Abstract
The osteogenic transcription factor, Runx2, is abnormally expressed in prostate cancer (PCa) and associated with metastatic disease. During bone development, Runx2 is activated by signals known to be hyperactive in PCa including the RAS/MAP kinase pathway, which phosphorylates Runx2 on multiple serine residues including S301 and S319 (equivalent to S294 and S312 in human Runx2). This study examines the role of these phosphorylation sites in PCa. Runx2 was preferentially expressed in more invasive prostate cancer cell lines (PC3 > C4-2B > LNCaP). Furthermore, analysis using a P-S319-Runx2-specific antibody revealed that the ratio of P-S319-Runx2/total Runx2 as well as P-ERK/total ERK was highest in PC3 followed by C4-2B and LNCaP cells. These results were confirmed by immunofluorescence confocal microscopy, which showed a higher percentage of PC3 cells staining positive for P-S319-Runx2 relative to C4-2B and LNCaP cells. Phosphorylated Runx2 had an exclusively nuclear localization. When expressed in prostate cell lines, wild type Runx2 increased metastasis-associated gene expression, in vitro migratory and invasive activity as well as in vivo growth of tumor cell xenografts. In contrast, S301A/S319A phosphorylation site mutations greatly attenuated these Runx2 responses. Analysis of tissue microarrays from 129 patients revealed strong nuclear staining with the P-S319-Runx2 antibody in primary prostate cancers and metastases. P-S319-Runx2 staining was positively correlated with Gleason score and occurrence of lymph node metastases while little or no Runx2 phosphorylation was seen in normal prostate, benign prostate hyperplasia or prostatitis indicating that Runx2 S319 phosphorylation is closely associated with prostate cancer induction and progression towards an aggressive phenotype. These studies establish the importance of Runx2 phosphorylation in prostate tumor growth and highlight its value as a potential diagnostic marker and therapeutic target.
Keywords: Prostate, Runx2, MAP kinase, phosphorylation, metastasis
Introduction
Prostate cancer (PCa), the most common male malignancy, is frequently associated with bone metastases(1). The metastatic process begins in the primary tumor with activation of genes that promote angiogenesis, tumor invasion and migration leading to colonization of peripheral tissues including bone(2). A major challenge for PCa treatment is to identify factors controlling tumor growth and metastasis.
Runx2, a transcription factor required for bone development, is abnormally elevated in PCa.(3, 4) The presence of Runx2 in PCa is positively correlated with increased Gleason score and metastasis.(5–7) Furthermore, Runx2 stimulates growth, migration and osteolytic activity of prostate and breast tumors.(6, 8, 9) Runx2 also directly induces genes associated with angiogenesis, invasiveness and metastasis including Vegf, Spp1, Mmp9 and Mmp13 and stimulates epithelial to mesenchymal transition of primary tumors.(5, 8, 10) Lastly, transgenic overexpression of Runx2 predisposes mice to T cell lymphomas, suggesting an oncogene function.(11, 12) Other runt domain transcription factors are also associated with cancers; Runx1 chromosomal translocations/mutations are commonly found in myeloid leukemias while Runx3 may function as a tumor suppressor in gastric cancers (for reviews, see(12, 13)).
MAP kinase (MAPK), PI3K/AKT and non-receptor tyrosine kinase signaling pathways are also elevated in PCa. Increased MAPK signaling due to RAS-RAF mutations is seen in 43 percent of primary tumors and 63 percent of metastases.(14, 15) Furthermore, RAS/MAPK activation positively correlates with disease progression.(16) Significantly, transgenic overexpression of RAS stimulates EMT and PCa formation in genetic models of PCa.(17) Similarly, targeted expression of mutant BRAF in prostate epithelium induces invasive carcinomas in mice.(18) PI3K/AKT and non-receptor kinases have also been related to PCa initiation and progression.(19, 20) However, there is currently no clear explanation for why kinase activation in PCa is associated with an invasive phenotype.
