Abstract
Background
Many studies have reported that the p53 codon 72 polymorphism is associated with acute myeloid leukemia (AML) susceptibility; however, the conclusions are inconsistent. Therefore, we performed this meta-analysis to obtain a more precise result.
Material/Methods
We searched PubMed to identify relevant studies, and 6 published case-control studies were retrieved, including 924 AML patients and 3832 controls. Odds ratio (OR) with corresponding 95% confidence interval (95%CI) was applied to assess the association between p53 codon 72 polymorphism and AML susceptibility. The meta-analysis was performed with Comprehensive Meta-Analysis software, version 2.2.
Results
Overall, no significant association between p53 codon 72 polymorphism and AML susceptibility was found in this meta-analysis (Pro vs. Arg: OR=0.94, 95%CI=0.81–1.10; Pro/Pro vs. Arg/Arg: OR=0.93, 95%CI=0.71–1.22; Arg/Pro vs. Arg/Arg: OR=0.79, 95%CI=0.55–1.13; (Pro/Pro + Arg/Pro) vs. Arg/Arg: OR=0.84, 95%CI=0.62–1.13; Pro/Pro vs. (Arg/Arg + Arg/Pro): OR=1.06, 95%CI=0.83–1.35). Similar results were also found in stratified analysis according to ethnicity and source of controls.
Conclusions
Our meta-analysis demonstrates that p53 codon 72 polymorphism may not be a risk factor for AML, which should be verified in future studies.
MeSH Keywords: Genes, p53; Leukemia, Myeloid; Meta-Analysis; Polymorphism, Genetic
Background
The tumor suppressor p53 (TP53) is a principal mediator of multiple cellular functions, including cell cycle arrest, senescence, and apoptosis in response to cellular stresses [1]. Located on chromosome 17p13, the TP53 gene has been considered as a significant determinant factor in human carcinogenesis [2]. The TP53 codon 72 polymorphism Arg72Pro (rs1042522), an amino acid substitution of arginine (Arg)→proline (Pro) at position 72 [3], is one of the most investigated polymorphisms. Published meta-analyses have indicated that TP53 Arg72Pro polymorphism is associated with increased risk of some malignancies, such as lung cancer [4], cervical cancer [5], bladder cancer [6], nasopharyngeal carcinoma [7], thyroid carcinoma [8], prostate cancer [9], and skin cancer [10]. However, other meta-analyses found no significant association between TP53 Arg72Pro polymorphism and certain malignancies, like head and neck cancer [11], oral squamous cell carcinoma [1], ovary cancer [12], and sarcoma [13]. Obviously, the associations between the polymorphism and tumors vary in different types of malignancies.
Acute myeloid leukemia (AML) is a hematological malignancy involving genetic alterations. Hence, much attention has been paid to the issue of whether TP53 Arg72Pro polymorphism is associated with AML risk. In 2000, Nakano et al. performed a case-control study and reported that this polymorphism might decrease the risk of AML in the Japanese population [14]. However, subsequent studies showed divergent results about TP53 Arg72Pro polymorphism and AML susceptibility. In this case, a meta-analysis is needed to pool these controversial outcomes for a more precise result [15].
Material and Methods
Literature search
A comprehensive search was conducted in PubMed for studies detecting the association between p53 gene polymorphism and ML susceptibility up to December 11, 2014. Keywords were combined with Boolean operators “OR” and “AND”, and contained the following MeSH or text words: (Tumor Suppressor Protein p53[MH] or “tumor protein p53” or genes, p53[MH] or “p53”) and (polymorphism[MH] or polymorph* or “SNPs” or “SNP” or mutation[MH] or mutat* or Genetic Variation[MH] or varian*) and (leukemia, myeloid[MH] or “myeloid leukemia”). The search strategy used English and Chinese languages, and the bibliographies of the included studies and recent reviews were checked for additional relevant publications.
