Abstract
Reconciliation methods explain topology differences between a species tree and a gene tree by evolutionary events other than speciations. However, not all phylogenies are trees: hybridization can occur and create new species and this results into reticulate phylogenies. Here, we consider the problem of reconciling a gene tree with a species network via duplication and loss events. Two variants are proposed and solved with effcient algorithms: the first one finds the best tree in the network with which to reconcile the gene tree, and the second one finds the best reconciliation between the gene tree and the whole network.
Keywords: tree reconciliation, gene evolution, phylogenetic, parsimony, phylogenetic networks
Background
Reconciliations explain topology incompatibilities between a species tree and a gene tree by evolutionary events - other than speciation - affecting genes [[1], for a review]. However, not all phylogenies are trees: indeed, hybridization can occur and create new species [2] and this results into reticulate phylogenies, i.e. species (phylogenetic) networks [3]. In [4], the authors presented a first contribution toward solving a problem similar to the reconciliation problem, namely the cophylogeny problem [5-8], on networks. In their article, they first propose a polynomial algorithm to solve this problem on dated host trees taking into account codivergence, duplication, host switching, and loss events. This model is similar to the DTL model in gene tree reconciliation - that takes into account speciation, duplication, transfer, and loss events [[9], among others]. However, when extending the cophylogeny problem to species networks, their model may not be the more pertinent one for the DTL problem. Indeed, in [4] the parasite tree that is "reconciled" with the host network can take any path in the latter, modeling the fact that some hybridization species can receive the parasites of both parents. In the problem of gene tree reconciliation, their model is more adapted to novel hybridizations, where the genes still keep trace of the polyploidy due to the hybridization. But, for ancient hybridizations, the polyploidy of the extant species being reduced, a model where each gene of an hybridization species can be inherited from at most one of its two parents is more pertinent. In other words, for solving the latter problem, we are interested in finding a tree that is "displayed" by the species network such that its reconciliation with a given gene tree is optimum. We propose an efficient algorithm that takes into account duplication and loss events whose complexity does not depend on the number of hybridization events in the species network but only on the level of the network, where the level is a measure of how much the network is "tangled". Moreover, we propose a faster algorithm solving the problem described in [4] when restricting to duplication and loss events (that is, host switching is not taken into account).
Basic notions
We start by giving some basic definitions that will be useful in the paper.
Definition 1 (Rooted phylogenetic network)A rooted phylogenetic network N on a label set is a rooted directed acyclic graph with a single root where each outdegree-0 node (the leaves) is labelled by an element of . The root of N, denoted by r(N), has indegree 0 and outdegree 2. All other internal nodes have either indegree 1 and outdegree 2 (speciation node), or indegree 2 and outdegree 1 (hybridization node).
Denote by V(N), I(N), E(N), L(N) and respectively the set of nodes, internal nodes (nodes with outdegree greater than 0), edges, leaves and leaf labels of N. The size of N, denoted by |N|, is equal to |V(N)| + |E(N)|. Given x in V(N), we denote by Nx the subnetwork of N rooted at x, i.e. the subgraph of N consisting of all edges and nodes reachable from x. If x is a leaf of N, we denote by s(x) the label of x in . If x is a speciation node, we denote by p(x) the only parent of x.
Given two nodes x and y of N, we say that x is lower or equal to y in N, denoted by x ≤N y (resp. lower, denoted by x <N y), if and only if there exists a path (possible reduced to a single node) in N from y to x (resp. and x ≠ y). If x ≤N y, then, for every path p from y to x, denote by length(p) the number of speciation nodes in N such that x <N z ≤N y. If N is a tree, then, for every two nodes u, v of N, LCAN (u, v) [10] is the lowest node of N that is above or equal to both u, v.
Given a speciation node x in N, two paths of N starting from x are said to be separated if each path contains a different child of x. Let x, y be two nodes of N. Denote by the set of nodes z of N such that there exist two separated paths in N from z to x and y. For example, in Figure 1(d), . Note that all nodes in are speciation nodes and, when N is a tree and x, y are not comparable, MN (x, y) contains exactly one node, which coincides with LCAN (x, y).
Figure 1.
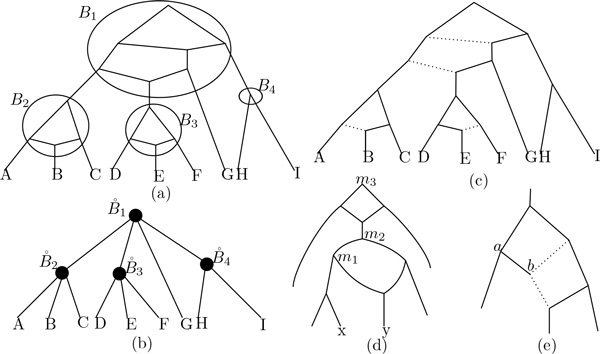
(a) An example of a level-2 network N, (b) the tree bc(N), (c) a switching of N where the switched-off edges are dotted, (d) a network N' such that , (e) another switching of a network. Note that the edge (a; b) will also be switched off since all out-going edges of b are off.
If every biconnected component of N has at most k hybridization nodes, we say that N is of level-k [11]. A rooted phylogenetic tree is a rooted phylogenetic network with no hybridization nodes, i.e. a level-0 network. In the following, we will refer to rooted phylogenetic networks and rooted phylogenetic trees simply as networks and trees, respectively. In this paper, we allow trees to contain artificial nodes, i.e. nodes with indegree and outdegree 1.
Let B be a biconnected component of a network N. Then B contains exactly one node r(B) without ancestors in B [[12], Lemma 5.3]; we call r(B) the root of B. If B is not trivial, i.e. B consists of more than one node, we can contract it by removing all nodes of B other than r(B), then connect r(B) to every node with indegree 0 created by this removal. Then the following definitions are well-posed.
Definition 2 (Tree bc(N)) Given a network N, the tree bc(N) is obtained from N by contracting all its biconnected components.
For example, Figure 1(a, b) shows respectively a level-2 network N and its associated tree bc(N).
Let denote by the node in bc(N) that corresponds to a biconnected component B in N. Given two biconnected component Bi, Bj , we say that Bi ≤ N Bj (resp. Bi <N Bj ) if and only if (resp. ). We say that Bi is the parent (resp. a child) of Bj if is the parent (resp. a child) of in bc(N). We also denote by LCAN(Bi, Bj) the biconnected component corresponding to in N.
Definition 3 (Elementary network) Given a network N, each biconnected component B that is not a leaf of N defines an elementary network, denoted by N(B), consisting of B and all cut-edges coming out from B.
Definition 4 (Switchings of a network [13]) Given a network N, a switching S of N is obtained from N by choosing, for each hybridization node, an incoming edge to switch on and the other to switch off. Once this is done, we also switch off all switched-on edges with the target node having only switched-off outgoing edges (see Figure 1(e) for an example). For each bi-connected component B of N, we also denote by S(B) the subgraph of S restricted to N(B).
