of the HCV genome. The reaction took place under stander thermal profile: incubation at 48°C for 30 minutes to transcribe viral RNA to cDNA by RT. This was followed by AmpliTaq gold activation at 95°C for 10 minutes. Denaturation was performed at 95°C for 15 seconds, followed by annealing and extension at 60°C for one minute with end point fluorescence detection.34 The fluorescence intensity increases proportionally with each amplification cycle in response to the increased amplicon concentration. This allows quantification of the template to be based on the fluorescent signal during the exponential phase of amplification, before limiting reagents, accumulation of inhibitors, or inactivation of the polymerase has started to have an effect on the efficiency of amplification. Software provided in the computer system should connect to the apparatus allowing real-time amplification plots to be viewed and to be analyzed during the PCR run.
HCV genotyping
Our samples were genotyped using both the RT-PCR hybridization fluorescence-based method and reverse hybridization line probe assay (INNO-LiPA).
RT-PCR, genotyping
HCV RNA-positive samples were genotyped using an HCV real-time genotype kit (AmpliSens® HCV-genotype-FRT PCR kit). It was able to detect HCV genotypes 1a, 1b, 2, 3, and 4, following the manufacturer’s instructions. Briefly, a 5 μL of a sample of cDNA, 4 μL of TaqF polymerase, and 6 μL of each PCR mix (PCR-mix-1-FRT HCV 1b/3, PCR-mix-1-FRT HCV 1a/2, and PCR-mix-1-FRT HCV 4/IC) were distributed on a MicroAmp® Optical 96-Well Reaction Plate. The PCR reactions were done in a 7500 Fast Real-Time PCR System (Applied Biosystems, Roche). Fluorescence curves were analyzed with Fast 7500 Sequence Detection Software v2.1 (Applied Biosystems, Roche) using the following cycling parameters: 50°C for two minutes; 95°C for 10 minutes; and 40 cycles at 95°C for 15 seconds, 50°C for 30 seconds, and 60°C for one minute. The total time required for performing this assay was only two hours, as previously described.35
INNO-LiPA II, genotyping
The line probe assay was to assess HCV genotypes using the versant HCV genotype 2.0 assay (LiPA), which is based on genotype-specific oligonucleotides from the 5′ UTR that are immobilized on a nitrocellulose strip. The 5′ UTR core region of HCV was amplified using RT-PCR, and the oligonucleotides were probed with a biotin-labeled 5′ UTR amplicon. The labeled amplicon was allowed to hybridize and mounted on a strip. After stringent washing, streptavidin labeled with alkaline phosphatase was used to trace the hybridized products, and BCIP/NBT chromogen was used as a substrate according to the manufacturer’s instructions. The probe reactivity patterns were interpreted using the chart provided by the manufacturer.36
Statistical analysis
A simple linear regression model was used to determine the relationship between RT-PCR and different clinical parameters. The Pearson product–moment correlation coefficient (r) was calculated to measure the degree of that relationship, and testing the significance of r was also carried out at P = 0.05. The analysis was applied using Sigma-Plot® 12.5 software. The analysis of variance model was used to test for the significance of resulting slopes of each regression line; and model of analysis of co-variance (ANCOVA) was applied to compare regression slopes of patients’ levels (Egyptian and Saudi) using adjusted (ie, least squares) means to determine if the slopes were significantly different from each other.37 Descriptive statistics for patients’ age (n = 20/level) were represented by a box plot constructed in Sigma-Plot® 12.5. To determine the differences in HCV genotypes between Egyptian and Saudi patients, as detected by RT-PCR and INNO-LiPA, a two independent-samples t-test, with two-tailed alternative hypothesis (Egyptian patients’ genotype ≠ Saudipatients’ genotype) was applied using SigmaPlot® 12.5 software extended with the statistical package. For any genotype that appeared once, a one-sample t-test, with single genotype as the hypothesized value, was applied to compare the single genotype with those detected in other patients.
Results
A total of 40 diagnosed HCV-positive cases, confirmed by RT-PCR for quantitative HCV RNA assay, were analyzed for HCV genotyping by RT-PCR and INNO-LiPA. Of the 40 patients included in the study, 20 (50%) were Egyptian and 20 (50%) were Saudi Arabian. The age was roughly symmetric around the median of Egyptian patients (49.50 years) with a minimum and maximum range of 32–76 years and Saudi patients (46 years) with a minimum and maximum range of 22–59 years (Fig. 1).
Figure 1.
