Abstract
Numerous studies indicate that p300 acts as a key transcriptional cofactor in vivo, at least, in part, through modulating activities of p53 by acetylation. Nevertheless, the regulation of the p53-p300 interplay is not completely understood. Here, we have identified the DEAD box RNA helicase DDX24 as a novel regulator of the p300-p53 axis. We found that DDX24 interacts with p300, and this interaction leads to suppression of p300 mediated acetylation of p53. Notably, RNAi-mediated knockdown of endogenous DDX24 significantly increases the acetylation levels of endogenous p53 in human cancer cells and subsequently promotes p53-mediated activation of its transcriptional targets such as p21 and PUMA. In contrast, DDX24 expression inhibits the p300-p53 interaction and suppresses p300-mediated acetylation of p53. Moreover, DDX24 is overexpressed in human cancer cells and reduction of DDX24 protein levels by RNAi induces cell cycle arrest and senescence in a p53 dependent manner. These results reveal DDX24 as an important regulator of p300 and suggest that the modulation of the p53-p300 interplay by DDX24 is critical in controlling p53 activities in human cancer cells.
Keywords: DDX24, p300, p53, senescence
Introduction
The p53 tumor suppressor protein provides powerful intrinsic defense mechanism against cancer, and mutations in p53 are the most frequent genetic alterations in human cancers29. p53 is kept in a latent form under normal condition until cells encounter stresses. The various stimuli from intrinsic and extrinsic environment lead to p53 activation and the activated p53 in turn controls cell cycle arrest, apoptosis, senescence, and other responses yet to be identified to obstruct tumor cell growth. Although the precise mechanisms of p53 activation are not fully understood, they are generally believed to involve post-translational modifications, of which p53 acetylation has been most studied and proved indispensable for p53 transcriptional activity8, 24. Most strikingly, when all the major acetylation lysine sites of p53 were mutated to acetylation-defective arginine, p53 mediated classic cellular functions were entirely abrogated39. Acetylation provides multifaceted mechanism for p53 activation. This modification impairs the interaction between p53 and MDM2, the main negative regulatory E3 ubiquitin ligase of p53, to relieve both ubiquitin-dependent and ubiquitin-independent repression mediated by MDM2 from p538, 35. p53 acetylation also augments DNA binding affinity and possibly recruits cofactors to enable promoter-specific p53 transcriptional activity4, 8, 38.
p53 acetylation occurs on multiple lysine sites and are catalyzed by Histone Acetyl Transferases(HATs) including structurally related p300 and CREB-Binding Protein (CBP), p300/CBP-Associated Factor (PCAF), Tat-Interactive Protein of 60 kDa (TIP60), and human males absent On the First (hMOF)8. p300/CBP, the first identified p53 acetyl transferase, has been proved a critical one in catalyzing multiple sites including six lysines (370, 372, 373, 381, 382, and 386) clustered at the C-terminus and lysine 164 in the DNA binding domain of p5318, 39. In addition, p300/CBP targets many other transcriptional factors, such as GATA-1 and c-Myc, and histones for acetylation to modulate global transcription activities20, 41. The critical role of p300/CBP in the regulation of global gene transcription was demonstrated by the embryonic lethality of conventional p300 or CBP knockout mice44. p300/CBP has been implicated in cancer development as many of its substrates are cancer molecules. However, the exact role has been under debate for quite a long time due to the limited genetic evidence. p300 promotes cancer cell proliferation by progressing G1/S transition and the downregulation of p300 results in growth inhibition21. Conversely, p300 mutations, albeit at low frequency, in colorectal and breast cancers, support p300 as a classic tumor suppressor gene 16, 20. Until recently, the tumor suppressor role of p300 has been substantiated by the evidence from large scale tumor sequencing projects. p300 and CBP gene mutations, especially mutations clustered around the sequence encoding the HAT domain have been frequently found in multiple types of human tumors including non-small cell lung cancers, acute lymphoblastic leukemia, and B-cell lymphoma27, 31, 33. Presumably, p300 behaves as a tumor suppressor protein through the acetylation of its HAT substrates involved in cancer development, particularly, tumor suppressor proteins such as p53. In fact, the molecular network surrounding the p300-p53 axis remains not fully understood as p300 often exists in multicomponent complex. It is critical to resolve the molecular complexity of p300 complex to allow comprehensive understanding of the tumor suppressive role of p300 and how it attributed to p53 function 36. Here we show that a novel interacting protein of p300, DDX24, identified from purified p300-complex, acts as a negative regulator of p53.
DDX24 is a family member of the DEAD box containing RNA helicases46. DEAD-box proteins are a large group of putative RNA helicases and have been characterized in a variety of organisms ranging from bacteria to humans46. The DEAD-box RNA helicases share nine conserved motifs in core region and one of the motifs bears the sequence Asp-Glu-Ala-Asp (DEAD) that inspires the family name19. The characteristic functions of this family are involved in RNA metabolic processes, such as transcription, pre-mRNA processing, RNA degradation, RNA export, ribosomal biogenesis, and translation5, 15. However, relatively few members have been documented to be truly RNA helicases13. The physiological functions and regulatory data are presently lacking for most of the DEAD box proteins13, 15. Emerging evidence has started to link DEAD box proteins to cancer development and progression1, 15, 19. Several DEAD box proteins appear aberrantly expressed in cancer tissues and are reported to be involved with other cancer proteins or pathways1, 7, 12, 14, 15.
In this report, we identified DEAD-box protein DDX24 as a negative regulator of tumor suppressor protein p53. Endogenous DDX24 and p300 co-immunoprecipitate from nuclear extracts and DDX24 antagonizes the acetylation activity of p300 towards p53. RNAi knockdown of DDX24 significantly elevates p53 acetylation level and subsequently p53 transcriptional activity with the most prominent effect on the p53 target gene p21. Consequently, DDX24 deficiency in human cancer cells leads to G1/S cell cycle arrest and ultimately triggers senescence. Of note, the frequent overexpression of DDX24 protein throughout breast tumor cell lines suggests a potential oncogenic role of DDX24 in breast cancer tumorigenesis.