Switchings will be used in the next section to model gene histories for genes evolving in a species network. For example, Figure 1 presents in (c) a switching of the level-2 network in (a). We denote by Von(S) the set of nodes of S that are not an endpoint of any switched-off edge. A path of S is a path of N that uses only switched-on edges.
Hereafter, G will denote a tree and N a network such that there is a bijection between L(N) and and . In the gene tree reconciliation problem, G represents a gene tree such that each leaf corresponds to a contemporary gene and is labeled by the species containing this gene, while N is a species network such that each leaf represents an extant species. In the cophylogeny problem, G represents a parasite tree such that each leaf corresponds to a parasite species that is labeled by the species that hosts it, while N is a host network such that each leaf represents an extant species.
Reconciliations
We will now extend the definition of reconciliation in [14] to networks. In a reconciliation, each node of G is associated to a node of S and an event - a speciation , a duplication or a contemporary event - under some constraints. A contemporary event associates a leaf u of G with a leaf x of S such that s(u) = s(x). A speciation in a node u of G is constrained to the existence of two separated paths from the mapping of u to the mappings of its two children, while the only constraint given by a duplication event is that evolution of G cannot go back in time. More formally:
Definition 5 (Reconciliation) Given a tree G and a network N such that , a reconciliation between G and N is a function α that maps each node u of G to a pair (αr(u), αe(u)) where αr(u) is a node of V(N) and αe(u) is an event of type or or, such that:
• if and only if u ∈ L(G), αr(u)∈L(N) and s(u) = s(αr(u));
• for every u ∈ I(G) with child nodes {u1, u2}, if , then ;
• for any two nodes u, v of V(G) such that v <G u, if , then αr (v) ≤N αr(u). Otherwise, αr (v) <N αr(u).
Note that, if N is a tree, this definition coincides with the one given in [14].
The number of duplications of α, denoted by d(α) is the number of nodes u of G such that . Since in a network there can be several paths between two nodes, we count the number of losses on shortest paths, as done in [4]. In more details, given two nodes x, y of N, the distance between x and y, denoted by dist(x, y), is defined as follows:
• If y ≤N x, then dist(x, y) = minp length(p) over all possible paths p from x to y;
• otherwise, dist(x, y) = +∞.
Then, for every u ∈ I(G) with child nodes {u1, u2}, the number of losses associated with u in a reconciliation α, denoted by lα(u), is defined as follows:
• if , then lα(u) = min{dist(x1, αr(u1)) + dist(x2, αr(u2)), dist(x1, αr(u2))+dist(x2, αr(u1))} where x1, x2 are the two children of αr(u);
• if , then lα(u) = dist(αr(u), αr(u1)) + dist(αr(u), αr(u2)).
The number of losses of a reconciliation α, denoted by l(α), is the sum of lα(·) for all internal nodes of G. Thus, the cost of α, denoted by cost(α), is d(α)·δ + l(α)·λ, where δ and λ are respectively the cost of a duplication and a loss event. We use cost(G,N) to denote the cost of the minimum reconciliations between G and N. A reconciliation having the minimum cost is called a most parsimonious reconciliation.
Let S be a switching of N, then a reconciliation between G and S is defined similarly to Definition 5 except that only switched-on edges are considered when defining paths, and only nodes in Von(S) are counted for calculating the length of the shortest path in the definition of dist. This is done to model the fact that, since each gene of an hybridization species is inherited from one of its two parents, we should not count as a loss the fact that the other parent does not contribute. Moreover, note that, for every two nodes x, y of S such that x ≤S y, there is a unique path from y to x in S.
When N is a tree, there is a unique reconciliation (the LCA reconciliation) between G and N which has minimum cost and this reconciliation can be found in O(|G|) time [15-18] as follows:
• for each node u in L(G), αr(u) is defined as the only node x of S such that s(u) = s(x), and ;
• for each node u in I(G) with child nodes {u1, u2}, αr(u) = LCAN (αr(u1), αr(u2)); if αr(u1) ≤N αr(u2) or αr(u2) ≤N αr(u1) then ; otherwise .
In the LCA reconciliation, the mapping αe is totally defined by αr, hence it can be omitted, and we will use α to refer to αr. Note that the algorithms used on trees to find the LCA reconciliation can also be used on switchings, which - when only switched-on edges are considered - are actually trees. Hereafter, when we refer to the reconciliation between a tree and a switching, we refer to the LCA reconciliation between them. The problems in which we are interested in here are defined as follows:
Problem 1 BEST SWITCHING FOR THE MODEL
Input A tree G, a network N such that , and positive costs δ and λ for respectively and events.
Output A switching S of N such that the cost(G, S) is minimum over all switchings of N.
Problem 2 MINIMUM RECONCILIATION ON NETWORKS
Input A tree G, a network N such that , and positive costs δ and λ for respectively and events.
Output A minimum reconciliation between G and N.
Remark 1 For the sake of simplicity, we suppose that G does not contain any internal node u such that (i.e. all nodes of Gu are mapped to a leaf of N). If it is not the case, we can simplify the instance by replacing in G each such subtree Gu by a leaf labeled by the unique label in and compute a reconciliation for the simplified tree G'. A parsimonious reconciliation for G can be easily obtained from a parsimonious one for G'.
Method
Best switching
We start by proving that finding the best switching for the model is NP-hard:
Theorem 1 Problem 1 is NP-hard.
Proof: To prove the theorem, we reduce Problem 1 to the TREE CONTAINMENT problem, which is NP-hard [19]. The TREE CONTAINMENT problem asks the following "Given a network N and a tree T, both with their leaf sets bijectively labeled by a label set , is there a switching S of N such that T can be obtained by S deleting all switched-off edges and nodes with indegree and outdegree 1?". Now, assume there is an algorithm to solve Problem 1 in polynomial time. Then, it is easy to see that, since λ, δ > 0, there is a solution of Problem 1 with cost 0 if and only if G is contained in N. Therefore, this method would provide a polynomial-time algorithm to solve the TREE CONTAINMENT problem, which is impossible.
In the following, we present a fixed-parameter tractable algorithm [20] in the level of the network to solve Problem 1. To do so, we need to introduce some notations.
Definition 6 (Mapping B) For every u ∈ V(G), B(u) is defined as the lowest biconnected component B of N such that ℒ(Nr(B)) contains ℒ(Gu).
Then the following remark holds:
Remark 2 If u ∈ L(G), then B(u) is the only leaf of N such that s(u) = s(B(u)). If u ∈ L(G), then B(u) = LCAN (B(u1), B(u2)) where u1, u2 are child nodes of u in G.
We define by GN the tree obtained from G by adding some artificial nodes on the edges of G and label each node of GN by a biconnected component of N via an extension of the labeling function B(·) as follows:
Definition 7 (Tree GN ) The tree GN is obtained from G as follows: for each internal node u in G with child nodes u1, u2 such that there exist k biconnected components strictly below B(u) and strictly above B(u1), we add k artificial nodes v1 > ... >vk on the edge (u, u1), and B(vj ) is fixed to . We do the same for u2.