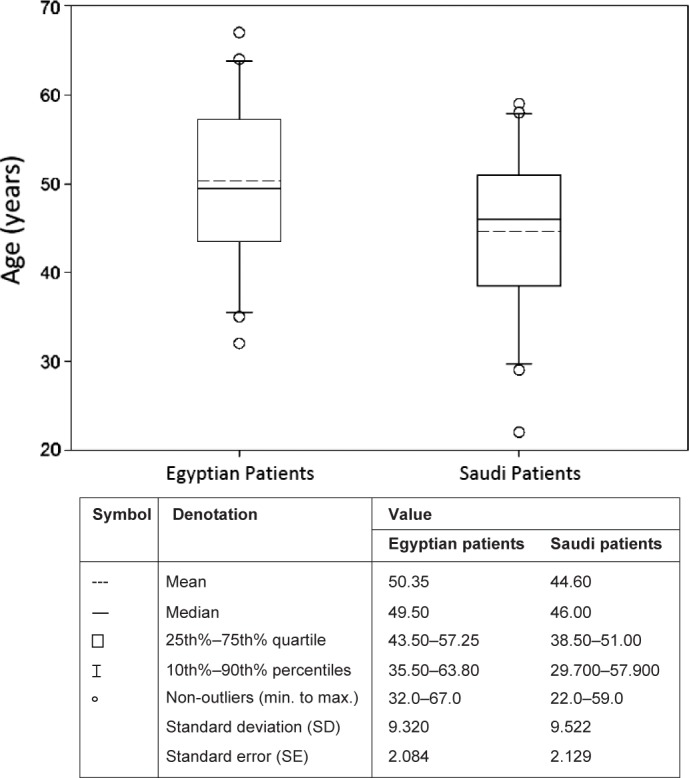
Shows the descriptive statistics for Egyptian and Saudi patients’ age (n = 20/level) represented by a box plot constructed in SigmaPlot® 12.5 software. The age was roughly symmetric around the median of Egyptian patients (49.50 years) with a minimum and maximum range of 32–76 years and Saudi patients (46 years) with a minimum and maximum range of 22–59 years.
Both RT-PCR and INNO-LiPA as genotyping methods have identified variations among Egyptian and Saudi Arabian patients for HCV genotypes and subtypes. HCV genotypes 4, mixed 4.1b, and 1 were detected in patients of both countries, while genotype 2 was only detected in Saudi Arabian patients. With regard to HCV subtypes, INNO-LiPA assay was a reliable test in HCV genotyping for the detection of major genotypes and subtypes, while RT-PCR-based assay was a good test at the genotype levels only. HCV subtypes 1a; 4a,4c,ad; and 4e were detected in Egyptian patients, while subtypes 1a; 1b; 4a,4c; 4c,4d; 4a,4c,4d; 4e; and 4a,4c,4d,1 were detected in Saudi Arabian patients (Table 1). The present study found that HCV genotype 4 was the predominant genotype among Egyptian and Saudi Arabian chronic patients. Of the Egyptian samples, 17/20 (85%), 2/20 (10%), and 1/20 (5%) were genotypes 4, mixed 4.1b, and 1, respectively. Of the Saudi Arabian samples 12/20 (60%), 5/20 (25%), 2/20 (10%), and 1/20 (5%) were genotypes 4, 1, mixed 4.1b, and 2, respectively (Fig. 2).
Table 1.
Shows both RT-PCR and INNO-LiPA as genotyping methods for HCV genotypes and subtypes among Egyptian and Saudi Arabian patients. Both methods showed variation in HCV genotypes and subtypes. HCV genotypes 4, mixed 4.1b, and 1 were detected in patients of both countries, while genotype 2 was only detected in Saudi Arabian patients. With regard to HCV subtypes, INNO-LiPA assay was a reliable test in HCV genotyping for the detection of major genotypes and subtypes, while RT-PCR-based assay was a good test at the genotype levels only. HCV subtypes 1a; 4a,4c,ad; and 4e were detected in Egyptian patients, while subtypes 1a; 1b; 4a,4c; 4c,4d; 4a,4c,4d; 4e; and 4a,4c,4d,1 were detected in Saudi Arabian patients.
| INNO-LiPA | RT-PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EGYPTIAN PATIENTS | TOTAL | SAUDI ARABIAN PATIENTS | TOTAL | |||||||
| 1 | 2 | 4 | 4,1b | 1 | 2 | 4 | 4,1b | |||
| 1 | – | – | – | – | – | 1 | – | – | – | 1 |
| 1a | 1 | – | – | – | 1 | 2 | – | – | – | 2 |
| 1b | – | – | – | – | – | 2 | – | – | – | 2 |
| 2 | – | – | – | – | – | – | 1 | – | – | 1 |
| 4 | – | – | 11 | – | 11 | – | – | 7 | – | 7 |
| 4a,c | – | – | – | – | – | – | – | 1 | – | 1 |
| 4c,d | – | – | – | – | – | – | – | 1 | – | 1 |
| 4a,c,d | – | – | 5 | – | 5 | – | – | 1 | – | 1 |
| 4e | – | – | 1 | – | 1 | – | – | 2 | – | 2 |
| 4,1b | – | – | – | 2 | 2 | – | – | – | 1 | 1 |
| 4,a,c,d,1 | – | – | – | – | – | – | – | – | 1 | 1 |
| Total | 1 | – | 17 | 2 | 20 | 5 | 1 | 12 | 2 | 20 |
Figure 2.