Results
Identification of DDX24 as a component in p300 protein complex
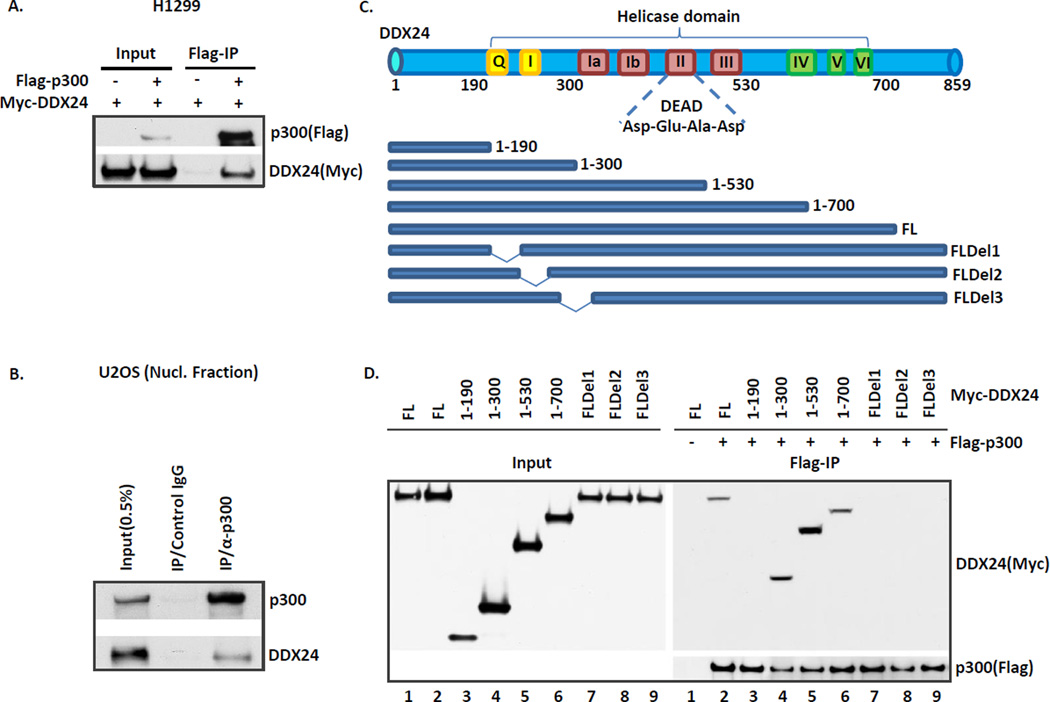
We previously made the initial efforts to explore endogenous p300 complex extracted from Hela cells and the following mass spectrometry analysis identified peptides specific for DDX24, an uncharacterized DEAD box RNA helicase, as one of the components in p300 protein complex 23. However, no further exploration has been made on this putative novel interaction. We first sought to confirm the interaction by performing co-immunoprecipitation (Co-IP) assays in H1299 cells transfected with Flag-p300 and/or Myc-DDX24 expression vectors. The western blot analysis of M2/Flag immunoprecipitates revealed that Myc-DDX24 specifically appeared in the Flag-p300 immunoprecipitates (Fig. 1A). To further examine the interaction of endogenous DDX24 and p300, we used U2OS osteosarcoma cells and performed p300 or control IgG immunoprecipitation in nucleic fractions as both p300 and DDX24 predominantly resides in the nucleus. As expected, DDX24 readily co-precipitated with p300 (Fig. 1B). Taken together, these data confirmed that DDX24 is a bona fide binding partner of p300.
Figure 1. Identification of DDX24 as a component in p300 protein complex.
(A) Myc-DDX24 co-immunoprecipitates with Flag-p300 in an overexpression system. H1299 cells were transfected with Flag-p300 and Myc-DDX24. The whole cell extracts or Flag-M2 immunoprecipitates from H1299 cells were subjected to Western blot analysis with α-Flag and α-Myc antibodies. (B) Endogenous co-immunoprecipitation of DDX24 with p300 from U2OS cells. U2OS nuclear extracts were immunoprecipitated with IgG or α-p300 antibody and subjected with lysate input to Western blot with α-p300 and α-DDX24 antibodies. (C) Schematic representation of DDX24 truncation and deletion mutants used in interaction domain mapping. Full length DDX24 and all the DDX24 mutants were subcloned into PCMV-Myc expression vector. FL, full length; FLDel1, Δ(193-214)aa; FLDel2, Δ(215-239)aa; FLDel3, Δ(240-264aa). (D) DDX24 interacts with p300 through its N-terminal fragment. H1299 cells were transfected with Myc-tagged DDX24 full length or mutant constructs. The cell lysates were incubated with Flag-M2 agarose beads bound with purified Flag-tagged p300 protein. The M2 immunoprecipitates were recovered by Flag peptide and analyzed by Western blot with α-Flag and α-Myc antibodies.
By sequence alignment, we characterized that the multiple-motif helicase domain of DDX24 clustered in a central core region and encompasses amino acids 200 to 700 (Fig.1C). Both amino- and carboxyl-terminal fragments flanking the core domain of DDX24 are highly variable and unique regions, which are presumably regulatory domains for post-translational modifications and protein-protein interaction19. We performed an extended analysis to map the p300 interacting domain on DDX24 by generating a series of Myc-tagged DDX24 truncation mutants as diagrammed in Figure 1C. Flag/M2 agarose-conjugated p300 were incubated with DDX24 mutants-expressing H1299 cell lysates. The first 300 amino acids in DDX24, which includes the N-terminal flexible sequence and the first two conserved motifs, Q-motif and motif I, in helicase domain of DDX24, is sufficient for p300 interaction (Fig. 1D). Furthermore, all three tandem deletion mutants (FLdel1(Δ193-214aa), FLdel2(Δ215-239aa) and FLdel3(Δ240-264aa)) disruptive in Q-motif or motif I of full length DDX24 completely abolished p300 interaction(Fig.1D). Taken together, the N-terminal end specific for DDX24 and the partial helicase domain containing Q-motif and motif I mediate the interaction with p300. Notably, DDX24 is a unique interacting partner of p300 as the majority of the proteins co-purified with p300 are either DNA damage response or repair proteins23.
