The apical scaffolding protein E3KARP, a paralogue of EBP50, is regulated by Ser-303 cell-cycle phosphorylation. This phosphorylation enhances its exchange rate in vivo and reduces its apical localization and function. The dynamics of EBP50 and E3KARP are regulated by sites in their tails, which are the most divergent regions between them.
Abstract
We examine the dynamics and function of the apical scaffolding protein E3KARP/NHERF2, which consists of two PDZ domains and a tail containing an ezrin-binding domain. The exchange rate of E3KARP is greatly enhanced during mitosis due to phosphorylation at Ser-303 in its tail region. Whereas E3KARP can substitute for the function of the closely related scaffolding protein EBP50/NHERF1 in the formation of interphase microvilli, E3KARP S303D cannot. Moreover, the S303D mutation enhances the in vivo dynamics of the E3KARP tail alone, whereas in vitro the interaction of E3KARP with active ezrin is unaffected by S303D, implicating another factor regulating dynamics in vivo. A-Raf is found to be required for S303 phosphorylation in mitotic cells. Regulation of the dynamics of EBP50 is known to be dependent on its tail region but modulated by PDZ domain occupancy, which is not the case for E3KARP. Of interest, in both cases, the mechanisms regulating dynamics involve the tails, which are the most diverged region of the paralogues and probably evolved independently after a gene duplication event that occurred early in vertebrate evolution.
INTRODUCTION
Polarized cells establish and maintain compositionally and morphologically distinct plasma membrane domains, the classic example being an epithelial cell, with its distinct apical and basolateral domains. The apical domain of epithelial cells is decorated by microvilli that contain a core of actin filaments linked to the plasma membrane in part by activated ezrin, a member of the ezrin/radixin/moesin (ERM) family. ERM proteins can bind directly to plasma membrane proteins and also associate with scaffolding proteins ezrin-binding phosphoprotein of 50 kDa (EBP50)/Na+-H+ exchanger-3 regulatory factor 1 (NHERF1) or its paralogue, exchanger 3 kinase A regulatory protein (E3KARP)/Na+-H+ exchanger-3 regulatory factor 2 (NHERF2; Fehon et al., 2010). Very little is known about the regulation of scaffolding proteins despite their importance in the functional organization of plasma membrane domains. Recent work has shown that EBP50’s function and dynamics are regulated by its ability to bind ligands (Garbett and Bretscher, 2014). In this study, we explore the biochemical properties of E3KARP and examine whether it is similarly regulated. We find that E3KARP’s function and dynamics are regulated during the cell cycle in a manner mechanistically different from those of EBP50.
EBP50 was identified as a binding partner of activated ezrin (Reczek et al., 1997) and as a factor necessary to confer cAMP regulation on NHE3 (Weinman et al., 1993), hence the alternate names. EBP50 has two postsynaptic density 95/discs large/zona occludens-1 (PDZ) domains, which mediate interactions with multiple PDZ ligands, including transporters and receptors in the plasma membrane (Weinman et al., 1995; Short et al., 1998; Cao et al., 1999; Hernando et al., 2002) and the cytoplasmic protein EBP50 PDZ interactor of 64 kDa (EPI64; Reczek and Bretscher, 2001). EBP50 binds ezrin through its C-terminal domain, and this interaction, as well as a functional PDZ1 domain, is required for microvillar assembly and maintenance (Garbett et al., 2010). EBP50 in microvilli is unexpectedly dynamic, exchanging in 5–10 s, and this dynamic behavior is strongly suppressed when ligand binding to its PDZ domains is inhibited (Garbett and Bretscher, 2012).
E3KARP was identified by its ability to bind the cytoplasmic tail of NHE3, recognized as being closely related to EBP50 (Yun et al., 1998), and subsequently renamed NHERF2 (Kurashima, 1999). E3KARP and EBP50 share 55% sequence identity, and both have two PDZ domains and an ezrin-binding domain (Reczek and Bretscher, 1998). The PDZ domains of E3KARP interact with transporters (PMCA 2b [ DeMarco et al., 2002] and SLC26A6 [ Lohi et al., 2003]), receptors (β-α-adrenergic receptor [ Hall et al., 1998] and parathyroid hormone receptor [ Mahon et al., 2002]), channels (cystic fibrosis transmembrane conductance regulator [ Sun et al., 2000] and ROMK [ Yun, 2002]), and a protein contributing to the glycocalyx (podocalyxin; Li et al., 2002), as well as with cytoplasmic proteins (EBP50 [ He et al., 2001] and EPI64 [ Reczek and Bretscher, 2001]). Of interest, E3KARP exhibits some clear differences from EBP50. For example, the expression of EBP50 is almost exclusively restricted to epithelial cells, whereas that of E3KARP is not (Ingraffea et al., 2002). Whereas both EBP50 and E3KARP can confer cAMP inhibition on NHE3, E3KARP is required for cGMP inhibition of NHE3, a function for which EBP50 cannot substitute (Cha et al., 2005; Yun et al., 1998; Weinman et al., 2003). In addition, E3KARP exhibits a much slower exchange rate than EBP50 in vivo in microvilli of interphase cells (Garbett and Bretscher, 2012). The region specifying the difference in dynamics between EBP50 and E3KARP is located in the region downstream of the PDZ domains (Garbett et al., 2013; Yang et al., 2013).
Here we explore the evolutionary divergence of EBP50 and E3KARP and then document E3KARP’s biochemical properties to show that, like EBP50, it exists as a monomer in solution. We then report that E3KARP is phosphorylated during mitosis on a conserved serine residue, not present in EBP50, and that this regulates its function and dynamics. We also identify A-Raf as a kinase necessary for E3KARP phosphorylation at the G2/M stage of the cell cycle. This study reveals that the function and dynamics of E3KARP are regulated but in a manner mechanistically different from those of EBP50, suggesting that after the gene duplication event that gave rise to these paralogues, different modes of regulation evolved.
RESULTS
E3KARP and EBP50 arose by gene duplication and evolution of their C-terminal regions
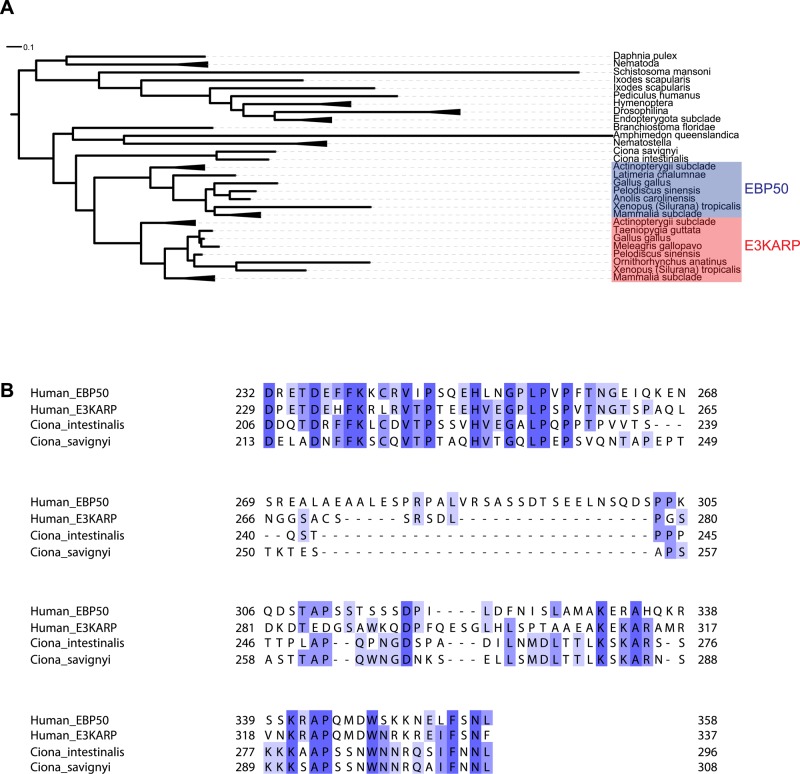
Mammalian E3KARP and EBP50 are closely related (Donowitz et al., 2005), so we conducted a phylogenetic analysis across metazoan genomes of these two proteins. E3KARP and EBP50 are derived from a unique root in early vertebrate evolution, whereas invertebrates have only one form (Figure 1A). Next we examined sequence homology between E3KARP and EBP50 in the vertebrate clade. Alignment of 18 E3KARP and EBP50 vertebrate species shows a high degree of conservation over the region of their PDZ domains (unpublished data), as well as over the C-terminal residues involved in binding ezrin (Supplemental Figure S1, A and B). Aligning these proteins on the basis of the PDZ regions alone clearly separates the EBP50 and E3KARP groups, suggesting a divergence soon after the gene duplication event that gave rise to them. The divergence between the consensus sequences of the full-length proteins reveals the presence of ∼20 amino acids upstream of the ezrin-binding domain that is present in all EBP50, but not in the E3KARP, homologues (Supplemental Figure S1C). To determine whether the ∼20 amino acids are a consequence of an insertion in EBP50 or a deletion in E3KARP, we compared their sequences with the species immediately outside the vertebrate clade. The tunicate Ciona species do not have these 20 amino acids (Figure 1, A and B). These data suggest that present-day E3KARP and EBP50 arose from a gene duplication event during vertebrate evolution, and soon thereafter EBP50 acquired a 20–amino acid insertion, followed by evolutionary divergence of the region between the PDZ domains and ezrin-binding site. This divergent region is partly responsible for the difference in dynamics between EBP50 and E3KARP (Garbett et al., 2013).
