In humans, homologues of Chlamydomonas ciliary docking complex subunit DC2 and assembly factor ODA10 are interacting axonemal proteins needed for outer dynein arm assembly. Surprisingly, Chlamydomonas ODA10 does not interact with DC2, localizes to a proximal axonemal domain, and is needed for cytoplasmic steps in dynein assembly, not for docking.
Abstract
Axonemal dyneins are multisubunit enzymes that must be preassembled in the cytoplasm, transported into cilia by intraflagellar transport, and bound to specific sites on doublet microtubules, where their activity facilitates microtubule sliding-based motility. Outer dynein arms (ODAs) require assembly factors to assist their preassembly, transport, and attachment to cargo (specific doublet A-tubule sites). In Chlamydomonas, three assembly factors—ODA5, ODA8, and ODA10—show genetic interactions and have been proposed to interact in a complex, but we recently showed that flagellar ODA8 does not copurify with ODA5 or ODA10. Here we show that ODA5 and ODA10 depend on each other for stability and coexist in a complex in both cytoplasmic and flagellar extracts. Immunofluorescence and immuno–electron microscopy reveal that ODA10 in flagella localizes strictly to a proximal region of doublet number 1, which completely lacks ODAs in Chlamydomonas. Studies of the in vitro binding of ODAs to axonemal doublets reveal a role for the ODA5/ODA10 assembly complex in cytoplasmic maturation of ODAs into a form that can bind to doublet microtubules.
INTRODUCTION
Dyneins are microtubule-associated, minus end–directed ATPase motor complexes. Extended amino-terminal tail domains of dyneins bind to cargoes, whereas globular motor heads built from AAA domains bind to and translocate along microtubules (Roberts et al., 2013). Cytoplasmic and related intraflagellar transport (IFT) dyneins transport cargoes such as proteins, RNAs, and vesicles in cells for fast and efficient transport when diffusion is too slow for the purpose. They appear to assemble spontaneously, and their functions may be regulated in part by their interactions with specific cargoes, to coordinate activation of their motor activity. Motile cilia have specialized axonemal dyneins, which bind to microtubules by both ends, by their tails to doublet A-tubule cargo attachment (docking) sites, and by their heads to doublet B-tubules. Evidence suggests that functional assembly of axonemal dyneins is a multistep process requiring not only the dynein subunits, but also cochaperones and dedicated transport factors (Kobayashi and Takeda, 2012). Axonemal dyneins have further diversified into inner dynein arms (IDAs), which have multiple subtypes and are largely responsible for ciliary waveform regulation, and outer dynein arms (ODAs), which provide power and beat frequency regulation (Kamiya and Yagi, 2014). Most inner dynein arm (IDA) isoforms are found once every 96 nm along all nine doublet microtubules, whereas the more abundant ODAs are usually found every 24 nm along eight of the nine doublets.
Most of the work to elucidate axonemal dynein assembly mechanisms has been done in the single-celled biflagellate alga Chlamydomonas reinhardtii using oda and ida mutants, most of which identify loci encoding conserved proteins that play similar roles in vertebrate axonemal dynein function. Here we focus on the role of ODA10 in the assembly of axonemal outer dynein arms in Chlamydomonas to shed light on the apparently complex assembly pathway required for axonemal dyneins. Chlamydomonas oda mutants all have a slow-swimming phenotype with reduced beat frequency, but they can be differentiated into functional assembly groups using cytoplasmic complementation after gamete fusion, termed dikaryon rescue (Kamiya, 1988). Based on this approach, available oda mutants were categorized into four genetic groups, each of which corresponds to proteins that physically interact in a complex or participate in a common mechanistic step in the dynein assembly process. One group consists of mutations in motor subunits, together with the cytoplasmic chaperones that are involved in forming an ODA complex in the cytoplasm (Freshour et al., 2007; Omran et al., 2008; Duquesnoy et al., 2009; Horani et al., 2012; Kott et al., 2012; Mitchison et al., 2012). These preassembled complexes are then proposed to interact with adaptor proteins that facilitate recognition of preassembled axonemal dyneins as cargoes for the IFT system. One putative adaptor protein, ODA16 (WDR69 in vertebrates), interacts with IFT46, and the oda16 mutation alone forms the second genetically defined group (Ahmed and Mitchell, 2005; Ahmed et al., 2008). Once transported into the ciliary compartment, ODAs dock onto axonemes through interaction with an independently assembled axonemal docking complex (ODA DC), and the two core ODA DC subunits form the third genetic group (Takada and Kamiya, 1994; Takada et al., 2002; Koutoulis et al., 1997; Wakabayashi et al., 2001; Casey et al., 2003).
A fourth group of assembly factors is defined by mutations at the ODA5, ODA8, and ODA10 loci, believed to encode subunits of a hypothesized accessory complex involved in docking of ODAs to doublet microtubules (Kamiya, 1988; Fowkes and Mitchell, 1998; Wirschell et at., 2004; Dean and Mitchell, 2013). Although initial characterizations of ODA5 (Wirschell et al., 2004), ODA8 (Desai et al., 2015), and ODA10 (Dean and Mitchell, 2013) show that all three gene products are present in flagella, support for their interaction in a complex is lacking, and their roles in ODA assembly are unclear. Immunoprecipitation of a dynein heavy chain showed that preassembly of ODA complexes in the cytoplasm of all three mutants was comparable to that of wild type, which suggests that these genes function after the initial cytoplasmic preassembly of ODAs (Fowkes and Mitchell, 1998). However, more recently, we showed that ODAs in oda8 cytoplasm dissociate readily and have greatly reduced affinity for axonemal docking sites (Desai et al., 2015), suggesting that these assembly factors may play a role in the cytoplasm rather than, or as well as, in flagella. ODA5 and ODA10 are coiled coil proteins that have predicted structural similarities and weak sequence homology with Chlamydomonas docking complex proteins ODA1 (ODA DC2) and ODA3 (ODA DC1), respectively (Dean and Mitchell, 2013). However, unlike DC1 and DC2, ODA10 and ODA5 are unnecessary for functional reconstitution of purified flagellar ODA complexes onto oda axonemes in vitro (Dean and Mitchell, 2013). Of interest, the vertebrate homologue of ODA10, CCDC151, is not expressed during Drosophila spermatogenesis, an IFT-independent process, whereas it is required for dynein assembly in other motile ciliated tissues in the same organism, all of which rely on IFT-based ciliary assembly (Jerber et al., 2014). Thus ODA10 may be involved in an IFT-dependent step of ODA assembly. To determine specific functions of these proteins in dynein assembly, we further characterized the ODA10 and ODA5 proteins and examined their interaction in both cytoplasmic and flagellar compartments. Our results reveal an unexpected distribution of ODA10 and ODA5 in flagella and a defect in ODA complexes in the cytoplasm of oda10 mutants. We hypothesize that these proteins form an assembly complex that acts after preassembly of dynein subunits in the cytoplasm but before transport and docking of dynein onto axonemes in flagella and supports a maturation step in the cytoplasm necessary to prepare ODAs as substrates for IFT-dependent transport and docking.
RESULTS
ODA5 and ODA10 interact in a complex
Because ODA5 and ODA10 both encode coiled-coil proteins with weak homology to Chlamydomonas docking complex subunits, which interact in an obligate heterodimer, we predicted that the ODA5 and ODA10 proteins would also form a heterodimer and might be dependent on each other for stability. We previously showed that the amount of ODA10 is significantly reduced in oda5 mutant cytoplasm. Based on the analysis of Wirschell et al. (2004), ODA5 does not assemble in oda10 flagella, but cytoplasmic abundance of ODA5 was not analyzed. To follow abundance, we used hemagglutinin (HA)- and FLAG epitope–tagged versions of ODA5 and ODA10 (Supplemental Figure S1). When we tested the abundance of ODA5HA in two independent meiotic products generated by crossing oda10 into the ODA5HA strain, we found that both strains almost completely lack ODA5HA in their flagella (Figure 1A) and show reductions in ODA5HA in deflagellated cell body samples (Figure 1B). To confirm that the reduction in ODA5HA level was due to the absence of ODA10, we introduced a plasmid encoding ODA10FL (ODA10 C-terminally tagged with 3xFLAG and hexahistidine [6xHIS]) into the meiotic product with the lowest level of ODA5HA expression, ODA5HA,oda10-1C. Cell body and flagellar samples of three independent transformants that showed varying levels of motility rescue were tested for the level of ODA5HA by Western blot. ODA5HA was restored upon expression of ODA10FL (Figure 1, A and B), and the levels of both ODA5HA and of ODA IC subunit IC2 that had assembled into flagella correlated with the flagellar abundance of ODA10FL. Only the highest ODA10FL expression levels observed supported assembly of dynein to wild-type levels.