DDX24 is a negative regulator of p53 transcriptional activity
Previous studies have demonstrated that p300 directly catalyzes lysine acetylation on p53 to boost its transcriptional activity. Moreover, p300 localizes to the promoter region of p53 target genes to acetylate histones in the vicinity of the basic transcription machinery to favor a more accessible chromatin structure, thus to facilitate the transcription activity of p5318, 39, 24, 25, 37. Since DDX24 apparently interacts with p300, we expect that endogenous DDX24 modulates p53 transcription activity. In Fig. 2A, RNAi mediated knockdown of DDX24 in U2OS cells significantly augmented expression level of p21 and PUMA, the classic p53 targets in regulating cell cycle arrest and apoptosis. Therefore, DDX24 acts as a negative modulator of p53 transcriptional activity. To explore further, both control and DDX24 siRNA treated cells were incubated with 20 µM etoposide for eight hours and the increased expression of p21 and PUMA was recapitulated in DDX24 depleted cells (Fig. 2A). This data suggests that DDX24 negatively regulates p53 activity not only under normal condition but also upon DNA damage stress. We further confirmed that DDX24 ablation affected p53 targets at the transcriptional level by examining the mRNA level of p53 targets using qRT-PCR (Fig. 2B). DDX24 deficiency has a broad impact on p53 target gene expression profile; we tested more p53 targets in various functional groups and all were affected in the same trend as p21 and PUMA, albeit, to different extents (Shi and Gu, unpublished data). Among all, the most drastic change occurs on p21. Additionally, p53+/+ and −/− isogenic HCT116 colon carcinoma cells were transfected with either control or DDX24 specific siRNA to induce DDX24 depletion. The considerable induction of p53 target genes was only present in HCT116 p53 wild type cells by DDX24 depletion and no perceptible differences of p21 and puma was observed in p53 null HCT116 cells (Fig. 2C). Therefore, the repressive effect of DDX24 on p21 and PUMA expression relies on an intact p53 pathway.
Figure 2. DDX24 is a negative regulator of p53 transcriptional activity.
(A) DDX24 siRNA effects on p21 and PUMA under normal and DNA damage conditions. U2OS cells were transiently transfected with either control or DDX24 specific siRNA and treated with or without 20 uM Etoposide (Etop) for 8 hours. Cell extracts were subjected to Western blot analysis of DDX24, p53, p21, PUMA and β-actin. (B) DDX24 siRNA effects on p21 and PUMA transcriptional level under normal and DNA damage conditions. Total RNA of the same samples presented in Fig. 2A was extracted, and cDNA was prepared by reverse transcription. The mRNA abundance for p21 and PUMA was assessed using quantitative real time PCR. (C) DDX24 siRNA effects on p53 dependent PUMA and p21 expression. Control or DDX24 siRNA were transfected into either p53+/+ or p53−/− HCT116 cells for three days, and the cell extracts were applied to Western blot analysis using α-DDX24, α-p53, α-p21, α-PUMA, and α- β-actin antibodies. (D) DDX24 siRNA effects on p53 target PUMA and p21 with multiple DDX24 specific siRNA oligos. U2OS cells were transiently transfected with control siRNA or four DDX24-specific siRNA oligos (#1, #2, #3, and #4). Total cell lysates were analyzed by Western blotting with α-DDX24, α-p53, α-p21, α-PUMA, and α- β-actin antibodies. (E) p53-ser15 phosphorylation and γ-H2AX in control and DDX24 siRNA treated U2OS cells. U2OS cells were transiently transfected with control or DDX24-specific siRNA, or U2OS cells were treated with 1 µM Doxorubicin (Dox) for 6 hours (6h). Total cell lysates were immunoblotted for p53 phosphoserine 15 (S15-phos-p53), γ-H2AX, p53, DDX24 and β-actin.
To exclude off-target effect of DDX24 siRNA, we ablated DDX24 in U2OS cells using four DDX24 siRNA oligos (RNAi/DDX24 #1, #2, #3, and #4) targeting different regions of DDX24 mRNA. Again, the dramatic up-regulation of p21 and PUMA were repeatedly shown up in all four samples with DDX24 protein level equivalently reduced (Fig. 2D). It is worth mentioning that the slight increase of p53 protein level was observed when DDX24 is depleted from both U2OS and HCT116 cells. However, the difference in p53 protein level is not comparable to the fold change of expression level of p53 target genes when control and DDX24 siRNA treated cells were compared. The very drastic increase in p53 transcriptional activity induced by DDX24 deficiency raised our concern of a potential p53-stabilizing stress response in DDX24 depleted cells 30. We investigated this possibility by detecting signals of p53 serine15 (S15) phosphorylation and γ-H2AX, markers of stress-induced p53 and global genome stress, in lysates of control and DDX24 siRNA treated U2OS cells. The cells had equivalently undetectable level of p53 S15 phosphorylation and γ-H2AX, compared to the robust induction of those in U2OS cells incubated with genotoxic drug doxorubicin (Fig. 2E). Therefore, a nonspecific stress mechanism was precluded to account for the hyperactivity of p53 in these cells. It is speculative that RNA helicase activity of DDX24, although not characterized yet, could possibly play a role in global transcription machinery. To this end, p53 mRNA level was evaluated by qRT-PCR and found unchanged in DDX24 depleted cells (Fig. S1). p53 overactivity in DDX24 deficient cells is not due to altered overall cellular transcription activities. Collectively, our data demonstrate that DDX24 serves as a strong negative regulator of p53 activity.