See Figure 2 for an example of GN . We have the following lemma:
Figure 2.
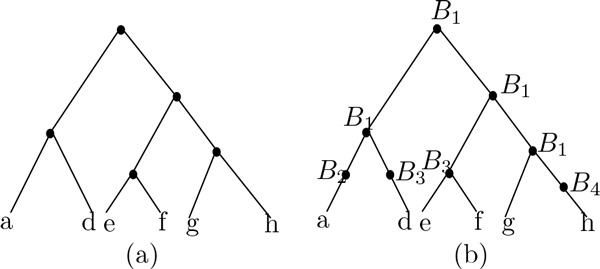
(a) A gene tree G and (b) the tree GN along with the labeling B(·) of its nodes (N here is the network in Figure 1(a)).
Lemma 1 Let u be a node of I(GN ), and u' be one of the children of u. If u is an artificial node, then B(u) is the parent of B(u') and a child of B(p(u)); otherwise, B(u') is either equal to B(u) or one of its children.
Proof: Suppose that u is an artificial node. Then, by Definition 7, B(p(u)), B(u), B(u') are three consecutive nodes of GN , thus B(u) is the parent of B(u') and a child of B(p(u)). Suppose now that u is not an artificial node, and let u" be the child of u in G such that u" ≤Gn u' ≤Gn u. If B(u") = B(u'), then by definition, u" = u' because no artificial node is added between u and u", and thus the claim holds. If B(u') ≠ B(u"), then by Definition 7, B(u') is the highest biconnected component of N that is below B(u) and above B(u"), which is then a child of B(u).
Definition 8 (GB ) Let B be a biconnected component of N different from a leaf, then GB is the set of all maximal connected subgraph H of GN such that B(u) = B for every u ∈ I(H).
For example, in Figure 3, consists of one binary tree, consists of one edge, and contains 1 tree and 1 edge.
Figure 3.
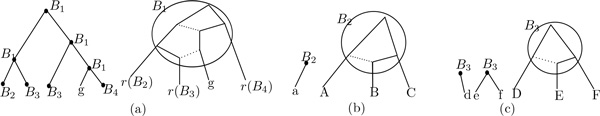
An example of execution of Algorithm 1 on the tree G in Figure 2(a) and the network N in Figure 1(a). (a) and a switching of N(B1) on which its reconciliation with contains 1 duplication and 2 losses. (b) and a switching of N(B2) on which the reconciliation with contains one loss. (c) and a switching of N(B3) on which the reconciliation with contains 2 losses.
Lemma 2 Let B be a biconnected component of N different from a leaf, then we have the following:
(i) for every H ∈ GB, H is either a binary tree or an edge whose upper extremity is an artificial node. Moreover, for every leaf u of H, B(u) is a child of B.
(ii) if B = B(r(G)), then GB consists of one binary tree.
Proof: (i) First, suppose that I(H) contains an artificial node u. Then this artificial node is the only internal node of H; indeed, by Lemma 1, the value of B(·) for the parent and the child of u are both different from B. Thus, H consists of only one edge whose upper extremity is u. If I(H) does not contain any artificial node, it follows that H is a binary tree. Moreover, by Lemma 1 and Definition 8, B(u) is a child of B for every leaf u of H.
(ii) Suppose now that B = B(r(G)), and GB contains at least two components H1, H2, rooted at two different nodes r1, r2 where B(r1) = B(r2) = B. Let , then, by Definition 6, B(v) = B for every node v on the two paths from r to r1 and to r2, because B(r(G)) = B(r1) = B(r2) = B and . But this contradicts the maximality of H1 and H2 since they can both be extended. Hence GB contains only one component. Suppose that this component is an edge; thus, its upper extremity is an artificial node that has been added on the path from a node u to a node v of G where u is strictly higher than r(G). But this is not possible, because r(G) is the highest node of G. Therefore, GB contains one binary tree.
Given a biconnected component Bi different from a leaf, denote by the set of binary trees of , and the set of edges of . Let Si be a switching of N(Bi), and let H be a tree in . By Lemma 2, for every u ∈ L(H), B(u) is a child of Bi and thus r(B(u)) is a leaf of N(Bi), which is also a leaf of Si. Hence, we can define a reconciliation between H and Si, denoted by , such that each leaf u of H is mapped to the leaf r(B(u)) of Si.
Lemma 3 Let S be a switching of N, and let α be the reconciliation between G and S. For every u ∈ I(G), there exists such that u ∈ I(H), and .
The proof of this lemma is deferred to the appendix. The following definition will be useful later on.
Definition 9 (cost(H,Si)) Let Bi be a biconnected component of N different from a leaf, and Si a switching of N(Bi); then cost(H,Si) is defined as follows:
• , if Bi = B(r(G)), and otherwise;
• , where u is the only leaf of H.
As we will see later, this cost corresponds to the contribution of H to a reconciliation between G and any switching of N that contains Si. For example, in Figure 3(b), H is the edge (B2, a) and, if Si is the switching on the left, then cost(H,Si) = λ.
The next lemma is a central one, and it permits to solve Problem 1 independently per each biconnected component of N :
Lemma 4 Let {B1,..., Bp} be the biconnected components of N that are not leaf nodes, and let S be a switching of N where each elementary network N(Bi) has Si as switching. Then
Proof: Let α be the reconciliation between G and S. Denote by dα(Si) the number of nodes in I(Si) associated to a duplication by α. By Remark 1, no duplication happens at leaves of S. Additionally, and the sets of internal nodes of are disjoint. Hence, we have because for every internal node u of H (Lemma 3).
Let us now consider the loss count. Note that a node/edge is on (resp. off) in Si if and only if it is also on (resp. off) in S. Let x, y be two nodes of S such that y ≤S x. Then we define as the number of nodes z in Von(Si) \ L(Si) such that y <S z ≤S x. Then, for each internal node u of G, we define the number of losses associated with u in Si, denoted by lα(u, Si), similarly to lα(u) but using the function instead of dist. Then, . Now, let u1, u2 be two children of u in G, then lα(u, Si) >0 only if the path from α(u) to either α(u1) or α(u2) in S contains at least a node of Bi. Therefore, we have three possible cases: (1) α(u) is in is in Bi, (3) α(u) is above r(Bi) while either α(u1) or α(u2) is in a biconnected component below Bi. In case (1), by Lemma 3, there exists a binary tree H of such that u∈I(H), and for every v∈I(Si), thus . Now, let suppose Si that case (2) holds for u1, then u1 must be the root of a binary tree H1 of Gt , and the contribution of u1 to lα(u, Si) is . Note that in this case, u1 ≠ r(G). Finally, let suppose that case (3) holds for u1 and let ua, ub be the artificial nodes added on the edge (u, u1) of G such that B(ua) = Bi and B(ub) is a child of Bi. Hence, , and the contribution of u1 to lα(u, Si) is dist(r(Si), r(B(ub))). Let call the set of nodes u in the first, second, and third case. By construction, V1 is disjoint from and . Moreover, and are disjoint because if a node u has two children u1, u2 such that u1 is in Bi, and u2 is below Bi, then u must be in Bi, i.e. cannot be above r(Bi). Thus,
Therefore,
As proved above (in case (3)), the first term between squared brackets is equal to by Definition 9. In the second term between squared brackets, the sum of the two first factors is exactly (as proved in case (1)), while the last factor is equal to (as proved in case (2)). Note that as mentioned in case (2), only nodes that are not the root of G are considered. Hence, the second term between squared brackets corresponds to by Definition 9.