FIGURE 1:
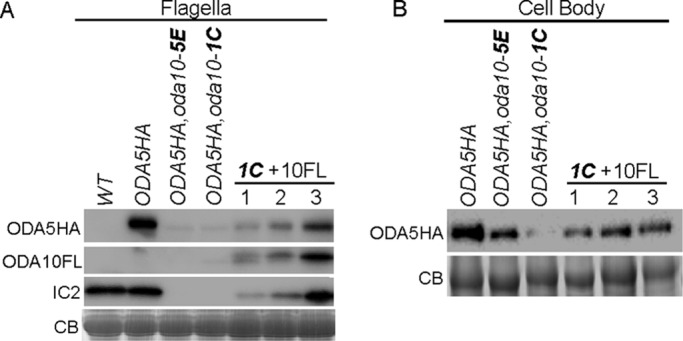
ODA5 stability is dependent on ODA10. (A) Blots of flagella from untagged wild -type cells (WT), pseudo-wild-type oda5,ODA5HA cells (ODA5HA), two meiotic products (5E and 1C) that introduce the oda10 mutation into the ODA5HA-tagged strain, and three transformants of meiotic product 1C with a construct that expresses FLAG-tagged ODA10 (1C+10FL), probed with anti-HA, anti-FLAG, and anti-IC2. (B) Blots of deflagellated cell body samples from the same strains, probed with anti-HA, show reduction of ODA5HA in the oda10 background and restoration from transgene expression of ODA10FL. CB, portions of Coomassie blue–stained gels showing equal protein loads.
Reduced abundance of ODA10 and ODA5 in cytoplasm could result from either reduced rates of protein synthesis (due to altered rates of transcription or translation) or increased rates of degradation. First, we tested whether protein synthesis was affected in mutant cells, using a simple measure of the time needed to reach a new steady-state concentration when turnover was inhibited. Levels of ODA10HA and ODA5HA were detected by Western blot in samples of whole cells treated with proteasomal protein degradation inhibiter MG132 for various times. In the wild-type background (Figure 2A, left), the abundance of ODA5HA increases rapidly after MG132 addition and reaches a new steady-state level within 1 h. The level of ODA5HA in the oda10 background, although much lower to begin with, also reaches a new steady state within 1 h, suggesting little or no defects in protein synthesis. A similar pattern of accumulation was seen for ODA10HA in wild-type and oda5 mutant backgrounds (Figure 2B), even though the ODA5HA protein is expressed from a tagged version of the endogenous gene, whereas the ODA10HA construct was introduced in a cassette that uses the promoter, 5′ untranslated region (UTR), and 3′ UTR from an unrelated, nonflagellar gene (Dean and Mitchell, 2013).
FIGURE 2:
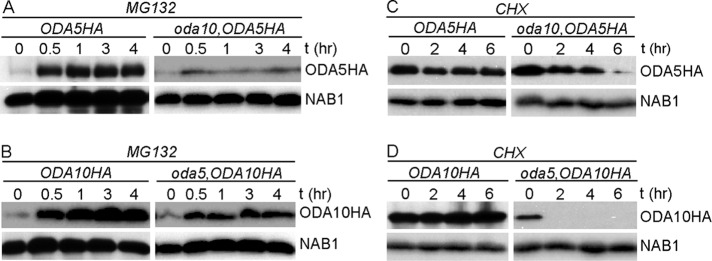
Half-lives of ODA5 and ODA10 are reduced by the oda10 and oda5 mutations. Blots of whole-cell samples probed with (top) anti-HA and (bottom) anti-NAB1 (loading control) isolated before and at indicated times after treatment with 10 μM MG132 (A, B) or 20 μg/ml cycloheximide (C, D). All strains expressing epitope-tagged transgenes also carry the mutant allele at the corresponding endogenous locus.
To test whether ODA5 and ODA10 are degraded faster than normal in mutant cytoplasm, we estimated half-lives of ODA5 and ODA10 by detecting the abundance of ODA10HA and ODA5HA in the cell body fraction of cells treated with cycloheximide for various times. Levels of ODA5HA (Figure 2C) and ODA10HA (Figure 2D) were unchanged in wild-type cells even after 6 h, whereas they were significantly reduced in mutant backgrounds by 2 h, supporting the hypothesis that mutual reduction in the amount of ODA5 and ODA10 is due to reduced half-lives of the proteins, not to changes in rates of protein synthesis. Apparent differences in the stability of each protein when expressed alone could result from real intrinsic differences in degradation rates or differential effects of the HA tag sequences introduced into each construct. These results show that ODA5 and ODA10 require each other for stability, as might be expected if they interact directly with each other in a complex.
To see whether ODA5 and ODA10 physically interact in a complex, we immunoprecipitated ODA10FL from a dialyzed axonemal salt extract of double-tagged oda5,oda10,ODA5HA,ODA10FL cells and probed the immunoprecipitate for the presence of ODA5HA. The result shows that ODA5HA coimmunoprecipitated with ODA10FL from flagellar extracts, supporting their interaction in a complex, whereas no ODA5HA appeared in control FLAG IPs from extracts of oda5,ODA5HA flagella (Figure 3). We were unable to perform similar single-step anti-FLAG immunoprecipitations (IPs) from cytoplasmic extracts because endogenous proteins similar in size to ODA10FL reacted with the anti-FLAG M2 antibody (Supplemental Figure S2). However, IP with anti-HA from cytoplasmic extracts of double-tagged cells shows that ODA10FL coimmunoprecipitates with ODA5HA (Figure 3A), which confirms that these proteins coexist in a complex in the cytoplasm. Based on these results, the low-abundance protein pool of ODA10 that remains in the cytoplasm of oda5 (and of ODA5 in the cytoplasm of oda10) represents protein that is both unstable and unable to support dynein assembly. Phenotypically, oda5 and oda10 mutants can be taken as essentially identical, and the functional depletion of both proteins in the cytoplasm of either mutant explains the lack of dynein assembly in temporary dikaryons between oda5 and oda10 mutant strains.
FIGURE 3:
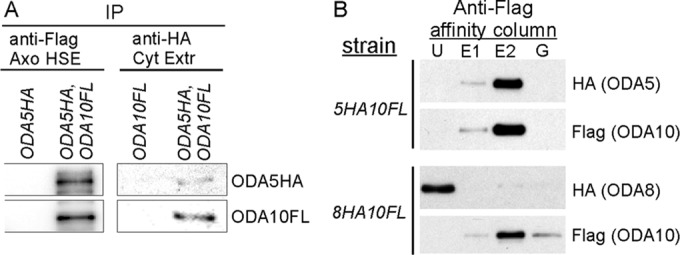
ODA5 and ODA10 interact with each other but not with ODA8. (A) Blots of immunoprecipitated proteins from a dialyzed axonemal salt extract (Axo HSE) of oda10,oda5,ODA10FL,ODA5HA strain flagella (left) probed with anti-HA and anti-FLAG show that ODA5HA coimmunoprecipitates with ODA10FL; blots of the reciprocal immunoprecipitate from a cytoplasmic extract of the same strain (right) shows that ODA10FL coimmunoprecipitates with ODA5HA. (B) Anti-FLAG affinity column purification of ODA10FL from cytoplasmic extracts of the indicated strains shows copurification of ODA5HA, but not ODA8HA, with ODA10FL. E1 and E2, eluted with 3xFLAG peptide; G, eluted with 0.1 M glycine, pH 3.5; U, unbound.
Although we previously showed that flagellar ODA8 does not copurify with flagellar ODA10 (Desai et al., 2015), the failure of dikaryon rescue between oda8 and oda5, and between oda8 and oda10, suggests that ODA8 might interact with ODA5 and ODA10 in the cytoplasm. To test this, we initially purified ODA10FL from cytoplasmic extracts of ODA5HA,ODA10FL and ODA8HA,ODA10FL strains based on the 6xHIS tag on ODA10FL, using a Ni-nitriloacetic acid (NTA) column. Peak fractions containing ODA10FL were subsequently purified on a FLAG M2 affinity column. As summarized in Figure 3B and Supplemental Figure S2, ODA5HA and ODA10FL quantitatively copurified from cytoplasmic extracts, but ODA8HA, although it coincidentally eluted from the Ni-NTA column in the same fractions as ODA10FL, failed to copurify with ODA10FL on the FLAG M2 column. Based on these results, ODA5 and ODA10 are obligate subunits of a complex, hereafter designated as the ODA assembly complex (ODA AC), whereas ODA8 does not appear to be associated with this complex under the conditions tested.