DDX24 inhibits p300 mediated acetylation of p53
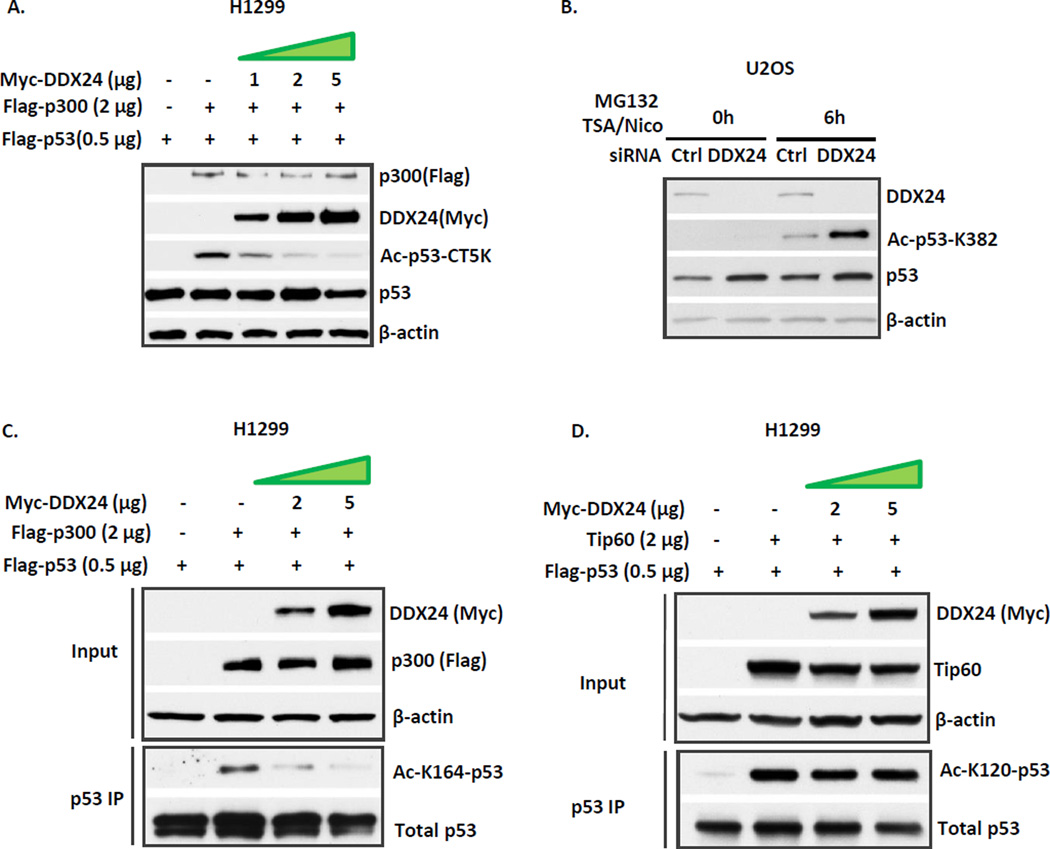
To gain mechanistic insight of DDX24 mediated repression of p53 transcription activity, we focused on the novel interaction between DDX24 and p300. Since p300 mediated p53 acetylation is critical for p53 transcription activity, we speculate that DDX24 suppresses p53 transcription activation through inhibiting p300-dependent acetyltransferase activity towards p53. To test this hypothesis, we assessed if the excessive expression of DDX24 modulates p300 mediated p53 acetylation. H1299 cells were transfected with p53 and p300 simultaneously, and the cell lysates were supplemented with HDAC inhibitor Trichostatin A (TSA) and Sirtuin inhibitor Nicotinamide to enrich for p53 acetylation. As expected, when p300 was co-expressed, p53 was readily acetylated on its five carboxyl-terminal lysines (Fig. 3A). The concomitant overexpression of DDX24 protein decreased p53 acetylation level in a DDX24 dose dependent manner (Fig. 3A). To further validate that DDX24 potentiates p300 mediated p53 acetylation under physiological settings, control and DDX24 siRNA transfected U2OS cells were incubated with TSA/ Nicotinamide to enrich for p53 acetylation and MG132 proteasome inhibitor to equalize p53 abundance. DDX24 knockdown substantially increased endogenous p53 acetylation level at the classic p53 C-terminal lysine 382 site, a representative of total p53 C-terminal acetylation lysine sites (Fig. 3B). Likewise, DDX24 inhibits p53 acetylation on other p300 mediated lysine site K164 (Fig. 3C). However, no apparent impact on K120 acetylation of p53 was observed when DDX24 is overexpressed (Fig.3D). Taken together, our data indicate that DDX24 modulates p53 transcriptional activity through impeding p300 mediated acetylation.
Figure 3. DDX24 inhibits p300 mediated acetylation of p53.
(A) DDX24 inhibits p300 mediated p53 acetylation in dose dependent manner. H1299 cells were transfected with plasmid DNA expressing Flag-p53, Flag-p300, and/or Myc-DDX24 as indicated. Cells were harvested in lysis buffer supplemented with 1 µM Trichostatin A (TSA) and 5 mM Nicotinamide. Cell lysates were subjected to Western blotting with α-Ac-p53-CT5K, α-Flag, α-Myc, α-p53, and α-β-actin antibodies. (B) Inactivation of DDX24 promotes p53 acetylation level. U2OS cells were transiently transfected with either control or DDX24 siRNA, and preincubated with 1 µM TSA, 5 mM Nicotinamide, and 10 µM MG132 for 6 hours (6h) prior to harvesting. Cell extracts were analyzed by western blot analysis using antibodies against DDX24, p53, Ac-p53-K382, and β-actin. (C) DDX24 inhibits p300 mediated p53 acetylation at lysine 164. H1299 cells were transfected with plasmid DNA expressing Flag-p53, Flag-p300, and/or Myc-DDX24 as indicated. Cells were harvested in lysis buffer supplemented with 1 µM TSA and 5 mM Nicotinamide. Cell extracts and p53 immunoprecipitates were subjected to Western blotting with α-acetyl-K164-p53, α-Flag, α-Myc, α-p53, and α-β-actin antibodies. (D) DDX24 does not inhibit Tip60 mediated p53 acetylation at lysine 120. H1299 cells were transfected with plasmid DNA expressing Flag-p53, Tip60, and/or Myc-DDX24 as indicated. Cells were harvested in lysis buffer supplemented with 1 µM TSA and 5 mM Nicotinamide. Cell extracts and M2 immunoprecipitates were subjected to Western blotting with α-acetyl-K120-p53, α-Tip60, α-Myc, α-p53, and α-β-actin antibodies.
