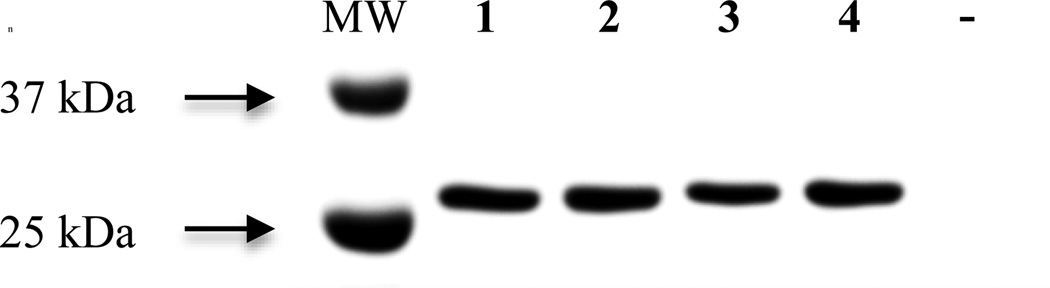Abstract
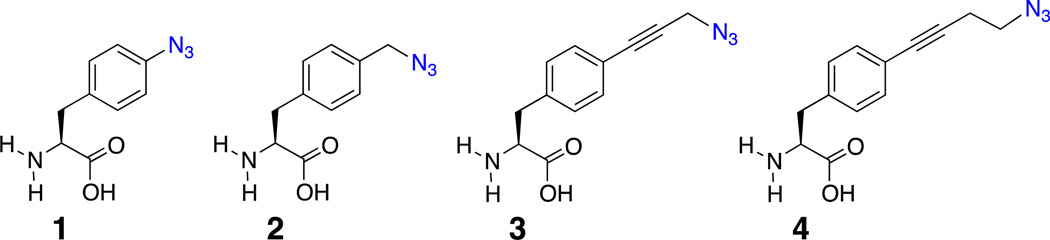
Two new azidophenylalanine residues (3 and 4) have been synthesized and, in combination with 4-azido-L-phenylalanine (1) and 4-azidomethyl-L-phenylalanine (2), form a series of unnatural amino acids (UAAs) containing the azide vibrational reporter at varying distances from the aromatic ring of phenylalanine. These UAAs were designed to probe protein hydration with high spatial resolution by utilizing the large extinction coefficient and environmental sensitivity of the azide asymmetric stretch vibration. The sensitivity of the azide reporters was investigated in solvents that mimic distinct local protein environments. Three of the four azido-modified phenylalanine residues were successfully genetically incorporated into a surface site in superfolder green fluorescent protein (sfGFP) utilizing an engineered, orthogonal aminoacyl-tRNA synthetase in response to an amber codon with high efficiency and fidelity. SDS-PAGE and ESI-Q-TOF mass analysis verified the site-specific incorporation of these UAAs. The observed azide asymmetric stretch in the linear IR spectra of these UAAs incorporated into sfGFP indicated that the azide groups were hydrated in the protein.
Keywords: Azide Vibrational Reporters, Unnatural Amino Acids, Infrared Spectroscopy, superfolder Green Fluorescent Protein
Introduction
Amino acids covalently modified with spectroscopic reporters offer the potential to probe local protein hydration with high spatial and temporal resolution when coupled with the appropriate spectroscopic technique. The development of effective unnatural amino acids (UAAs) to serve as sensitive, site-specific probes rests on several key principles. First, the UAAs must have a readily measurable spectroscopic observable that is sensitive to local environment. Secondly, the synthesis of the UAA should be efficient, cost-effective, and amenable to a large scale. Thirdly, the UAA must be able to be incorporated in either peptides or proteins in a site-specific manner with high efficiency and fidelity. Lastly, the UAA needs to be stable and minimally invasive to the native protein structure.
A number of unnatural amino acids containing a variety of vibrational probes have been utilized to study protein structure and dynamics. For instance, the nitrile (-CN), 1–12 azide (-N3),13–21 thiocyanate (-SCN),22–26 selenocyanate (-SeCN)27–29 and metal carbonyl30–32 vibrational reporters have been utilized for this purpose. These reporter moieties have been incorporated into peptides and/or proteins by standard solid-phase peptide synthesis,1 post-translational modifications,22, 31 and/or genetic techniques.33 Each probe has a vibrational signature that occurs in a relatively clear region of the infrared spectrum and is sensitive to local environment. Additionally, the relative small size of most of these reporters helps to minimize any perturbation to the native protein structure.
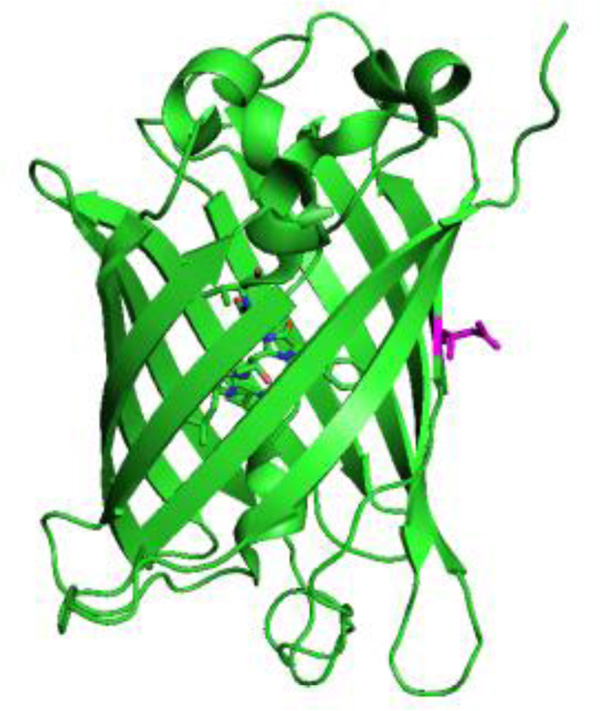
Recent work in our laboratory has focused on unnatural amino acids containing the azide vibrational reporter to take advantage of the relatively large oscillator strength of the azide asymmetric stretch vibration and the high sensitivity of this vibration to local environment. This work has included the development of 4-azidomethyl-L-phenylalanine (2), which is a stable analogue of the photo-reactive UAA, 4-azido-L-phenylalanine (1).21 As expected, the azide asymmetric stretch of 2 was sensitive to local protein environment as demonstrated through the incorporation of this UAA at multiple sites in the 247 residue, β-barrel superfolder green fluorescent protein (sfGFP, Figure 2)34 utilizing an engineered, orthogonal aminoacyl-tRNA synthetase.