The ODA AC has a restricted proximal flagellar localization
Localization patterns of the ODA AC could constrain its possible functions in ODA assembly. By immunoblot, the ODA AC is present primarily in axonemal fractions of flagella, with only trace amounts seen in detergent-soluble membrane/matrix fractions, and therefore it interacts in some way with axonemal microtubules (Wirschell et al., 2004; Dean and Mitchell, 2013). If this interaction is involved directly in promoting the attachment of ODAs to docking sites, then the ODA AC should be doublet associated and evenly distributed along the entire length of the axoneme. To determine its localization pattern, we used confocal immunofluorescence (IF) microscopy with an anti-HA antibody. Compared with the nonspecific background seen in untagged wild-type cells, ODA10HA appears only in a restricted region on the flagella of oda10,ODA10HA cells, limited to ∼2–3 μm near the base (Figure 4A); note that faint fluorescence seen along the length of these flagella is equivalent to the signal often seen on negative controls and is therefore considered nonspecific. This proximal localization of ODA10HA was not altered by the absence of the ODA complex itself, as seen in oda9,oda10,ODA10HA cells (oda9 is a mutation in the outer dynein IC1 subunit and is essential for ODA assembly) or by the absence of ODA8, as seen in oda8,oda10,ODA10HA cells.
FIGURE 4:
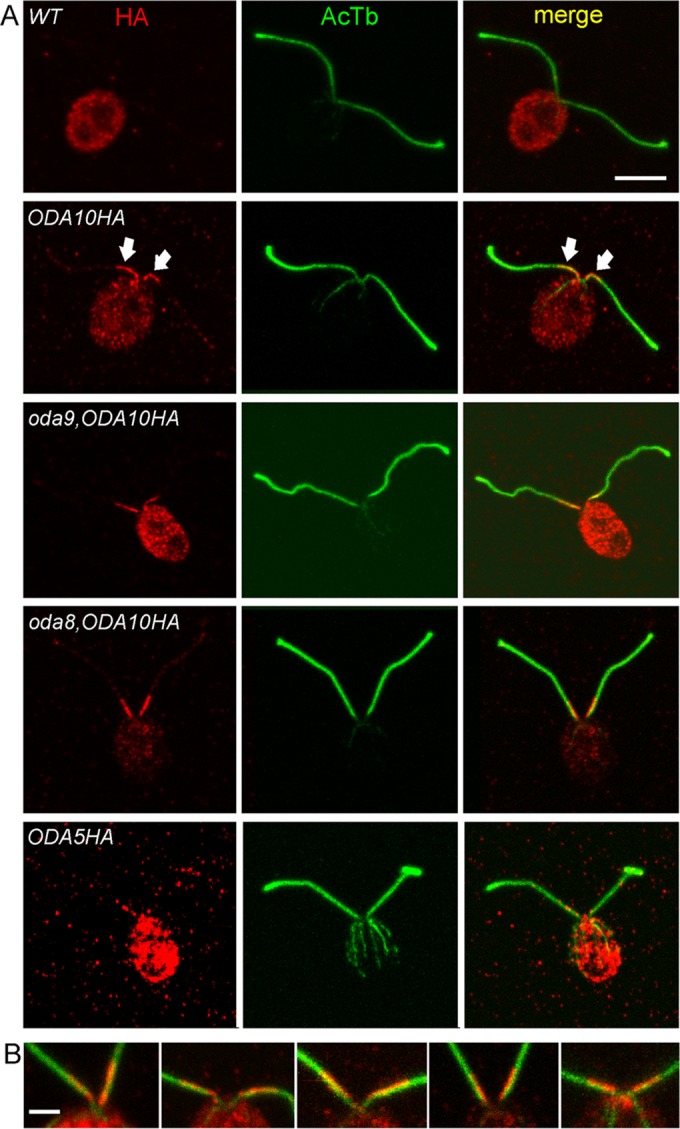
ODA10 localizes to a unique proximal flagellar domain. (A) IF localization of the HA epitope (red) and acetylated tubulin (green) reveals ODA10HA only in the first 2–3 μm at the proximal end of each flagellum. This localization is unaffected by the absence of ODAs in oda9,oda10,ODA10HA cells or the combined absence of ODAs and ODA8 in oda8,oda10,ODA10HA cells. A similar distribution of signal is seen in oda5,ODA5HA cells, although the signal is weaker due to the lower expression level of ODA5HA. (B) Enlarged views of the anterior end of cells and proximal portion of flagella of ODA10HA-expressing strains stained with anti-HA (red) and anti-acetylated tubulin (green). The HA signal does not overlap symmetrically with the tubulin signal but consistently labels a region shifted toward the midline of the cell. All images are maximum intensity projections of z-stacks from confocal analysis. Bar, 5 μm (A), 2 μm (B).
Further IF localization with unfixed, isolated oda10,ODA10HA axonemes confirmed the proximal localization seen in fixed whole cells; however, no signal was visible on ∼20% of axonemes, and, when present, it did not always extend to the extreme proximal end of the severed axoneme (Supplemental Figure S3). Lack of signal on apparently full-length axonemes likely results from problems of antibody access to axonemes that have already adhered in random orientations to the poly-l-lysine–coated coverslip before antibody incubation, blocking the limited region of ODA10 distribution. Absence of a signal at the extreme proximal end suggests that the complex preferentially dissociates from this region during axoneme purification or that the domain specifying an attachment site for this complex does not extend to the transition zone where flagellar severing occurs. A similar pattern was observed for ODA5HA, although the lower expression level of this construct resulted in a weaker signal than that typically seen for ODA10HA (Figure 4A and Supplemental Figure S3).
Enlarged images of ODA10 localization consistently displayed a biased distribution toward the side of each flagellum that faced the midline between the two flagella on each cell (Figure 4B). This distribution could not be due to misalignment of signals from different confocal channels, as it was apparent on multiple cells within a field of view, regardless of their orientation. Given that Chlamydomonas flagella have a fixed orientation with doublet microtubule 1 (DM1) facing the midline (Hoops and Witman, 1983), this fluorescence signal distribution suggests that the ODA AC is concentrated on the DM8-9-1-2-3 side of the axoneme.
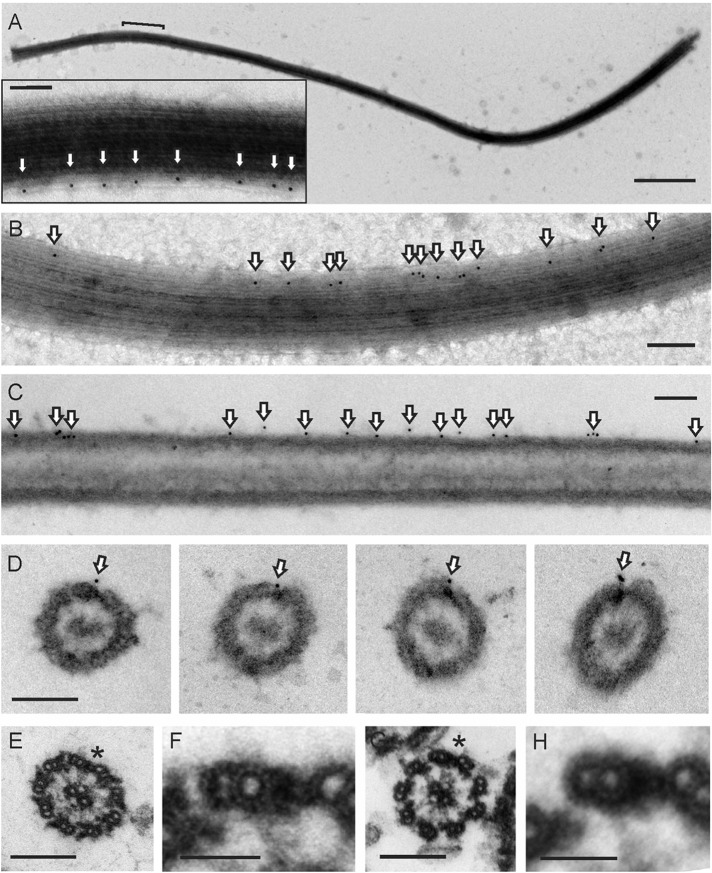
To see whether the proximal flagellar localization of ODA10 corresponded to a specific ultrastructural feature, we performed whole-mount negative-stain immunogold electron microscopy using secondary 10-nm gold-labeled antibodies to detect anti-HA on unfixed oda10,ODA10HA axonemes. Gold particles appeared on only a single microtubule doublet toward one end of any given axoneme (Figure 5, A and B). This end was identified as the base by the orientation of ODAs, which tilt toward the base, and by the appearance of the doublets, which all end together at the severed base but appear uneven in length and are reduced to A-tubules alone at the distal tip. The overall distribution of gold particles (Figure 6A) was similar to that seen by immunofluorescence in isolated axonemes and intact cells. Longitudinal thin sections of embedded axonemes labeled with 10-nm gold displayed an identical gold particle distribution, with single rows of gold particles visible along the margin of axonemes that were in a favorable orientation (Figure 5C). The spacing between gold particles in both negatively stained and longitudinal thin-section images peaks around 24 nm (Figure 6B).