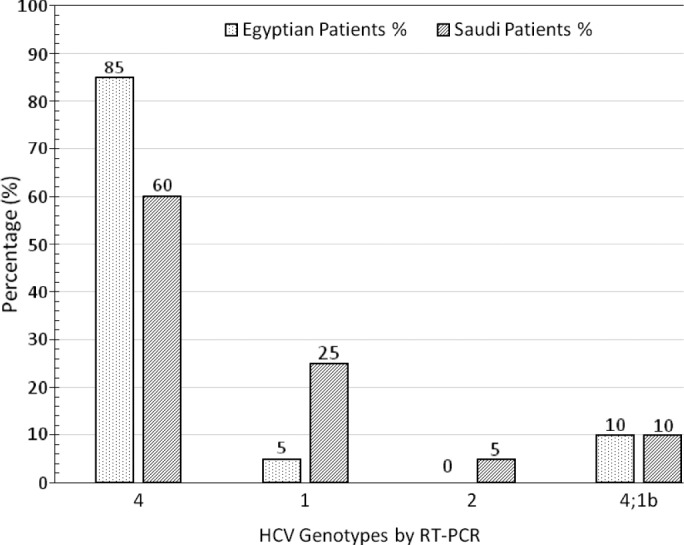
Shows variation of HCV genotypes among Egyptian and Saudi Arabian patients identified by RT-PCR. Our study showed that HCV genotype 4 was found to be the predominant genotype among Egyptian and Saudi Arabian chronic patients. Of these samples, 17/20 (85%), 2/20 (10%), and 1/20 (5%) of Egyptian patients were genotypes 4, mixed 4.1b, and 1, respectively. Of the Saudi Arabian patients, 12/20 (60%), 2/20 (25%), 2/20 (10%), and 1/20 (5%) were genotypes 4, 1, mixed 4.1b, and 2, respectively.
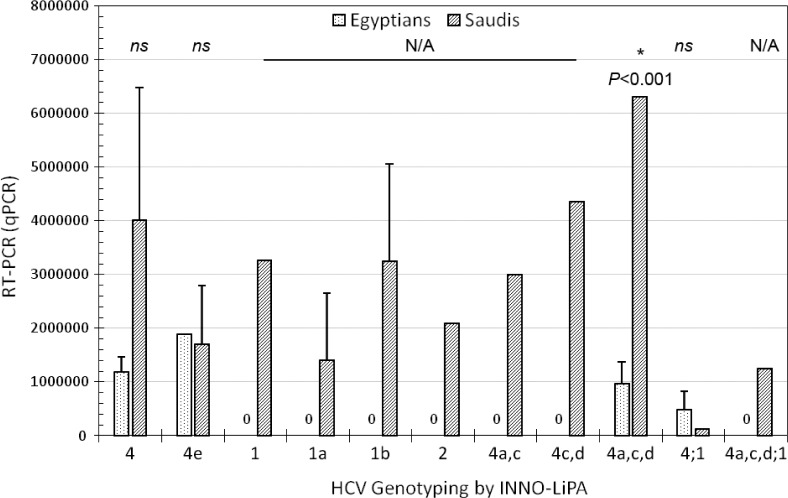
Figures 3 and 4 show the prevalence of HCV infection identified by quantitative RT-PCR among Egyptian and Saudi Arabian patients in correlation with different HCV genotypes. Our results showed that the viral load of genotype 4 in Egyptian patients was significantly lower than in Saudi patients (P = 0.0426). No significant differences were detected between the two patients’ levels at other HCV genotypes (Fig. 3). The viral load for HCV subtypes 4a, 4c, and 4d in Egyptian patients was significantly lower than in Saudi patients (P < 0.001). No significant differences were detected between the two patients’ levels in other HCV subtypes (Fig. 4).
Figure 3.
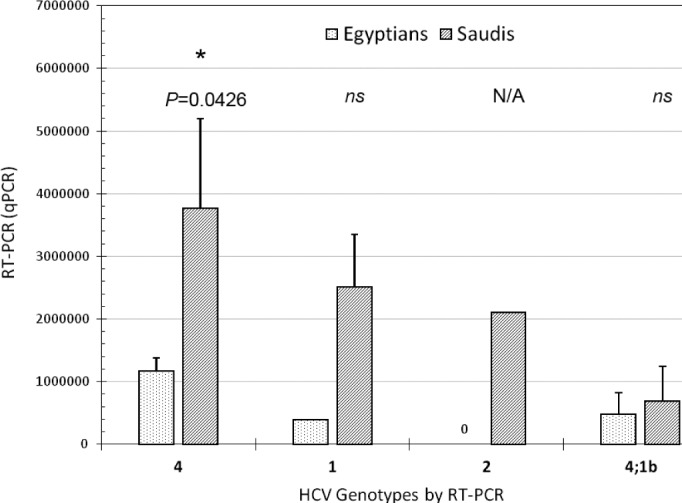
Shows the prevalence of HCV-infection identified by quantitative RT-PCR among Egyptian and Saudi Arabian patients and HCV genotypes detected by RT-PCR. Our results showed that the viral load of genotype 4 in Egyptian patients was significantly lower than in Saudi patients (P = 0.0426). No significant differences were detected between the two patients’ levels at other genotypes. Stars represent the significance level between Egyptian and Saudi patients; ns denotes not significant. Error bars represent standard error (SE). (*p < 0.05, n.s. p > 0.05).
Figure 4.
Shows the prevalence of HCV-infection identified by quantitative RT-PCR among Egyptian and Saudi Arabian patients and HCV genotypes and subtypes detected by INNO-LiPA. The viral load for HCV subtypes 4a,4c,4d in Egyptian patients was significantly lower than in Saudi patients (P < 0.001). No significant differences were detected between the two patients’ levels at other HCV subtypes. Stars represent the significance level between Egyptian and Saudi patients; ns denotes not significant. Error bars represent standard error (SE). (*p < 0.05, n.s. p > 0.05).