Here, we have extended the toolkit of azido-modified amino acids to serve as sensitive, site-specific vibrational probes of local protein hydration. The series of azido-modified UAAs (shown in Figure 1) were designed to systematically alter the distance between the azide group and the phenyl ring of the modified phenylalanine residues. The alkyne group in 3 and 4 was designed as a rigid spacer that limits the conformational flexibility between the azide group and aromatic ring. In concert, this series has the potential to probe protein hydration systematically with high spatial resolution thus creating a vibrational hydration ruler. The synthesis of 3 and 4, the genetic incorporation of these UAAs into sfGFP at site 150 (Figure 2), and the subsequent infrared analysis of the resulting sfGFP constructs is discussed in addition to future applications of these UAAs.
Figure 1.
Structure of the series of the azido-modified unnatural amino acids.
Figure 2.
Structure of wt-sfGFP (PDB ID 2B3P) with site 150 (magenta) highlighted.
Experimental
General Information
Chemical reagents were purchased from Sigma-Aldrich, Strem, Gelest, Alfa Aesar, Peptech, and Tokyo Chemical Industry and used without further purification. Deuterated chloroform (98.8% D enrichment) and deuterated dimethyl sulfoxide (99.5% D enrichment) were purchased from Cambridge Isotope Laboratories. DH10B cells and pBadA were purchased from Invitrogen. 4-Azido-L-phenylalanine (1) was purchased from Bachem. The synthesis of Boc-4-iodo-L-Phe methyl ester (6) was carried out following a literature procedure.35 All aqueous solutions were prepared using 18 MΩ cm water.
Reactions were carried out under a dry argon atmosphere and stirred with a magnetic stir bar. An ice bath was utilized for reactions performed below ambient temperature. Analytical thin layer chromatography (TLC) was carried out using 0.2 mm silica plastic coated sheets (Selecto Scientific) with F254 indicator. Flash column chromatography was carried out using 230−00 mesh silica gel.
NMR spectra were obtained with a Varian INOVA 500 multinuclear Fourier transform NMR spectrometer at the following frequencies: 1H (499.7 MHz) and 13C (125 MHz). Chemical shifts are reported in parts per million (ppm) while all coupling constants are reported in hertz (Hz). 1H NMR spectra in CDCl3 were referenced to the residual solvent peak at 7.26 ppm while 13C NMR spectra in CDCl3 were referenced to the solvent peak at 77.0 ppm. 1H NMR spectra in DMSO were referenced to the residual solvent peak at 2.49 ppm while 13C NMR spectra in DMSO were referenced to the solvent peak at 39.5 ppm. All IR characterization for synthetic purposes were carried out as ATR thin film analyses with all peaks reported in wavenumbers (cm−1). Mass spectral (MS) analyses of the free UAAs (3, 4) for synthetic purposes were carried out on an Agilent 1100 series LC/MSD SL ion trap mass spectrometer with electrospray ionization and MS/MS capabilities. Protein constructs were analyzed by electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass analysis at the University of Illinois at Urbana-Champaign.
Boc-4-(3-hydroxy-1-propynyl)-L-Phe methyl ester (7a)
Pd(PPh3)2Cl2 (693 mg, 0.0987 mmol, 1 mol%) and CuI (376 mg, 0.19 mmol, 2 mol%) were suspended in triethylamine (40 mL) and 6 (4.00g, 9.87 mmol) was added, followed by propargyl alcohol (632 µL, 610 mg, 10.9 mmol, 1.1 equiv), which caused immediate darkening of the solution to a brown color. The reaction was stirred at room temperature for 5 h. The crude mixture was filtered through a glass frit and the solid residue was washed with Et3N. The filtrate was concentrated under reduced pressure and the residue was purified by flash column chromatography (hexanes/ethyl acetate, 3:2) to give 7a as a yellow-orange oil (2.73 g, 8.18 mmol, 83% yield). IR 3379.5 (br, w), 2977.5 (w), 1741.2 (s), 1693.3 (s), 1509.2 (s), 1437.6 (m), 1366.4 (s), 1252.0 (m), 1218.6 (m), 1164.2 (s), 1027.1 (s), 951.9 (w), 823.6 (w), 781.1 (w).1H NMR (CDCl3) 7.34 (d, J = 8.5, 2H), 7.06 (d, J = 8.5, 2H), 4.96 (d, J = 7.5, 1H), 4.56 (m, 1H), 4.47 (d, J = 6.0, 2H), 3.68 (s, 3H), 3.05 (m, 2H), 1.39 (s, 9H). 13C NMR (CDCl3) 172.09, 155.01, 136.45, 131.65, 129.12, 121.26, 87.60, 84.87, 79.97, 54.17, 52.13, 51.18, 38.05, 28.12.
Boc-4-(4-hydroxy-1-butynyl)-L-Phe methyl ester (7b)
7b was synthesized in the same manner as 7a using the following reagents: 3-butyn-1-ol (626 µL, 583 mg, 8.22 mmol, 1.1 equiv), Pd(PPh3)2Cl2 (524 mg, 0.075 mmol, 1 mol%), CuI (285 mg, 0.149 mmol, 2 mol%) and 6 (3.03 g, 7.47 mmol) in Et3N (30 mL). Purification by flash column chromatography (hexanes/ethyl acetate, 3:2) yielded 7b as a light orange oil (2.02 g, 5.79 mmol, 78% yield). IR 3374.3 (br, w), 2977.3 (w), 1694.0 (s), 1509.9 (s), 1436.1 (m), 1366.0 (s), 1249.9 (m), 1216.3 (m), 1163.9 (s), 1053.6 (s), 1021.4.4 (m), 729.8 (m). 1H NMR (CDCl3) 7.28 (d, J = 7.9, 2H), 7.01 (d, J = 7.9, 2H), 5.03 (m, 1H), 4.52 (m, 2H), 3.75 (t, J = 6.4, 2H), 3.65 (s, 3H), 3.05 (m, 2H ), 2.62 (t, J = 6.4, 2H), 1.37 (s, 9H). 13C NMR (CDCl3) 172.11, 154.98, 135.88, 131.68, 129.11, 122.01, 86.55, 81.90, 79.92, 61.00, 54.20, 52.15, 38.07, 28.16. 23.70.