FIGURE 5:
ODA10 is restricted to DM1. (A, B) Negative-stain electron micrograph of ODA10HA localized with 10-nm gold particles (arrows) on unfixed axonemes. Bracket in A indicates the region enlarged in the inset. (C, D) Pre-embedding, labeled oda10,ODA10HA axonemes, thin section. (E) Wild-type axoneme (proximal). (F) Enlarged averaged view of wild- type DM1–DM2 bridge. (G) oda10 axoneme (proximal). (H) Enlarged averaged view of oda10 DM1–DM2 bridge. The DM1–DM2 bridge is marked by an asterisk in E and G. Scale bar, 1 μm (A; inset, 100 nm); 200 nm (B–E, G), 50 nm (F, H).
FIGURE 6:
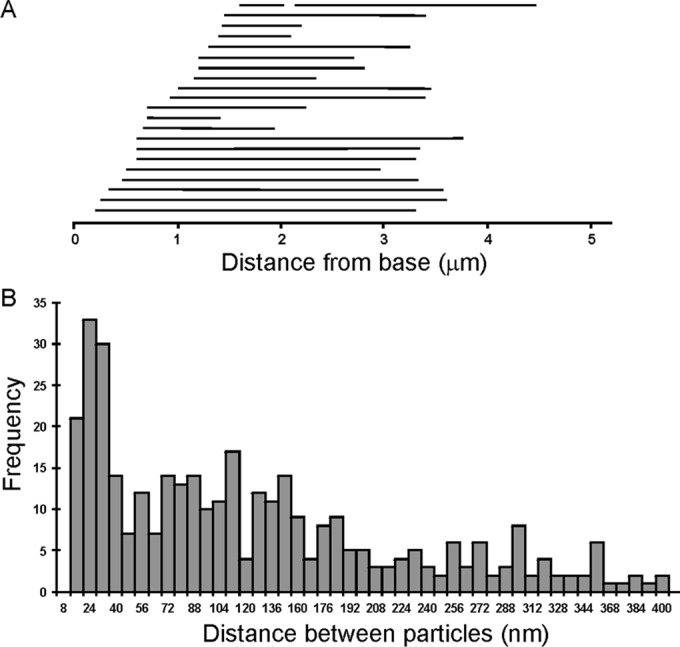
Gold particle distribution on oda10,ODA10HA axonemes labeled with anti-HA and 10-nm gold secondary antibodies. (A) Extent of labeled region on each of 22 negatively stained axonemes arranged in order of the distance from the base to the first gold particle. (B) Distance between adjacent gold particles binned in 8-nm increments (357 particles, from images of 47 axonemes).
Cross sections of pre-embedding, gold antibody–labeled axonemes show that ODA10HA is located exclusively on DM1 (Figure 5D), which was identified by the absence of electron density that corresponds to ODAs and by the presence of the DM1–DM2 bridge, an electron-dense structure that connects DM1 to DM2 in the proximal region of Chlamydomonas axonemes (Hoops and Witman, 1983; Bui et al., 2009; Lin et al., 2012). To control for the possibility that gold labeling of ODA10HA was only observable on DM1 because ODA complexes blocked antibody access on DM2-9, we also examined gold labeling on axonemes from an oda strain (oda9,oda10,ODA10HA; Supplemental Figure S4), as well as on wild-type (oda10,ODA10HA) axonemes that were fixed with formaldehyde (similar to those used for immunofluorescence analysis in Figure 5A). Both of these controls generated identical labeling limited to a single microtubule doublet, supporting the conclusion that ODA10HA is restricted to the proximal region of DM1. Previous ultrastructural analyses of oda5 and oda10 noted that axonemes of both mutants lacked ODAs but retained the ODA-DC (Kamiya, 1988; Takada and Kamiya, 1994); however, the appearance of the DM1–DM2 bridge was not analyzed. We compared bridge structures using thin sections of traditionally fixed, unlabeled wild-type and oda10 axonemes. No difference between wild-type (Figure 5, E and F) and oda10 (Figure 5, G and H) bridge structures was detected using this method.
To gain further insight into the relationship between a proximally localized ODA AC and the overall assembly of ODAs, which occurs primarily at the flagellar tip during all phases of flagellar elongation, we observed ODA10HA in vegetative cells that were regenerating their flagella after a pH shock–induced deflagellation. By IF microscopy, ODA10HA was seen at the proximal end of the growing flagella as soon as they were observable, but this ODA10HA signal never extended beyond the proximal 2–3 μm over the 90 min required for flagella to reach 10 μm in length (Figure 7).
FIGURE 7:
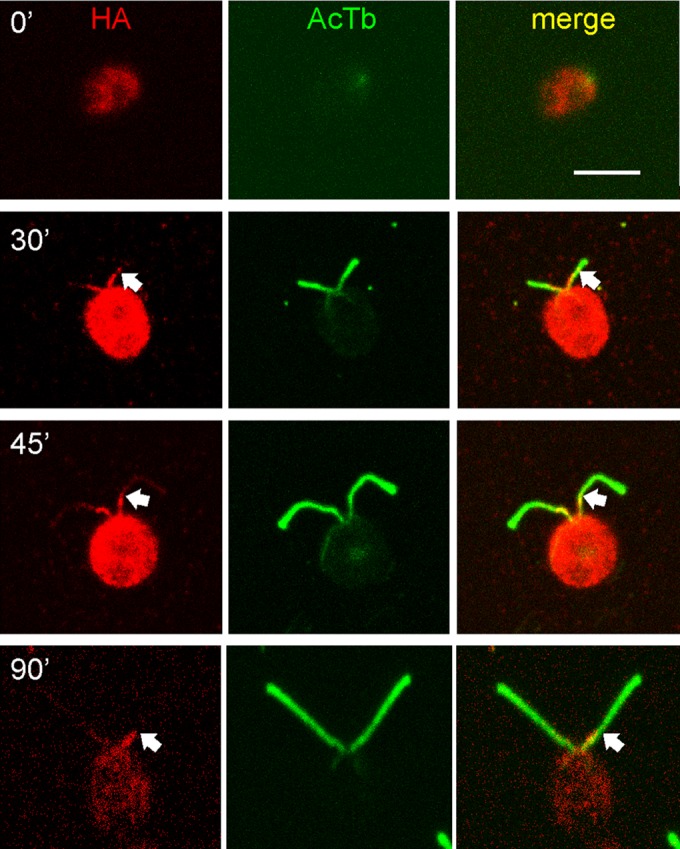
Localization of the ODA AC during flagellar growth. oda10,ODA10HA cells deflagellated by pH shock and fixed at the indicated times during flagellar assembly were stained for HA (red) and acetylated tubulin (green). HA signal appears at the earliest stages of assembly but does not extend beyond the proximal ∼3 μm (arrows). Bar, 5 μm.