Study selection criteria
Every study included in this analysis had to meet the following criteria: (1) with case-control or cohort design; (2) investigating the association between TP53 gene Arg72Pro polymorphism and the susceptibility to AML; (3) cases were enrolled from patients with ML, and controls were from healthy population. Both diagnosed cases and controls accorded with laboratory medicine and clinical criteria, and their details were clearly reported; (4) with sufficient data for estimating the odds ratios (ORs) and 95% confidence intervals (95%CIs).
In addition, articles were excluded if they satisfied any of the following exclusion criteria: (1) abstracts or unpublished records; (2) studies in which the genotype frequencies were not reported and could not be calculated. As for overlapped publications, the most comprehensive one was selected.
Data extraction
Two reviewers were responsible for data extraction separately following the same standard. The principal information of included studies to be extracted included first author, publication year, country, ethnicity, source of controls, numbers of cases and controls, genotype distribution, genotyping method, and Hardy-Weinberg equilibrium (HWE). All discrepancies during this work were solved by discussion between the 2 reviewers.
Statistical analysis
The OR and its 95%CI were used to assess the association under 5 genetic models: Pro vs. Arg, Pro/Pro vs. Arg/Arg, Arg/Pro vs. Arg/Arg, Pro/Pro vs. (Arg/Arg + Arg/Pro), and (Pro/Pro + Arg/Pro) vs. Arg/Arg. Comprehensive Meta Analysis software (version 2.2; Biostat, Englewood, N.J., USA) [16,17] was used for forest plots, heterogeneity test, and other data analyses. Heterogeneity was evaluated by the Cochran’s Q statistic [18] and the I2 statistic [19]. If heterogeneity was significant (P<0.1 or I2 >25%), the random-effects model was used, otherwise, the fixed-effects model was employed. Subgroup analysis was also conducted. In addition, the influence of every single study on the overall results was investigated by removing each study in turn so as to test the robustness of the main results. Potential publication bias was assessed by visual inspection of the funnel plots, and Egger’s regression method provided corresponding statistical evidence (P<0.05 represented statistical significance) [20,21].
Results
Study characteristics
Of the 579 records found initially, 6 case-control studies [14,22–26] were ultimately included involving 924 cases and 3832 controls. A detailed flowchart of the selection process is shown in Figure 1. Table 1 exhibits the major characteristics of the 6 case-control studies [14,22–26]. Four studies were conducted in Asian populations [14,23–25] and 2 in white populations [22,26]. In terms of source of controls, 2 studies recruited controls from hospital (HB) [23,26] and 4 from general population (PB) [14,22,24,25]. The genotype distributions of controls from all included studies were consistent with HWE.
Figure 1.
Flowchart of study selection.
Table 1.
Characteristics of the studies included in the meta-analysis.
| Reference | Country (Ethinicity) | Source of control | Case | Control | Genotype method | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | AA | AP | PP | Total | AA | AP | PP | |||||
| Nakano 2000 | Japan (Asian) | PB | 200 | 82 | 93 | 25 | 188 | 59 | 95 | 34 | PCR-SSCP | 0.77 |
| Ellis 2008 | USA/UK (Caucasian) | PB | 171 | 95 | 66 | 10 | 3022 | 1714 | 1127 | 181 | PCR-RFLP | 0.85 |
| Xiong 2009 | China (Asian) | HB | 231 | 52 | 127 | 52 | 128 | 39 | 64 | 25 | PCR-RFLP | 0.99 |
| Chauhan 2012 | India (Asian) | PB | 131 | 38 | 71 | 22 | 199 | 51 | 112 | 36 | PCR-RFLP | 0.06 |
| Dunna 2012 | India (Asian) | PB | 141 | 64 | 44 | 33 | 245 | 79 | 123 | 43 | PCR-RFLP | 0.68 |
| El-Danasouri 2014 | Egypt (Caucasian) | HB | 50 | 20 | 20 | 10 | 50 | 14 | 31 | 5 | PCR-RFLP | 0.24 |
AA represents individuals who do not inherit a mutant allele; AP represents individuals who are heterozygote for the mutant allele; PP represents individuals who are homozygote for the mutant allele; HB – hospital based; PB – population based; HWE – Hardy-Weinberg equilibrium.