FIGURE 1:
E3KARP and EBP50 diverged after gene duplication and insertion of 22 amino acids in the EBP50 tail. (A) Phylogenetic tree of EBP50 and E3KARP homologues. The scale bar indicates the number of substitutions. Mammalia, Actinopterygii, Nematoda, Drosophilina, Endopterygota, and Nematostella species have each been collapsed into a unique branch. The shaded boxes indicate the E3KARP and EBP50 groups in vertebrate clades. The tree was generated using the EggNOG database and iTOL software. (B) Sequence alignment of C-terminal tails of human EBP50 and E3KARP with two species outside the vertebrate clade, Ciona intestinalis and Ciona savignyi. Conserved residues are highlighted.
E3KARP is a slightly elongated monomer
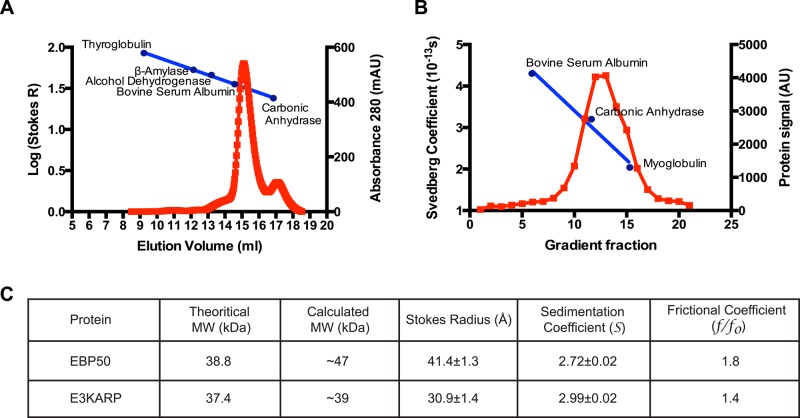
Biochemical and biophysical studies on EBP50 and E3KARP have yielded contradictory results on the capacity of EBP50 and E3KARP to oligomerize through their PDZ domains (Lau and Hall, 2001). To interpret subsequent results, it was necessary to first document the properties of E3KARP in vitro. Pure EBP50 has been shown to exist as an elongated monomer with no tendency to oligomerize (Li et al., 2007; Garbett et al., 2010). To examine the properties of E3KARP, recombinant E3KARP tagged with hexahistidine (6xHis)-SUMO was expressed in bacteria and purified using Ni2+ resin, and the 6xHis-SUMO tag was cleaved to generate free E3KARP (Supplemental Figure S2A). E3KARP migrated as a single species on a Superdex 200 gel filtration column and on a sucrose gradient. By gel filtration, E3KARP has a Stokes radius of 31 Å, corresponding to a molecular weight of 40 kDa for a globular protein (Figure 2, A and C). E3KARP sedimented at 2.99S (Figure 2, B and C, and Supplemental Figure S2B). These two physical parameters can be combined to predict a native molecular weight of ∼39 kDa (Siegel and Monty, 1966), close to the predicted monomeric mass, indicating that E3KARP exists as a monomer in solution with no tendency to oligomerize. Using a protein partial specific volume of 0.74 cm3/g, we calculate a frictional ratio (f/f0) of 1.4, indicating that E3KARP is a slightly elongated monomer in solution. When compared with the physical constants for EBP50 (Garbett et al., 2010), this shows that EBP50 is more elongated than E3KARP in solution (Figure 2C). This difference is presumably due to the divergence in the tail region between the two proteins.
FIGURE 2:
E3KARP is a slightly elongated monomer. (A) The Stokes radius of untagged recombinant E3KARP was analyzed by gel filtration on a Superdex 200 10/300GL column and (B) the sedimentation coefficient on a sucrose velocity gradient. The migration of protein standards is shown in blue. (C) Summary of the results for E3KARP compared with those obtained for EBP50 (from Garbett et al., 2010).
E3KARP can complement EBP50’s function in microvilli biogenesis
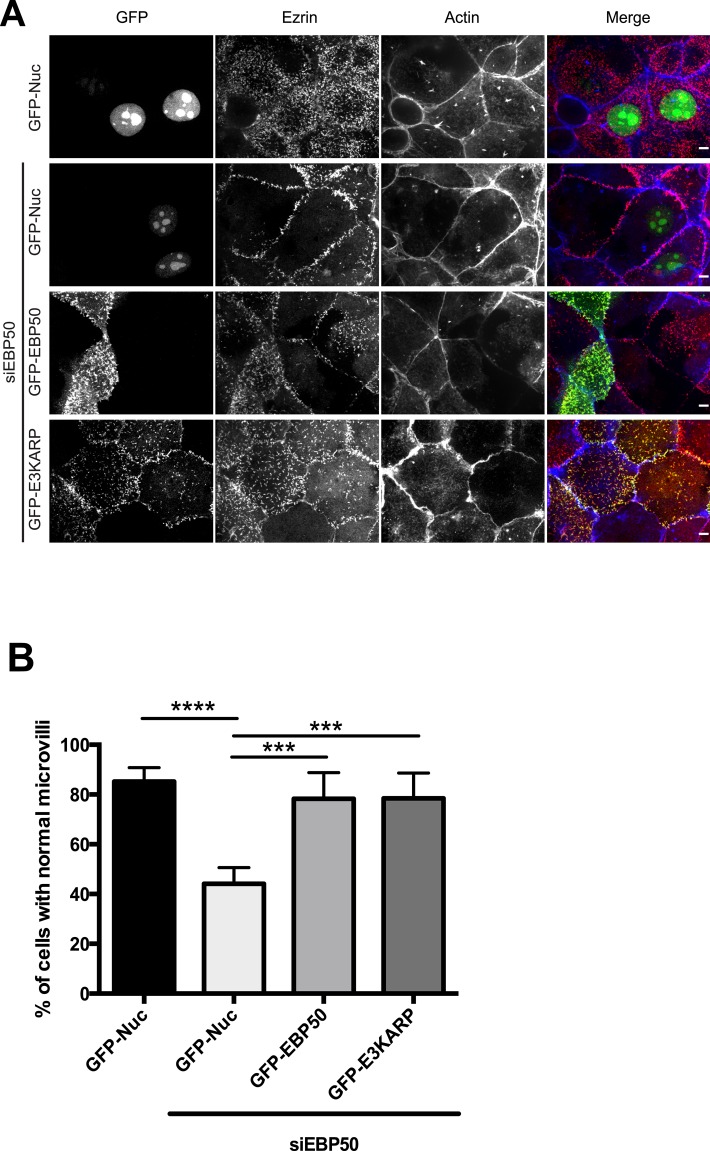
To explore whether E3KARP has similar functions to EBP50, we used the human choriocarcinoma JEG-3 cell line, as these cells normally display abundant microvilli. JEG-3 cells express EBP50 but have no detectable E3KARP. Depletion of EBP50 by small interfering RNA (siRNA) causes a significant loss of microvilli (Garbett et al., 2010; Figure 3A). When cells are depleted of EBP50 by siRNA and transfected to express green fluorescent protein (GFP) targeted to the nucleus (GFP-Nuc), only 44% of the cells show normal microvilli (Figure 3B). However, expression of RNA interference–resistant GFP-EBP50 restores microvilli in 78% of the cells. Expression of GFP-E3KARP also restores normal microvilli in the cells to the same extent as EBP50 (Figure 3, A and B). These data suggest that E3KARP shares a similar function to EBP50 that is necessary to assemble microvilli.
FIGURE 3:
E3KARP can rescue the loss of microvilli induced by siRNA against EBP50. (A) Maximum projection images of the apical surface from the microvillar rescue assay. Cells were transfected with GFP-tagged construct (green) and stained for ezrin (red) and actin (blue). The contrast for F-actin was increased for clarity in the panel representing siRNA-treated cells transfected with GFP-E3KARP. Scale bar, 5 μm. (B) Results from scoring cells for the presence of microvilli; errors bars indicate mean ± SD; n = 3. ***p < 0.001, ****p < 0.0001.
E3KARP is phosphorylated at the G2/M transition of the cell cycle
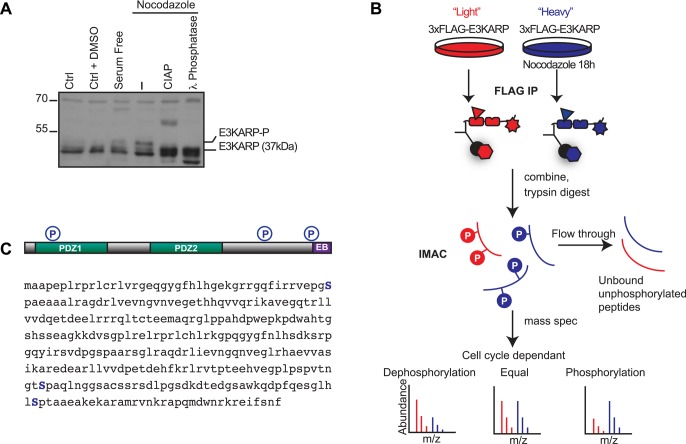
Phosphorylation of EBP50 during both interphase and mitosis can regulate the formation of microvilli (Hall et al., 1999; He et al., 2001; Garbett et al., 2010). To investigate the potential regulation of E3KARP by phosphorylation during the cell cycle, we used epithelial colorectal adenocarcinoma Caco-2 cells, which express endogenous E3KARP. Treatment of Caco-2 cells with nocodazole for 18–20 h arrested the cells in mitosis and resulted in a shift in the mobility of E3KARP, as seen by SDS–PAGE (Figure 4A). Because the shift can be reversed by in vitro phosphatase treatment, this suggests that E3KARP is phosphorylated during mitosis.