Based on previous work in bone, we propose that Runx2 and the RAS/MAPK pathway cooperatively interact in PCa to regulate metastasis-related gene expression. During osteoblast differentiation, ERK1/2 and p38 MAPKs phosphorylate Runx2 on several serine and threonine residues.(21–24) Of these, Ser 301 and Ser 319 (murine type I Runx2 sequence) are particularly important for Runx2-dependent transcriptional activity.(21) ERK phosphorylates Runx2 directly on the chromatin of target genes.(21, 25, 26) Phosphorylated Runx2 then stimulates epigenetic changes including histone acetylation and transcription leading to induction of gene expression.(25, 27)
In the present study, we show that Runx2 is phosphorylated in PCa cells and that the same phosphorylation sites previously demonstrated to be important for osteoblast gene expression are also required for Runx2-dependent stimulation of metastasis-associated gene expression, in vitro cell migration, invasion and in vivo tumor growth. Furthermore, the presence of P-Runx2 as measured with a P-S319-Runx2-specific antibody is correlated with PCa onset and severity in a patient population.
Results
Runx2 is preferentially phosphorylated in metastatic PCa cell lines
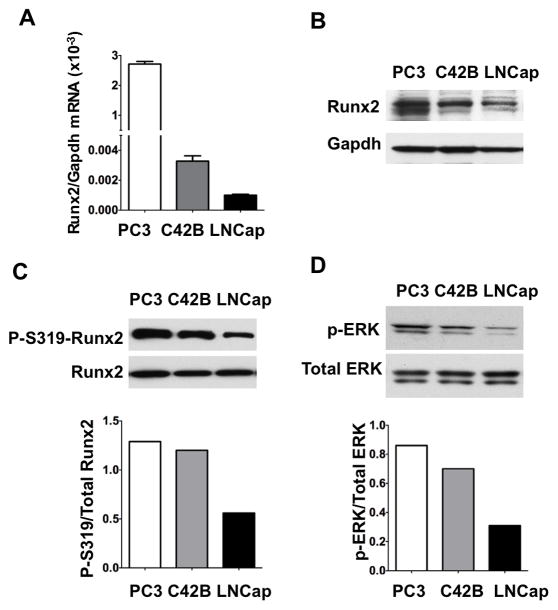
Runx2 expression was previously compared between different human prostate cancer cell lines.(6, 7) PC3 cells have high metastatic potential while LNCaP cells have little or no activity. C4-2B cells are a metastatic subclone derived from LNCaP cells.(28, 29) These cell lines were compared to determine if there is a correlation between MAPK activity, Runx2 phosphorylation and metastatic potential (Figure 1). Phosphorylated Runx2 was detected using an anti-S319-phospho-Runx2 specific antibody.(26) Although MAPK phosphorylates Runx2 at additional sites including S301, phosphorylation at S319 is closely correlated with Runx2 transcriptional activity in osteoblasts and is likely to reflect phosphorylation at other MAPK sites for which immune reagents are not currently available.(21, 24, 26) Runx2 mRNA and protein were highest in PC3 cells followed by C4-2B and LNCaP (Fig 1AB). Panels C and D compare the S319-phospho-Runx2/total Runx2 ratio with the level of MAPK activation (P-ERK/total ERK) in each cell line. For P-Runx2 analysis, protein loading was adjusted to give the same amount of total Runx2 in each lane. C4-2B and PC3 cells had high MAPK activity and Runx2 phosphorylation. S319-phospho-Runx2/total Runx2 and P-ERK/total ERK ratios in the metastatic cells were more than twice the ratios seen in LNCaP cells.
Figure 1. Comparison of total Runx2, phospho-S319-Runx2 and MAPK activity in human PCa cell lines with high (PC3, C4-2B) and low (LNCaP) metastatic potential.