Meta-analysis and sensitivity analysis
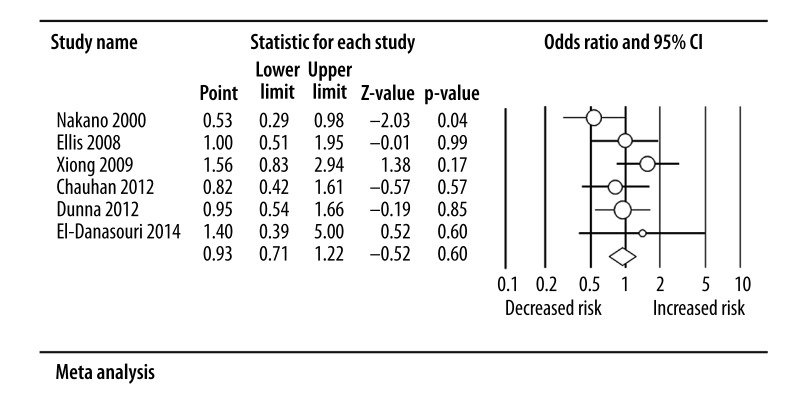
Table 2 shows the main results of meta-analysis. Overall, no significant association was observed between TP53 Arg72Pro polymorphism and AML risk [Pro vs. Arg: OR=0.94, 95%CI=0.81–1.10; Pro/Pro vs. Arg/Arg: OR=0.93, 95%CI=0.71–1.22, Figure 2; Arg/Pro vs. Arg/Arg: OR=0.79, 95%CI=0.55–1.13; (Pro/Pro+Arg/Pro) vs. Arg/Arg: OR=0.84, 95%CI=0.62–1.13; Pro/Pro vs. (Arg/Pro+Arg/Arg): OR=1.06, 95%CI=0.83–1.35]. Similarly, in the succeeding stratified subgroup analysis, we also did not find any significant association (Table 2).
Table 2.
Pooled ORs and 95% CIs for the association between p53 genetic polymorphism and AML susceptibility.
| Overall and subgroups | N | Pro vs. Arg | ProPro vs. ArgArg | ArgPro vs. ArgArg | (ProPro + ArgPro) vs. ArgArg | ProPro vs. (ArgPro + ArgArg) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | I2 (%) | OR (95%CI) | I2 (%) | OR (95%CI) | I2 (%) | OR (95%CI) | I2 (%) | OR (95%CI) | I2 (%) | ||
| Overall | 6 | 0.94 (0.81–1.10) | 30 | 0.93 (0.71–1.22) | 21.55 | 0.79 (0.55–1.13) | 69.2 | 0.84 (0.62–1.13) | 60 | 1.06 (0.83–1.35) | 23 |
| Source of controls | |||||||||||
| PB | 4 | 0.88 (0.77–1.02) | 6.37 | 0.80 (0.58–1.09) | 0 | 0.74 (0.51–1.08) | 67.3 | 0.78 (0.58–1.04) | 51.36 | 0.97 (0.69–1.36) | 30.46 |
| HB | 2 | 1.17 (0.90–1.54) | 0 | 1.53 (0.87–2.69) | 0 | 0.87 (0.27–2.78) | 80.87 | 1.00 (0.40–2.52) | 72.94 | 1.34 (0.82–2.17) | 0 |
| Ethnicity | |||||||||||
| Asian | 4 | 0.91 (0.73–1.14) | 53.73 | 0.89 (0.58–1.38) | 48.92 | 0.79 (0.49–1.27) | 74.75 | 0.82 (0.54–1.24) | 69.29 | 1.02 (0.72–1.44) | 36.67 |
| Caucasian | 2 | 1.02 (0.81–1.28) | 0 | 1.07 (0.59–1.94) | 0 | 0.77 (0.34–1.72) | 68.05 | 0.89 (0.54–1.49) | 39.59 | 1.29 (0.59–2.78) | 34.33 |
HB – hospital based; PB – population based; OR – odds ratio; CI – confidence interval; I2 – test for heterogeneity.