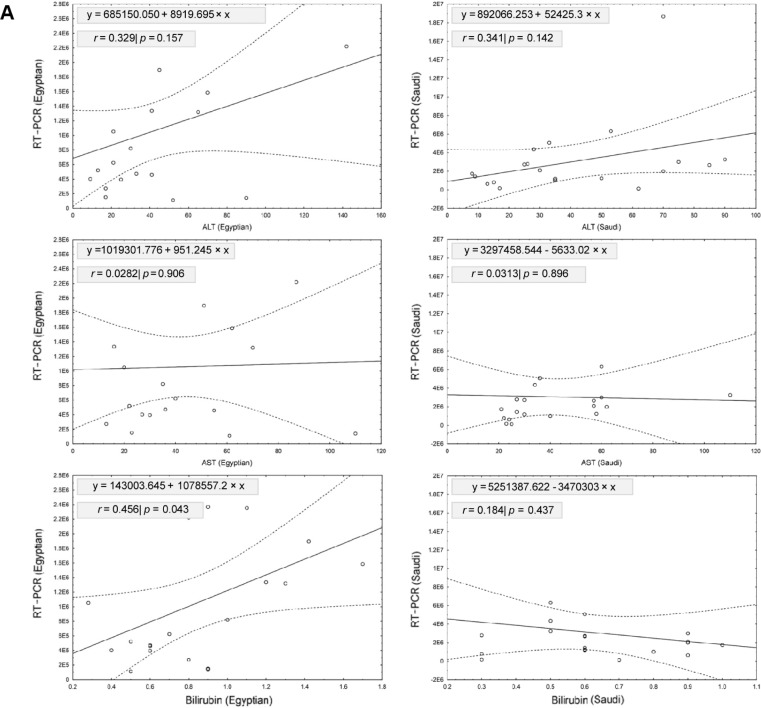
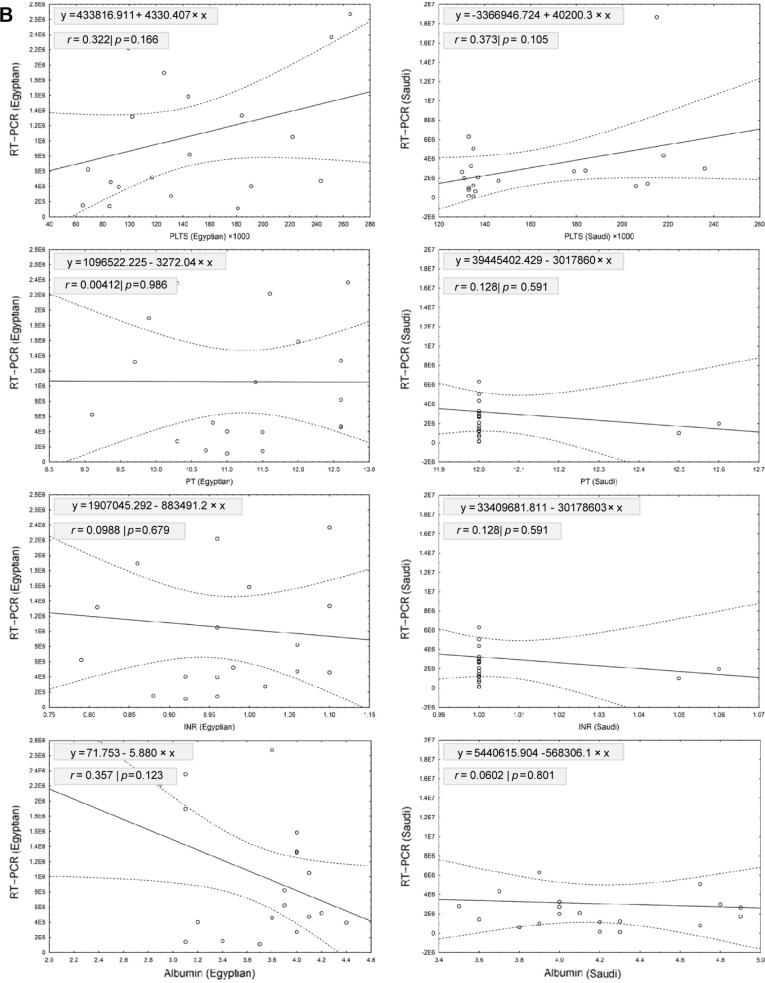
Table 2 shows the linear regression model for relationship between RT-PCR tested for HCV RNA viral load and the other biochemical analyses for patients included in our study, which were ALT, AST, bilirubin, PLTS, PT, INR, and albumin. Our results showed that a significant positive relationship was detected between RT-PCR and bilirubin in Egyptian patients (r = 0.4560; slope = 1078557.2; P = 0.043). However, no significant relationships were detected between RT-PCR and other biochemical tests from Egyptian and Saudi patients (Fig. 5). Testing the variations between slopes of regression lines by ANCOVA revealed no significant differences between slopes of age (F2,36 = 0.636, P = 0.535), ALT (F2,36 = 2.353, P = 0.109), AST (F2,36 = 0.017, P = 0.983), bilirubin (F2,36 = 0.778, P = 0.467), PLTS (F2,36 = 2.875, P = 0.069), PT (F2,36 = 0.287, P = 0.752), INR (F2,36 = 0.294, P = 0.747), and albumin (F2,36 = 0.219, P = 0.804) among the two levels of patients.