FIGURE 4:
Identification of cell cycle–dependent E3KARP phosphorylation sites. (A) Western blot of total lysates from Caco-2 cells treated without or with nocodazole as indicated and then subjected to treatment with calf intestinal alkaline phosphatase (CIAP) or λ phosphatase. (B) Experimental scheme using SILAC for identifying phosphorylation sites in E3KARP during mitosis. JEG-3 cells stably expressing 3xFLAG-E3KARP were labeled with “light” or “heavy” arginine and lysine. The cells growing in the “heavy” medium were arrested in mitosis by treatment with 50 ng/ml nocodazole overnight. FLAG immunoprecipitations were then performed on the two samples, which were then combined and subjected to trypsin digestion. Peptides were applied to a fresh IMAC resin, and the enriched phosphopeptides were eluted with IMAC elution buffer and analyzed by mass spectrometry. (C) Location of the three serine residues that were found to be phosphorylated in mitosis.
To identify the residues phosphorylated during mitosis, we subjected JEG-3 cells stably expressing 3xFLAG-tagged E3KARP to stable isotope labeling of amino acids in cell culture (SILAC) in normal (“light”) or 13C-arginine and 13C-lysine (“heavy”) medium (Figure 4B). 3xFLAG-E3KARP was separately immunoprecipitated from untreated cells grown in light medium or from cells grown in heavy medium, after treatment with nocodazole to arrest them in mitosis. The immunoprecipitates were mixed and subject to trypsin digestion. To enrich for phosphopeptides, we subjected the peptide mixture to immobilized metal affinity chromatography (IMAC) followed by mass spectrometry (Figure 4B). Analysis of tryptic peptides enriched from the cells grown in heavy medium identified three serine residues whose phosphorylation was enhanced at least threefold during mitosis (indicated in blue in Figure 4C). Serine S43 is localized in the PDZ1 domain, and, of interest, serines S261 and S303 are conserved residues in the C-terminal region of E3KARP and divergent from EBP50 (Figure 1A and Supplemental Figure S1B).
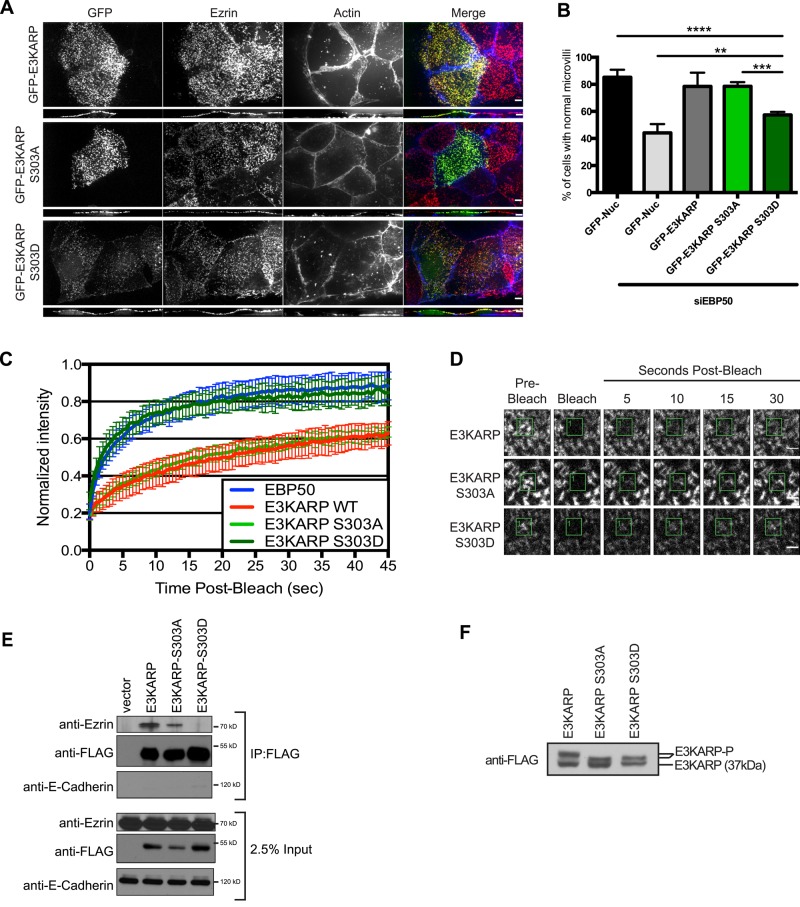
Phosphorylation of Ser-303 regulates the localization, function, and dynamics of E3KARP
To investigate the potential role of S261 and S303 phosphorylation in vivo, we generated phosphomimetic and phosphodeficient mutants. The localization and dynamics of expressed GFP-E3KARP S261A or GFP-E3KARP S261D were indistinguishable from those of GFP-E3KARP, and both constructs were able to rescue the loss of microvilli after knockdown of EBP50, indicating that phosphorylation of E3KARP at S261 has no detectable effect on E3KARP’s functions in microvilli assembly (Supplemental Figure S3). By contrast, phosphomimetic GFP-E3KARP S303D is weakly localized to microvilli in JEG-3 cells, being mostly cytosolic. The nonphosphorylatable mutant GFP-E3KARP S303A localized well to microvilli (Figure 5A). Expression of GFP-E3KARP or GFP-E3KARP S303A restored microvilli in cells depleted of EBP50, whereas GFP-tagged E3KARP S303D was impaired in its ability to rescue microvilli (Figure 5B). To determine whether these mutations affect the exchange rate of E3KARP, we used fluorescence recovery after photobleaching (FRAP) on interphase JEG-3 cells expressing GFP-E3KARP, GFP-E3KARP S303A, or GFP-E3KARP S303D. Mutation of Ser-303 to alanine (S303A) has no effect on the recovery rate, whereas the phosphomimetic S303D mutant increased the dynamics of E3KARP dramatically to a level similar to that seen for EBP50 (Figure 5, C and D). Because E3KARP is not normally present in JEG-3 cells, we repeated our analysis on Caco-2 cells that express endogenous E3KARP and found that each construct behaves the same as in JEG-3 cells (Supplemental Figure S4). We have shown a biochemical correlation between the ability to coimmunoprecipitate EBP50 and ezrin and the in vivo dynamics of EBP50, which is low when EBP50 is highly dynamic and enhanced when it is less dynamic (Garbett and Bretscher, 2012). We used the same assay with cells expressing the 3xFLAG-E3KARP variants. 3xFLAG-E3KARP S303A coprecipitated about as much ezrin as 3xFLAG-E3KARP, whereas 3xFLAG-E3KARP S303D showed very minimal recovery of ezrin (Figure 5E). We also explored whether phosphorylation affected the ability of E3KARP to bind the PDZ1 ligand EPI64 (Reczek and Bretscher, 2001). JEG-3 cells were transfected to express 3xFLAG-E3KARP, 3xFLAG-E3KARP S303A, or 3xFLAG-E3KARP S303D, and then FLAG immunoprecipitates were probed for the presence of EPI64 (Supplemental Figure S4C). EPI64 was able to bind E3KARP wild type, as well as the phosphodeficient mutant S303A. The S303D phosphomimetic mutation may slightly reduce the binding of EPI64, but this difference is much more modest than the effect on ezrin coimmunoprecipitation. Thus the major effects of the S303D phosphomimetic mutation are to enhance E3KARP’s dynamics and reduce its in vivo interaction with ezrin.
FIGURE 5:
Phosphorylation of Ser-303 regulates the localization, function, and dynamics of E3KARP. (A) Maximum projection images of the apical surface of JEG-3 cells. Cells were transfected to express GFP-tagged constructs (green) and then stained for ezrin (red) and actin (blue). A vertical cross section through the cells is shown under each maximum projection. The contrast for F-actin was increased for clarity in the panel representing siRNA-treated cells transfected with GFP-E3KARP S303D. Scale bar, 5 μm. (B) Ability of GFP-E3KARP constructs to restore microvilli in cell treated with siRNA against EBP50. Results from scoring individual cells for the presence of microvilli; errors bars indicate mean ± SD; n = 3, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) FRAP curves of for JEG-3 cells expressing GFP-E3KARP (n = 19), GFP-E3KARP S303A (n = 12), and GFP-E3KARP S303D (n = 29). Error bars show SD. (D) Representative time points from FRAP experiments of GFP-E3KARP, E3KARP S303A, and E3KARP S303D in JEG-3 cells. Scale bars, 2 μm. (E) FLAG-tagged E3KARP, E3KARP S303A, and E3KARP S303D were immunoprecipitated and blotted for FLAG, endogenous ezrin, and E-cadherin. (F) Western blot of total lysates from JEG-3 cells stably expressing FLAG-tagged E3KARP, E3KARP S303A, and E3KARP S303D arrested in mitosis by treatment with nocodazole.