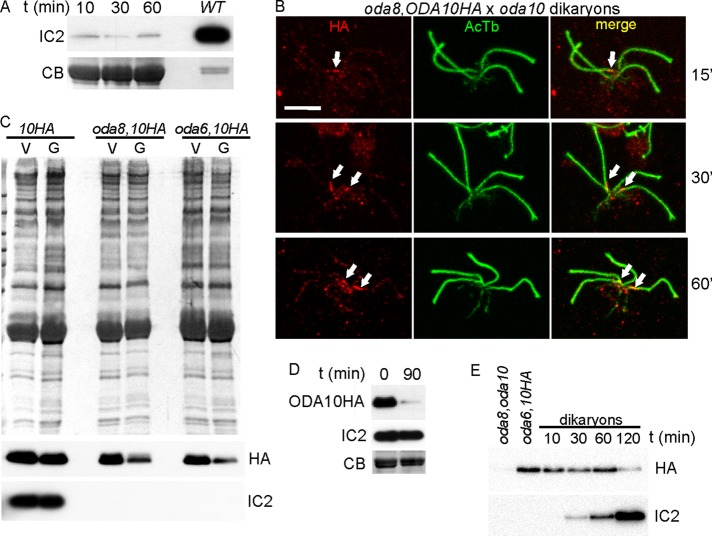
The genetic interaction between oda8 and oda10 was originally identified through lack of cytoplasmic complementation in temporary dikaryons formed by fusion of oda8 and oda10 gametes (Kamiya, 1988), where complementation was scored as rescue of beat frequency from the oda level of ∼24 Hz to the wild-type dikaryon level of ∼45 Hz. To understand this phenomenon, we first tested whether lack of beat frequency restoration was due to lack of dynein assembly or to assembly of a defective dynein that could not contribute to motility. Blot analysis of flagella isolated at various times after gamete mixing (Figure 8A) confirms that there is no increase in dynein abundance above the level contributed by the oda10 gametes (∼1% of the wild-type level). Dikaryons from a cross in which ODA10HA was expressed in the oda8 strain (oda10 × oda8,oda10,ODA10HA) were then analyzed by IF to determine whether the ODA10 protein present in oda8 mutant cytoplasm could assemble into the oda10 mutant flagella. Surprisingly, ODA10HA only appeared in one pair of flagella in dikaryons fixed at 15, 30, or 60 min after mixing (Figure 8B), and some dikaryons completely lacked an HA signal in all four flagella. In addition, the HA signal consistently appeared attenuated in gametes compared with vegetative cells and in dikaryons compared with unmated gametes in the same samples. To provide a more quantitative analysis, we isolated flagella from vegetative and gametic cells and blotted them to determine the relative levels of ODA10HA. ODA10 is maintained at the same levels in vegetative and gametic flagella of wild-type cells but is reduced by about half in gametes of ODA assembly mutants regardless of whether dynein assembly was blocked by a mutation in the interacting oda8 locus or in an ODA subunit locus, oda6 (Figure 8C). Next we compared ODA10 levels during an oda10 × ODA10HA cross, in which one pair of flagella is already fully populated with ODA complexes and with ODA10HA, before and 90 min after mixing (Figure 8D) and observed a nearly complete loss of ODA10HA from the dikaryon flagella. To see whether this loss also occurred in dikaryons during a dikaryon rescue in which one set of gamete flagella initially has ODA10 assembled but all of the gamete flagella lack dynein, we isolated flagella from an oda8,oda10 × oda6,oda10,ODA10HA cross. As seen in Figure 8E, the levels of ODA10HA decreased, whereas the levels of IC2 increased, over a 2-h period. We conclude that the ODA AC is mobilized in gametes and during a dikaryon rescue and that steady-state concentrations of this complex in flagella are not required for ODA assembly. Thus, although ODA10 fails to assemble into oda10 mutant flagella during a cross between oda8 and oda10 mutant cells, this failure may not be related to the failure of ODA complexes to assemble into the dikaryon flagella. We therefore turned to an analysis of the role of the ODA AC in formation of assembly-competent ODA complexes in the cytoplasm.
FIGURE 8:
Flagellar ODA10 is dynamic in dikaryons. (A) Coomassie-stained gel (CB, bottom) and Western blot (IC2, top) of flagella samples isolated from a mating mixture of oda8 and oda10 gametes at various times. WT lane, flagellar sample of vegetative cells, loaded five times less than the gamete flagella. Dynein fails to rescue in these oda8 × oda10 dikaryons. (B) IF analysis of dikaryons between oda8,oda10,ODA10HA and oda10 gametes fixed at the indicated times, showing ODA10HA in only one pair of flagella (arrows). Scale bar, 5 μm. (C) Amido black–stained blot (top) and immunoblots (bottom) of flagellar samples from vegetative (V) and gametic (G) cells demonstrates reduction of flagellar ODA10HA in oda-strain gametes. (D) Flagella isolated from a mating mixture of oda10 and oda10,ODA10HA gametes immediately after and 90 min after mixing of gametes. Blots show loss of ODA10HA but no loss of IC2. (E) Blots of flagella from gametes and dikaryons formed between oda8,oda10 and oda6,oda10,ODA10HA show that ODA10 is lost from flagella during the time when ODAs (IC2) are assembling.
ODA complexes in oda10 mutant cytoplasm are defective
Because axonemal binding sites for ODAs appear normal in both oda8 (Desai et al., 2015) and oda10 (Dean and Mitchell, 2013), whereas ODA complexes in the cytoplasm of oda8 do not appear normal, we hypothesized that the genetic interaction between oda8 and oda10 involved a shared role in maturation of ODA complexes in the cytoplasm, after preassembly of dynein but before transport of ODA complexes into flagella. We therefore tested whether preassembled dynein in the cytoplasm of oda10 mutants was defective in its ability to bind to outer doublet microtubules. Dynein binding assays were performed by incubating cytoplasmic extracts with axonemes lacking outer arm dyneins, and the pellets were analyzed for the presence of dynein, using Western blots. The results show that ODAs from oda10 mutant cytoplasm are highly defective for binding to axonemes (Figure 9A). ODA subunits are present at wild-type abundance in oda10 cytoplasm, and their assembly state is comparable to that in wild-type cytoplasm, based on coimmunoprecipitation with a monoclonal antibody to ODA heavy chain β (Fowkes and Mitchell, 1998). Recently, however, we observed that ODA complexes in the cytoplasm of another ODA assembly mutant, pf22, could not be detected using our monoclonal for immunoprecipitates, even though ODA heavy chain β is abundant in the pf22 cytoplasm by blot (Mitchison et al., 2012), leading us to question the accuracy of IP analysis based on this anti-β monoclonal. Subsequently, IP with a Myc-epitope tagged IC (ODA IC2) revealed reduced structural integrity of ODA complexes in oda8 compared with wild-type cytoplasm (Desai et al., 2015). With this anti-Myc IP approach, when ODA complexes in oda10 cytoplasm were compared with those in wild-type cytoplasm, <50% of the wild-type level of dynein heavy chains α, β, and γ was seen to coimmunoprecipitate with IC2 (Figure 9B), indicating comparable or greater defect in stability of ODA complexes in oda10 cytoplasm and oda8 cytoplasm. Because the Chlamydomonas ODA5/ODA10 complex appears to function in cytoplasmic steps needed for correct ODA assembly rather than as an accessory to the docking complex for ODA attachment to doublet microtubules, we suggest that its continued designation as the ODA AC should serve to indicate ODA assembly complex rather than ODA accessory complex.