(A, B) Runx2 mRNA and protein levels. Runx2 mRNA was measured by real-time RT/PCR. Total Runx2 protein was measured by Western blotting (10 μg protein/lane) using an anti-Runx2 antibody (MBL monoclonal antibody). Loading efficiency was assessed by reprobing each blot with a Gapdh antibody. (C) Comparison of P-S319-Runx2/total Runx2 ratio. Based on the results shown in panel B, sample loading from each cell line was adjusted to give an equivalent amount of total Runx2 protein (Note increased GAPDH in C4-2B and LNCaP lanes). Blots were then probed with a P-S319-Runx2-specific antibody or anti-Runx2 antibody and the ratio of P-S319-Runx2/total Runx2 was determined by densitometry (lower panel). (D) MAPK activity. Cell extracts (10 μg protein/lane) were probed with P-ERK and total ERK antibody and the P-ERK/total ERK ratio was determined by densitometry (lower panel).
Differences between cell lines were also noted when total and S319-P-Runx2 staining was examined by confocal microscopy (Figure 2, Suppl. Figure S1). Most cells were positively stained with total Runx2 antibody (Suppl. Fig S1). However, clear differences in cellular distribution were noted; Runx2 was mainly nuclear in PC3 and C4-2B cells while a diffuse cytoplasmic and nuclear distribution was observed in LNCaP cultures (Fig. 2). These differences could not be explained by non-specific staining of LNCaP cells with the total Runx2 antibody (Suppl. Fig. S2). In contrast, S319-P-Runx2 staining was exclusively nuclear in all 3 lines with strongest staining observed in PC3 followed by C4-2B and LNCaP cells. The percentage of Runx2-positive cells also staining for P-Runx2 (yellow/orange staining in merged images, Suppl Fig. S1) was as follows: PC3, 72.5 ± 0.8; C4-2B, 39.5 ± 0.2; LNCaP, 16.5 ± 0.5.
Figure 2. Cellular distribution of total Runx2 and phospho-S319-Runx2 immunofluorescence in PCa cell lines.
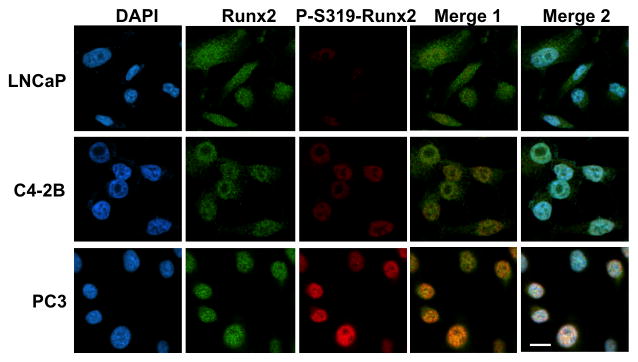
LNCaP, C4-2B and PC3 cells were plated on glass cover slips, fixed, permeablized and stained for DNA (DAPI), total Runx2 (Abcam primary Mab, secondary antibody Alexa Fluor 488 green) and P-S319-Runx2 (rabbit anti-P-S319-Runx2 primary polyclonal antibody, secondary antibody Alexa Fluor 555 red) as indicated in Methods. High magnification (100x oil objective) Z stack confocal images are shown. Merge 1 contains total Runx2 and P-S319-Runx2 images. Merge 2 combines total and P-S319-Runx2 with the DAPI image to facilitate visualization of the cytoplasmic Runx2 staining in LNCaP cells. Scale bar = 5 μm. Lower power images and IgG controls are shown in Supplementary Figures S1 and S2.
These results indicate that nuclear P-S319-Runx2 staining is increased in more metastatic PCa cell lines.