Table 2.
Shows the linear regression model for relationship between RT-PCR tested for HCV RNA viral load and different clinical data for patients included in the study. Our results showed that a significant positive relationship was detected between RT-PCR and bilirubin in Egyptian patients (r = 0.4560; slope = 1078557.2; P = 0.043). However, no significant relationships were detected between RT-PCR and other measurements from Egyptian and Saudi patients.
| PATIENTS | EGYPTIANS | SAUDIS | ||||
|---|---|---|---|---|---|---|
| r | SLOPE | P-VALUE | r | SLOPE | P-VALUE | |
| Age | 0.0939 | 8573.807 | 0.694 | 0.1880 | 79099.6 | 0.428 |
| ALT | 0.3290 | 8919.695 | 0.157 | 0.3410 | 52425.3 | 0.142 |
| AST | 0.0282 | 951.2450 | 0.906 | 0.0313 | −5633.02 | 0.896 |
| Bilirubin | 0.4560 | 1078557.2 | 0.043* | 0.1840 | −3470303 | 0.437 |
| PLTS | 0.3220 | 4330.407 | 0.166 | 0.3730 | 40200.30 | 0.105 |
| PT | 0.0041 | −3272.040 | 0.986 | 0.128 | −33017860 | 0.591 |
| INR | 0.0988 | −883491.2 | 0.679 | 0.128 | −30178603 | 0.591 |
| Albumin | 0.3570 | −5.880 | 0.123 | 0.0602 | −568306.1 | 0.801 |
Note:
Represent the significance at P < 0.05.
Figure 5.
The linear regression model for relationship between RT-PCR tested for HCV RNA viral load and different biochemical analyses for patients included in our study; these biochemical analyses are ALT, AST, bilirubin, PLTS, PT, INR, and albumin. (A) Shows the linear regression model for relationship between RT-PCR tested for HCV RNA viral load and different liver enzymes (ALT, AST, bilirubin). Our results showed that a significant positive relationship was detected between RT-PCR and bilirubin in Egyptian patients (r = 0.4560; slope = 1078557.2; P = 0.043). However, no significant relationships were detected between RT-PCR and ALT or AST from Egyptian and Saudi patients. (B) Shows the linear regression model for relationship between RT-PCR tested for HCV RNA viral load and different biochemical tests. Our results showed no significant relationships were detected between RT-PCR and these biochemical tests from Egyptian and Saudi patients. Scattered plots showed the relationship between RT-PCR and different clinical parameters for Egyptian (left) and Saudi (right) patients. Solid lines represent the linear regression with ±95% confidence intervals (dashed line).
Discussion
Genotyping of HCV has been regarded as a fundamental factor for treatment, prognosis, and prediction in clinical management,38 so that to begin the treatment and then follow-up are in accordance with the virus genotype.39 The length of the therapy is also influenced by genotypes.5,11 Recent studies have shown that the distribution of HCV genotypes varies in different geographical regions. Thus, determination of the predominant and less prevalent genotypes and subtypes of HCV in various areas has a fundamental role. The interest for identifying HCV genotypes is increasing worldwide.39 The identification of different HCV genotypes, subtypes, and isolates is helpful in understanding the evolution and the epidemiology of the virus in relation to patient’s age, area, risk factors, and degree of liver disease.40 Although sequence analysis is considered the gold standard for HCV genotype determination, it is expensive, time consuming, and inconvenient for routine use. Analysis of the 5′ UTR by the RT-PCR hybridization fluorescence-based method and reverse hybridization line probe assay (INNO-LiPA) provides a fast, cheap, and easy method for determination of the HCV genotype and subtypes.41 To date, there are only a few comparison studies of these two tests and most of them are reported from Europe and North America.42,43 Our study was the first comparison between these two methods using clinical samples from Egypt and Saudi Arabia. This study analyzed 40 HCV-positive samples for genotyping. The results indicated that HCV genotypes 4, mixed 4.1b, and 1 were detected in patients of both countries, while genotype 2 was only detected in Saudi Arabian patients. HCV genotype 4 was found to be the predominant genotype among Egyptian and Saudi Arabian chronic patients. With regard to HCV subtypes, INNO-LiPA assay was a reliable test in HCV genotyping for the detection of major genotypes and subtypes, while RT-PCR-based assay was a good test at the genotype levels only. This finding was consistent with recent studies performed in Egypt and Saudi Arabia, all of which reported that the distribution of HCV genotypes varies in different geographical regions, and HCV genotype 4 was found to be the predominant genotype among Egyptian and Saudi Arabian chronic patients.29,30,43–47 In contrast, other results suggest that HCV genotyping of the 5′ UTR cannot accurately distinguish genotypes 1a and 1b.41,48 Our results showed that HCV subtypes 1a; 4a,4c,4d; and 4e were detected in Egyptian patients, while subtypes 1a; 1b; 4a,4c; 4c,4d; 4a,4c,4d; 4e; and 4a,4c,4d,1 were detected in Saudi Arabian patients. The lack of agreement reported in this study at the subtype level is not surprising considering the high diversity of genotype 4, especially in the Egyptian and Saudi Arabian populations as previously shown by others.49,50 Comparing serotyping and genotyping methods, the detection of a large number of individual mutated strains by hybridization assay such as INNO-LiPA in a single test was accurate and reliable for HCV genotypes and subtypes.42 Overall, the RT-PCR hybridization fluorescence-based method was also accurate and correctly identified all samples belonging to HCV genotypes 1–4. These results are similar to those reported by another study using the 5′ UTR with a type agreement of 99.5%.48,51