Finally, we asked whether the phosphorylation on Ser-303 is responsible for the gel shift observed in mitotic cells (Figure 4A). JEG-3 cells stably expressing 3xFLAG-E3KARP, 3xFLAG-E3KARP S303A, or 3xFLAG-E3KARP S303D were treated with nocodazole for 18 h to arrest them in mitosis, lysed in 2× sample buffer, separated by gel electrophoresis, and blotted for FLAG. Of interest, a similar mobility shift is observed for 3xFLAG-E3KARP S303A and 3xFLAG-E3KARP S303D, suggesting that both are phosphorylated at another site during mitosis, most likely at one of the other sites we identified. However, wild-type E3KARP is subjected to a greater gel shift than E3KARP S303A, showing that S303 is indeed responsible for a mitosis-specific gel shift (Figure 5F).
Phosphorylation of S303 regulates the dynamics of the E3KARP tail region
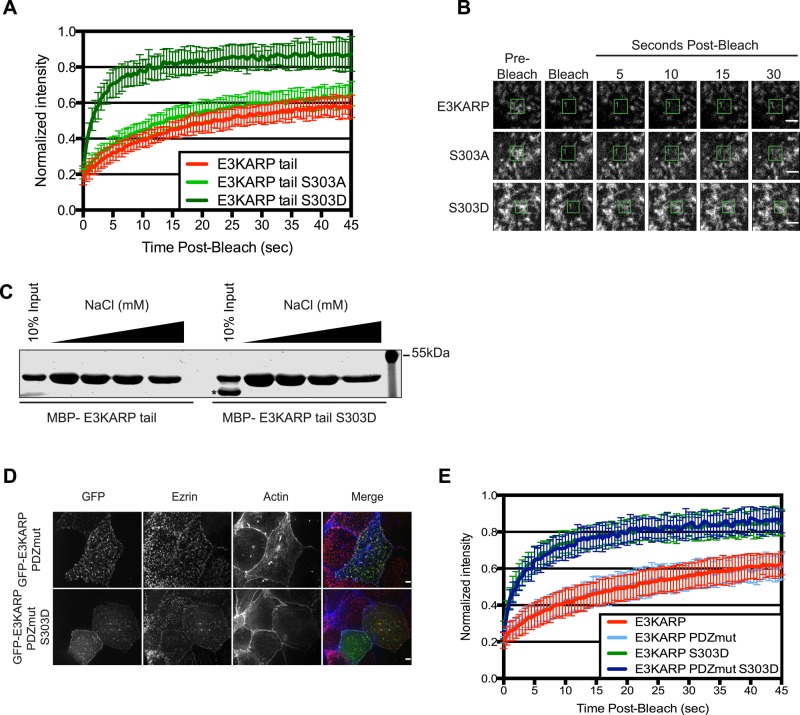
The very different dynamics of full-length EBP50 and E3KARP have been traced to the tail regions, and the tail region of EBP50 alone shows the same dynamics as the full-length protein when the PDZ domains are occupied (Garbett et al., 2013). We therefore examined the dynamics of the tail of E3KARP. FRAP experiments on cells transfected with GFP-tagged E3KARP tail wild type or S303A or S303D mutants showed that the phosphomimetic S303D mutation enhances the dynamics, whereas the nonphosphorylatable mutant has no effect (Figure 6, A and B). Of importance, the regulation of the dynamics of the tail closely reflects the dynamics of the corresponding full-length proteins.
FIGURE 6:
S303 phosphorylation of the E3KARP tail regulates its exchange rate, whereas mutation of the PDZ domains in the full-length protein does not. (A) FRAP curves of GFP-tagged E3KARP tail (n = 11), E3KARP tail S303A (n = 14), and E3KARP tail S303D (n = 14) expressed in JEG-3 cells. Errors bars show SD. (B) Representative time points from FRAP experiments of GFP-tagged E3KARP tail, E3KARP tail S303A, and E3KARP tail S303D in JEG-3 cells. Scale bars, 2 μm. (C) MBP-tagged E3KARP tails (WT or S303D) were incubated with ezrin FERM domain beads over a range of 150–1000 mM NaCl. The retained MBP-E3KARP tails were recovered and analyzed by SDS–PAGE and the gel stained with IRDye Blue. The asterisk indicates the MBP tag that was cleaved during MBP-E3KARP tail S303D purification. (D) Maximum projection images of the apical surface of JEG-3 cells expressing the indicated GFP-tagged constructs (green) and then stained for ezrin (red) and actin (blue). Scale bar, 5 μm. (E) FRAP curves of GFP-tagged E3KARP (n = 19), E3KARP PDZmut (n = 15), E3KARP S303D (n = 29), and E3KARP PDZmut S303D (n = 16) expressed in JEG-3 cells. Errors bars show SD.
The simplest explanation for the S303D mutation enhancing the dynamics of the E3KARP tail would be that it reduces the affinity of the tail for active ezrin. We therefore examined the ability of the E3KARP wild-type tail and the corresponding S303D mutant to bind immobilized ezrin FERM domain in which the E3KARP binding site is fully accessible. Maltose-binding protein (MBP) fusions of both tails bound immobilized FERM beads equivalently over a range of 150–1000 mM NaCl (Figure 6C). We conclude that the S303D mutation has no effect on the ability of the tail to bind active ezrin, and so the different dynamics seen in vivo must be due to some additional factor, most likely one involved in the binding to the S303D tail, thus destabilizing its interaction with ezrin.
The high dynamics of full-length EBP50 is regulated by occupancy of its PDZ domains: the EBP50 tail is intrinsically dynamic, and this is suppressed in the full-length protein by the presence of the PDZ domains when they cannot bind ligand; this suppression is relieved in the wild-type protein upon occupancy of the PDZ domains (Garbett and Bretscher, 2012). To see whether a similar situation exists for E3KARP, we mutated both PDZ domains to inhibit ligand binding in the context of either wild-type E3KARP or the S303D mutant. Surprisingly, mutating both PDZ domains of wild-type E3KARP had no effect on its dynamics, nor did mutating the PDZ domains of the dynamic S303D phosphomimetic mutant (Figure 6, D and E). Thus, in contrast to the situation with EBP50, E3KARP dynamics is not regulated by PDZ domain occupancy but only by phosphorylation.
In cells arrested in mitosis, E3KARP shows a fast exchange rate due to S303 phosphorylation
Our data indicate that E3KARP is phosphorylated on S303 during mitosis and that GFP-E3KARP S303D expressed in interphase cells is much more dynamic than the corresponding wild-type construct. We therefore investigated the localization and dynamics of E3KARP in mitotic cells. JEG-3 cells were transfected to express GFP-E3KARP or the S303A or S303D mutants and then arrested in mitosis by nocodazole treatment. In the rounded mitotic cells, GFP-E3KARP S303A shows a strong cortical localization. However, both GFP-E3KARP and GFP-E3KARP S303D are both largely cytoplasmic in mitotic cells, implying that S303 phosphorylation alters the localization of E3KARP (Figure 7A). FRAP analysis on these constructs shows that GFP-E3KARP S303A has a relatively slow recovery rate, similar to that seen for the wild-type protein in interphase cells, whereas E3KARP S303D and GFP-E3KARP in mitotic cells are both very dynamic with fast recovery rates (Figure 7B). We then used cells expressing 3xFLAG-E3KARP or an empty vector to examine whether the interaction between E3KARP and ezrin is compromised in mitotic cells. Immunoprecipitates of 3xFLAG-E3KARP coprecipitated endogenous ezrin, whereas when the cells were arrested in mitosis, almost no ezrin was recovered (Figure 7C). Thus E3KARP phosphorylated in mitosis behaves very similarly to the expression phosphomimetic E3KARP in interphase cells (Figure 5E). We conclude that E3KARP is phosphorylated on S303 in mitotic cells and this alters its localization and binding to ezrin and greatly enhances its dynamics.