Figure 2.
Overall ORs for AML susceptibility and p53 genetic polymorphism under Pro/Pro vs. Arg/Arg model with random-effects model.
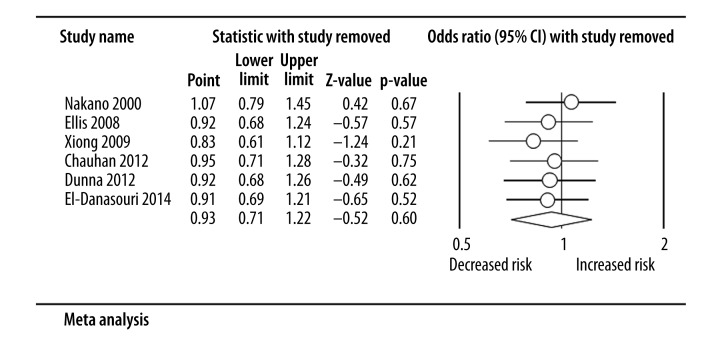
No substantial alterations occurred in results during sensitivity analysis through omitting 1 included study every time (Figure 3 shows the result for the Pro/Pro vs. Arg/Arg model), implying the robustness of the results.
Figure 3.
Forest plot of sensitivity analysis (Pro/Pro vs. Arg/Arg model).
Publication bias
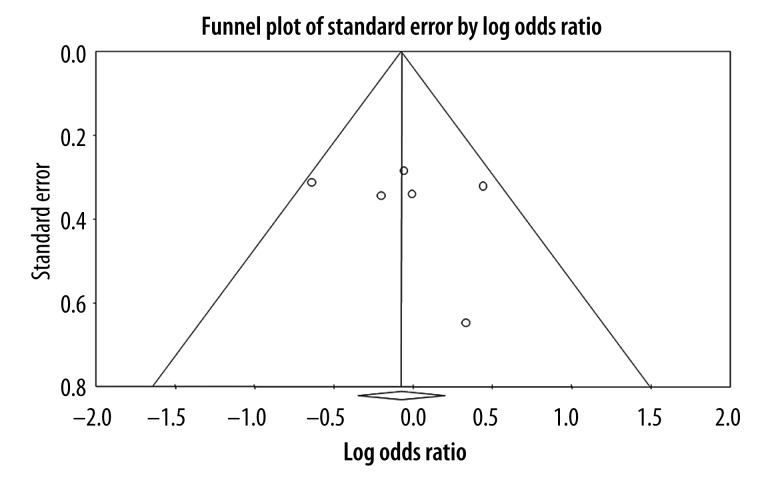
Begg’s funnel plot seemed symmetric for each genetic model, showing no significant publication bias (Figure 4 for Pro/Pro vs. Arg/Arg model), which was confirmed with Egger’s test [Pro vs. Arg, P=0.99; Pro/Pro vs. Arg/Arg, P=0.61; Arg/Pro vs. Arg/Arg, P=0.42; (Pro/Pro+Arg/Pro) vs. Arg/Arg, P=0.60; Pro/Pro vs. (Arg/Pro+Arg/Arg), P=0.50].
Figure 4.
Funnel plot for publication bias (Pro/Pro vs. Arg/Arg mode).
Discussion
AML is a multifactorial and complex disease, in which genetic effect has been considered as an important element. Many studies reported the effects of TP53 Arg72Pro (rs1042522) polymorphism on the susceptibility of myeloid leukemia. In 2004, for the first time, Bergamaschi et al. [27] reported that allele A1 (proline residue, Pro72) was more frequent in patients with CML than in controls, and among CML patients who had no cytogenetic response than among responders. However, the subsequent studies did not achieve the same or similar results, and the association between TP53 Arg72Pro polymorphism and AML susceptibility is still controversial. This meta-analysis of 6 case-control studies was performed to assess the relationship between TP53 Arg72Pro polymorphism and AML susceptibility, but no significant association was found in overall analysis. Furthermore, similar results were also found in stratified analysis according to ethnicity and source of controls.