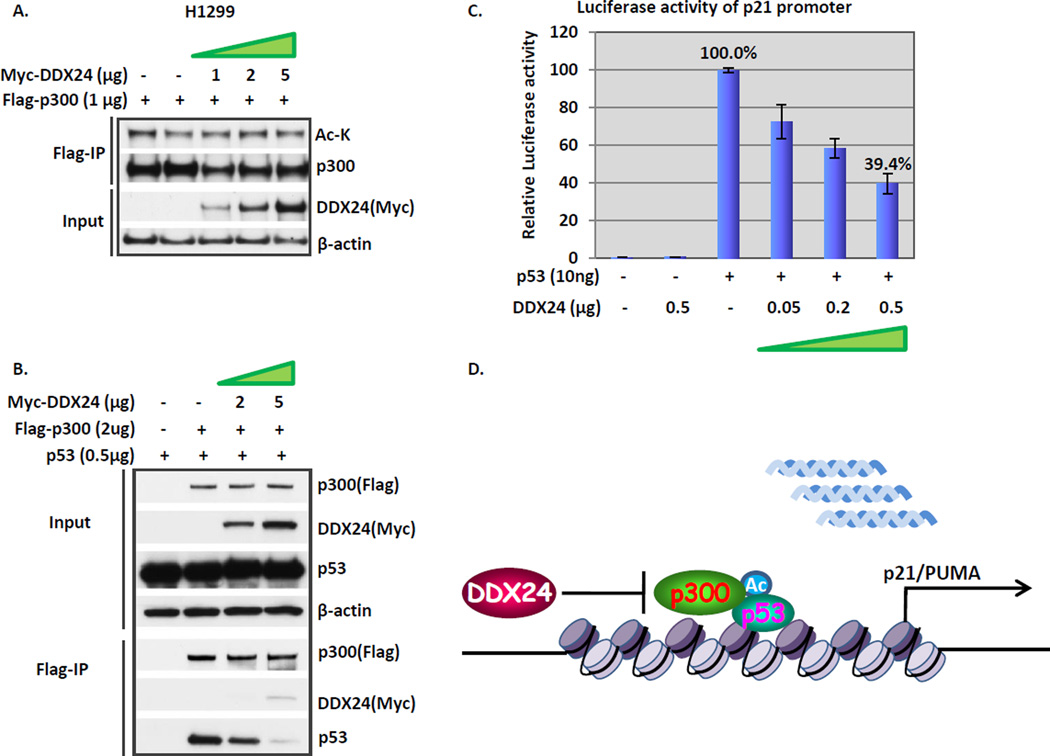
Next we asked whether DDX24 influences the intrinsic HAT activity of p300, which is reflected by its autoacetylation level9, 34. Increasing amount of Myc-tagged DDX24 plasmids were transfected with equal amount of Flag-tagged p300 and the cell lysates were then subjected to Flag/M2 IP. The autoacetylation level on p300 was prominent when p300 was transfected alone, and the addition of DDX24 does not change the autoacetylation level even slightly (Fig.4A). Evidently, DDX24 does not exert direct impact on the intrinsic enzymatic activity of p300. Previous studies ascertain that p300 exists in the same complex with p5317. We hypothesize that DDX24 suppresses p300 dependent p53 acetylation by interfering with this p300-p53/enzyme-substrate interaction. To this end, we first confirmed the binding between p300 and p53 in an overexpression system in H1299 cells (Fig. 4B). Upon DDX24 expression, the amount of p300-bound p53 decreased drastically (Fig. 4B). As a consequence, DDX24 strongly antagonized p53 transactivation of p21 promoter in a luciferase essay (Fig. 4C). Our data gradually unraveled the mechanism underlying the repressive role of DDX24 towards the p53 activity. As diagramed in Fig. 4D, DDX24 interferes with p300 catalyzed p53 acetylation by blocking p300-p53/HAT-substrate interaction.
Figure 4. DDX24 negatively regulates p53 activity by interfering with p300-p53 interaction.
(A) DDX24 has no effect on p300 autoacetylation level. Increasing amount of plasmid DNA expressing Myc-DDX24 with identical amount of plasmid DNA expressing Flag-p300 were co-transfected into H1299 cells. Cells were then incubated with1 µM TSA and 5 mM Nicotinamide for 6 hours. Cell lysates and Flag-M2 immunoprecipitates were subjected to Western blot analysis with α-acetyl lysine (Ac-K), α-p300, α-Myc, and α-β-actin antibodies. (B) DDX24 inhibits p300-p53 interaction. H1299 cells were transfected with expressing vectors for Flag-p300 and p53, in combination with Myc-tagged DDX24. Total cell lysates and Flag M2 immunoprecipitates were analyzed by Western blot with antibodies against Myc, Flag, p53, and β-actin. (C) DDX24 antagonizes p53 transactivation of p21 promoter. Luciferase reporter activity of p21 promoter from H1299 cells co-transfected with wild type p53 and DDX24 expression vectors. Relative luciferase activity was a ratio of firefly (p21 promoter) and renilla luciferase activity for each sample. (D) A diagram for the mechanism underlying the repressive role of DDX24 towards p53 activity. See text for details.
DDX24 negatively regulates p53 mediated cell cycle arrest and senescence
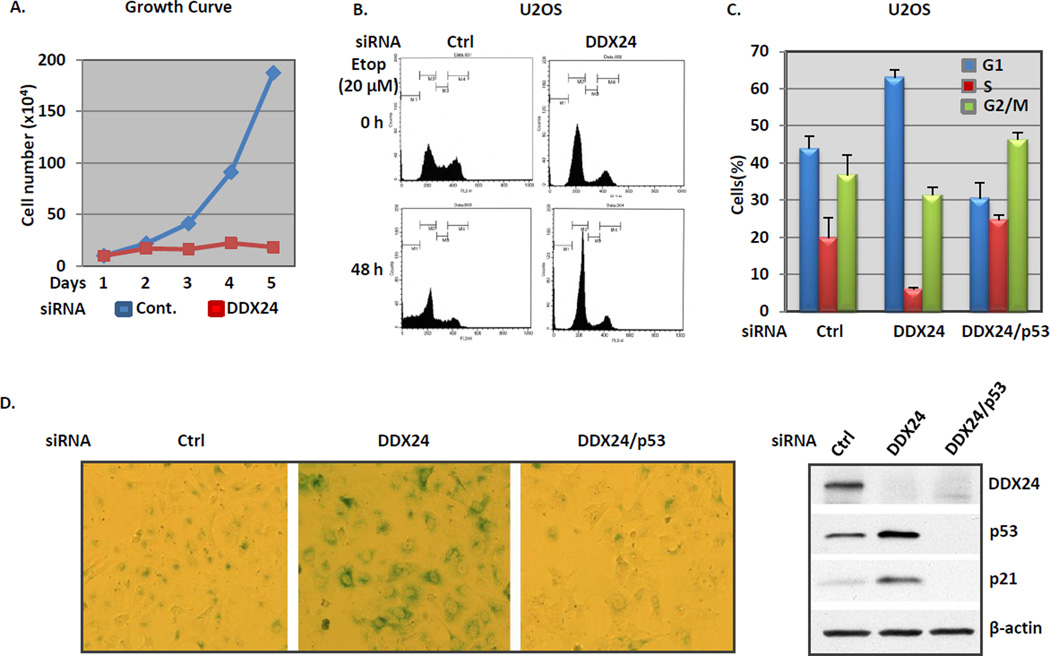
To further investigate the physiological significance of DDX24-p300 interaction, we tested the effect of endogenous DDX24 in modulating p53 mediated cellular functions. U2OS cells, bearing wild type p53, were treated with either control or DDX24 specific siRNA and incubated for 48 hours to achieve satisfactory knockdown efficiency. Cells were then reseeded at identical numbers and allowed to grow for another four days, and cell proliferation rate were measured by recording cell number on each successive day. It is evident that the proliferation of DDX24 depleted cells is significantly impeded while control cells double every 24 hours (Fig. 5A). As p21 is the most affected p53 targets in DDX24 deficient cells, it is speculated that the altered cell growth rate in same cells is due to p21 induced G1/S cell cycle arrest. As presented in Fig. 5C, 43% of the total control cells arrested at G1 phase, and DDX24 knockdown led to the significant increase of G1 population to 62%. In accordance, the reduction of S phase is drastic when DDX24 is depleted from cells (Fig. 5C). Apparently, DDX24 deficient cells underwent G1/S cell cycle arrest, presumably dependent on amplified p21 expression. We further knockdown p53 in DDX24 depleted cells, and the cell cycle arrest phenotype was reversed (Fig.5C). The DDX24 depletion induced cell cycle arrest is also prominent under DNA damage condition. In Fig. 5B, control siRNA treated cell, when challenged with etoposide for 48 hours, underwent apoptosis reflected by the subG1 peak in the histogram of cell cycle profile. DDX24 siRNA treated cells, under same treatment, did not undergo apoptosis but showed even more predominant G1/S arrest (Fig.5B). Interestingly, prolonged culturing of DDX24 siRNA treated U2OS cells ultimately resulted in enlarged and flattened cell shape, the phenotypes typical of senescence45. Most strikingly, DDX24 knockdown cells appeared nearly 100% positive for β-galactosidase staining, which proves the occurrence of senescence in cells lacking DDX24 protein (Fig. 5D). Since p21 was originally discovered as a senescence associated gene42, it is not surprising that concomitant DDX24 and p53 knockdown conferred resistance to senescent phenotype (Fig. 5D). The siRNA knockdown efficiency was examined by western blotting analysis of individual targeted genes (Fig. 5D, right panel). Taken together, DDX24 negatively regulates p53 dependent G1/S cell cycle arrest and senescence.