Mixed HCV with more than one genotype was observed in some studies with higher prevalence rates in multiple-exposure groups, such as hemophiliacs, patients on chronic hemodialysis (HD), and injection drug users.47,52,53 In the present study, the prevalence of mixed HCV genotype was seen in our patients from both countries, indicating the contribution and association of various risk factors to the clinical profile and disease progression in different patients. This finding was consistent with studies performed in Egypt,52 Saudi Arabia,46 Serbia,53 Hong Kong, and China.54
The prevalence of HCV infection identified by quantitative RT-PCR among Egyptian and Saudi Arabian patients was assessed. Our results showed that the viral load for genotype 4 and subtypes 4a,c,d in Egyptian patients was significantly lower than in Saudi patients (P = 0.0426 and P < 0.001, respectively). No significant differences were detected between the two patient groups’ levels for other genotypes and subtypes. These results are similar to those reported by another study, which showed a high prevalence of genotype 4 for HCV infection amongst a Saudi sample group, with high viral load.55 In that study, 22% of chronic hepatitis C (CHC) patients progressed to cirrhosis and another 22% had treatment; liver-related mortality was more common in patients with advanced cirrhosis. In the same context, there are current findings confirming the corrected prevalence of HCV-4 in Saudi Arabian to be at least 97.6%, greater than the previously reported prevalence observed among the Saudi population. Further, it was found that most Saudi patients are infected with a single subtype of HCV 4, without genetic variability. This suggests a recent circulation of a single HCV 4 strain among Saudi patients.44 The linear regression model for the relationship between RT-PCR testing for HCV RNA viral load and different clinical data was assessed. Our results showed a significant positive relationship between RT-PCR and bilirubin in Egyptian patients (r = 0.4560; slope = 1078557.2; P = 0.043). However, no significant relationships were detected between RT-PCR and other measurements from Egyptian and Saudi patients. This finding is in agreement with another study that showed elevation in the level of bilirubin in Egyptian HCV patients, in comparison with HCV RNA identified by RT-PCR.56
In conclusion, HCV genotyping employed methods showed no difference in the classification at the genotype level. The employed methods were accurate overall and provided an attractive alternative for HCV genotyping. HCV genotypes 4, mixed 4.1b, and 1 were detected in patients of both countries, while genotype 2 was only detected in Saudi Arabian patients. HCV genotype 4 is found to be the predominant genotype among HCV-infected Egyptian and Saud Arabian patients. The data analysis for detecting and genotyping HCV provides precise genotype and subtype identification and an accurate epidemiological representation of circulating viral strains among Egyptian and Saudi Arabian chronic patients.
Footnotes
ACADEMIC EDITOR: Wen-Zhe Ho, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,207 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: MMSF, MRA, and MAA. Analyzed the data: MMSF. Wrote the first draft of the manuscript: MMSF. Contributed to the writing of the manuscript: MMSF and MRA. Agree with manuscript results and conclusions: MMSF, ARS, AAM, MAA, and MRA. Jointly developed the structure and arguments for the paper: MMSF, ARS, AAM, MAA, and MRA. Made critical revisions and approved final version: MMSF. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Perz JF, Alter MJ. The coming wave of HCV related liver disease: dilemmas and challenges. J Hepatol. 2006;44:441–443. doi: 10.1016/j.jhep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Sherlock S, Dooley J. Diseases of Liver and Biliary System. 11th ed. Oxford, UK: Wiley-Blackwell; 2002. pp. 305–320. [Google Scholar]
- 3.The C, Everett Koop Institute; Dartmouth College . Hepatitis C: An Epidemic for Anyone. Worldwide Prevalence. Hanover, NH: The C. Everett Koop Institute; 2009. [Google Scholar]
- 4.Perz JF, Armstrong GL, Farrington LA, Hutin YI. The contributions of hepatitis B virus and hepatitis C virus infections tocirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Simmonds P. Genetic diversity and evolution of hepatitis C virus. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 6.Bukh J, Miller R, Purcell R. Genetic heterogenicity of hepatitis C virus: quasi-species and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 9.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 10.Smith DB, Mellor J, Jarvis LM, et al. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. The International HCV Collaborative Study Group. J Gen Virol. 1995;76:1749–1776. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 11.Lindh M, Hannoun C. Genotyping of hepatitis C virus by Taqman real-time PCR. J Clin Virol. 2005;34:108–114. doi: 10.1016/j.jcv.