Therefore, and this concludes the proof.
Algorithm 1 computes the switching of N for which its reconciliation with G has the smallest cost, by analyzing each biconnected component of N independently. See Figure 3 for an example of application of this algorithm to the species network in Figure 1(a) and the gene tree in Figure 2(a).
Algorithm 1 Solving Problem 1
1: Input: A species network N and a gene tree G such that , and positive costs δ, λ for duplication and loss events, respectively.
2: Output: A switching S of N that is optimal in the sense of Problem 1.
3: Compute the tree GN and its labeling function B(·);
4: Compute for each biconnected component Bi of N that is not a leaf;
5: for each biconnected component Bi of N do
6: for each switching of N(Bi) do
7: ;
8: the switching of N(Bi) with the lowest value of costj over all j;
9: Return the switching S of N in which each elementary network N(Bi) has as switching.
Theorem 2 Let N be a level-k network with p biconnected components. Algorithm 1 runs in O(|N| + 2k·p·|G|) time and returns a switching S of N such that cost(G,S) is minimum.
Proof: Complexity: It is well-known that all biconnected components of N can be computed in linear time, i.e. O(|N|), by using depth-first-search [10]. After a linear preprocessing, LCA operations on a tree can be performed in constant time [21,22]. Thus, from Remark 2, the mapping B(·) can be computed in O(|GN|). Hence, the tree GN can be constructed in times O(|GN | + |N|).
By a simple traversal of GN which takes time O(|GN |), we can compute for all Bi. Each biconnected component Bi of N has at most k hybridization nodes, then N(Bi) has at most 2k switchings. For each switching of N(Bi), it takes O(|GB |) to compute costj [23,24]. The overall size of all GBi is the size of GN . Therefore, the total complexity of the loop at lines 5 - 8 is O(2k ·|GN |). Each edge of G can have at most p artificial nodes added to it. Hence in the worst case O(|GN |) = O(|G|·p), i.e. the total complexity of Algorithm 1 is O(|N| + 2k·p·|G|).
Correctness: Let S be a switching of N where each elementary network N(Bi) has Si as the switching. By Lemma 4, cost(G,S) = . Hence cost(G,S) is minimum if and only if is minimum for every Si. Lines 5-8 in Algorithm 1 search, for each network N(Bi), the tree such that is minimum, which implies the correctness of the algorithm.
Minimum Reconciliation on Networks
Given a tree G and a network N, in [4] the authors prove that reconciling G on N can be solved in polynomial time, when host switchings (or transfer events, in the DTL reconciliation terminology) are also accounted for. In their model, each node of N is dated by a single date while each node of G is dated by a set of dates, and they search for a parsimonious reconciliation between G and N, i.e. one that has minimum cost, under the constraint that an event associated to a node u of G can only happen at a node/edge of N whose date is contained in the set of possible dates of u. Although the algorithm complexity stays polynomial, it is very high: O(τ3·|G|·|N|5) for a binary tree and a binary network, where τ is the number of possible dates of the nodes of G and N, which is at most O(|G| + |N|). A drawback of this model is that it requires information on the dates that is often not available. Moreover, transfers are not always possible in all parts of Tree of Life. Here, we take into account only speciation, duplication and loss events, and consider G and N as undated (see Problem 2 and Definition 5 for more details). Using a similar dynamic algorithm on this simpler model, and by some further analyses, we provide an algorithm that is a generalization of the LCA algorithm to networks that has a much smaller complexity than the algorithm in [4], namely O(h2·|G|·|N|), where h is the number of hybridization nodes of N.
Let x, y be two nodes of N. Denote by the subset of such that, for every , there does not exist any such that every path from z to x and to y passes through z'. For example, in Figure 1(d), m1, m2, m3 are in but only m1 and m2 are in because every path from m3 to x, y must pass m2.
For the sake of simplicity, we consider only reconciliations without duplications on hybridization nodes: indeed, since losses are not counted at hybridization nodes, a duplication on such nodes can be moved to its only child without changing the total cost of the reconciliation.
Lemma 5 Let α be a reconciliation of minimum cost between G and N, then for every node u of G that has two children u1, u2, we have:
(i) if , then ;
(ii) if , then either αr (u1) ≤N αr(u2) and αr(u) = αr(u2), or αr(u2) ≤N αr(u1) and αr(u) = αr(u1).
Proof: (i) Suppose that , then by definition αr(u) must be a node of . Let x1, x2 be two children of αr(u) such that lα(u) = dist(x1, αr(u1))+dist(x2, αr(u2)). Suppose that , then there exists a node y in such that every path from αr(u) to αr(u1) (resp. to αr(u2)) passes for y. Let y1, y2 be the two children of y.
First, we suppose that the shortest path from x1 to αr(u1) passes through y, y1, while the one from x2 to αr(u2) passes y, y2. Consider the reconciliation α' such that and for every v ≠ u, while and . It is easy to see that α' respects Definition 5, and that d(α) = d(α'). Denote by f = dist(αr(u), y), then f >0. The numbers of losses in α and α' are different on those associated with u and p(u) (if u is not the root of G). We thus have lα(p(u)) ≥ lα'(p(u)) - f if u ≠ r(G). Moreover, lα(u) = dist(x1, αr(u1)) + dist(x2, αr(u2)) = dist(x1, y) + 1 + dist(y1, αr(u1)) + dist(x2, y) + 1 +dist(y2, αr(u2)) ≥ dist(αr(u), y) + dist(y1, α' (u1) + dist(αr(u), y)+dist(y2, α' (u2))) ≥ 2·f +lα' (u). Hence, whether u coincides with r(G) or not, l(α) > l(α'), i.e. cost(α) > cost(α'), a contradiction.
Suppose now that both the shortest paths from x1 to αr(u1) and to αr(u2) pass through y, and then pass through y1 (or y2). This means that y1 is above both αr(u1) and αr(u2). Let y' be one of the lowest nodes below or equal to y1 that is above both αr(u1) and αr(u2). Let , be the two children of y', and sup-1 2 pose, without loss of generality, that is above or equal to αr(u1) and is above or equal to αr(u2). Hence, the shortest path from x1 to αr(u1) must pass, in the order, through y, y1, y', and y', while the shortest path from x2 to αr(u2) must pass through y, y1, y', and . By defining the reconciliation α'as done above apart for α'(u), which is fixed to y', we arrive at a contradiction by a similar argument.