FIGURE 7:
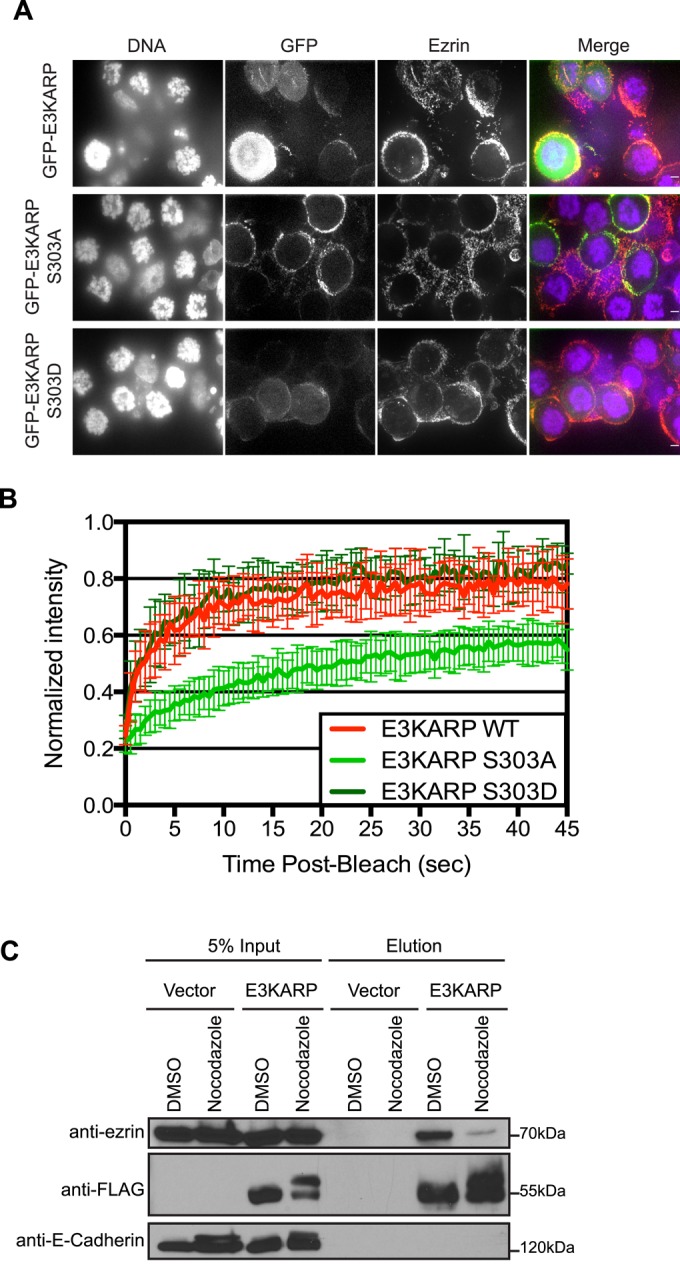
Cell cycle–dependent phosphorylation of E3KARP regulates its dynamics. (A) Maximum projection of a few slices through JEG-3 cells expressing the indicated GFP-tagged constructs (green) and arrested in mitosis by 50 ng/ml nocodazole overnight. Cells were fixed and stained for ezrin (red) and DNA (blue). Scale bar, 5 μm. (B) FRAP curves of GFP-tagged E3KARP (n = 13), E3KARP S303A (n = 16), and E3KARP S303D (n = 7) in JEG-3 cells arrested by nocodazole treatment. Errors bars show SD. (C) Empty vector FLAG or FLAG-tagged E3KARP were immunoprecipitated and blotted for FLAG and endogenous ezrin and E-cadherin in control cells or treated with nocodazole.
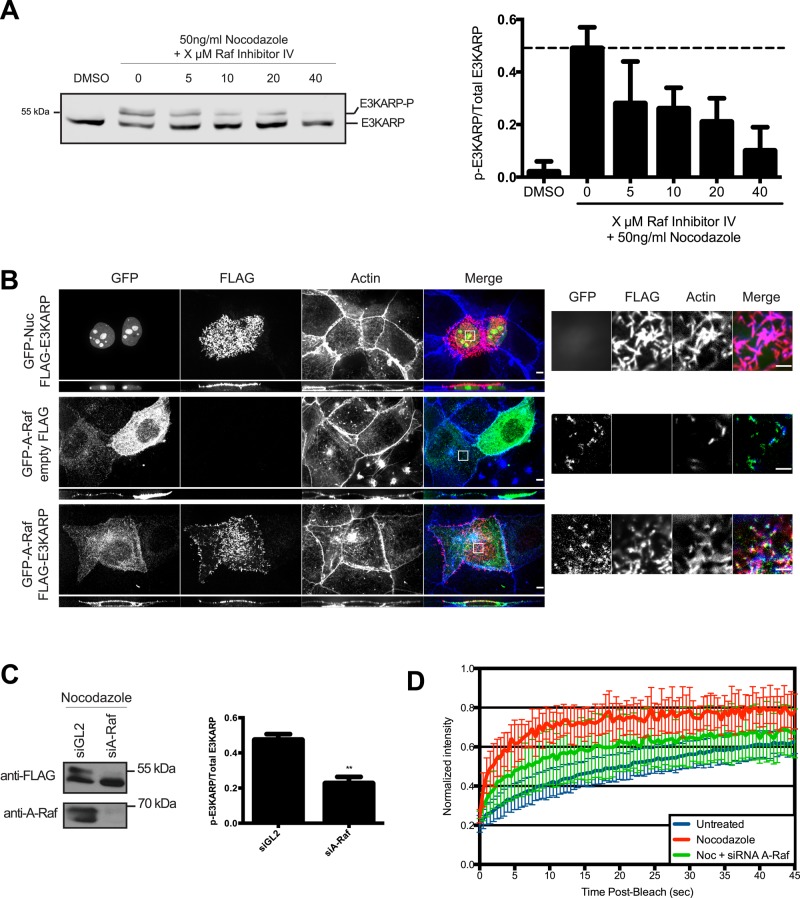
Mitotic phosphorylation of E3KARP depends on A-Raf
The sequence surrounding S303 in E3KARP does not provide a clear indication of the identity of a relevant kinase important for phosphorylation at this residue, although the presence of an adjacent proline suggests that it might be in the CDK1 family. Therefore cells were arrested in mitosis with nocodazole and the effect of kinase inhibitors on E3KARP phosphorylation assessed. Roscovitine, an inhibitor of CDK1 that phosphorylates EBP50 (Planchais et al., 1997), had no effect on E3KARP phosphorylation under conditions that eliminate EBP50 cell cycle–dependent phosphorylation (Garbett et al., 2010) Similarly, preliminary experiments with other inhibitors, including ones that affect the c-Jun amino-terminal kinase, mitogen-activated protein kinase (MAPK) kinase (MEK), and MAPK families, had no effect (Supplemental Figure S5A). However, a selective Raf kinase inhibitor reduced E3KARP phosphorylation in a dose-dependent manner (Figure 8A). The mechanism of phosphorylation is conserved among cell types, as Raf inhibitor IV treatment of Caco-2 also inhibited mitotic phosphorylation of endogenous E3KARP (Supplemental Figure S5B). Raf kinases are serine/threonine kinases downstream of Ras and upstream of the MEK/extracellular signal–regulated kinase (ERK) pathway (Cseh et al., 2014). There are three Raf kinases: A-Raf, B-Raf, and Raf-1. The Raf inhibitor IV targets A-Raf and Raf-1 kinases (Shelton et al., 2003; White, 2003). We next examined the localization of expressed GFP–A-Raf and found that it is mostly cytoplasmic but also exhibited a weak enrichment in microvilli (Figure 8B), whereas Raf-1 was cytoplasmic (unpublished data). To explore whether A-Raf might affect E3KARP phosphorylation in mitosis, we knocked down A-Raf using siRNA treatment, arrested the cells in mitosis with nocodazole, and examined the effect on E3KARP phosphorylation. We observed a strong reduction in mitotic phosphorylation of E3KARP in the absence of A-Raf (Figure 8C). Finally, we asked whether knockdown of A-Raf would affect the exchange rate of E3KARP in mitosis. First, we treated JEG-3 cells expressing GFP-E3KARP with siRNA against A-Raf for 24 h. We then arrested the cells in mitosis by incubation in nocodazole for 18 h. FRAP analysis showed that A-Raf knockdown partially reduced the exchange rate of GFP-E3KARP in mitotic cells (Figure 8D). Whether the inability to completely reverse the effect on dynamics is due to incomplete knockdown of A-Raf or another kinase that phosphorylated S303 is not clear. Nevertheless, the data suggest that A-Raf kinase contributes to cell-cycle phosphorylation of E3KARP, which enhances its dynamics.
FIGURE 8:
A-Raf contributes to the cell cycle–dependent phosphorylation of E3KARP. (A) JEG-3 cells stably expressing 3xFLAG-tagged E3KARP were treated with DMSO, nocodazole (50 ng/ml), and/or Raf inhibitor IV at the indicated concentrations overnight at 37°C. Cells were lysed, E3KARP was detected by Western blotting, and the level of phosphorylation was quantitated using Odyssey software (n ≥ 3). Errors bars show SD. (B) Maximum projection images of the apical surface of JEG-3 cells expressing the indicated constructs. Cells were stained for FLAG (red) and actin (blue). Scale bar, 5 μm. White box indicates area magnified on the right, in which a single z-plane is shown. Scale bar, 2 μm. (C) Cells stably expressing 3xFLAG-tagged E3KARP were treated with 10 nM control (siGL2) or A-Raf (siA-Raf) siRNA for 72 h. At 18 h before lysing of the cells, 50 ng/ml nocodazole was added. Cell extracts separated by SDS–PAGE were Western blotted for FLAG and A-Raf. The level of phosphorylated 3xFLAG-tagged E3KARP was determined using Odyssey software (n = 3). Errors bars show SD. **p < 0.01 (D) FRAP curves of GFP-tagged E3KARP in interphase JEG-3 cells (n = 19), cells arrested by nocodazole treatment (n = 13), and cells treated with siRNA against A-Raf and then arrested by nocodazole treatment (n = 20). Errors bars show SD.
DISCUSSION
Scaffolding proteins are generally believed to bring together two or more proteins to enhance a specific biochemical pathway. Although it is likely that scaffolding proteins are subject to dynamic regulation, little attention has been paid to this aspect of their biology (Garbett and Bretscher, 2014). In this study, we show that the apical scaffolding protein E3KARP is subject to cell-cycle phosphorylation and that this regulates its function and dynamics.