Requirement for Runx2 S301, S319 phosphorylation in metastasis-related gene expression
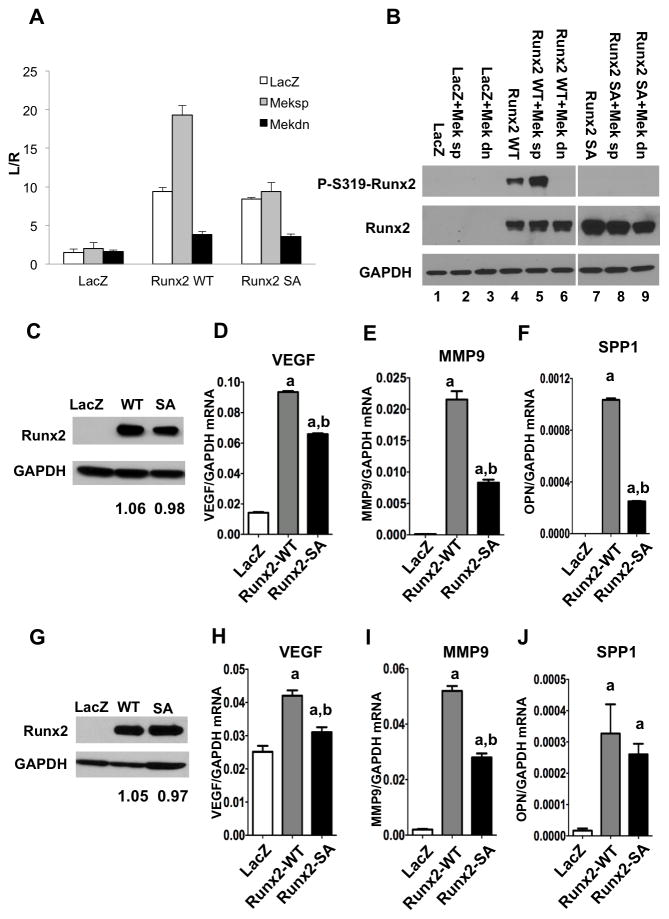
As previously shown, MAPK activation of Runx2 transcriptional activity requires phosphorylation at Ser 301 and Ser 319.(21) Two approaches were used to determine if these same two phosphorylation sites are also required for induction of metastasis-associated gene expression in prostate cells (Fig. 3). First, PC3 cells were transfected with a pOPN-luc reporter plasmid containing the proximal promoter of Spp1, a gene previously associated with metastatic activity(30), and Runx2 phosphorylation and transcriptional activity were examined with or without MAPK activation (Fig 3A, B). Spp1 contains a previously characterized, functional Runx2 binding site between -136 to -130 bp of the proximal promoter.(31) Wild type Runx2 strongly stimulated promoter activity and this response was further increased with Meksp, a constitutively-active MEK1 mutant.(32) Meksp also increased Runx2 phosphorylation as measured with anti-S319-P-Runx2 antibody (Fig 3B). In contast, dominant-negative MEK1 strongly inhibited Runx2 phosphorylation and activity. To assess whether increased Runx2 phosphorylation was necessary for MAPK-dependent transcriptional activity, cells were transfected with Runx2 containing S301A, S319A mutations (Runx2 SA). Although basal stimulation with Runx2 SA was similar to wild type, mutant Runx2 was resistant to activation by Meksp, as would be expected if the two phosphorylation sites were necessary for MAPK activation. These results cannot be explained by differences in levels of wild type versus Runx2 SA. Also immunoreactivity of the anti-Runx2 antibody was not affected by Runx2 phosphorylation since similar band intensities were obtained with or without Meksp or phosphorylation site mutation (Fig. 3B).
Figure 3. Requirement for Runx2-S301, S319 phosphorylation for induction of metastasis-associated genes.
(A, B) Spp-1 promoter activity. PC3 cells were transfected with pOPN-Luc reporter, pRL-SV40 renilla luciferase normalization plasmid, lacZ (control), wildtype Runx2 (Runx2WT) or Runx2-S301A, S319A mutant (Runx2 SA) and either constitutively-active (Meksp) or dominant-negative (Mekdn) MEK1 expression vectors as indicted. Firefly luciferase activity was normalized to renilla luciferase. Levels of P-S319-Runx2 and total Runx2 protein are shown in panel B. Samples in lanes 7–9 were run on a separate gel from those in lanes 1–6, but were blotted and exposed under identical conditions. (C–J) mRNA induction. PC3 (C–F) or RWPE1 cells (G–J) were transduced with LacZ, Runx2-WT or Runx2-SA adenovirus expression vectors and total protein and RNA was isolated after 48 h. Viral titers were adjusted to give equivalent levels of Runx2 protein as measured on Western blots; values below each lane indicate the Runx2/Gapdh protein ratio as determined by densitometry (C, G). VegfA (D, H), Mmp9 (E, I) and Spp-1 (F, J) mRNAs were measured by real-time RT/PCR. Statistics (D-F, H-J): aSignificantly different from LacZ control (p < 0.01), bSignificantly different from Runx2-WT (p < 0.01); N = 3/group.