(ii) Suppose that , and αr(u1), αr(u2) are not comparable. Hence, is not empty. If , then there exists a node such that every path from αr(u) to αr(u1) and to αr(u2) must pass through y. Similarly to case (i), we have that the reconciliation α' that coincides with α apart for the fact that and has smaller cost than α, a contradiction. Hence, αr(u) 2 ℳinN (αr(u1), αr(u2)). Considering now the reconciliation α' that coincides with α but for α' (u), which is fixed to . Hence d(α) = d(α')+1, and if u ≠ r(G). Let x1, x2 be the two children of αr(u). If the shortest path from αr(u) to αr(u1) passes through x1 while the shortest path from αr(u) to αr(u2) passes through x2, then . Therefore, cost(α) > cost(α'), a contradiction. Thus, both the two shortest paths from αr(u) to αr(u1) and αr(u2) must pass through x1 (or x2). Let y be one of the lowest nodes located on both of these paths. Then, y ∈ ℳinN (αr(u1), αr(u2)). By the same argument as in the previous case, the reconciliation α" coinciding with α but for and must have smaller cost than α, a contradiction.
Hence, either αr(u1) ≤N αr (u2) or αr(u2) ≤N αr (u1). Suppose that the first case holds (the second case is similar), but αr(u) ≠ αr(u2), i.e. αr(u2) <N αr (u). Let x1, x2 be two children of αr(u). If the shortest path from αr(u) to αr(u1) passes through x1 while the shortest path from αr(u) to αr(u2) passes through x2, then by replacing αe(u) by an event, we obtain a reconciliation with a smaller cost. Thus, both shortest paths from αr(u) to αr(u1) and αr(u2) must pass through x1 (or x2). Let y be a node that is located on both paths such that there is no other node below y on these two paths. Then, y ≠ ℳinN (αr(u1), αr(u2)). By the same argument as in the case (i), the reconciliation α' such that , for every v ≠ u, , must have smaller cost than α, a contradiction.
Now, we are ready to describe a dynamic algorithm to compute a reconciliation of minimum cost between G and N. Let α be a reconciliation between G and N. For every u ∈ V(G), denote by costα(u) the cost of the reconciliation of α restricted to Gu. Hence, if α is a most parsimonious reconciliation, then costα(u) is the minimum cost among all reconciliations between Gu and N that maps u to αr(u). Algorithm 2 aims at computing, for each u, the set (u) containing all pairs (x,c) such that c is the minimum cost among all reconciliations between Gu and N mapping u to x. It is straightforward to see that the cost of a most parsimonious reconciliation between G and N is the minimum cost involved in a pair in (r(G)).
The function merge(L1, L2) used in Algorithm 2 takes as input two lists of pairs (x,c) - where x is a node of N and c is a real number - and merges them keeping, for each x, the pair (x,c) with the smallest value of c. The method computeMin(y, z) used in Algorithm 2 is detailed in Algorithm 3. This method computes, for two nodes y, z of N, the set by using two breath-first-searches (BFS) starting respectively from y and z up to the root of N (note that, to perform the breath-first-searches, the edges are considered as directed in the inverse order). For this, it labels each node v in such a way that, if v is not strictly above y and z, then label (v) = ∅. Otherwise, label(v) is the lowest node such that every path from v to y and z passes through it. This method also computes the value of the function dist between y (resp. z) and each node visited in the corresponding breath-first-search.
Algorithm 2 Solving Problem 2
1: Input: A network N and a tree G such that , and positive costs δ, λ for duplication and loss events, respectively.
2: Output: The set (u) of pairs (x,c) for every .
3: for each node u of G in post-order do
4: (u) ← ∅;
5: if u is a leaf then
6: Let x be the leaf of S such that s(x) = s(u);
7: (u) ← {(x, 0)};
8: else
9: Let u1, u2 be the two children of u;
10: for each (y, c1) ∈ (u1) and each (z, c2) ∈ (u2) do
11: computeMin(y, z);
12: C ← ∅;
13: for each do
14: Let x1, x2 be the two children of x in N ;
15: c = c1 + c2 + λ·min{dist(x1, y) + dist(x2, z), dist(x2, z) + dist(x1, y)};
16: C ← C ∪ {(x, c)};
17: if y ≤N z then
18: c = δ + λ·dist(z, y)+ c1 + c2;
19: C ← C ∪ {(z, c)};
20: else if z ≤N y then
21: c = δ + λ·dist(y, z) + c1 + c2;
22: C ← C ∪ {(y, c)};
23: (u) = merge;
24: Return .
The following theorem proves the correctness of Algorithm 2:
Theorem 3 Algorithm 2 returns a matrix such that, for every u ∈ V(G), (x,c) is contained in (u) if and only if there exists a most parsimonious reconciliation between Gu and N mapping u to x with cost c.
Algorithm 3 computeMin(y, z)
1: Proceed a BFS from y, store the ordered list of visited nodes in BFS(y) and compute, for each u in BFS(y), dist(y,v);
2: Do the same from z;
3: for each node v ∈ BFS(y) do
4: if v = y or v = z or v is not in BFS(z) then label(v) = ∅
5: else if v has only one child v1 that is in BFS(z) or BFS(y) then
6: label(v) = label(v1);
7: else
8: Let v1, v2 be the two children of v;
9: if label(v1) = label(v2) ≠ ∅ then
10: label(v) = label(v1);
11: else
12: label(v) = {v}; Add v into ;
Proof: For each u ∈ V(G), we need to prove that, if α' is a reconciliation of minimum cost between Gu on N, then (α'(u), costα') is contained in (u). This is obviously true for every leaf of G (by lines 5-7). Let now u be an internal node having two children u1, u2. Then, following Lemma 5, (u) can be computed from (u1) and (u2) by using the information contained in and dist. Lines 10-23 in Algorithm 2 computes (u) following this lemma.
It remains now to prove that Algorithm 3 correctly computes . For every node v that is above y, z, denote by low(v) the lowest node such that every path from v to y, z must pass through this node. There is only one such node. Indeed, suppose that there are two such nodes m, m', then every path from v to y must pass through both m, m', i.e. either m <N m' or m' <N m, contradicting the lowest property of m, m'. To prove the claim, we need to show that if v is a node of BFS(y), then:
(i) if v is not strictly above y and z, then label(v) = ∅;
(ii) otherwise, label(v) = low(v) and line 12 of Algorithm 3 is performed if and only if v is actually in .