Boc-4-(3-azido-1-propynyl)-L-Phe methyl ester (8a)
A solution of 7a (2.1813 g, 6.81 mmol), dimethyl formamide (5.4 mL) and triethylamine (1.41 mL, 10.21 mmol, 1.5 equiv) was cooled to 0°C under argon. Methanesulfonyl chloride (686 µL, 8.85 mmol, 1.3 equiv) was added over approximately 6 min causing the solution to become cloudy. After an additional 6 min at 0 °C, the solution was allowed to stir at room temperature for 30 min. The reaction was then quenched with water, extracted with diethyl ether (2 × 30 mL), and the combined organic layers were washed with H2O (50 mL) and brine (50 mL) before drying over MgSO4, filtering through a glass frit, and concentrating under reduced pressure. This crude product was immediately moved forward to the next step. The residue was dissolved in dimethyl formamide (5.4 mL) and sodium azide (664 mg, 10.21 mmol, 1.5 equiv) was added using appropriate precautions. The solution was allowed to stir at temperature for 72–96 h, or until TLC confirmed that the reaction was complete. Water (20 mL) was added and the product was extracted with ethyl acetate (3 × 25 mL). The combined organic layers were washed with water (1 × 25 mL), saturated aqueous NH4Cl (1 × 25 mL), water (1 × 25 mL), and brine (1 × 25 mL). The organic layer was dried over MgSO4, filtered through a glass frit, and concentrated under reduced pressure. Purification by flash column chromatography (hexanes/ethyl acetate 3:1) yielded 8a as a pale yellow oil (1.523 g, 4.42 mmol, 65% yield). IR 3405.5 (br, w), 2976.8 (w), 2118.8 (m), 1743.3 (m), 1701.1 (s), 1508.0 (s), 1437.1 (m), 1366.2 (s), 1247.8 (m), 1164.5 (s), 1108.5 (w), 1058.0 (m), 1020.8 (m), 860.5 (w). 1H NMR (CDCl3) 7.34 (d, J = 8.1, 2H) 7.05 (d, J = 8.1, 2H), 5.03 (d, J = 8.3, 1H), 4.53 (m, 1H), 4.08 (s, 2H), 3.66 (s, 3H), 3.03 (m, 1H), 1.36 (s, 9H). 13C NMR (CDCl3) 171.97, 154.90, 137.05, 131.88, 129.25, 120.50, 86.99, 81.03, 79.86, 54.14, 52.15, 42.65, 40.42, 38.13, 28.13.
Boc-4-(4-azido-1-butynyl)-L-Phe methyl ester (8b)
8b was synthesized in the same manner as 8a using the following reagents: 7b (1.1788 g, 3.39 mmol), dimethyl formamide (2.7 mL), triethylamine (704 µL, 5.09 mmol, 1.5 equiv), and methanesulfonyl chloride (342 µL, 4.41 mmol, 1.3 equiv). Following work up and concentration, the product was combined with dimethyl formamide (2.7 mL) and NaN3 (331 mg, 5.09 mmol, 1.5 equiv). Purification yielded 8b as a clear oil (706 mg, 1.90 mmol, 56% yield) IR 3354.0 (br, w), 2978.2 (w), 2108.6 (m), 1743.7 (m), 1712.1 (s), 1508.7 (m), 1438.0 (m), 1364.9 (s), 1249.6 (m) 1165.6 (s), 1056.7 (m), 1016.5 (m), 778.3 (m). 1H NMR (CDCl3) 7.30 (d, J = 8.1, 2H), 7.02 (d, J = 8.1, 2H), 4.97 (d, J = 7.3, 2H), 4.53 (m, 1H). 3.66 (s, 3H), 3.42 (t, J = 6.9, 2H), 3.03 (m, 2H), 2.67 (t, J = 6.9, 2H), 1.38 (s, 9H). 13C NMR (CDCl3) 172.05, 154.95, 136.11, 131.65, 129.17, 121.81, 85.85, 82.17, 79.89, 54.21, 52.14, 49.85, 38.15, 28.19, 20.53.
4-(3-azido-1-propynyl)-L-Phe (3)
8a (1.34 g, 3.87 mmol) was dissolved in a mixture of THF:H2O (3:1, 15.5 mL) and LiOH monohydrate (244 mg, 5.80 mmol, 1.5 equiv) was added and the mixture was stirred at room temperature for 4 h. 0.5 M NaHSO4 was added to adjust the pH of the solution to ~2.5. The mixture was extracted with ethyl acetate (2 × 25 mL) and the combined organic layers were washed with water (30 mL) and brine (30 mL), dried over MgSO4, filtered through a glass frit, and concentrated under reduced pressure. The crude product was immediately taken to the next step. The residue was dissolved in a solution of dry HCl in dioxane (2.5 M, 11 mL) and stirred for 4 h at room temperature. The solution was concentrated under reduced pressure to about a third of its initial volume, and pentane was added to precipitate the solid product. The product was isolated by filtration to obtain 3 as an off white to yellow powder (817 mg, 3.34 mmol, 86%). 1H NMR (DMSO) 7.43 (d, J = 8.5, 2H), 7.30 (d, J = 8.5, 2H), 4.36 (s, 2H), 4.13 (t, J = 6.2, 1H), 3.13 (d, J = 6.5, 2H).13C-NMR (DMSO) 170.24, 136.24, 131.78, 129.99, 120.18, 86.16, 82.60, 52.89, 39.00, 35.51. MS(ESI) 245.0 (M+, 100), 171.0 (40), 145.1 (20), 488.8 (15).