It should be noted that there are some limitations in the present study. Significant heterogeneity, for example, appeared among most of the genetic models. Inter-study heterogeneity may be frequent in the meta-analysis of studies on genetic association, but its occurrence also has certain relevance to some aspects, such as different enrollment criteria for study subjects, diverse environmental circumstances, multiple interactions among genes and environment factors, and various genotyping methods [28]. After stratification analyses by ethnicity, and source of control, the significance of heterogeneity still could not be eliminated completely. In addition, the number of included studies was limited, and the sample size was relatively small. Therefore, the evidence about the association in this meta-analysis may be less powerful. Furthermore, AML onset involves multiple genetic and environmental factors, and although p53 polymorphism showed no independently significant association with the risk of the disease, it may influence AML risk in combination with other elements, which was not analyzed in our study due to the lack of sufficient data. Despite the above limitations, the results in the present meta-analysis are reliable. First, there was no significant publication bias among selected studies. Second, no single included study had a crucial impact on the whole results, indicating the stability of the outcomes. Lastly, the meta-analysis itself presents a more powerful tool compared with any single study.
Conclusions
Although p53 gene polymorphism has been confirmed to be associated with increased risk of some malignancies, our meta-analysis suggests that p53 gene polymorphism may not be independently associated with AML risk. In the future, larger-scale case-control studies are needed to further investigate the exact correlation of the TP53 codon 72 polymorphism with AML susceptibility.
Footnotes
Conflict of interest
All authors declared there was none conflict of interest.
Source of support: This work was supported by the Foundation of Wuhan University (2042015kf0156)
References
- 1.Zeng XT, Luo W, Geng PL, et al. Association between the TP53 codon 72 polymorphism and risk of oral squamous cell carcinoma in Asians: a meta-analysis. BMC Cancer. 2014;14:469. doi: 10.1186/1471-2407-14-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsui IF, Poh CF, Garnis C, et al. Multiple pathways in the FGF signaling network are frequently deregulated by gene amplification in oral dysplasias. Int J Cancer. 2009;125:2219–28. doi: 10.1002/ijc.24611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ara S, Lee PS, Hansen MF, Saya H. Codon 72 polymorphism of the TP53 gene. Nucleic Acids Res. 1990;18:4961. doi: 10.1093/nar/18.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye XH, Bu ZB, Feng J, et al. Association between the TP53 polymorphisms and lung cancer risk: a meta-analysis. Mol Biol Rep. 2014;41:373–85. doi: 10.1007/s11033-013-2871-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Gu Y, Zhang SL. Association between p53 codon 72 polymorphism and cervical cancer risk among Asians: a HuGE review and meta-analysis. Asian Pac J Cancer Prev. 2012;13:4909–14. doi: 10.7314/apjcp.2012.13.10.4909. [DOI] [PubMed] [Google Scholar]
- 6.Xu T, Xu ZC, Zou Q, et al. P53 Arg72Pro polymorphism and bladder cancer risk – meta-analysis evidence for a link in Asians but not Caucasians. Asian Pac J Cancer Prev. 2012;13:2349–54. doi: 10.7314/apjcp.2012.13.5.2349. [DOI] [PubMed] [Google Scholar]
- 7.Zhuo XL, Cai L, Xiang ZL, et al. TP53 codon 72 polymorphism contributes to nasopharyngeal cancer susceptibility: a meta-analysis. Arch Med Res. 2009;40:299–305. doi: 10.1016/j.arcmed.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Wang P, Wang B, et al. Association between TP53 Arg72Pro polymorphism and thyroid carcinoma risk. Tumour Biol. 2014;35:2723–28. doi: 10.1007/s13277-013-1359-x. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, Liu Y, Zeng J, et al. Association of p53 codon 72 polymorphism with prostate cancer: an update meta-analysis. Tumour Biol. 2014;35:3997–4005. doi: 10.1007/s13277-014-1657-y. [DOI] [PubMed] [Google Scholar]