Our data from mass spectrometry indicate that there are three sites phosphorylated in mitotically arrested cells: Ser-43, -261, and -303. This parallels the finding that EBP50 is also phosphorylated in mitosis (He et al., 2001; Raghuram et al., 2003; Li et al., 2007; Garbett et al., 2010). The identification of novel phosphorylation sites in mitotically arrested cells has allowed us to investigate their functional significance. Our initial experiments led us to focus on phosphorylation of Ser-303, as the S303D mutation dramatically increased E3KARP’s dynamics in interphase cells. Ser-303 lies just upstream of the C-terminal ezrin-binding site, so our initial thought was that it might reduce the affinity of E3KARP for active ezrin.
We found that the E3KARP tail alone, like the full-length protein, is intrinsically not dynamic but becomes dynamic upon S303 phosphorylation, with the PDZ domains playing little or no role in regulating the dynamics. This is in contrast to EBP50, in which the tail is intrinsically dynamic, with the unoccupied PDZ domains suppressing the dynamics, and this suppression is relieved by PDZ ligand binding (Garbett and Bretscher, 2012). Furthermore, in vitro analyses of E3KARP revealed that the interaction with active ezrin is unperturbed even when E3KARP is highly dynamic, indicating that in vivo dynamics is mediated by unknown cytoplasmic proteins. It is important to note that in vitro, pure EBP50 and E3KARP both exist as monomers in solution (Garbett et al., 2010; this study) and do not oligomerize through their PDZ domains as had been suggested (Lau and Hall, 2001), so the regulation of their dynamics conferred by the tail regions is unlikely to be explained by oligomerization status.
Phylogenetic analysis reveals that EBP50 and E3KARP arose by gene duplication followed by sequence divergence in their tails. The regions in the tail responsible for regulation of their dynamics are especially divergent, but in the case of E3KARP, for which only one residue is involved, this residue is highly conserved among vertebrate homologues. Therefore it appears that the regulation of the dynamics of EBP50 and E3KARP evolved independently, which is consistent with the mechanistic differences in regulation. We found that the longer EBP50 tail is a result of an insertion of 20 amino acids and that this insertion is in the region identified as necessary for conferring the higher dynamic of EBP50. Because EBP50 and E3KARP show specific and mutually exclusive tissue distributions (Ingraffea et al., 2002), the modes of regulation are presumably reflective of their different functions.
Interphase cells, in which EBP50 has been depleted, had greatly reduced microvilli, which were restored upon expression of GFP-E3KARP. Thus E3KARP can provide a function normally performed by EBP50, despite the differences in their dynamics. Expression of GFP-E3KARP with the phosphomimetic S303D mutation displayed an impaired ability to restore microvilli in EBP50-depleted cells. It is likely that the S303D mutation is a good mimic for S303 phosphorylation, as its dynamics is indistinguishable in mitotic cells from that of E3KARP phosphorylated in vivo on S303. Thus phosphorylation of S303 enhanced the dynamics of E3KARP and compromised its function in microvilli biogenesis. By contrast, GFP-EBP50 was highly dynamic after expression in EBP50-depleted cells, restoring microvilli, thereby indicating that enhanced dynamics alone does not necessarily impair the microvillar functions of these scaffolding proteins but may reflect the mechanistically different ways the dynamics is regulated.
Using several different approaches, we found that A-Raf contributes to S303 phosphorylation of E3KARP. A-Raf is the least-studied member of the three members of the Raf serine/threonine kinase family, with much more known about B-Raf and Raf-1 (Roskoski, 2010; Cseh et al., 2014). These kinases are upstream of the MEK-ERK cascade and are regulated by phosphorylation and subsequent binding to 14-3-3 proteins (Fischer et al., 2009; Roskoski, 2010). For example, 14-3-3 proteins can bind and activate Raf-1 at the G2/M transition (Hayne et al., 2000). We do not know whether A-Raf is selectively activated during mitosis, which will be an interesting topic for future studies. Our results with MEK-ERK inhibitors suggest that they are not involved in E3KARP phosphorylation; thus E3KARP may be a direct substrate of A-Raf or of an unknown downstream kinase. However, knockdown of A-Raf only partially eliminated the enhanced dynamics of E3KARP seen in mitotic cells. It is therefore likely that another kinase contributes to S303 phosphorylation in mitosis to regulate E3KARP’s dynamics. It will be interesting to understand the implication of E3KARP phosphorylation during the cell cycle. Because the actin cytoskeleton has been involved in membrane tension regulation during the cell cycle to prevent endocytosis (Boulant et al., 2011; Kaur et al., 2014), it will also be interesting to examine how E3KARP is involved in this mechanism.
In summary, we have uncovered a new mechanism regulating the apical scaffolding protein E3KARP during the cell cycle. The phosphoregulation of E3KARP brings specific properties to the protein that have not been explored before. Together with our earlier studies on EBP50, our results highlight how two homologous proteins can provide similar functions despite having unique mechanisms of regulation.
MATERIALS AND METHODS
Antibodies and reagents
Antisera and affinity-purified antibodies against human ezrin, E3KARP and EPI64 have been described (Bretscher, 1989; Ingraffea et al., 2002; Hokanson and Bretscher, 2012). The mouse anti-FLAG, mouse anti–E-cadherin, and rabbit anti–A-Raf (C20) antibodies were purchased from Sigma-Aldrich (St. Louis, MO), BD Biosciences (Franklin Lakes, NJ), and Santa Cruz Biotechnology (Dallas, TX), respectively. Goat anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidase were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and MP Biomedicals (Solon, OH), respectively. Hoechst 32258 DNA stain, Alexa Fluor 568 donkey anti-rabbit antibody, and Alexa Fluor 660–conjugated phalloidin were purchased from Invitrogen (Life Technologies, Grand Island, NY). IRDye 680– and 800–conjugated secondary antibodies were from LI-COR Biosciences (Lincoln, NE). Dimethyl sulfoxide (DMSO) and roscovitine were obtained from Sigma-Aldrich. Raf Inhibitor IV was obtained from Santa Cruz Biotechnology. Sorafenib, PLX 4720, U0126, and doramapimod were obtained from Cayman Chemical (Ann Arbor, MI). PD 98058 was obtained from Calbiochem (EMD Millipore, Billerica, MA). The siRNAs targeting human EBP50 (5′-CGGCGAAAACGTGGAGAAG-3′) and luciferase GL2 (5′-CGUACGCGGAAUACUUCGA-3′) were obtained from Thermo Fisher Scientific and Applied Biosystems (Lafayette, CO). The siRNA targeting human A-Raf (5′-ACCGAGAUCUCAAGUCUAAUU-3′) was obtained from Dharmacon (GE Healthcare, Pittsburgh, PA).
Cell culture and transfection
JEG-3 cells, Caco-2 cells, and Phoenix-AMPHO (American Type Culture Collection, Manassas, VA) were maintained in a 5% CO2 humidified atmosphere at 37°C in MEM with 10% fetal bovine serum (FBS) or DMEM with 10% FBS (MEM and DMEM from Thermo Fisher Scientific; FBS from Invitrogen). JEG-3 cells were transfected with polyethylenimine (PE; Polysciences, Warrington, PA) and DNA as described previously (Hanono et al., 2006). Caco-2 were transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. For microvillar rescue assays, cells were first transfected with 1–2 μg of DNA at low confluence, allowed to recover for 24 h, and then transfected again with 10 nM of siRNA using Lipofectamine RNAiMAX (Life Technologies). Cells were then allowed to grow for another 48 h and then processed for immunofluorescence and counting. For mitotic arrest studies, JEG-3 and Caco-2 cells were incubated with 50 ng/ml and 1 μg/ml nocodazole (Sigma-Aldrich), respectively, for 18–20 h before preparation for analysis. For siRNA A-Raf assays, cells were transfected with 10 nM siRNA using Lipofectamine RNAiMAX at low confluence, allowed to recover for 48 h, and then treated with 50 ng/ml nocodazole. At 72 h after siRNA transfection, cells were lysed in 2× sample buffer. To analyze the effect of A-Raf knockdown on the dynamics of E3KARP in mitosis, JEG-3 cells were transfected with PE and 1 μg of DNA to express GFP-E3KARP and with 10 nm siRNA to A-Raf using Lipofectamine RNAiMAX for 24 h. The cells were then arrested in mitosis by 18-h nocodazole treatment and subjected to FRAP analysis.
For generation of stable JEG-3 cell lines expressing 3xFLAG-E3KARP, Phoenix-AMPHO cells were cotransfected with the foregoing construct in pQCXIP in addition to a plasmid encoding VSV-G using PE. The resulting retroviruses were then used to infect JEG-3 cells, which were then selected and maintained with 2μg/ml puromycin (Sigma-Aldrich).
DNA constructs
All EBP50 and E3KARP constructs were created in pEGFP-C2 (Takara Bio, Otsu, Japan) and pE-SUMO (LifeSensors, Malvern, PA). The 3xFLAG tagged E3KARP construct was created using PCR and inserted into pQCXIP (BD Biosciences; Garbett et al., 2013). All point mutations in pEGFP-E3KARP were generated using the mutagenesis kit (QuikChange; Agilent Technologies, Santa Clara, CA).