4-(4-azido-1-butynyl)-L-Phe (4)
4 was synthesized by the same method as 3 using the following reagents: 8b (1.961 g, 5.27 mmol), THF:H2O (3:1, 21.1 mL), and LiOH monohydrate (331 mg, 7.90 mmol). Following reaction completion and isolation, 2.5 M HCl in dioxane (13.9 mL). Filtration yielded 4 as a pale pink powder (1.353 g, 4.59 mmol, 87%). 1H NMR (DMSO) 7.34 (d, J = 8.3, 2H), 7.27 (d, J = 8.3, 2H), 4.13 (t, J = 5.9, 1H), 3.49 (t, J = 6.4, 2H), 3.14 (d, J = 6.4, 2H), 2.73 (t, J = 6.4, 2H). 13C NMR (DMSO) 170.18, 135.30, 131.79, 129.86, 121.59, 87.57, 81.67, 52.99, 49.24, 35.43, 19.89. MS(ESI) 259.1 (M+, 100), 213.1 (5), 517.1 (2M, 25).
Expression and Purification of sfGFP Constructs
A codon-optimized gene coded with a C-terminal 6-His affinity tag for wild-type sfGFP (wt-sfGFP) 10, 21, 34, 36, 37 was inserted into pBadA, generating pBad-sfGFP. The N150 codon was replaced through site-directed mutagenesis with the amber stop codon (TAG) generating pBad-sfGFP-150TAG. The engineered, orthogonal aminoacyl-tRNA synthetase for the incorporation of 1, 2, 3, and 4, was inserted into pDule, generating pDule-pN3Phe.10, 21, 35 These plasmids were obtained from Dr. Ryan A. Mehl (Oregon State University).
pBad-sfGFP was transformed into DH10B E. coli cells and pBad-sfGFP-150TAG was co-transformed with pDule-pN3Phe into DH10B E. coli cells. Five milliliters of noninducing media were inoculated with the transformed cells, which grew to saturation while shaking (250 rpm) at 37 °C. A portion of the cultured cells (2.5 mL aliquot) was used to inoculate 250 mL of autoinduction media containing the appropriate UAA at 1 mM. The negative control experiment excluded the UAAs from the autoinduction media. After shaking at 37 °C for 24−30 h, the cells from the autoinduction media were collected by centrifugation and the expressed protein was purified using TALON cobalt ion-exchange chromatography (Clontech) similar to previous procedures.10, 21
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass analysis were used to verify the incorporation of the UAAs into sfGFP at site 150 with high efficiency and fidelity. The sfGFP constructs containing 1, 2, or 4 yielded between 5 – 150 mg of purified protein per liter of autoinduction media. The yields for protein expression were calculated using the extinction coefficient of sfGFP at 488 nm.34
Equilibrium FTIR Measurements
A Bruker Vertex 70 FTIR spectrometer, equipped with a globar source, KBr beamsplitter, and a liquid nitrogen cooled mercury cadmium telluride (MCT) detector, was used to record all equilibrium FTIR absorbance spectra. The spectra were recorded using a temperature-controlled transmission cell with an embedded thermocouple to measure the temperature of the cell. The dual-compartment transmission cell consisted of two calcium fluoride windows with a path length of ~100 µm. The spectra were the result of 1024 scans recorded at a resolution of 1.0 cm−1. The spectra were recorded at 25°C, baseline corrected, and intensity normalized. Igor Pro was used for the analysis of all spectra (Wavemetrics).
Results and Discussion
Synthesis of Azido-Modified UAAs
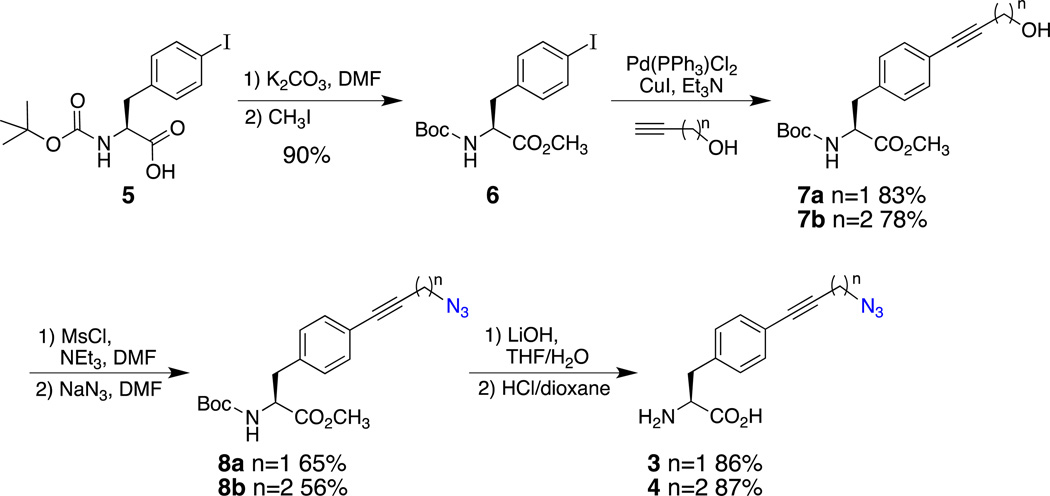
The syntheses of 3 and 4 were accomplished in gram quantities in just four steps from the commercially available Boc-4-iodo-L-Phe (5) in overall yields of 42% and 34%, respectively (Scheme 1). The first step for both syntheses was the protection of the carboxylic acid of 5 as its methyl ester (6), which was accomplished in 90% yield by the alkylation of the carboxylate with methyl iodide following a known procedure.35 The syntheses diverged in the Sonogashira coupling step; coupling with propargyl alcohol led to alcohol 7a (83%), whereas coupling with 3-butyn-1-ol provided alcohol 7b (78%). Azides 8a and 8b were then prepared in 65% and 56% yields, respectively, by a two-step procedure of mesylate formation followed by displacement by sodium azide. Finally, the well-established deprotection method35 of saponification with lithium hydroxide, Boc-removal with HCl-dioxane, and pentane precipitation provided UAA 3 (86%) and UAA 4 (87%) in excellent yields.