Let v be a node of BFS(y), then (i) holds by line 4. Now consider the case where v is strictly above y and z. We will prove (ii) by recursion on v following the order of the nodes in BFS(y). The recursion begins from the set of lowest nodes that are strictly above y and z, i.e. the set of nodes of v in such that there is not any node in that is below v. Let v1, v2 be two children of v, then by hypothesis v1, v2 are not strictly above y, z, i.e. label(v1) = label(v2) ≠ ∅; and low(v) = v. Thus, due to line 12, label(v) = v = low(v). Now let v be a node strictly above y, z such that (ii) is correct for each node below v which is strictly above or equal to y, z. If v is a hybridization node, then it is evident that low(v) = low(v1) where v1 is the only child of v. Moreover, since label(v1) = low(v1) (by the hypothesis of recurrence), then label(v) = label(v1) = low(v1) = low(v). If v is a speciation node having two children v1, v2 such that v2 is not in either BFS(y) or BFS(z), then we also have low(v) = low(v1). Hence, due to lines 5 - 6, we have label(v) = label(v1) = low(v1) = low(v). Now consider the last case, i.e. v is a speciation node having two children v1, v2 that are both in BFS(y) and BFS(z). If there exists a node q = low(v1) = low(v2), then low(v) = q because every from v to y, z must pass either through v1 or v2, i.e. always pass through q. Following line 10, we fix label(v) = label(v1) = q = low(v). Moreover, in this case v can not be in because every path from v to y, z passes through a node q below v. In the last case, we have . Since both v1, v2 are above y, z, then there exists a node q1 = label(v1), and a different node q2 = label(v2). We will prove that . Indeed, suppose otherwise, then there is a node q such that every path from v to y, z must pass through q. Hence, every path from v1 (resp. v2) to y, z must also pass through q, so q1 ≤N q (resp. q2 ≤N q). It means that there is a path from v1 to q to q2 and then to y, z that does not pass through q1, a contradiction. Hence v is in , and thus by definition the only node that every path from v to y, z must pass through is v itself (line 12).
We now present some intermediate results that will be useful to prove the complexity of Algorithm 2.
We extend the definition of to a subset of leaves. Let L be a subset of L(N ). If |L| = 1, then . Otherwise, is the set of nodes m of N such that m is above all leaves in L and there exist at least two separated paths from m to two distinct leaves of L.
Given a node u of G, LN (u) is defined as the set of leaves of N to which α maps the leaf set of Gu, i.e. and s(u) = s(x)}.
Lemma 6 Let α be a most parsimonious reconciliation between G and N, then, for every node u of .
Proof: It is true for every leaf u of G. Let u now be an internal node having two children u1, u2.
If u1, u2 ∈ L(G), then following Remark 1, LN (u) consists of two distinct nodes αr(u1), αr(u2). By Lemma 5, and , i.e. there exist two separated paths from αr(u) to αr(u1) and αr(u2). It means that .
Let u1 ∉ L(G), then following Remark 1, |LN (u1)| ≥ 2, then there always exist two distinct leaves x, y of LN (u) such that x ∈ LN(u1), y ∈ LN(u2), i.e. x is below αr(u1) and y is below αr(u2). If , then following Lemma 5, , i.e. there exist two separated paths from αr(u) to αr(u1), αr(u2). By extending these two paths from αr(u1) to x, and from αr(u2) to y, we have to two separated paths from αr(u) to x, y. In other words, αr(u) ∈ ℳN (LN (u)). If αe(u) = and suppose that αr(u2) ≤N αr (u1), then αr(u) = αr(u1) following Lemma 5, and αr(u) is not a leaf of N following Remark 1. Let u' be the highest node such that u' ≤G u1 and . Then following Lemma 5, , and there exists two separated paths from αr(u'), i.e. from αr(u), to two distinct leaves of LN(u'), i.e. two distinct leaves of LN (u). We can prove the claim similarly for the case when αr(u1) ≤N αr (u2).
Lemma 7 If N is a network that contains h hybridization nodes, then for every subset L of holds.
The proof of this lemma is deferred to the appendix. We are now ready to state the complexity of Algorithm 2.
Theorem 4 The time complexity of Algorithm 2 is O(h2·|G|·|N|) where h is the number of hybridization nodes of N.
Proof: For every u ∈ V(G), is equal to the possible nodes of N that u can be mapped to, which is bounded by by Lemma 6, and so by O(h) following Lemma 7.
The for loop at lines 3 - 23 is performed |G| times, and, at each iteration, the for loop at lines 10-23 is performed O(h2) times. In each iteration of the second loop, the operation computeMin, as detailed in Algorithm 3, requires two breath-first-search traversals, which can be performed in time O(|N|). Moreover, for every node x of , by definition there exists two separated paths from x to y, z, which can be extended to be two separated paths from x to two distinct leaves l1, l2 of LN (u) where l1 is a leaf below y of and l2 is a leaf below z. This is always possible because, by Remark 1, |LN (u)| > 1. Hence, x must be in by definition, i.e. , and thus . Therefore, the loop at lines 13 - 16 can be performed in time O(h). The operation merge(L1, L2) at lines 23 for two lists of size O(h) can be implemented in times O(h), if we know that the resulting list is also of size O(h). Hence, it takes O(|N| + h) = O(|N|) times for each iteration of the loop 10 - 23. Therefore, the total complexity is O(h2· |G|·|N|).
Finally, a reconciliation of minimum cost between G and N can be then obtained by a standard back-tracking of the matrix , starting from any pair (x,c) of (r(G)) such that c is the minimum value over all pairs in (r(G)).
Conclusions
In this paper, we have studied two variants of the reconciliation problem between a gene tree and a species network. In particular, for the problem of finding the "most parsimonious" switching of the network, even though the number of switchings can be exponential with respect to the number of hybridization nodes, we proposed an algorithm that is exponential only with respect to the level of the network, which is often low for biological data. Moreover, the problem of finding a reconciliation between a gene tree and a network, which was solved in [4] for a more general model but with a very high complexity, was re-studied here for a simpler model, which is more pertinent for same parts of the Tree of Life, and an algorithm with a much smaller complexity was provided. In a further work, we intend to implement the algorithms presented in this paper and apply them to biological data.
Appendix
Proof of lemma 3
By Definition 8, for every u ∈ I(G), there must exist H ∈ GB(u) such that u ∈ I(H). If H is an edge, then u is the only internal node of H, which must be an artificial node by Lemma 2. But this is not possible because nodes of G cannot be artificial. Hence, H must be a binary tree. Let denote Bi = B(u), and Si = S(B(u)). We will prove now that by recursion on the height of u.
Let u be an internal node of G that has two children u1, u2 in G, and let H be the binary tree of such that u ∈ I(H). Denote , , and , . For j = 1, 2, if uj is a leaf, let Hj be equal to uj , otherwise Hj is the binary tree of such that uj ∈ I(Hj). For the sake of convenience, if uj is a leaf, we also denote by the reconciliation that maps the only leaf uj to the only leaf x of N such that s(x) = s(uj ). Note that s(x) = α(uj ).
We now suppose that for j = 1, 2 (which is evidently true if uj is a leaf), and we will show that this implies that the claim is true for u.
Let (resp. ) be the child of u in GN such that . We respectively denote and . By definition of the LCA reconciliation, we have .
(i) If , then , and H = H1 = H2. This implies that , because otherwise H will contain an artificial node. The same holds for and u2. Thus,
(ii) If , then , and H = H2. As in point (i), this implies and thus . If , then we have . Otherwise, is an artificial node which is a leaf of H that is mapped to r by . By Lemma 1, , so all nodes of Bi are above all nodes of and above . This implies that . Similarly for the case where .