Immunoprecipitation and Western blotting
JEG-3 cells transiently expressing 3xFLAG-E3KARP, 3xFLAG-E3KARP S303A, and 3xFLAG-E3KARP S303D constructs were lysed in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1% IGEPAL-630, 50 mM NaF, 0.1 mM Na3VO4, 10 mM β-glycerol phosphate, and 2.5% glycerol) and incubated with anti-FLAG M2 affinity resin (Sigma-Aldrich) while nutating at 4°C for 2 h. The resin was then washed three times in lysis buffer with 0.1% IGEPAL-630, and the remaining bound material was eluted using 3xFLAG-peptide (Sigma-Aldrich) for 30 min at room temperature. Then protein samples were boiled in reducing sample buffer. Boiled protein samples were then separated by SDS–PAGE, transferred to Immobilon-FL (EMD Millipore) for Western blotting, and then visualized using ECL (GE Healthcare) or an Odyssey infrared imaging system (LI-COR Biosciences).
SILAC and mass spectrometry
For SILAC, JEG-3 stable cell lines expressing 3xFLAG-E3KARP were grown for ∼3 wk in MEM (Thermo Fisher Scientific) containing dialyzed FBS (Invitrogen) and either C12-arginine and lysine or C13-arginine and lysine (Sigma-Aldrich), respectively. FLAG immunoprecipitations were as described, with slight modifications for mass spectrometry processing, as described previously (Smolka et al., 2007; Viswanatha et al., 2012). Briefly, after immunoprecipitation, bound protein was eluted in 50 mM Tris (pH 8.0) and 1% SDS and then precipitated with 50% ethanol, 49.9% acetone, and 0.1% acetic acid. Protein samples were then mixed, trypsin digested (Promega, Madison, WI), and desalted in a C18 column (Waters, Milford, MA). The tryptic peptides were dehydrated in a speed vacuum and then resuspended in 1% acetic acid. Phosphopeptides were enriched by IMAC as previously described (Smolka et al., 2007; Albuquerque et al., 2008; Ohouo et al., 2013), reconstituted in 85 μl of solution containing 80% acetonitrile and 1% formic acid, and fractionated by hydrophilic interaction liquid chromatography. The resulting fractions were injected into a mass spectrometer (Qexactive LC-MS/MS; Thermo Fisher Scientific) and the data analyzed using Proteome Discoverer (Thermo Fisher Scientific).
Immunofluorescence
For immunofluorescence, cells grown on glass coverslips were fixed in 3.7% formaldehyde/phosphate-buffered saline (PBS) for 10 min at room temperature. Cells were permeabilized in 0.2% Triton X-100/PBS for 5 min at room temperature, rinsed in PBS, and incubated in primary antibodies in PBS containing 3% FBS for 1 h at room temperature. After washing in PBS, secondary antibodies and additional markers (phalloidin, DNA counterstain) were added in 3% FBS in PBS for 1 h at room temperature. After further washing in PBS, coverslips were mounted onto glass slides using Vectashield (H-1000; Vector Laboratories, Burlingame, CA), and images were then acquired on a CSU-X spinning disk microscope (Intelligent Imaging Innovations, Santa Monica, CA) with spherical aberration correction device, 100×/1.46 numerical aperture (NA) objective on an inverted microscope (DMI6000B), and an HQ2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) using SlideBook 5.5 (Intelligent Imaging Innovations, Denver, CO). Images were also processed using SlideBook 5.5 and Photoshop (Adobe, San Jose, CA).
Scoring the apical surface phenotype of transfected cells
Transfected JEG-3 cells were scored as described previously (Hanono et al., 2006; Garbett et al., 2010). In brief, cells were stained for ezrin and F-actin to indicate the apical membrane phenotype, whereas the GFP-tagged constructs marked transfected cells. At least 250 cells were counted for each condition in each of three replicates. Cells were scored as having normal microvilli or no microvilli.
Live imaging and FRAP
Transfected cells grown in 35-mm glass-bottom dishes (MatTek, Ashland, MA) were washed in PBS and then maintained in low–sodium bicarbonate, phenol red–free MEM (Sigma-Aldrich) supplemented with 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.4), 10% FBS, and GlutaMAX (Invitrogen). Live cells were imaged by time-lapse microscopy on a spinning disk (CSU-X; Yokogawa, Tokyo, Japan) with a spherical aberration correction device, a 100×/1.46 NA objective (Leica, Wetzlar, Germany) on an inverted microscope (DMI6000B; Leica), and an electron-multiplied CCD camera (QuantEM; Photometrics) at 37°C in an environmental chamber (Okolab, Ottaviano, Italy) controlled by SlideBook 5.5. Regions selected for FRAP were illuminated with a point-scanner galvanometer-based system (Vector; Intelligent Imaging Innovations) coupled to a 473-nm, 50-mW diode-pumped solid-state laser. For all FRAP experiments, an independent region was also monitored to control for photobleaching during the observation period. Movies were processed using SlideBook and analyzed using Excel (Microsoft, Redmond, WA) and Prism (GraphPad, La Jolla, CA). The normalized data from multiple biological replicates were subjected to a two-way analysis of variance using Prism.
Purification of recombinant proteins
SUMO-E3KARP constructs were purified as described previously (LaLonde et al., 2010). In brief, induced bacterial pellets were lysed in binding buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4, and 1% Triton X-100) by sonication, centrifuged, and run over a His GraviTrap column (GE Healthcare), washed, and eluted in elution buffer (20 mM sodium phosphate, 500 mM NaCl, and 500 mM imidazole, pH 7.4). Eluate was dialyzed into binding buffer, cleaved with 6xHis-Ulp1 for 40 min at 30°C to remove the SUMO tag, and then run over another His GraviTrap column, and flowthrough containing untagged EBP50 was collected and dialyzed into 150 mM NaCl and 10 mM Tris, pH 7.4, with 1 mM dithiothreitol (DTT).
MBP fused to the last 38 amino acids of the E3KARP tails with various mutations was purified as described previously (Finnerty et al., 2004). In brief, induced cells were lysed in PBS with 1 mM DTT and complete protease inhibitor by sonication, centrifuged, and run over amylose resin column (New England Biolabs, Ipswich, MA), washed with PBS, and then eluted in PBS with 10 mM maltose.
Gel filtration and sucrose gradients
From 1 to 2 mg of untagged full-length E3KARP was run over a Superdex 200 10/300GL column on an AKTA FPLC (GE Healthcare) in 150 mM NaCl and 10 mM Tris, pH 7.4, with 1 mM DTT. Stokes radii were calculated as described previously (Begg et al., 2001). Full-length E3KARP was run on 5–20% sucrose gradients with size standards bovine serum albumin (4.3S), carbonic anhydrase (3.2S), and myoglobin (2.04S) in a rotor (SW60Ti; Beckman Coulter, Brea, CA) at 50,000 rpm for 26 h at 4°C. Fractions were collected manually, run out by SDS–PAGE, and stained with IRDye blue protein stain (LI-COR Biosciences) for band densitometry and sedimentation coefficient calculation. Frictional ratios were calculated from ƒ/ƒ0 = RS(4πN/3vMr)1/3, where RS is the experimentally determined Stokes radius, N is Avogadro’s number, v is the partial specific volume (taken to be 0.74 cm3/g for protein), and Mr is the calculated molecular mass.
In vitro binding assays
For binding of MBP-tagged E3KARPtail to ezrin-FERM, 20 μg of MBP-E3KARP tail constructs was incubated with 7 μl of ezrin FERM-CNBr bead slurry (coupled to be 2 μg/μl on the beads) in a range of 150–1000 mM NaCl, 0.1% Triton X-100, 5% glycerol, and 20 mM Tris, pH 7.4, for 1 h at 4°C. After washing, the bound protein was eluted by boiling in SDS sample buffer. Samples were run on SDS–PAGE, stained with IRDye blue protein stain, and scanned using an Odyssey infrared imaging system.
Phylogenetic analysis
The phylogenic tree was generated using the database EggNOG on the metazoan clades of E3KARP (Powell et al., 2012) and the online software iTol (Letunic and Bork, 2011). All alignments were generated using Clustal Omega and processed for publication using Jalview (Waterhouse et al., 2009).
Supplementary Material
Acknowledgments
We are very grateful to Francisco Bastos de Oliveira and Marcus Smolka (Cornell University) for help with the mass spectrometry. We also thank members of the Bretscher laboratory for critically reading the manuscript and Benoît Castandet (Boyce Thompson Institute, Ithaca, NY) for help with phylogenetic analysis. This work was supported by National Institutes of Health Grant GM036552.
Abbreviations used:
- E3KARP
NHE3 kinase A regulatory protein
- EBP50
ERM-binding protein of 50 kDa
- EPI64
EBP50 PDZ interactor of 64 kDa
- ERM
ezrin/radixin/moesin
- FERM
4.1 ERM
- FRAP
fluorescence recovery after photobleaching
- NHERF
Na+-H+ exchanger-3 regulatory factor
- PDZ
postsynaptic density 95/disks large/zona occludens-1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0498) on August 26, 2015.