Scheme 1.
Solvent Dependence of the Azide Asymmetric Stretch of the UAAs
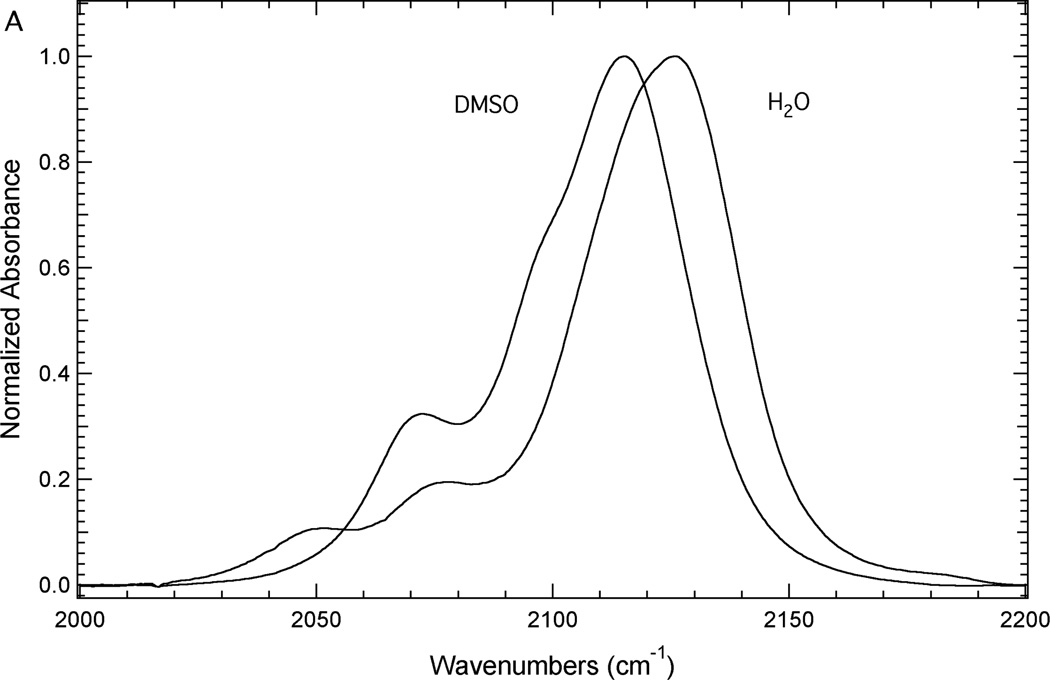
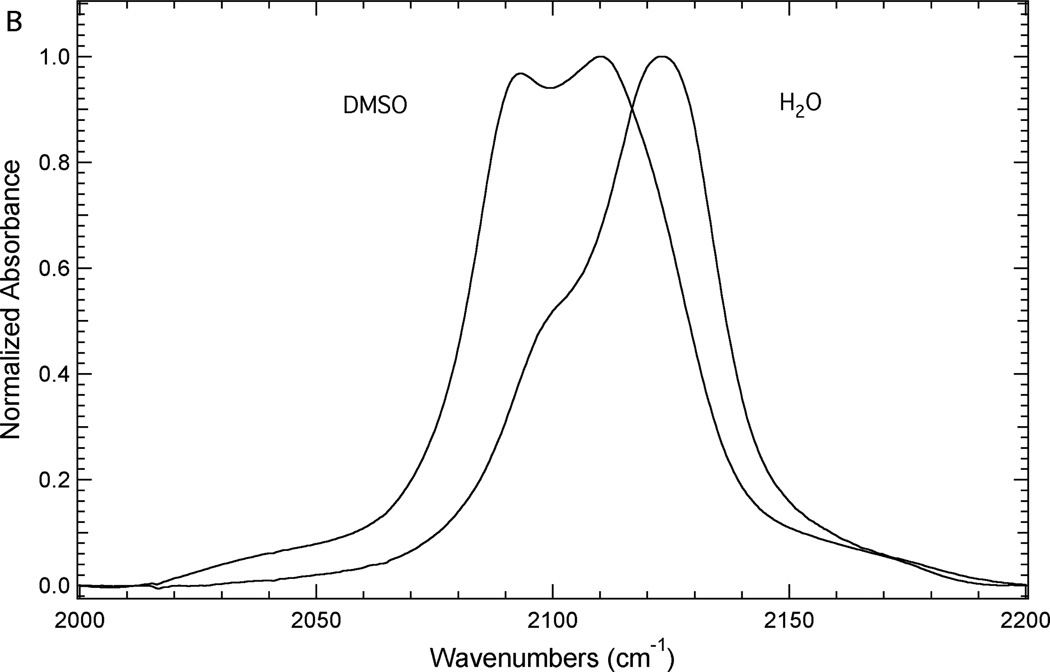
Figure 3 illustrates the sensitivity of the azide asymmetric stretch of 3 and 4 to local environment. Figure 3A shows the FTIR spectra of 3 dissolved in either DMSO or water to mimic a buried or a solvated position in a protein, respectively. Each spectrum contains at least three components. The most intense high frequency component is assigned as resulting from the azide asymmetric stretch vibration based upon literature precedent.14, 16, 18, 21, 38 The other components of this absorption band are likely the result of anharmonic coupling such as accidental Fermi resonance.39, 40 Alternatively, this complexity could be the result of the azide group sampling multiple distinct environments.14 Based upon this assignment, the azide asymmetric stretch vibration of 3 shifts from 2115.1 cm−1 in DMSO to 2126.2 cm−1 in water. This 11.1 cm−1 blue shift is mainly the result of hydrogen bonding interactions between water and the azide group of 3 in the aqueous solution.13, 28, 41
Figure 3.
FTIR absorbance spectra of 3 (Panel A) and 4 (Panel B) dissolved in either DMSO or an aqueous basic solution (200 mM NaOH) at a concentration of ~50 mM.