FIGURE 9:
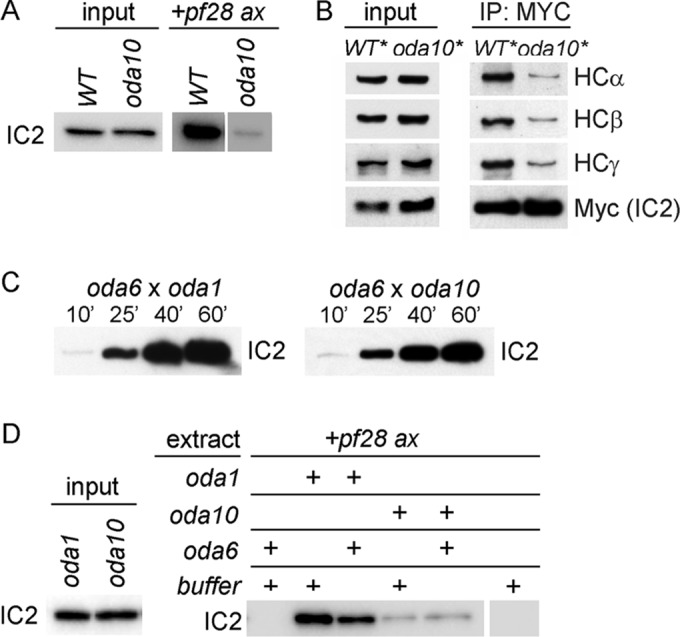
Defective ODA complexes in oda10 mutant cytoplasm. (A) Axonemal binding of ODA complexes from wild-type cytoplasm (WT) and oda10 cytoplasm to pf28 axonemes (+pf28 ax lanes). Blots of pelleted axonemes were probed for IC2 to reveal bound ODA complexes. Input shows equal amounts of ODA IC2 in the extracts. (B) Blot of total cytoplasmic proteins to show relative abundance (left) and anti-Myc IP of ODA complexes (right) from extracts of wild- type (oda6,IC2-Myc) and oda10 cells expressing IC2-MYC (oda10,oda6,IC2-Myc). (C) Comparison of the rates of ODA assembly into fully formed flagella of dikaryons between oda6 and oda1 (left) and between oda6 and oda10 (right). Flagella isolated at the indicated times were blotted for IC2. (D) Binding assays comparing the ability of ODA complexes from oda10 and oda1, with and without preincubation with an oda6 cytoplasmic extract, to bind to pf28 axonemes. Input lanes show equal amounts of IC2 in the extracts.
The defective ODAs in oda10 cytoplasm can apparently be repaired and assembled into the flagella of dikaryons formed between oda10 gametes and gametes harboring mutations in either intermediate chains (oda6 or oda9) or heavy chains (oda2 or oda4; Kamiya, 1988; Figure 8E). To see whether this repair required a measurably slow process such as protein synthesis, as reported recently for rescue between two I1 inner arm dynein mutants (Viswanadha et al., 2014), we measured the relative rate of repair in a cross between oda6 (missing IC2) and oda10 (missing the ODA AC) and in a cross between oda6 and oda1 (missing the ODA DC). As seen in Figure 9C, the repair in oda6 × oda10 dikaryons does not display any lag indicative of the need for synthesis of new proteins during dikaryon rescue. A successful replication of this cytoplasmic complementation phenomenon in vitro could allow us to biochemically isolate the components in oda6 cytoplasm that modified the defective dynein into an assembly-competent form. Thus we tested whether the axonemal binding defect in ODAs from oda10 cytoplasm could be restored by mixing together cytoplasmic extracts of oda6 and oda10 cells and then preincubating the mixture to allow any necessary enzymatic activity in vitro before adding axonemes to test binding. No repair was observed in this experiment (Figure 9D) or in several others with variations of the time, temperature, and ATP concentration during preincubation. In contrast, ODAs in the cytoplasm of docking complex mutant oda1 bound to axonemes with high affinity, which was unaffected by preincubation with oda6 extracts.
DISCUSSION
ODA assembly is demonstrably a nonspontaneous process requiring multiple complexes and proteins that do not function as part of the motor itself. Preassembly of ODA motor subunits (heavy chains and ICs) begins in the cytoplasm with the assistance of cytoplasmic assembly factors (Freshour et al., 2007; Omran et al., 2008; Duquesnoy et al., 2009; Horani et al., 2012; Mitchison et al., 2012). Transport of preassembled ODAs into cilia uses an ODA-specific IFT adaptor protein (Ahmed and Mitchell, 2005; Ahmed et al., 2008), and, once in cilia, ODAs assemble onto axonemes at locations specified by the independently assembled ODA DC (Takada and Kamiya, 1994; Takada et al., 2002; Koutoulis et al., 1997; Wakabayashi et al., 2001; Casey et al., 2003; Ide et al., 2013). There is at least one additional step required for ODA assembly in Chlamydomonas, which requires three proteins—ODA5, ODA8, and ODA10—after cytoplasmic preassembly and before docking of ODAs onto axonemes in flagella (Kamiya, 1988; Fowkes and Mitchell, 1998; Wirschell et al., 2004; Dean and Mitchell, 2013; Desai et al., 2015). Here we showed that ODA5 and ODA10 exist together in a complex, the ODA AC, in both the cytoplasm and in flagella and that these two proteins depend on each other for stability. Thus lack of cytoplasmic complementation in dikaryons between oda5 and oda10 follows from an absence of the ODA AC in both strains, but the exact role of the ODA AC is unclear. Sequence and predicted structure comparisons suggest that these proteins share a common ancestor with ODA DC subunits (Dean and Mitchell, 2013). Most organisms with motile flagella powered by ODAs only retain a single homologue of ODA10 and a single homologue (or two close paralogues) of Chlamydomonas DC2, and evidence has been presented that the vertebrate ODA10 homologues (Hjeij et al., 2014; Jerber et al., 2014) and DC2 homologues (Knowles et al., 2013; Onoufriadis et al., 2014) are interacting axonemal proteins important for ODA assembly. We hypothesize that ancestrally a single heterodimer performed the functions of both the Chlamydomonas AC and DC complexes, but through gene duplication in the green algal lineage, two separate complexes emerged with separate functions, one dedicated to assembly and the other to forming an axonemal docking site. If so, then the single complex found in mammals formed through interaction of DC2 homologue CCDC114 and ODA10 homologue CCDC151 (Hjeij, 2014) may still perform both assembly and docking functions. In tissues in which IFT-dependent assembly is not required, such as Drosophila testis, the need for an ODA10 homologue appears to have been lost (Jerber et al., 2014). It remains to be seen whether a docking complex is still required for attachment of ODA complexes in fly sperm and whether a DC2 homologue participates in its formation.
Of interest, immunofluorescence and immunogold electron microscopy showed that ODA5 and ODA10 are localized to DM1 in the proximal 2–3 μm of flagella (Figures 4 and 5). DM1 in Chlamydomonas lacks outer arms, and ODA10 localization was not affected by lack of ODAs; thus binding sites on DM1 for the ODA AC are assembled independently of ODAs (Figure 4). The only other structures with a similar localization to ODA10 are bridges with 8-nm periodicity that connect DM1 and DM2 in the proximal region of Chlamydomonas flagella (Hoops and Witman, 1983; Bui et al., 2009; Lin et al., 2012). The function of these bridges is unclear, but similar structures are present in sea urchin sperm axonemes (Lin et al., 2012), and they are hypothesized to be responsible for creating asymmetric planar waveforms typical of ciliary motility by restraining sliding between DM1 and DM2. ODA10 localization did not reach the extreme proximal end of axonemes (Figures 5A and 6A), a region that lacks the DM1–DM2 bridge (Bui et al., 2012). This pattern is not the same as the pattern of another coiled-coil protein that has been implicated in dynein assembly in vertebrates, CCDC11, which extends along the entire cilium in some zebrafish tissues but is restricted to the basal body region (coincident with γ-tubulin) in nodal cilia, where it may be needed for outer arm dynein assembly (Narasimhan et al., 2015).
Our data suggest that some DM1-specific proteins direct assembly of the DM1–DM2 bridge and also favor interaction of the AC in this region, whereas the remaining eight doublets show higher affinity for the DC at the same relative A-tubule position. DM1 distal to the bridge lacks any electron density at the position occupied by the DC (and outer arms) on the remaining eight DMs and fails to bind outer arms in vitro in the presence of the DC (Takada and Kamiya, 1994). Thus the AC does not play a role in docking ODA onto axonemes, consistent with our dynein reconstitution assay, which showed that ODA10 is unnecessary for dynein to bind to axonemes in vitro (Dean and Mitchell, 2013). The structural and implied evolutionary relationship between the heterodimeric DC1/DC2 core of the docking complex and the heterodimeric ODA5/ODA10 assembly complex would be consistent with a 24-nm periodicity for the interaction of both complexes with doublet microtubules. Although the peak of ODA10 immunogold particle separation distance was ∼24 nm, the size of gold particle–tagged immune complexes precludes accurate measurement of closer spacing. Thus AC complexes could have true periodicities of 8 or 16 rather than 24 nm.
Immunofluorescence microscopy performed on cells fixed at various times during ciliogenesis showed that ODA10 assembles from base to tip coincident with assembly of the proximal bridge-associated domain, suggesting that ODA10 and ODA5 function in the proximal region of flagella to facilitate ODA assembly in flagella (Figure 7). However, reductions in flagellar ODA10 during a dikaryon rescue, at a time when ODAs are still being assembled (Figure 8E), suggest that the proximal location of the ODA AC is not essential for dynein assembly and that ODAs no longer need the ODA AC once they have been assembled. However, because we were unable to distinguish HA signals at the single-molecule level from the background IF signal seen along the flagella of control (untagged) cells, we cannot eliminate the possibility that low levels of ODA10 are transported along with dynein to the flagellar tip during the assembly process.