In a second approach, we examined the effect of phosphorylation site mutation on the ability of Runx2 to stimulate endogenous expression of three metastasis-associated genes; Vegf, Mmp9 and Spp-1.(6, 33) PC3 cells were transduced with adenovirus expressing LacZ (control), wild type Runx2 or phosphorylation-deficient Runx2 (Fig. 3C–F). To assess whether Runx2 could also stimulate gene expression in non-tumorigenic cells, we examined RWPE1 cells, a non-transformed, immortalized cell line derived from normal human prostate epithelium (Fig. 3G–J).(34) Adenovirus vectors were toxic to LNCaP cells, preventing them from being included in this study. In these experiments, Runx2 was activated by endogenous MAPK rather than by added Meksp. Viral titers were adjusted to produce equivalent amounts of wild type or mutant Runx2 protein as measured on Western blots (panels C, G). Note that due to the high level of Runx2 expression, Western blots were exposed for a shorter time than in Fig. 1 so endogenous Runx2 in PC3 cells was not visible in the LacZ group, but was detected when blots were deliberately over-exposed (Supplementary Figure S3). We were not able to detect Runx2 protein in RWPE1 cells (result not shown). Wild type Runx2 strongly induced Vegf, Mmp9 and Spp1 mRNAs in both PC3 and RWPE1 cells. In all cases, the S301A, S319A Runx2 mutant had considerably less activity than wild type Runx2.
Role of Runx2 phosphorylation in cell migration and invasion
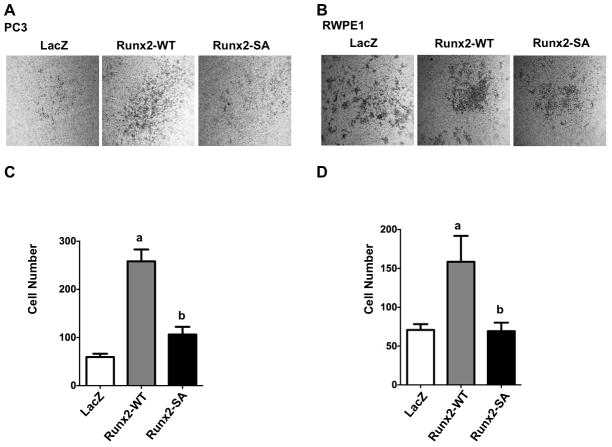
Runx2 is required for in vitro PCa cell migration and invasion.(5, 33) The following experiments assessed the requirement for Runx2 phosphorylation in these two tumor-related processes using LacZ, Runx2-WT or Runx2-SA transduced PC3 and RWPE1 cells.
Migration assays measured the movement of cells into an initially cell-free area (Fig. 4). Panels A and B show representative images of cells at the beginning of the experiment and after a specific migration period while panels C and D show the percent occupancy of the initially cell-free area over time. Consistent with their metastatic phenotype, PC3 cells migrated faster than the non-transformed RWPE1 cell line. For example, after 6 h, control PC3 cells filled approximately 30 percent of the initially cell-free area versus only a 10 percent fill for RWPE1 cells. However, wild type Runx2 stimulated migration of both cell types with PC3 cells filling 80 percent of the cell-free area after 6 h and nearly the entire area after 8 h and RWPE1 cells filling 18 percent of the cell-free area after 6 h and 38 percent after 12 h. In contrast, equivalent levels of phosphorylation-deficient Runx2 had approximately half the activity of wild type Runx2 in stimulating migration of PC 3 cells and no significant stimulatory activity on RWPE1 cells.
Figure 4. Requirement for Runx2-S301, S319 phosphorylation in stimulation of prostate cell migration.