(iii) Suppose now that , are not comparable and that and (the other cases can be shown reusing the arguments of point (ii)). Then, similarly as in point (ii), , and , are leaves of H mapped respectively to and by . Since is a node of , is a node of , then
Therefore, in all cases we always have , and thus the same is true for every u ∈ I(G) by recursion.
Proof of lemma 7
We will prove this lemma by recursion on the number of hybridization nodes. See the figure 4 for an example.
Figure 4.
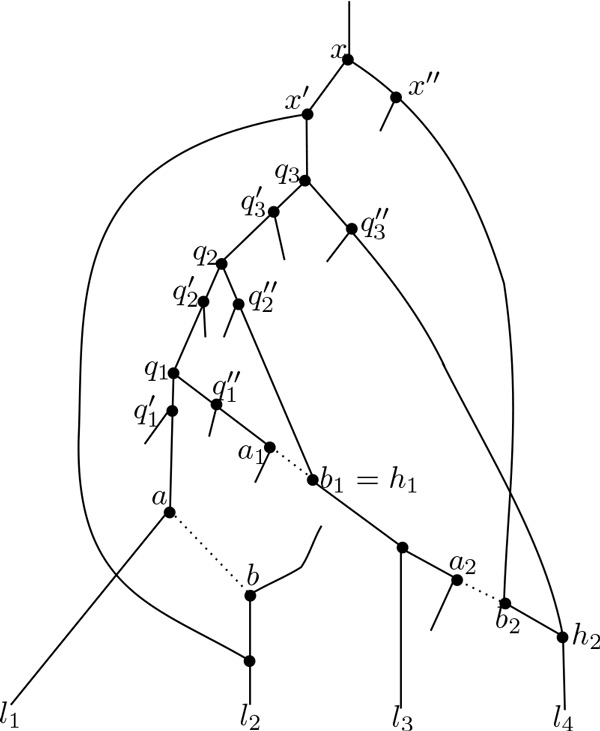
An illustration for the proof of Lemma 7. In this example: L = {l1, l2, l3, l4}, Q = {q1, q2, q3}, L* = {l1, l3, l4}, L3 = {l4}, L2 = {l3}, and L1 = ∅. For i = 1, let l(q1) = l3; thus, we have c(1) = 2 and the path p1 starts from q1, passes through , b1 to l3, while the path starts from q2, passes through , b1 to l3. In this case, we have h1 = b1. For i = 2, let l(q2) = l3; thus we have c(2) = 3 and the path p2 starts from q2, passes through , b1, b2, h2 to l4, while the path starts from q3 passes through to l4. Let x be a node in , then x is also in where N" is obtained from N' by removing all edges (ai, bi).
If there is no hybridization node, then N is a tree, and it is evident that contains exactly one node.
Suppose that the claim is correct for every network having h hybridization nodes. Let N now be a network that has h + 1 hybridization nodes. Let (a, b) be an edge of N having as target a hybridization node (namely, b) such that it does not exist any hybridization node above a (such a node always exists because N is a directed acyclic graph). Let N' be the network obtained by removing (a, b) from N (and also removing all nodes of indegree 1 and outdegree 1 created by this removal), then N' has exactly h hybridization nodes. Let , then every node q in Q must be above a. Indeed, if q is not above a, then every path from q to every node of L does not contains (a, b), thus q is in , a contradiction. Moreover, by hypothesis, there is no hybridization node above a, hence all nodes of Q must be contained in a path leading a, and this path does not contain any hybridization to node. Let enumerate the nodes in Q as q1,... qm from the lowest to the highest one.
If , we are done.
Suppose now that |Q| = m >1. In the following, we will define m - 1 edges of N having as target a hybridization node such that if N" is the network obtained from N' by removing these edges, then .
Denote by L* the set of nodes in L that are below qm in N'. Hence L \ L* is not empty since otherwise qm would be in , a contradiction. For every qi, i = 1,..., m, let , be the two children of qi such that is above or equal to a. Hence, , is not above or equal to a since there is no hybridization node above a, thus every path from qi to L \ L* must pass through and a, b. By definition, there must exist at least two separated paths from qi to two leaves of L. Hence, for every i, there exists always a path from qi to a node of L* that passes through . Denoted this node by l(qi).
Denote by Lm the set of nodes in L* such that, for every l ∈ Lm, there is a path from q to l that passes through . As explained above, Lm is not empty. Recursively, for every i< m, let Li be the set of nodes in L* \ Lm \... Li+1 such that, for every l ∈ Li, there is a path from qi to l that passes . Note that this set may be empty if i< m, and for every i ≠ j, Li ∩ Lj = ∅.
We will define, for each i < m, an index c(i) that is strictly greater than i such that Lc(i) ≠ ∅, together with two paths pi (resp. ) from qi (resp. qc(i)) to a node of Lc(i) as follows: if l(qi) is not in Li, then by the definition, there exists a unique j such that Lj contains l(qi), and j > i. We fix c(i) = j. Next, we define pi (resp. ) as a path from qi (resp. qc(i)) to l(qi) that passes (resp. ). If l(qi) is in Li, then let c(i) be the smallest number that is greater than i and Lc(i) ≠ ∅ (such an index always exists because Lm ≠ ∅). Let l' be a node of Lc(i). Since qi is in MN (L), then there must exist a path from qi to l', and we define pi as this path. The path is defined as the one from qc(i) to l' that passes through . Note that pi, must contain at least one common hybridization node since they start from different nodes and end at a same leaf of L. Denote by hi the highest common hybridization node of pi and . Hence, all his are distinct since each pi starts at a different node qi, and each starts at a node qc(i) that is strictly greater than qi. We define (ai, bi) recursively in increasing order of i from 1 to m - 1 as follows. If i = 1, then bi is the highest hybridizatition node on pi. If i >1, then bi is the highest hybridization node on pi and different from all bk for every k < i. There exists always such a node bi, for example hi. Therefore, all bis are distinct. Denote by ai the parent of bi on the path pi.
Denote by N" the network obtained from N' by removing all edges (ai, bi). For every node x in , we will prove that x is also in . Denote by x', x" the two children of x. By definition, for every l ∈ L, there exists a path, denoted by f'(l), in N' from x to l such that at least one path among them passes through x' and one other passes through x". To prove that x is in , we will now construct another set of paths in N" (i.e. in N' and does not contain any (ai, bi)) from x to each leaf l of L, denoted by f"(l), such that at least one path among them passes through x'and one other passes through x".