Figure 5. DDX24 negatively regulates p53 mediated cell cycle arrest and senescence.
(A) Cell growth rate analysis of normal and DDX24 inactivated U2OS cells. U2OS cells were treated with control or DDX24 siRNA for 48 hours. A total of 10x104 cells from each sample were seeded into 6-well plates at day 0 and counted for four successive days. (B) FACS analysis of DDX24-inactivated U2OS cells treated with and without etoposide. U2OS cells transiently transfected with either control or DDX24 siRNA were treated with 20 µM etoposide (Etop) for 0 or 48 hours. Cells were fixed in cold 80% methanol, stained with propidium iodide, and subjected to DNA content analysis by flow cytometry. (C) DDX24 RNAi induces p53-dependent G1/S cell cycle arrest. Percentages of cells in G1, S, and G2/M cell cycle phases from control, DDX24, and both DDX24 and p53 (DDX24/p53) siRNA treated U2OS cells are presented. Values are an average of three independent experiments. Error bars, +/−1 standard deviation. (D) Images of senescence-associated β-galactosidase staining of control, DDX24, and both DDX24 and p53 (DDX24/p53) siRNA treated U2OS cells. Cells treated with siRNA as indicated were cultured for six days and were fixed and stained for β-galactosidase activity. The right panel is the Western blotting analysis of cells three days after siRNA treatment as indicated by DDX24, p53, p21, and β-actin antibodies.
DDX24 antagonizes p53 function to promote tumor cell growth
The primary role of p53 is tumorigenesis is to suppress tumor cell growth. To address whether DDX24 influences the suppressive function of p53 in tumor cell growth, a colony formation assay was carried out in p53 null H1299 cells11. Cells were transfected with the combinations of plasmids expressing DDX24 or p53 protein (Fig. 6A). The transfected cells were reseeded at the same number, incubated for 14 days, and stained with crystal violet. As expected, the colonies formed on singly p53 expressed plate dramatically decreased to 5% in comparison to the empty vector transfected plates (Fig.6A). Significantly, the concomitant expression of DDX24 reversed the growth suppressive effect of p53 in a dose dependent manner (Fig.6A). Therefore DDX24 antagonizes the function of p53 to promote tumor cell growth. Interestingly, DDX24 is highly expressed in a wide range of breast cancer cell lines, while the expression level of DDX24 in normal breast cells (MCF-10A) is low (Fig.6B), suggesting a potential role of DDX24 in breast cancer tumorigenesis presumably attributed to its negative regulatory effect on p53. To strengthen this notion, we analyzed the correlation between p53 mutation status and the expression level of DDX24 in breast tumor samples acquired from the TCGA database. As presented in Figure S2, the transcriptional abundance of DDX24 is lower in TP53-mutant (mut) samples than in its wild-type (wt) samples in TCGA breast cancer data-sets. The difference of DDX24 expression level is statistically significant in the two groups, with a p-value of 0.0002282. The correlation of higher DDX24 expression level with the functional integrity of wild type p53 pathway in human breast tumor data supports the significant impact of DDX24 in the p53 signaling pathway. MCF7 was selected at random to test DDX24 effect on p53 and the result validated that DDX24 repression of p53 on target p21, not tissue or cell context specific (Fig.6C). Taken together, the counteractive role of DDX24 in p53 biology and the frequently aberrant expression of DDX24 protein throughout breast tumor cells suggest a putative oncogenic role of DDX24 in breast cancer tumorigenesis.
Figure 6. DDX24 antagonizes p53 function to promote tumor cell growth.
(A) Colony-formation assay of H1299 cells transfected with empty vector(Empt-Vec), Flag-p53, and Flag-DDX24 expression plasmids as indicated. The transfected cells were incubated for 48 hours, split, and subjected to colony-formation assay visualized by crystal violet staining (Left panel). Western blot analysis of cell extracts from H1299 cells transfected as indicated for colony formation assay (Right upper panel). Quantification of colonies formed in H1299 cells transfected with empty vector, Flag-p53, and Flag-DDX24 expression plasmids after 14 days culture in the presence of G418 (1mg/ml). Graphs are presented as percentage of colonies formed of Flag-DDX24 plasmid transfected cells, and values are shown as average percentage ±SD of three different experiments (Right lower panel). (B) DDX24 expression in breast cancer cell lines. Western blot analysis of cell extracts from a number of breast cancer cell lines and normal breast cell line MCF-10A by α-DDX24 and α- β-actin antibodies. (C) DDX24 siRNA effects on p53 and p21 in breast cancer MCF7 cells. Control or DDX24 siRNA were transfected into MCF7 cells for three days and the cell extracts were applied to Western blot analysis using α-DDX24, α-p53, α-p21, and α- β-actin antibodies.