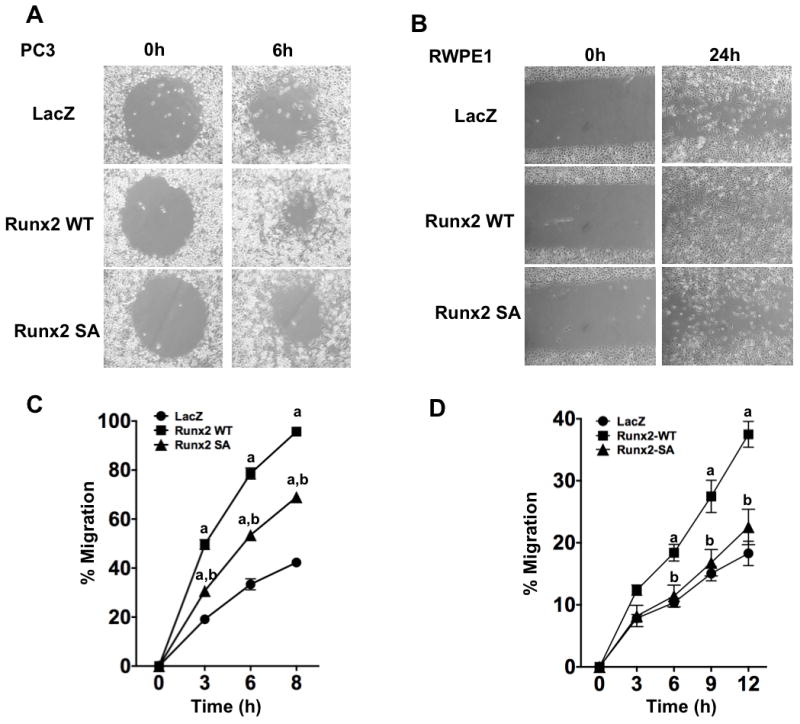
PC3 and RWPE1 cells were transduced with LacZ, Runx2-WT or Runx2-SA adenovirus vectors. After 48 h, cells were trypsinized and cell migration was measured as described in Methods. (A, C) PC3 cell migration. Cell images after 0 and 6 h (A). Percent occupancy of the initially cell-free area was determined after the indicated times (C). (B, D) RWPE1 cell migration. Images of cells after 0 or 24 h (B). Percent occupancy of the initially cell-free area was determined after the indicated times (D). Statistics: C, aSignificantly different from LacZ control (p < 0.001), bSignificantly different from Runx2-WT (p < 0.001); D, aSignificantly different from LacZ control (p < 0.05), bSignificantly different from Runx2-WT (p < 0.01), N = 4/group.
To measure cell invasion, PC3 or RWPE1 cells were plated in the upper well of a Transwell apparatus precoated with MatrigelTM. The lower chamber contained 10% fetal bovine serum and SDF-1, a chemoattractant. Migration of cells through the MatrigelTM and Millipore membrane was scored after 24 h (Fig. 5). Representative images are shown in panels A and B while invasion is quantified in panels C and D. Wild type Runx2 increased invasion activity of PC3 cells 5-fold while phosphorylation-deficient Runx2 only increased activity 2-fold. Similarly, wild type Runx2 stimulated activity of RWPE1 cells 2.3-fold while the phosphorylation-deficient mutant was inactive. These results cannot be explained by differences in proliferation since cell numbers in each group were identical over the time frame of these experiments (result not shown).
Figure 5. Requirement for Runx2-S301, S319 phosphorylation in stimulation of prostate cell invasion.
PC3 (A, C) and RWPE1 cells (B, D) were transduced with LacZ, Runx2-WT or Runx2-SA adenovirus vectors, trypsinized and cells were assayed for invasive activity using MatrigelTM containing transwells as described in Methods. (A, B) Representative fields of stained cells migrating to bottom side of transwell membrane are shown. Cells were counted in 5 fields/insert and averaged, 3 inserts/group (C, D). Statistics: Panel C, aSignificantly different from LacZ control (p < 0.0001), bSignificantly different from Runx2-WT (p < 0.0001); Panel D, aSignificantly different from LacZ control (p < 0.05), bSignificantly different from Runx2-WT (p < 0.05), N = 4/group.