Consider first the case that x is above qm (as shown in the figure 4). Without loss of generality, suppose that x' is above or equal to qm while x" is not. Suppose that f'(l) contains qm, then l ∈ L* and f'(l) must pass through x' because there is no hybridization node above qm. Suppose that . Let k be the index such that l ∈ Lk, then we can choose a path f"(l) in N' from x to l that does not contain any (ai, bi) as follows. This path starts from x, passes through x', goes down to qk , then takes the path from qk to l that contains q". Note that this path does not include any pi since, by construction, every path pi starts from qi and goes to a node in Lc(i) that is different from Li, while this path passes through qk and goes to a node in Lk . Moreover, this path and pi can not have any common hybridization node above ai because bi is the highest hybridization node on pi. Hence, it can not pass through (ai, bi) for any i. If l is not in , it means that every path from qm to l must pass through , then we take the path starting from x, going down to , then continuing to l. It is evident that this path does not include any pi, and it cannot have with pi any common hybridization node above ai because bi is the highest hybridization node on pi. Hence it does not pass through any (ai, bi). If f'(l) does not contain qm, then we fix f"(l) = f'(l). It is easy to see that f'(l) does not contain any edge (ai, bi) because otherwise f'(l) and pi must have at least a common hybridization node above ai (since there is no hybridization node above qi). But this is not possible because bi is the highest hybridization node on pi. Remark that at least one of the paths f'(l) in this case must pass through x" since all paths in the first case must pass through x'. Hence at least two of the paths f"(l) are separated, thus x is in by definition.
Consider now the case that x is not above qm, then similarly as in the previous case where f'(l) does not contain qm, we deduce that f'(l) does not contain any (ai, bi) for every l. Hence, by choosing f"(l) = f'(l) for every l, we are done.
Therefore, we have .
The network N" contains h - |Q| + 1 hybridization nodes, then following the hypothesis of recurrence, . This implies that , thus .
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Both authors contributed to design the models, algorithms and to write the paper.
Acknowledgements
This work was partially funded by the French Agence Nationale de la Recherche Investissements d'Avenir/ Bioinformatique (ANR-10-BINF-01-02, Ancestrome). The publication charges of this article were funded by the French Grant Agence Nationale de la Recherche: Investissements d'Avenir/Bioinformatique (ANR-10-BINF-01-02, Ancestrome).
This article has been published as part of BMC Genomics Volume 16 Supplement 10, 2015: Proceedings of the 13th Annual Research in Computational Molecular Biology (RECOMB) Satellite Workshop on Comparative Genomics: Genomics. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenomics/supplements/16/S10.
References
- Doyon J-p, Ranwez V, Daubin V, Berry V. Models, algorithms and programs for phylogeny reconciliation. Briefings in bioinformatics. 2011;12(5):392–400. doi: 10.1093/bib/bbr045. doi:10.1093/bib/bbr045. [DOI] [PubMed] [Google Scholar]
- Maddison WP. Gene trees in species trees. http://sysbio.oxfordjournals.org/content/46/3/523.full.pdf+html. Systematic Biology. 1997;46(3):523–536. doi:10.1093/sysbio/46.3.523. [Google Scholar]
- Huson D, Rupp R, Scornavacca C. Phylogenetic Networks. Cambridge University Press; 2010. [Google Scholar]
- Libeskind-Hadas R, Charleston MA. On the computational complexity of the reticulate cophylogeny reconstruction problem. JCB. 2009;16(1):105–117. doi: 10.1089/cmb.2008.0084. [DOI] [PubMed] [Google Scholar]
- Page RDM. Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics. 1994;10(2):155–173. [Google Scholar]
- Ronquist F. Reconstructing the history of host-parasite associations using generalized parsimony. Cladistics. 1995;11(1):73–89. doi: 10.1111/j.1096-0031.1995.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Charleston M. Jungles: a new solution to the hostparasite phylogeny reconciliation problem. Math Biosci. 1998;149(2):191–223. doi: 10.1016/s0025-5564(97)10012-8. [DOI] [PubMed] [Google Scholar]
- Merkle D, Middendort M. Reconstruction of the cophylogenetic history of related phylogenetic trees with divergence timing information. Theory Biosci. 2005;123(4):277–299. doi: 10.1016/j.thbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Doyon JP, Scornavacca C, Gorbunov KY, Szöllősi GJ, Ranwez V, Berry V. Research in Computational Molecular Biology: Proceedings of the 14th International Conference on Research in Computational Molecular Biology (RECOMB) Vol. 6398. LNCS, Springer, Berlin/Heidelberg, Germany; 2010. An effcient algorithm for gene/species trees parsimonious reconciliation with losses duplications and transfers; pp. 93–108. Software downloadable at http://www.atgc-montpellier.fr/Mowgli/ [Google Scholar]
- Cormen TH, Stein C, Rivest RL, Leiserson CE. Introduction to Algorithms. 2. McGraw-Hill Higher Education; 2001. [Google Scholar]
- Choy C, Jansson J, Sadakane K, Sung W-K. Computing the maximum agreement of phylogenetic networks. Theoretical Computer Science. 2005;335(1):93–107. [Google Scholar]
- Fischier M, van Iersel L, Kelk S, Scornavacca C. On Computing the Maximum Parsimony Score of a Phylogenetic Network. SIAM Journal on Discrete Mathematics - SIDMA. 2015. in press .
- Kelk S, Scornavacca C, Van Iersel L. On the elusiveness of clusters. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 2012;9(2):517–534. doi: 10.1109/TCBB.2011.128. [DOI] [PubMed] [Google Scholar]
- Doyon J, Chauve C, Hamel S. Space of gene/species trees reconciliations and parsimonious models. Journal of Computational Biology. 2009;16(10):1399–1418. doi: 10.1089/cmb.2009.0095. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17(9):821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- Goodman M, Czelusniak J, Moore GW, Romero-Herrera AE, Matsuda G. Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Systematic Biology. 1979;28(2):132–163. [Google Scholar]
- Page RDM. Maps between trees and cladistic analysis of historical associations among genes, organisms, and areas. Systematic Biology. 1994;43(1):58–77. [Google Scholar]
- Chauve C, El-Mabrouk N. New perspectives on gene family evolution: Losses in reconciliation and a link with supertrees. Research in Computational Molecular Biology, 13th Annual International Conference, RECOMB 2009, Tucson, AZ, USA, May 18-21, 2009. Proceedings. 2009. pp. 46–58.
- Kanj Ia, Nakhleh L, Than C, Xia G. Seeing the trees and their branches in the network is hard. Theoretical Computer Science. 2008;401:153–164. doi:10.1016/j.tcs.2008.04.019. [Google Scholar]
- Downey RG, Fellows MR. Fundamentals of Parameterized Complexity. Springer. 2013;4 [Google Scholar]
- Gabow HN, Tarjan RE. A linear-time algorithm for a special case of disjoint set union. Proceedings of the Fifteenth Annual ACM Symposium on Theory of Computing. 1983. pp. 246–251.
- Harel D, Tarjan RE. Fast algorithms for finding nearest common ancestors. SIAM Journal on Computing. 1984;13(2):338–355. [Google Scholar]
- Zhang L. On a mirkin-muchnik-smith conjecture for comparing molecular phylogenies. Journal of Computational Biology. 1997;4:177–187. doi: 10.1089/cmb.1997.4.177. [DOI] [PubMed] [Google Scholar]
- Bender M, Farach-Colton M. The lca problem revisited. LATIN 2000: Theoretical Informatics. Lecture Notes in Computer Science Springer. 2000;1776:88–94. [Google Scholar]