Discussion
In this report, we described the identification of DDX24 as a novel and negative regulatory factor of p53. We have demonstrated that DDX24 plays an inhibitory role in p53 transcriptional activity and subsequently in p53 controlled cell growth arrest and senescence. The underlying mechanism by which DDX24 regulates p53 lies in the Mass Spectrometry identified original DDX24-p300 interaction. DDX24 competes with p53 for p300 interaction, and therefore exhibits an adverse effect on p300 catalyzed p53 acetylation. DDX24 is often aberrantly overexpressed in breast cancer cells, suggesting a potential role in tumorigenesis by antagonizing the tumor suppressor function of p53.
Our data strongly support the role of DDX24 as a negative regulator of p53. However, the precise mechanism by which DDX24 suppresses p53 function requires further elucidation. It is well accepted that acetylation and ubiquitination are counteracting processes that compete for the same lysine residues on p53. Since the forced expression of DDX24 represses p53 acetylation, p53 ubiquitination will be promoted on the same p53 lysines. This hypothesis was confirmed as DDX24 overexpression promotes p53 ubiquitination in in vivo ubiquitination assay, adding one more layer of p53 regulation via DDX24 (Fig. S3). When U2OS and HCT116 cells were treated with three rounds of DDX24 siRNA, knockdown efficiency was maximized and the DDX24 protein was completely depleted from cells. Under this condition, the increased p53 protein level in DDX24 depleted cells becomes more perceptible, as a result of prolonged half-life of p53 protein (Fig. S4). Altogether, DDX24 acts as a double-edged sword, repressing p53 acetylation and simultaneously promoting p53 ubiquitination. Of note, another group recently showed that DDX24 knockdown activates p53 through a distinctive mechanism43. It was presented that the depletion of DDX24 induces nucleolar stress, which in turn stabilizes p53 protein and activates p53 activity 43. To what extent these two mechanisms contribute to the regulation of p53 activity is of great interest for further exploration. In fact, as shown in Figure 4C, p53-mediated transactivation of p21 promoter can be severely inhibited by DDX24 overexpression, indicating that DDX24-mediated effect on p53 is direct, rather than indirectly through nucleolar stress. Moreover, DDX24-mediated repression of p53 activity is significantly abrogated when the acetylation-defective mutant p53-6KR was used, suggesting that DDX24-mediated effects act at least, in part, through modulating p53 acetylation (Fig. S5).
The discovery of DDX24 as a novel modulator of p300-p53 axis adds extra complexity to an already complicated molecular network centered on p300 and p53 proteins. p300-p53 axis is regulated through multiple mechanisms including p53 serine15 phosphorylation via ATM, MDM2/MDMX competition for p53 transactivation domain, and the interaction with other co-factors6. DDX24 may interact with numerous co-factors of p300/p53 to complement or to antagonize their functions. For example, we have observed that DDX24 was linked to MDM2. As shown in Figure S6, the inhibitory effect of DDX24 on p53-mediated transcriptional activation is significantly abrogated when the cells were pretreated with Nutlin-3a, which effectively blocks p53-Mdm2 interaction, indicating that DDX24 may act in part through modulating the p53-Mdm2 interaction to weaken p53 activity. A recent study found that MDM2 catalyzes polyubiquitin chains onto DDX24 but does not lead to proteasome degradation of DDX24 protein43. It is intriguing to investigate what signaling cascade this polyubiquitinated form of DDX24 involves and how it affects p300-p53 complex composition and p53 activity. Furthermore, DDX24 is expected to antagonize the function of positive regulators of p300-p53 axis, such as JMY and Strap, interacting with p300 and promoting p53 acetylation6. We hypothesize that DDX24 functions as a component in a multi-protein complex to control p53 acetylation and the regulatory role of DDX24 might be extended to other p300 substrates. Until now, little is known about modification or regulation of DDX24 protein itself. The elucidation of how DDX24 is regulated at the transcriptional, translational, and even post-translational level under various contexts will certainly help to gain a greater understanding of the multiple regulatory levels that p53 is subjected to.
Recently, the Barber group demonstrated DDX24 as a negative modulator of innate immune signaling26. DDX24 is induced in response to interferon treatment, disrupting the association between FADD and RIP1 and causing inhibition of the NF-κB pathway required for optimal interferon production26. This observation, combined with our discovery of DDX24 as a negative modulator of p53 pathway, indicates that DDX24 potentially bridge the interplay between NF-κB and p53 signaling pathways. It is well accepted that the crosstalk between p53 and NF-κB pathways plays important roles in tumorigenesis22. DDX24 will be another example of signaling molecules, like MDM2 and Wip1, to link p53 and NF-κB pathway2, 40, 47. A detailed study of the interplay between these two signaling pathways through DDX24 will undoubtedly uncover new complexities in p53 biology.
The DEAD box protein group has 36 identified family members throughout the human genome and they are presumably involved in RNA metabolism 19. Emerging evidence links this family of proteins to cancer development and progression1, 15, 19. Dysregulation of DEAD box proteins were identified in diverse cancer types, for example, DDX3 in Squamous cell carcinoma (SCC) and DDX6 in colon cancer3, 12, 15, 28, 32. In addition, several DEAD box proteins are reported to form complexes with cancer proteins: DDX3 with Casein Kinase 1 in regulating β-Catenin signaling pathway, for instance7. Besides, DDX5 and DDX17 have been established as positive regulators of p53 through the direct contact with transcriptional machinery at p53 target promoters14. In contrast, DDX24 is the first identified negative regulator of p53 from the DEAD box protein family and modulates p53 activity with distinctive mechanisms. In this report, we also propose that DDX24 might be involved in the tumorigenesis of breast cancer. Further study is required to determine the extent of contribution and the exact role of DDX24 in neoplastic transformation in animal models. DDX24 is overexpressed in breast cancer cells but at low levels in non-cancerous cells, making DDX24 a potentially attractive cancer therapeutic target.