- 10.Ye J, Li XF, Wang YD, Yuan Y. Arg72Pro polymorphism of TP53 gene and the risk of skin cancer: a meta-analysis. PLoS One. 2013;8:e79983. doi: 10.1371/journal.pone.0079983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia LY, Zeng XT, Li C, et al. Association between p53 Arg72Pro polymorphism and the risk of human papillomavirus-related head and neck squamous cell carcinoma: a meta-analysis. Asian Pac J Cancer Prev. 2013;14:6127–30. doi: 10.7314/apjcp.2013.14.10.6127. [DOI] [PubMed] [Google Scholar]
- 12.Alqumber MA, Akhter N, Haque S, et al. Evaluating the association between p53 codon 72 Arg >pro polymorphism and risk of ovary cancer: a meta-analysis. PLoS One. 2014;9:e94874. doi: 10.1371/journal.pone.0094874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Z, Yu X. Association between p53 codon 72 polymorphism and sarcoma risk among Caucasians. Tumour Biol. 2014;35:4807–12. doi: 10.1007/s13277-014-1631-8. [DOI] [PubMed] [Google Scholar]
- 14.Nakano Y, Naoe T, Kiyoi H, et al. Poor clinical significance of p53 gene polymorphism in acute myeloid leukemia. Leuk Res. 2000;24:349–52. doi: 10.1016/s0145-2126(99)00187-3. [DOI] [PubMed] [Google Scholar]
- 15.Zeng X, Zhang Y, Kwong JS, et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med. 2015;8:2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Zeng XT, Ruan XL, et al. Association between XPD Lys751Gln polymorphism and bladder cancer susceptibility: an updated and cumulative meta-analysis based on 6,836 cases and 8,251 controls. Mol Biol Rep. 2014;41:3621–29. doi: 10.1007/s11033-014-3226-2. [DOI] [PubMed] [Google Scholar]
- 17.Zeng XT, Liu DY, Kwong JS, et al. Meta-analysis of association between interleukin-1beta C-511T polymorphism and chronic periodontitis susceptibility. J Periodontol. 2015;86(6):812–19. doi: 10.1902/jop.2015.140698. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 19.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan XL, Li S, Zeng XT, et al. No association between cytochrome P450 2D6 gene polymorphism and risk of acute leukemia: evidence based on a meta-analysis. Chin Med J (Engl) 2013;126:3750–53. [PubMed] [Google Scholar]
- 21.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis NA, Huo D, Yildiz O, et al. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–49. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong X, Wang M, Wang L, et al. Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk Res. 2009;33:1454–58. doi: 10.1016/j.leukres.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan PS, Ihsan R, Mishra AK, et al. High order interactions of xenobiotic metabolizing genes and P53 codon 72 polymorphisms in acute leukemia. Environ Mol Mutagen. 2012;53:619–30. doi: 10.1002/em.21723. [DOI] [PubMed] [Google Scholar]
- 25.Dunna NR, Vure S, Sailaja K, et al. TP53 codon 72 polymorphism and risk of acute leukemia. Asian Pac J Cancer Prev. 2012;13:347–50. doi: 10.7314/apjcp.2012.13.1.349. [DOI] [PubMed] [Google Scholar]
- 26.El-Danasouri NM, Ragab SH, Rasheed MA, et al. MDM2 SNP309 and p53 codon 72 genetic polymorphisms and risk of AML: an Egyptian study. Ann Clin Lab Sci. 2014;44:449–54. [PubMed] [Google Scholar]
- 27.Bergamaschi G, Merante S, Orlandi E, et al. TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica. 2004;89:868–69. [PubMed] [Google Scholar]
- 28.Prejzner W. Relationship of the BCR gene breakpoint and the type of BCR/ABL transcript to clinical course, prognostic indexes and survival in patients with chronic myeloid leukemia. Med Sci Monit. 2002;8(5):BR193–97. [PubMed] [Google Scholar]