Similarly the FTIR spectra of 4 in either DMSO or water (Figure 3B) show a complex absorption profile in the region of the azide asymmetric stretch likely due to anharmonic effects.39, 40 The spectrum in DMSO contains at least two components although the two most prominent components have nearly the same intensity in this spectrum. The high frequency component (2110.2 cm−1) is the most intense component and is assigned as arising from the azide asymmetric stretch vibration of 4. The spectrum in water also contains at least two components where the more intense high frequency component (2123.3 cm−1) is assigned as resulting from the azide asymmetric stretch. Thus, this vibration blue shifts 13.1 cm−1 upon moving from DMSO to water. This shift is slightly larger than the solvent-induced shift in this vibration of 3, although the difference could be the result of variations in the strength of the anharmonic-coupling present in these spectra.28, 30, 42–44
The solvent-dependence of the azide asymmetric stretch of 3 and 4 is similar to the solvent dependence of this vibration in 1 and 2. We have previously shown that the azide asymmetric stretch vibration of 1 shifts from 2115.5 cm−1 to 2128.6 cm−1 upon moving from DMSO to water, respectively, resulting a 13.1 cm−1 blue shift.21 The azide asymmetric stretch vibration of 2 was also shown to blue shift upon going from DMSO to water. This vibration shifted from 2097.7 cm−1 to 2111.2 cm−1 upon moving from DMSO to water, respectively, resulting in a 13.5 cm−1 blue shift.21 These results illustrate that the azide groups of 1−4 are effective vibrational reporters of local environment where the azide asymmetric stretch of each of these UAAs has a similar sensitivity to local environment.
Genetic Incorporation of Azido-Modified UAAs into sfGFP
Site 150 in sfGFP was selected since it is located on the edge of the β-barrrel where the side chain is directed away from the protein and into the solvent.34 Thus, the solvation of the protein can be studied at varying distances from the protein surface and the incorporated UAAs should be minimally perturbative to the native protein structure compared with the UAA side chains directed into the interior of the protein.
The azido-modified phenylalanine residues 1, 2, and 4 were genetically incorporated into site 150 of sfGFP in response to an amber codon in an efficient, site-specific manner with high fidelity utilizing an engineered, orthogonal aminoacyl-tRNA synthetase. The incorporation was verified by SDS-PAGE (see Figure 4) and ESI-Q-TOF mass analysis (see Supporting Information). The incorporation of 1, 2, or 4 into site 150 resulted in the production of the protein constructs sfGFP-150-1, sfGFP-150-2 and sfGFP-150-4, respectively. The high fidelity of the UAA incorporation was demonstrated by the lack of protein expression when the UAAs were not included in the expression media (see lane 6 of Figure 4).
Figure 4.
Coomassie blue stained tris-glycine SDS-PAGE illustrating UAA incorporation into sfGFP at site 150 with high fidelity. The protein constructs were expressed from pBad-sfGFP-150TAG and pDule- pN3Phe (lanes 2 – 6) in the presence (lanes 2, 3, 4, and 5) or the absence (lane 6) of 1, 2, 3, or 4, respectively.
Although a band corresponding to a sfGFP construct containing an UAA was observed in the SDS-PAGE (Figure 4, Lane 4) when 3 was present in the expression media, further analysis by ESI-Q-TOF mass spectroscopy and FTIR spectroscopy revealed this sfGFP construct did not contain an azide group. The expected mass was not observed in the ESI-Q-TOF mass spectrum and the FTIR spectrum of this construct did not contain an azide asymmetric stretch vibration. This was surprising because azides in biomolecules are known to be quite stable.45 The loss of the azide group from 3 is likely due to the reactivity propargyl linker as the methylene protons are somewhat acidic and the benzylic position can act as an electrophile in a SN2′ mechanism. These mechanistic pathways are not likely for the other three UAAs, consistent with the loss of the azide group only with 3. It is unclear whether 3 was incorporated into the protein before or after the azide group was lost.
IR Characterization of sfGFP Constructs Containing UAAs
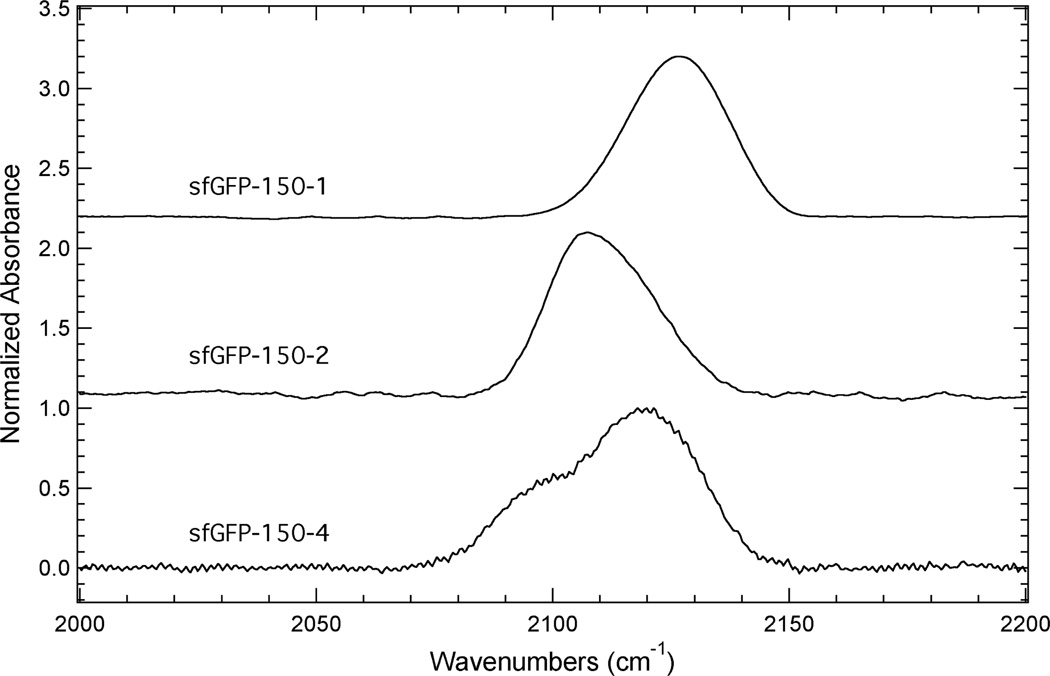
The FTIR spectra, in the region 2000 – 2200 cm−1, of sfGFP-150-1, sfGFP-150-2 and sfGFP-150-4 dissolved in an aqueous buffer are shown in Figure 5. The IR spectrum of sfGFP-150-1 shows a fairly symmetrical absorption band resulting from the azide asymmetric stretch vibration, while the absorption band for sfGFP-150-2 in this region is asymmetric towards higher frequency. The IR spectrum of sfGFP-150-4 contains at least two spectral components in this region with the high frequency component having the largest intensity, which is assigned as resulting from the azide asymmetric stretch vibration. The presence of more than one spectral component in this region of the spectrum is, once again, likely the result of anharmonic effects.39, 40
Figure 5.