Materials and Methods
Colony Formation Assay
H1299 cells were transfected with either empty or PCIN4-Flag-p53 and -DDX24 expression plasmids for 48 hr. Cells were then seeded at the density of 1,000 cells per dish and cultured under the selection of G418 (1 mg/ml) for 14 days. Cells were stained with crystal violet solution and colonies were counted.
Senescence-Associated-β-Galactosidase (SA-β-gal) Staining
Cells were fixed in 3% formaldehyde for 5 min RT, washed three times with PBS, and then incubated at 37°C for 16 hr with fresh Senescence-Associated-β-Galactosidase (SA-β-gal) Stain solution (1 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactosidase (X-Gal), 40 mM citric acid/sodium phosphate, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2)10.
All reagents, antibodies and other Materials and Methods are described in the Supplementary Information.
Supplementary Material
Acknowledgements
This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5RO1 CA172023, 5RO1 CA166294, 5RO1CA085533 and 2P01CA080058 to W. G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. D. Shi is also supported by NIH cancer biology training grant T32-CA09503. We especially thank Dr. Ruping Sun for his help with the analysis of TCGA database.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Abdelhaleem M. Do human RNA helicases have a role in cancer? Bba-Rev Cancer. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G, et al. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci U S A. 2010;107:18061–18066. doi: 10.1073/pnas.1006163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer research. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 4.Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, et al. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Molecular cell. 2011;42:584–596. doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Coutts AS, La Thangue NB. The p53 response: emerging levels of co-factor complexity. Biochemical and biophysical research communications. 2005;331:778–785. doi: 10.1016/j.bbrc.2005.03.150. [DOI] [PubMed] [Google Scholar]
- 7.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, et al. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 8.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends in molecular medicine. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nature structural & molecular biology. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 10.Dimri GP, Lee XH, Basile G, Acosta M, Scott C, Roskelley C, et al. A Biomarker That Identifies Senescent Human-Cells in Culture and in Aging Skin in-Vivo. P Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nature protocols. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 12.Fukawa T, Ono M, Matsuo T, Uehara H, Miki T, Nakamura Y, et al. DDX31 regulates the p53-HDM2 pathway and rRNA gene transcription through its interaction with NPM1 in renal cell carcinomas. Cancer research. 2012;72:5867–5877. doi: 10.1158/0008-5472.CAN-12-1645. [DOI] [PubMed] [Google Scholar]
- 13.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic acids research. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochemical Society transactions. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 15.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA biology. 2013;10:121–132. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 17.Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Molecular cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Gustafson EA, Wessel GM. DEAD-box helicases: posttranslational regulation and function. Biochemical and biophysical research communications. 2010;395:1–6. doi: 10.1016/j.bbrc.2010.02.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 21.Iyer NG, Xian J, Chin SF, Bannister AJ, Daigo Y, Aparicio S, et al. p300 is required for orderly G1/S transition in human cancer cells. Oncogene. 2007;26:21–29. doi: 10.1038/sj.onc.1209771. [DOI] [PubMed] [Google Scholar]
- 22.Johnson RF, Perkins ND. Nuclear factor-kappaB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends in biochemical sciences. 2012;37:317–324. doi: 10.1016/j.tibs.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Jung SY, Malovannaya A, Wei J, O'Malley BW, Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Molecular endocrinology. 2005;19:2451–2465. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- 24.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Molecular cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 26.Ma Z, Moore R, Xu X, Barber GN. DDX24 negatively regulates cytosolic RNA-mediated innate immune signaling. Plos Pathog. 2013;9:e1003721. doi: 10.1371/journal.ppat.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa Y, Morikawa H, Hirata I, Shiozaki M, Matsumoto A, Maemura K, et al. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. British journal of cancer. 1999;80:914–917. doi: 10.1038/sj.bjc.6690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor perspectives in biology. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oren M, Damalas A, Gottlieb T, Michael D, Taplick J, Leal JF, et al. Regulation of p53: intricate loops and delicate balances. Annals of the New York Academy of Sciences. 2002;973:374–383. doi: 10.1111/j.1749-6632.2002.tb04669.x. [DOI] [PubMed] [Google Scholar]
- 31.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV, et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood. 2011;118:903–915. doi: 10.1182/blood-2010-11-318022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44 doi: 10.1038/ng.2396. 1104-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi D, Gu W. Dual Roles of MDM2 in the Regulation of p53: Ubiquitination Dependent and Ubiquitination Independent Mechanisms of MDM2 Repression of p53 Activity. Genes & cancer. 2012;3:240–248. doi: 10.1177/1947601912455199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, et al. A novel cofactor for p300 that regulates the p53 response. Molecular cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 37.Szerlong HJ, Prenni JE, Nyborg JK, Hansen JC. Activator-dependent p300 acetylation of chromatin in vitro: enhancement of transcription by disruption of repressive nucleosome-nucleosome interactions. The Journal of biological chemistry. 2010;285:31954–31964. doi: 10.1074/jbc.M110.148718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Y, Luo JY, Zhang WZ, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Molecular cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomasova D, Mulay SR, Bruns H, Anders HJ. p53-independent roles of MDM2 in NF-kappaB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia. 2012;14:1097–1101. doi: 10.1593/neo.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO reports. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Blandino G, Givol D. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene. 1999;18:2643–2649. doi: 10.1038/sj.onc.1202632. [DOI] [PubMed] [Google Scholar]
- 43.Yamauchi T, Nishiyama M, Moroishi T, Yumimoto K, Nakayama KI. MDM2 mediates nonproteolytic polyubiquitylation of the DEAD-Box RNA helicase DDX24. Molecular and cellular biology. 2014;34:3321–3340. doi: 10.1128/MCB.00320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhao H, Darzynkiewicz Z. Biomarkers of cell senescence assessed by imaging cytometry. Methods in molecular biology. 2013;965:83–92. doi: 10.1007/978-1-62703-239-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Yu L, Fu Q, Chen W, Jiang J, Gao J, et al. Cloning and characterization of human DDX24 and mouse Ddx24, two novel putative DEAD-Box proteins, and mapping DDX24 to human chromosome 14q32. Genomics. 2000;67:351–355. doi: 10.1006/geno.2000.6255. [DOI] [PubMed] [Google Scholar]
- 47.Zhu YH, Bulavin DV. Wip1-dependent signaling pathways in health and diseases. Progress in molecular biology and translational science. 2012;106:307–325. doi: 10.1016/B978-0-12-396456-4.00001-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.