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Manns MP, McHutchison GJ, Gordon SC, Rustgi VK, Shiffman M, Reindollar R. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 13.Choo OL, Richman KH, Han JH, Berger K, Lee C, Dong C. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germer JJ, Rys PN, Thorvilson IN, Persing DH. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J Clin Microbiol. 1999;37:2625–2630. doi: 10.1128/jcm.37.8.2625-2630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halfon P, Trimoulet P, Bourliere M, Khiri H, de Ledinghen V, Couzigou P. Hepatitis C virus genotyping based on 5_noncoding sequence analysis (Trugene) J Clin Microbiol. 2001;39:1771–1773. doi: 10.1128/JCM.39.5.1771-1773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross RS, Viazov SO, Holtzer CD, Beyou A, Monnet A, Mazure C. Genotyping of hepatitis C virus isolates using CLIP sequencing. J Clin Microbiol. 2000;38:3581–3584. doi: 10.1128/jcm.38.10.3581-3584.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y. Typing hepatitis C virus by polymerase chain reaction with typespecific primers: application to clinical surveys and tracing infectious sources. Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 18.McOmish F, Yap PL, Dow BC, Follett EA, Seed C, Keller AJ. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32:884–892. doi: 10.1128/jcm.32.4.884-892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacherjee V, Prescott LE, Pike I, Rodgers B, Bell H, El-Zayadi AR. Use of NS-4 peptides to identify type-specific antibody to hepatitis C virus genotypes 1, 2, 3, 4, 5 and 6. J Gen Virol. 1995;76:1737–1748. doi: 10.1099/0022-1317-76-7-1737. [DOI] [PubMed] [Google Scholar]
- 20.Dixit V, Quan S, Martin P, Larson D, Brezina M, DiNello R. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978–2983. doi: 10.1128/jcm.33.11.2978-2983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 22.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forns X, Maluenda MD, Lopez-Labrador FX, Ampurdanes S, Olmedo E, Costa J. Comparative study of three methods for genotyping hepatitis C virus strains in samples from Spanish patients. J Clin Microbiol. 1996;34:2516–2521. doi: 10.1128/jcm.34.10.2516-2521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau JY, Mizokami M, Kolberg AJ, Davis GL, Prescott LE, Ohno T. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J Infect Dis. 1995;171:281–289. doi: 10.1093/infdis/171.2.281. [DOI] [PubMed] [Google Scholar]
- 25.Schroter M, Zollner B, Schafer P, Laufs R, Feucht HH. Comparison of three HCV genotyping assays: a serological method as a reliable and inexpensive alternative to PCR based assays. J Clin Virol. 2001;23:57–63. doi: 10.1016/s1386-6532(01)00186-x. [DOI] [PubMed] [Google Scholar]
- 26.Bullock GC, Bruns DE, Haverstick DM. Hepatitis C genotype determination by melting curve analysis with a single set of fluorescence resonance energy transfer probes. Clin Chem. 2002;48:2147–2154. [PubMed] [Google Scholar]
- 27.Schroter M, Zollner B, Schafer P, Landt O, Lass U, Laufs R. Genotyping of hepatitis C virus types 1, 2, 3, and 4 by a one-step LightCycler method using three different pairs of hybridization probes. J Clin Microbiol. 2002;40:2046–2050. doi: 10.1128/JCM.40.6.2046-2050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zekri AR, Bahnassy AA, Shaarawy SM, et al. Hepatitis C virus genotyping in relation to neu-oncoprotein overexpression and the development of hepatocellular carcinoma. J Med Microbiol. 2000;49:89–95. doi: 10.1099/0022-1317-49-1-89. [DOI] [PubMed] [Google Scholar]
- 29.Alzahrani AJ, Obeid OE, Al-Ali A, Imamwardi B. Detection of hepatitis C virus and human immunodeficiency virus in expatriates in Saudi Arabia by antigen-antibody combination assays. J Infect Dev Ctries. 2009;3:235–238. doi: 10.3855/jidc.42. [DOI] [PubMed] [Google Scholar]
- 30.Karkar A. Hepatitis C in dialysis units: the Saudi experience. Hemodial Int. 2007;11:354–367. doi: 10.1111/j.1542-4758.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- 31.Jarvis LM, Ludlam CA, Simmonds P. Hepatitis C virus genotypes in multitransfused individuals. Haemophilia. 1995;1:3–7. doi: 10.1111/j.1365-2516.1995.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 32.Eyster ME, Sherman KE, Goedert JJ. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. The Multi-center Hemophilia Cohort Study. J Infect Dis. 1999;179:1062–1069. doi: 10.1086/314708. [DOI] [PubMed] [Google Scholar]
- 33.Hu YW, Balaskas E, Kessler G. Primer specific and mispair extension analysis (PSMEA) as a simple approach to fast genotyping. Nucleic Acids Res. 1998;26:5013–5015. doi: 10.1093/nar/26.21.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleiber J, Walter T, Haberhausen G, Tsaung S, Babeil R, Rosenstaus M. Performance characteristics of a quantitative, homogeneous TaqMan RT-PCR test for HCV RNA. J Mol Diagn. 2000;2(3):158–166. doi: 10.1016/S1525-1578(10)60632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handbook “Sampling, transportation, storage of clinical material for PCR diagnostic”, developed by Federal Budget Institution of Science (Central Research Institute for Epidemiology) of Federal Service for Surveillance on Consumers’ Rights Protection and Human Well-Being. 3A Novogireevskaya Street Moscow 111123 Russia. 2008.