FTIR absorbance spectra of sfGFP constructs containing 1, 2, or 4 incorporated at site 150 dissolved in a pH 7.3 aqueous buffer containing 50 mM sodium phosphate and 150 mM sodium chloride at a concentration of ~1 mM.
Based upon these assignments, the azide asymmetric stretch for sfGFP-150-1, sfGFP-150-2 and sfGFP-150-4 was observed at 2126.6 cm−1, 2107.3 cm−1, and 2119.1 cm−1, respectively. These frequencies suggest that the azide group in each of these protein constructs is solvated since these frequencies are similar to the frequencies of the respective free unnatural amino acids dissolved in water (see above). The exact nature of the hydration state is partially complicated due to the anharmonic effects present in these spectra. However, the position and profile of the absorption band of sfGFP-150-2 suggests that the hydration state of the azide group at site 150 is slightly different than the corresponding hydration state of this reporter incorporated at site 134 of sfGFP, which is in a fully solvated site in a loop region of the protein. Specifically, the absorption band resulting primarily from the azide asymmetric stretch vibration for 2 incorporated at site 134 occurs at 2109.8 cm−1 and is fairly symmetrical.21 This 2.5 cm−1 blue shift compared to sfGFP-150-2 and different line shape are indicators that the azide vibrational reporter can probe different hydration states and dynamics, although complementary techniques, such as 2D IR spectroscopy, would be needed to extract out the finer details of this difference.28, 30, 42–44
UAAs 1, 2, and 4 place the azide in different locations relative to the protein backbone in sfGFP. In a more general sense, when coupled with 2D IR spectroscopy, these probes hold great potential to investigate protein hydration dynamics with high spatial and temporal resolution near the protein-water interface.
Conclusion
In this study, we have successfully synthesized two novel azidophenylalanine residues, 3 and 4, through multi-step syntheses. Similar to 4-azido-L-phenylalnine (1) and 4-azidomethyl-L-phenylalanine (2), the azide asymmetric stretch vibration for these new UAAs was found to be sensitive to local environment. The position of this vibration for 3 and 4 blue shifted 11.1 cm−1 and 13.1 cm−1, respectively, upon going from DMSO to water, which is similar to the previously observed shift for 1 and 2 in these solvents. 1, 2, and 4 were genetically incorporated with high efficiency and fidelity into site 150 of sfGFP, which is located on the edge of the β-barrel where the side chain is at the protein-water interface. On the other hand, the incorporation of 3 into sfGFP resulted in a protein construct that did not contain an azide group.
Linear infrared spectroscopy revealed that the azide groups of 1, 2, and 4 were hydrated in the protein at site 150 as illustrated by the frequency of the azide asymmetric stretch vibration. This series of UAAs and the ability to incorporate these probes site-specifically into proteins provides the ability to systematically investigate the protein-water interface with high spatial resolution. In combination with 2D IR spectroscopy, this series has great potential to examine protein hydration dynamics at this interface. This is significant because of the known relationship between the structure and dynamics of waters of hydration and protein structure, dynamics, and function.31, 46–51
These UAAs have the added benefit of potentially serving as conduits for the post-translational incorporation of a variety of other spectroscopic reporters since azides can undergo bioorthogonal click cycloaddition reactions with terminal or strained alkynes.52–56 This series permits the controlled placement of the azide group in the protein thus modulating its solvent accessibility to help promote an efficient cycloaddition reaction.
Supplementary Material
Acknowledgements
We thank Lisa Mertzman for obtaining materials and supplies; Beth Buckwalter for acquiring NMR spectra; and Kenneth Hess for technical assistance with the mass spectral analysis of 3 and 4. This work was supported by F&M Hackman, Leser, and Snavely funds to EMT; NSF (CHE-1053946) to SHB; and NIH (R15GM093330) to SHB/EEF.
Footnotes
Electronic supplementary information (ESI) available: Sequence of wt-sfGFP and ESI-Q-TOF mass analysis of the sfGFP protein constructs.