Dynein binding assays show that ODAs in oda10 cytoplasm, like those in oda8 cytoplasm (Desai et al., 2015), do not bind to axonemes with the same affinity as ODAs in wild-type or oda1 cytoplasm (Figure 9), suggesting that preassembled ODAs are in a different state in the absence of ODA8 or the AC. Cytoplasmic complementation analysis showed that ODAs in oda10 cytoplasm can be assembled to rescue motility in dikaryons formed between oda10 and oda6 (ODA-IC2 mutant), suggesting that ODAs in oda10 cytoplasm can be restored, given the right conditions. However, we could not replicate this restoration in vitro. Preincubating oda10 cytoplasmic extract with oda6 cytoplasmic extract, which lacks dynein but has ODA8, ODA10, and ODA5, did not restore the defective ODA in oda10 cytoplasm to wild-type affinity (Figure 9D). ODAs in oda10 cytoplasm may require an intact cellular architecture, such as the presence of basal body/centrosome–associated factors that are not present in these extracts, in order to be repaired or transitioned to a mature state.
In summary, we hypothesize that ODA10 and ODA5 function in an assembly complex required for dynein maturation in the cytoplasm after preassembly of most ODA subunits and that ODA8 is also important in this maturation process. In this instance, maturation could involve a posttranslational modification of a dynein subunit, such as a lipidation, phosphorylation, or the isomerization of a proline, or a change in dynein interaction with a specifically activated transport factor, such as a G-protein, as seen for other ciliary cargoes (Sung and Leroux, 2013). This function and the traditional doublet docking function may be dual roles of the docking complex in metazoan organisms. In Chlamydomonas, where these functions are separate, the function of the ODA AC in the cytoplasm may be essential to form ODA complexes that can be recognized by the IFT machinery as substrates for transport and that will interact productively with axonemal docking sites on outer doublet microtubules.
MATERIALS AND METHODS
Strains and culture conditions
The following strains of Chlamydomonas reinhardtii can be obtained from the Chlamydomonas Genetics Center (University of Minnesota, Minneapolis, MN): wild-type 137c,mt- (CC-124), 137c,mt+ (CC-125), pf28,mt- (CC-1877), pf28,mt+ (CC-3661; an allele of oda2), oda1,mt+ (CC-2228), oda6,mt+ (CC-2239), oda5,mt+ (CC-2236), oda8,mt+ (CC-2242), oda9,mt+ (CC-2244), oda9,mt- (cc-2245), oda10-1,mt+ (CC-2246), and oda10-1,mt- (CC-2247). Strains oda5,ODA5HA (CC-4030), insertional mutant oda10-2,mt-, and oda10-2,mt-,arg7 (Wirschell et al., 2004) were obtained from George Witman (University of Massachusetts Medical School, Worcester, MA). oda5,oda10, oda8,oda10, oda5,oda10,ODA10HA, oda8,oda10,ODA10HA, and oda10-2,ODA10HA (Dean and Mitchell, 2013), oda6, ODA6MYC, and oda8,ODA8HA (Desai et al., 2015), and oda10-2,ODA10FLAG, oda5,oda10,ODA5HA,ODA10FLAG, oda10,oda5,ODA5HA, and oda9,oda10,ODA10HA were constructed in our laboratory. oda10-2,ODA10FLAG was created by cotransformation of FLAG-tagged ODA10 cDNA with ARG2 gene plasmid pJD67 into an arginine-dependent oda10-2,arg7 strain. oda6,ODA6HA has been previously described (Mitchison et al., 2012). The double-mutant strain oda10,oda5 was crossed with oda5,ODAHA to create oda10,oda5,ODA5HA from nonparental ditype tetrads. oda mutant strains that expressed ODA10HA and ODA10FLAG were selected using whole-cell and flagellar sample Western blots probed with anti-HA and anti-FLAG antibodies, respectively. oda10-2,ODA10HA was crossed with oda9 to create oda9,oda10,ODA10HA from nonparental ditype tetrads. oda10-2,ODA10FLAG and oda5,oda10,ODA5HA,ODA10FLAG were created by cotransformation of FLAG-tagged ODA10 cDNA with ARG2 gene plasmid pJD67 and paromomycin-resistant gene plasmid pChlamiRNA3int (Chlamydomonas Genetics Center), respectively, into arginine-dependent oda10-2,arg7 and oda10,oda5,ODA5HA strains. Mating and tetrad analyses were performed according to standard protocols (Dutcher, 1995). Expression of all tagged proteins was in the background of a cell that was null at the endogenous locus. Expression levels are documented in Supplemental Figure S1.
Cell fractionation
Whole-cell protein samples were prepared by acetone precipitation, dissolved in 1× high-SDS buffer, boiled for 2 min, and spun in a microcentrifuge to remove cell ghosts. β-Mercaptoethanol was added, and supernatants were boiled for 5 min. Cell body samples were prepared identically to whole-cell samples after deflagellation by pH shock. Cytoplasmic extracts for binding assays and immunoprecipitations were prepared as previously described (Dean and Mitchell, 2013) and adjusted to equalize abundance of ODA IC2 by Western blot before use. For column purification, extracts of cells that had not been pretreated with gametic lysine were prepared from 32-l cultures using a commercial bead beater (Biospec, Bartlesville, OK) and an extraction buffer of 140 mM NaCl, 20 mM Tris, pH 7.8, and 5 mM imidizole. Up to 10 ml of extract was fractionated on a 1-ml His Bind column (Novagen). Peak fractions from the His Bind column were applied directly to a 0.1-ml anti-FLAG M2 affinity column (Sigma-Aldrich) and eluted with 3xFLAG peptide (APE Bio).
Flagella, axonemes, and axonemal salt extracts were prepared from vegetative cells using standard methods as detailed previously (Dean and Mitchell, 2013). For binding assays, axonemes were incubated with cytoplasmic extracts on ice for 1 h and pelleted in a microcentrifuge at 4°C for 5 min. Pellets were made directly into gel samples or washed as detailed in Results. For dikaryon analysis, cells were washed into medium lacking nitrogen and incubated overnight under constant illumination. Gametes were resuspended at equal densities before mixing. At the indicated times, samples were removed for pH shock deflagellation for fixation and immunofluorescence analysis.
Antibodies and immunodetection
Antibodies used include rat monoclonal anti-HA epitope 3F10 (Roche), mouse monoclonal anti–ODA-IC2 (C11.4; Mitchell and Rosenbaum, 1986), mouse monoclonal anti-FLAG epitope M2 (Sigma-Aldrich, Cell Signaling), anti-NAB1 (Agrisera AB, Vannas, Sweden), and mouse anti–acetylated-tubulin 6B11 (Sigma-Aldrich). The anti-Myc 9E 10 monoclonal antibody, developed by J. Michael Bishop, was obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). For Western blots, horseradish peroxidase–labeled secondary antibodies were detected using Clarity Western ECl Substrate (Bio-Rad). Chemiluminescence signals were detected using either radiography or Chemidoc MP Imager (Bio-Rad). The blots were quantified using Chemidoc software (Bio-Rad).
For immunoprecipitation, extracts were preincubated with 15 μl of a 50% suspension of protein A–Sepharose (PA-Seph) for 1 h, and spun, and supernatants were transferred to fresh tubes. For IP with anti-HA and anti-FLAG, antibodies were added and, after 2 h, 15 μl of PA-Seph suspension was added and incubated for 1 h while rotating. For IP with anti-Myc, beads with covalently bound anti-Myc (Sigma-Aldrich) were used. The mixtures were spun in a microcentrifuge, and pellets were washed three times with 0.5 ml of HMDEK50 buffer (30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM ethylene glycol-bis (β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid [EGTA], 50 mM potassium acetate) for 10 min while rotating. Finally, the pellets were resuspended in SDS-sample buffer and boiled for 2 min.