REFERENCES
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg GE, Harper SL, Speicher DW. Characterizing recombinant proteins using HPLC gel filtration and mass spectrometry. Curr Protoc Protein Sci. 2001;Chapter 7, Unit 7:10. doi: 10.1002/0471140864.ps0710s16. [DOI] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh J-C, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TT, Deacon HW, Reczek D, Bretscher A, von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- Cha B, Kim JH, Hut H, Hogema BM, Nadarja J, Zizak M, Cavet M, Lee-Kwon W, Lohmann SM, Smolenski A, et al. cGMP inhibition of Na+/H+ antiporter 3 (NHE3) requires PDZ domain adapter NHERF2, a broad specificity protein kinase G–anchoring protein. J Biol Chem. 2005;280:16642–16650. doi: 10.1074/jbc.M500505200. [DOI] [PubMed] [Google Scholar]
- Cseh B, Doma E, Baccarini M. “RAF” neighborhood: protein-protein interaction in the Raf/Mek/Erk pathway. FEBS Lett. 2014;588:2398–2406. doi: 10.1016/j.febslet.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco SJ, Chicka MC, Strehler EE. Plasma membrane Ca2+ ATPase isoform 2b interacts preferentially with Na+/H +exchanger regulatory factor 2 in apical plasma membranes. J Biol Chem. 2002;277:10506–10511. doi: 10.1074/jbc.M111616200. [DOI] [PubMed] [Google Scholar]
- Donowitz M, Cha B, Zachos NC, Brett CL, Sharma A, Tse CM, Li X. NHERF family and NHE3 regulation. J Physiol. 2005;567:3–11. doi: 10.1113/jphysiol.2005.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117:1547–1552. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- Fischer A, Baljuls A, Reinders J, Nekhoroshkova E, Sibilski C, Metz R, Albert S, Rajalingam K, Hekman M, Rapp UR. Regulation of RAF activity by 14-3-3 proteins: RAF kinases associate functionally with both homo- and heterodimeric forms of 14-3-3 proteins. J Biol Chem. 2009;284:3183–3194. doi: 10.1074/jbc.M804795200. [DOI] [PubMed] [Google Scholar]
- Garbett D, Bretscher A. PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J Cell Biol. 2012;198:195–203. doi: 10.1083/jcb.201204008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Bretscher A. The surprising dynamics of scaffolding proteins. Mol Biol Cell. 2014;25:2315–2319. doi: 10.1091/mbc.E14-04-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, LaLonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Sauvanet C, Viswanatha R, Bretscher A. The tails of apical scaffolding proteins EBP50 and E3KARP regulate their localization and dynamics. Mol Biol Cell. 2013;24:3381–3392. doi: 10.1091/mbc.E13-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Premont RT, Chow CW, Blitzer JT, Pitcher JA, Claing A, Stoffel RH, Barak LS, Shenolikar S, Weinman EJ, et al. The beta2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H +exchange. Nature. 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- Hall RA, Spurney RF, Premont RT, Rahman N, Blitzer JT, Pitcher JA, Lefkowitz RJ. G protein-coupled receptor kinase 6A phosphorylates the Na(+)/H(+) exchanger regulatory factor via a PDZ domain-mediated interaction. J Biol Chem. 1999;274:24328–24334. doi: 10.1074/jbc.274.34.24328. [DOI] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayne C, Tzivion G, Luo Z. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J Biol Chem. 2000;275:31876–31882. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- He J, Lau AG, Yaffe MB, Hall RA. Phosphorylation and cell cycle-dependent regulation of Na+/H +exchanger regulatory factor-1 by Cdc2 kinase. J Biol Chem. 2001;276:41559–41565. doi: 10.1074/jbc.M106859200. [DOI] [PubMed] [Google Scholar]
- Hernando N, Déliot N, Gisler SM, Lederer E, Weinman EJ, Biber J, Murer H. PDZ-domain interactions and apical expression of type IIa Na/P(i) cotransporters. Proc Natl Acad Sci USA. 2002;99:11957–11962. doi: 10.1073/pnas.182412699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Bretscher AP. EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol Biol Cell. 2012;23:701–715. doi: 10.1091/mbc.E11-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraffea J, Reczek D, Bretscher A. Distinct cell type-specific expression of scaffolding proteins EBP50 and E3KARP: EBP50 is generally expressed with ezrin in specific epithelia, whereas E3KARP is not. Eur J Cell Biol. 2002;81:61–68. doi: 10.1078/0171-9335-00218. [DOI] [PubMed] [Google Scholar]
- Kaur S, Fielding AB, Gassner G, Carter NJ, Royle SJ. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. Elife. 2014;3:e00829. doi: 10.7554/eLife.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurashima K. The apical Na+/H+ exchanger isoform NHE3 is regulated by the actin cytoskeleton. J Biol Chem. 1999;274:29843–29849. doi: 10.1074/jbc.274.42.29843. [DOI] [PubMed] [Google Scholar]
- LaLonde DP, Garbett D, Bretscher A. A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol Biol Cell. 2010;21:1519–1529. doi: 10.1091/mbc.E10-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AG, Hall RA. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry. 2001;40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Poulikakos PI, Dai Z, Testa JR, Callaway DJ, Bu Z. Protein kinase C phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J Biol Chem. 2007;282:27086–27099. doi: 10.1074/jbc.M702019200. [DOI] [PubMed] [Google Scholar]
- Li Y, Li J, Straight SW, Kershaw DB. PDZ domain-mediated interaction of rabbit podocalyxin and Na(+)/H(+) exchange regulatory factor-2. Am J Physiol Renal Physiol. 2002;282:F1129–F1139. doi: 10.1152/ajprenal.00131.2001. [DOI] [PubMed] [Google Scholar]
- Lohi H, Lamprecht G, Markovich D, Heil A, Kujala M, Seidler U, Kere J. Isoforms of SLC26A6 mediate anion transport and have functional PDZ interaction domains. Am J Physiol Cell Physiol. 2003;284:C769–C779. doi: 10.1152/ajpcell.00270.2002. [DOI] [PubMed] [Google Scholar]
- Mahon MJ, Donowitz M, Yun CC, Segre GV. Na(+)/H(+) exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature. 2002;417:858–861. doi: 10.1038/nature00816. [DOI] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Liu Y, Ma CJ, Smolka MB. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature. 2013;493:120–124. doi: 10.1038/nature11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S, Glab N, Tréhin C, Perennes C, Bureau JM, Meijer L, Bergounioux C. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram V, Hormuth H, Foskett JK. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc Natl Acad Sci USA. 2003;100:9620–9625. doi: 10.1073/pnas.1633250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J Cell Biol. 2001;153:191–206. doi: 10.1083/jcb.153.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399:313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- Shelton JG, Moye PW, Steelman LS, Blalock WL, Lee JT, Franklin RA, McMahon M, McCubrey JA. Differential effects of kinase cascade inhibitors on neoplastic and cytokine-mediated cell proliferation. Leukemia. 2003;17:1765–1782. doi: 10.1038/sj.leu.2403052. [DOI] [PubMed] [Google Scholar]
- Short DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, Stutts MJ, Milgram SL. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen S, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Hug MJ, Lewarchik CM, Yun C-HC, Bradbury NA, Frizzell RA. E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells. J Biol Chem. 2000;275:29539–29546. doi: 10.1074/jbc.M004961200. [DOI] [PubMed] [Google Scholar]
- Viswanatha R, Ohouo PY, Smolka MB, Bretscher A. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J Cell Biol. 2012;199:969–984. doi: 10.1083/jcb.201207047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman EJ, Steplock D, Shenolikar S. CAMP-mediated inhibition of the renal brush border membrane Na+-H+ exchanger requires a dissociable phosphoprotein cofactor. J Clin Invest. 1993;92:1781–1786. doi: 10.1172/JCI116767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman EJ, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na(+)-H +exchanger. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman EJ, Wang Y, Wang F, Greer C, Steplock D, Shenolikar S. A C-terminal PDZ motif in NHE3 binds NHERF-1 and enhances cAMP inhibition of sodium-hydrogen exchange. Biochemistry. 2003;42:12662–12668. doi: 10.1021/bi035244l. [DOI] [PubMed] [Google Scholar]
- White MK. Small-molecule inhibitors of signal transduction pathways in leukemia therapeutics: how to assess selectivity for malignant signals. Leukemia. 2003;17:1759–1761. doi: 10.1038/sj.leu.2403053. [DOI] [PubMed] [Google Scholar]
- Yang J, Singh V, Cha B, Chen T-E, Sarker R, Murtazina R, Jin S, Zachos NC, Patterson GH, Tse CM, et al. NHERF2 protein mobility rate is determined by a unique C-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells. J Biol Chem. 2013;288:16960–16974. doi: 10.1074/jbc.M113.470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CC. The serum and glucocorticoid-inducible kinase SGK1 and the Na+/H+ exchange regulating factor NHERF2 synergize to stimulate the renal outer medullary K+ channel ROMK1. J Am Soc Nephrol. 2002;13:2823–2830. doi: 10.1097/01.asn.0000035085.54451.81. [DOI] [PubMed] [Google Scholar]
- Yun CH, Lamprecht G, Forster DV, Sidor A. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J Biol Chem. 1998;273:25856–25863. doi: 10.1074/jbc.273.40.25856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.