- 36.Bouchardeau F, Cantaloube J, Chevaliez S, Portal CH, Razer A, Lefrère JJ. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol. 2007;45:1140–1145. doi: 10.1128/JCM.01982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zar JH. Biostatistical Analysis. 4th ed. Upper Saddle River, NJ, USA: Prentice Hall; 1999. p. 663. [Google Scholar]
- 38.Nakatani SM, Santos CA, Riediger IN, Krieger MA, Duarte CA, do Carmo Debur M. Comparative performance evaluation of hepatitis C virus genotyping based on the 5′ untranslated region versus partial sequencing of the NS5B region of Brazilian patients with chronic hepatitis C. Virol J. 2011;8:459. doi: 10.1186/1743-422X-8-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MirDavood O, Khadem AM. Hepatitis c virus genotyping by melting curve analysis in west Azerbaijan, Northwest of Iran. Hepat Mon. 2009;9:133–136. [Google Scholar]
- 40.Icardi G, Bonanni P, Wicks R, et al. Characterization of hepatitis C virus genotypes in Italian subjects at different risk infection. Viral Hepatitis and Liver Disease. 1997;68:598–600. [Google Scholar]
- 41.Chen Z, Weck K. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J Clin Microbiol. 2002;40:3127–3134. doi: 10.1128/JCM.40.9.3127-3134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ansaldi F, Torre F, Bruzzone B, Piciotto A, Crovari P, Icardi G. Evaluation of new hepatitis C virus sequencing assay as a routine method for genotyping. J Med Virol. 2001;63:17–21. [PubMed] [Google Scholar]
- 43.Zheng X, Pang M, Chan A, Roberto A, Warner D, Yen-Liberman B. Direct comparison of hepatitis C virus genotypes tested by INNO-LiPA HCV II and TRUGENE HCV genotyping methods. J Clin Virol. 2003;28:214–216. doi: 10.1016/s1386-6532(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed AS, Mohammad BS, Gaber SM, Abdel-Aziz AS, Fayssal FM. HCV infection among Saudi population: high prevalence of genotype 4 and increased viral clearance. PLoS One. 2012;7:29781–29787. doi: 10.1371/journal.pone.0029781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Faleh F, Huraib S, Sbeih F, Al-Karawi M, Al-Rashed R. Hepatitis C virus genotypes in patients with chronic liver disease and haemodialysis patients from Saudi Arabia. J Viral Hepat. 1995;2:293–296. doi: 10.1111/j.1365-2893.1995.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 46.Mohammed AA. Prevalence of mixed hepatitis C virus (HCV) genotypes among recently diagnosed dialysis patients with HCV infection. Saudi J Kidney Dis Transpl. 2011;22:712–716. [PubMed] [Google Scholar]
- 47.Osoba O, Ibrahim M, Abdelaal A, Al-Mowallad A, Al Shareef B. Hepatitis C virus genotyping by polymerase chain reaction and DNA enzyme immunoassay among Saudi patients in the Western Province, Saudi Arabia. Ann Saudi Med. 2000;20:394–397. doi: 10.5144/0256-4947.2000.394. [DOI] [PubMed] [Google Scholar]
- 48.Nolte FS, Green AM, Fiebelkorn KR, et al. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5′ noncoding region. J Clin Microbiol. 2003;4:1558–1564. doi: 10.1128/JCM.41.4.1558-1564.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zekri AR, Bahnassy A, Ramadan A, El-Bassuoni A, Badran A, Madwar MA. Hepatitis C virus genotyping versus serotyping in Egyptian patients. Infection. 2001;29:24–26. doi: 10.1007/s15010-001-0010-8. [DOI] [PubMed] [Google Scholar]
- 50.Zekri AR, El-Din HM, Bahnassy AA, et al. TRUGENE sequencing versus INNO-LiPA for sub-genotyping of HCV genotype-4. J Med Virol. 2005;75:412–420. doi: 10.1002/jmv.20293. [DOI] [PubMed] [Google Scholar]
- 51.Nakatani SM, Santos CA, Riediger IN, et al. Comparative performance evaluation of hepatitis C virus genotyping based on the 5′ untranslated region versus partial sequencing of the NS5B region of Brazilian patients with chronic hepatitis C. Virol J. 2011;8:459–465. doi: 10.1186/1743-422X-8-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Omran MH, Youssef SS, El-Garf WT. Phylogenetic and genotyping of hepatitis C virus in Egypt. Aust J Basic Appl Sci. 2009;3:1–8. [Google Scholar]
- 53.Stamenkovic G, Zerjav S, Velickovic ZM. Distribution of HCV genotypes among risk groups in Serbia. Eur J Epidemiol. 2000;16:949–954. doi: 10.1023/a:1011060505152. [DOI] [PubMed] [Google Scholar]
- 54.Zhang YY, Lok AS, Chan DT, Widell A. Greater diversity of hepatitis C virus genotypes found in Hong Kong than in Mainland China. J Clin Microbiol. 1995;33:2931–2934. doi: 10.1128/jcm.33.11.2931-2934.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hisham AO, Ahmad AG, Faten Q, Hind FI, Maha RA. Chronic hepatitis C in Saudi Arabia: three years local experience in a University Hospital. Hepat Mon. 2012;12:5810–5812. doi: 10.5812/hepatmon.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahib AA, Seif MS, Mangoud AM, El Shazly AM, Morsy AT. The liver function profile in PCR-RNA Egyptian HCV-patients and normal controls. J Egypt Soc Parasitol. 2005;35:451–466. [PubMed] [Google Scholar]