References
- 1.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 2.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 3.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J. Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 4.Weeks CL, Polishchuk A, Getahun Z, DeGrado WF, Spiro TG. J. Raman Spectrosc. 2008;39:1606–1613. doi: 10.1002/jrs.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxer SG. J. Phys. Chem. B. 2009;113:2972–2983. doi: 10.1021/jp8067393. [DOI] [PubMed] [Google Scholar]
- 6.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 7.Urbanek DC, Vorobyev DY, Serrano AL, Gai F, Hochstrasser RM. J. Phys. Chem. Lett. 2010;1:3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waegele MM, Culik RM, Gai F. J. Phys. Chem. Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung JK, Thielges MC, Fayer MD. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazewicz CG, Lipkin JS, Smith EE, Liskov MT, Brewer SH. J. Phys. Chem. B. 2012;116:10824–10831. doi: 10.1021/jp306886s. [DOI] [PubMed] [Google Scholar]
- 11.Bagchi S, Boxer SG, Fayer MD. J. Phys. Chem. B. 2012;116:4034–4042. doi: 10.1021/jp2122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhikary R, Zimmermann J, Dawson PE, Romesberg FE. Chemphyschem. 2014;15:849–853. doi: 10.1002/cphc.201400017. [DOI] [PubMed] [Google Scholar]
- 13.Oh KI, Lee JH, Joo C, Han H, Cho M. J. Phys. Chem. B. 2008;112:10352–10357. doi: 10.1021/jp801558k. [DOI] [PubMed] [Google Scholar]
- 14.Ye SX, Huber T, Vogel R, Sakmar TP. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taskent-Sezgin H, Chung JA, Banerjee PS, Nagarajan S, Dyer RB, Carrico I, Raleigh DP. Angew Chem Int Edit. 2010;49:7473–7475. doi: 10.1002/anie.201003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye SX, Zaitseva E, Caltabiano G, Schertler GFX, Sakmar TP, Deupi X, Vogel R. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 17.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gai XS, Coutifaris BA, Brewer SH, Fenlon EE. Phys. Chem. Chem. Phys. 2011;13:5926–5930. doi: 10.1039/c0cp02774j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagarajan S, Taskent-Sezgin H, Parul D, Carrico I, Raleigh DP, Dyer RB. J. Am. Chem. Soc. 2011;133:20335–20340. doi: 10.1021/ja2071362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JH, Raleigh D, Cho M. J. Phys. Chem. Lett. 2011;2:2158–2162. doi: 10.1021/jz200980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazewicz CG, Liskov MT, Hines KJ, Brewer SH. J. Phys. Chem. B. 2013;117:8987–8993. doi: 10.1021/jp4052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. J. Am. Chem. Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh KI, Choi JH, Lee JH, Han JB, Lee H, Cho M. J. Chem. Phys. 2008;128:154504. doi: 10.1063/1.2904558. [DOI] [PubMed] [Google Scholar]
- 24.McMahon HA, Alfieri KN, Clark CAA, Londergan CH. J. Phys. Chem. Lett. 2010;1:850–855. doi: 10.1021/jz1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo H, Culik RM, Korendovych IV, DeGrado WF, Gai F. Biochemistry. 2010;49:10354–10356. doi: 10.1021/bi101711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker DM, Hayes EC, Webb LJ. Phys. Chem. Chem. Phys. 2013;15:12241–12252. doi: 10.1039/c3cp51284c. [DOI] [PubMed] [Google Scholar]
- 27.Park KH, Jeon J, Park Y, Lee S, Kwon HJ, Joo C, Park S, Han H, Cho M. J. Phys. Chem. Lett. 2013;4:2105–2110. [Google Scholar]
- 28.Kim H, Cho M. Chem Rev. 2013;113:5817–5847. doi: 10.1021/cr3005185. [DOI] [PubMed] [Google Scholar]
- 29.Maj M, Oh Y, Park K, Lee J, Kwak KW, Cho M. J. Chem. Phys. 2014;140:235104. doi: 10.1063/1.4883505. [DOI] [PubMed] [Google Scholar]
- 30.Baiz CR, Mcrobbie PL, Anna JM, Geva E, Kubarych KJ. Acc. Chem. Res. 2009;42:1395–1404. doi: 10.1021/ar9000263. [DOI] [PubMed] [Google Scholar]
- 31.King JT, Kubarych KJ. J. Am. Chem. Soc. 2012;134:18705–18712. doi: 10.1021/ja307401r. [DOI] [PubMed] [Google Scholar]
- 32.Woys AM, Mukherjee SS, Skoff DR, Moran SD, Zanni MT. J. Phys. Chem. B. 2013;117:5009–5018. doi: 10.1021/jp402946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, Schultz PG. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 34.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 35.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 36.Miyake-Stoner SJ, Refakis CA, Hammill JT, Lusic H, Hazen JL, Deiters A, Mehl RA. Biochemistry. 2010;49:1667–1677. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 37.Smith EE, Linderman BY, Luskin AC, Brewer SH. J. Phys. Chem. B. 2011;115:2380–2385. doi: 10.1021/jp109288j. [DOI] [PubMed] [Google Scholar]
- 38.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, Schultz PG, Fayer MD. J. Phys. Chem. B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nydegger MW, Dutta S, Cheatum CM. J. Chem. Phys. 2010;133:134506. doi: 10.1063/1.3483688. [DOI] [PubMed] [Google Scholar]
- 40.Lipkin JS, Song R, Fenlon EE, Brewer SH. J. Phys. Chem. Lett. 2011;2:1672–1676. doi: 10.1021/jz2006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi JH, Oh KI, Cho MH. J. Chem. Phys. 2008;129:174512. doi: 10.1063/1.3001915. [DOI] [PubMed] [Google Scholar]
- 42.Zheng J, Kwak K, Fayer MD. Acc. Chem. Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- 43.Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A. Acc. Chem. Res. 2008;41:432–441. doi: 10.1021/ar700188n. [DOI] [PubMed] [Google Scholar]
- 44.Kim YS, Hochstrasser RM. J. Phys. Chem. B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 46.Otting G, Liepinsh E, Wuthrich K. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 47.Nandi N, Bhattacharyya K, Bagchi B. Chem. Rev. 2000;100:2013–2045. doi: 10.1021/cr980127v. [DOI] [PubMed] [Google Scholar]
- 48.Raschke TM. Curr. Opin. Struc. Biol. 2006;16:152–159. doi: 10.1016/j.sbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Moilanen DE, Levinger NE, Spry DB, Fayer MD. J. Am. Chem. Soc. 2007;129:14311–14318. doi: 10.1021/ja073977d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindquist BA, Furse KE, Corcelli SA. Phys. Chem. Chem. Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 51.Sharp KA, Vanderkooi JM. Acc. Chem. Res. 2010;43:231–239. doi: 10.1021/ar900154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brase S, Gil C, Knepper K, Zimmermann V. Angew Chem Int Edit. 2005;44:5188–5240. doi: 10.1002/anie.200400657. [DOI] [PubMed] [Google Scholar]
- 53.van Dongen SFM, Teeuwen RLM, Nallani M, van Berkel SS, Cornelissen JJLM, Nolte RJM, van Hest JCM. Bioconjugate Chem. 2009;20:20–23. doi: 10.1021/bc8004304. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, Chin JW. J. Am. Chem. Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 55.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson H, Pedersen DS. Eur. J. Org. Chem. 2012:4267–4281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.