For immunofluorescence analysis, cells were simultaneously fixed and permeabilized by mixing with equal volumes of 10 mM HEPES/6% paraformaldehyde/3% Triton X-100, and the mixture was immediately applied to 0.1% poly-l-lysine (Sigma-Aldrich)–coated coverslips and allowed to settle for 12 min. Slides were plunged into methanol for 5 min at −20°C and air dried. After rehydration in phosphate-buffered saline (PBS), samples were blocked for 1 h in 100% block solution (PBS/5% bovine serum albumin/1% cold-water fish gelatin), incubated with primary antibodies diluted in 20% block solution for 1 h at room temperature, washed 3 × 5 min in PBS, and incubated with secondary antibodies diluted in 20% block solution for 1 h at room temperature. Coverslips were washed as before and mounted in 50% glycerol/50% Tris-saline with 2% N-propyl-gallate as an anti-fade reagent. Slides were stored at 4°C until imaged. For multiple staining, the slides were incubated with one antibody at a time, followed by its secondary, with 30-min blocks before the next primary antibody, to avoid cross-reaction between antibodies. Alexa Fluor 488 goat anti–mouse and Alexa Fluor 555 goat anti–rat secondary fluorescent antibodies (Invitrogen) were used. Images were captured on a Leica SP5 scanning confocal microscope with an HCX PL APO 63×/1.40–0.60 Oil λ BL objective (Leica, Bannockburn, IL) with Leica LAS AF 2.6 software. Maximum Z-stack projections were made in ImageJ (National Institutes of Health, Bethesda, MD). Images were cropped, and brightness and contrast levels were adjusted using Photoshop (Adobe).
Electron microscopy
For traditional thin-section analysis, axonemes were pelleted and fixed in 1% glutaraldehyde, 1% tannic acid, and 100 mM sodium phosphate buffer, pH 7.0, rinsed, postfixed in osmium tetroxide, dehydrated, and embedded in Epon. Thin sections were stained with ethanolic uranyl acetate and lead citrate and imaged in a JEOL JEM1400 transmission electron microscope with a Gatan 832 charge-coupled device camera. Median density of images was adjusted with the Levels function in Photoshop. For immunogold analysis, axonemes adhering to carbon/Formvar-coated nickel grids or to poly-l-lysine–coated glass cover slips were incubated in 10% block in HMDEK50 for 10 min, treated with 3F10 (rat monoclonal anti-HA) diluted 1:10 in 10% block for 1 h at room temperature, rinsed in 10% block for 10 min, treated with 10 nm gold goat anti-rat (Electron Microscopy Sciences) diluted 1:20 in 10% block for 1 h at room temperature, and rinsed 2 × 10 min in HMDEK50. Preparations on grids were negatively stained with 2% aqueous uranyl acetate. Preparations on coverslips were fixed, dehydrated, embedded, and sectioned as described, except that HMDEK50 was substituted for phosphate buffer during glutaraldehyde fixation. Sections were lightly stained with uranyl acetate and lead citrate before viewing. Point-to-point distances between adjacent gold particles and from the abscission zone at the base of the axoneme to each gold particle were measured using Gatan Digital Micrograph DM3 software.
Supplementary Material
Acknowledgments
We thank Judy Freshour for technical support and Nickolas Deakin for help with IF microscopy. This work was supported by National Institutes of Health Grant R01 044228 to D.R.M.
Abbreviations used:
- AC
assembly complex
- DC
docking complex
- ODA
outer dynein arm.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0317) on August 26, 2015.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Mitchell DR. ODA16p, a Chlamydomonas flagellar protein needed for dynein assembly. Mol Biol Cell. 2005;16:5004–5012. doi: 10.1091/mbc.E05-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey DM, Inaba K, Pazour GJ, Takada S, Wakabayashi K, Wilkerson CG, Kamiya R, Witman GB. DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol Biol Cell. 2003;14:3650–3663. doi: 10.1091/mbc.E03-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AB, Mitchell DR. Chlamydomonas ODA10 is a conserved axonemal protein that plays a unique role in outer dynein arm assembly. Mol Biol Cell. 2013;24:3689–3696. doi: 10.1091/mbc.E13-06-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PB, Freshour JR, Mitchell DR. Chlamydomonas axonemal dynein assembly locus ODA8 encodes a conserved flagellar protein needed for cytoplasmic maturation of outer dynein arm complexes. Cytoskeleton (Hoboken) 2015;72:16–28. doi: 10.1002/cm.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy P, Escudier E, Vincensini L, Freshour J, Bridoux A-M, Coste A, Deschildre A, de Blic J, Legendre M, Montantin G, et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am J Hum Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 1995;47:531–540. doi: 10.1016/s0091-679x(08)60857-2. [DOI] [PubMed] [Google Scholar]
- Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9:2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour J, Yokoyama R, Mitchell DR. Chlamydomonas flagellar outer row dynein assembly protein ODA7 interacts with both outer row and I1 inner row dyneins. J Biol Chem. 2007;282:5404–5412. doi: 10.1074/jbc.M607509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjeij R, Onoufriadis A, Watson CM, Slagle CE, Klena NT, Dougherty GW, Kurkowiak M, Loges NT, Diggle CP, Morante NFC, et al. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, Van Arendonk LG, Thornton KC, Giacalone JC, Albee AJ, Wilson KS, et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Owa M, King SM, Kamiya R, Wakabayashi K. Protein-protein interactions between intermediate chains and the docking complex of Chlamydomonas flagellar outer arm dynein. FEBS Lett. 2013;587:2143–2149. doi: 10.1016/j.febslet.2013.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerber J, Baas D, Soulavie F, Chhin B, Cortier E, Vesque C, Thomas J, Durand B. The coiled-coil domain containing protein CCDC151 is required for the function of IFT-dependent motile cilia in animals. Hum Mol Genet. 2014;23:563–577. doi: 10.1093/hmg/ddt445. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, Yagi T. Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zool Sci. 2014;31:633–44. doi: 10.2108/zs140066. [DOI] [PubMed] [Google Scholar]
- Knowles MR, Ostrowski LE, Loges NT, Hurd T, Leigh MW, Huang L, Wolf WE, Carson JL, Hazucha MJ, Yin W, et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am J Hum Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Takeda H. Ciliary motility: the components and cytoplasmic preassembly mechanisms of the axonemal dyneins. Differentiation. 2012;83:S23–S29. doi: 10.1016/j.diff.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kott E, Duquesnoy P, Copin B, Lagandre M, Dastot-Le Moal F, Montantin G, Jeanson L, Tamalet A, Papon J-F, Siffroi J-P, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am J Hum Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Heuser T, Song K, Fu X, Nicastro D. One of the nine doublet microtubules of eukaryotic flagella exhibits unique and partially conserved structures. PLoS One. 2012;7:e46494. doi: 10.1371/journal.pone.0046494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. Protein-protein interactions in the 18S ATPase of Chlamydomonas outer dynein arms. Cell Motil Cytoskeleton. 1986;6:510–520. doi: 10.1002/cm.970060510. [DOI] [PubMed] [Google Scholar]
- Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, O’Callaghan C, Blau H, Al Dabbagh M, Olbrich H, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–382. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V, Hjeij R, Vij S, Loges NT, Wallmeier J, Koerner-Rettberg C, Werner C, Thamilselvam SK, Boey A, Choksi SP, et al. Mutations in CCDC11, which encodes a coiled-coil containing ciliary protein, causes situs inversus due to dysmotility of monocilia in the left-right organizer. Hum Mutat. 2015;36:307–318. doi: 10.1002/humu.22738. [DOI] [PubMed] [Google Scholar]
- Omran H, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoufriadis A, et al. Combined exome and whole-genome sequencing identifies mutations in ARMC4 as a cause of primary ciliary dyskinesia with defects in the outer dynein arm. J Med Genet. 2014;51:61–67. doi: 10.1136/jmedgenet-2013-101938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Leroux MR. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol. 1994;126:737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell. 2002;13:1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanadha R, Hunter EL, Yamamoto R, Wirschell M, Alford LM, Dutcher SK, Sale WS. The ciliary inner dynein arm, I1 dynein, is assembled in the cytoplasm and transported by ift before axonemal docking. Cytoskeleton (Hoboken) 2014;71:573–586. doi: 10.1002/cm.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Takada S, Witman GB, Kamiya R. Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil Cytoskeleton. 2001;48:277–286. doi: 10.1002/cm.1015. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Pazour G, Yoda A, Hirono M, Kamiya R, Witman GB. Oda5p, a novel axonemal protein required for assembly of the outer dynein arm and an associated adenylate kinase. Mol Biol Cell. 2004;15:2729–2741. doi: 10.1091/mbc.E03-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.