Role of Runx2 phosphorylation in tumor growth in vivo
In previous studies, Runx2 stimulated in vivo tumor formation by PC3 cell xenografts.(9) To determine if this response requires Runx2 phosphorylation, PC3 cells stably expressing a firefly luciferase reporter (PC-luc cells) were transduced with LacZ, Runx2-WT or Runx2-SA adenovirus and subcutaneously implanted into immunodeficient mice. Tumor development was monitored over a 16-day period by measuring whole-body luciferase activity. Mice were then sacrificed at day 17 and tumors were weighed and used for RNA analysis or immunohistochemistry. Wild type Runx2 significantly increased tumor growth as measured by in vivo luminescence (Fig 6A, C) and tumor size/weight at sacrifice (Fig. 6D, E). The increased growth of Runx2 transduced cells was first detected at day 4 with the most dramatic increase observed between day 9 and day 16 (Suppl. Fig. S4). In contrast, tumors formed by the Runx2-SA group were not significantly different from LacZ controls. Similar to cell culture results, tumors from Runx2-WT-treated cells expressed higher levels of VEGF, MMP9 and SPP1 mRNAs relative to LacZ controls while levels were lower with Runx2-SA transduction (Panel F).
Figure 6. Requirement for Runx2-S301, S319 phosphorylation in stimulation of in vivo tumor growth.
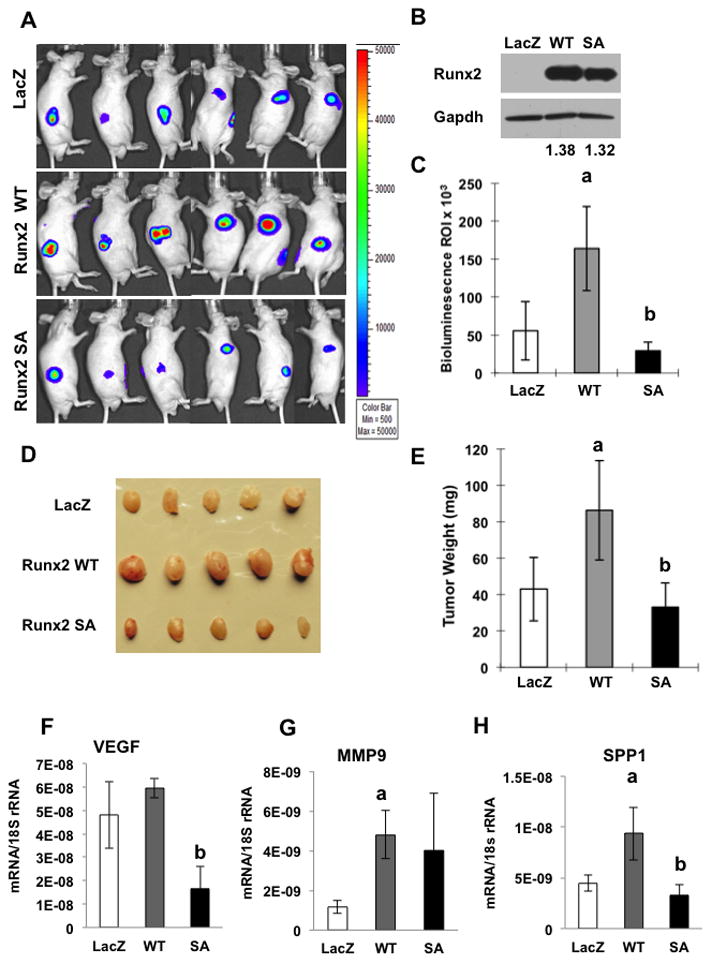
PC3-luc cells were transduced with LacZ, Runx2-WT or Runx2-SA adenovirus vectors, trypsinized, suspended in MatrigelTM and subcutaneously implanted into immunodeficient mice. Equal levels of wild type and mutant Runx2 protein expression were verified on Western blots (B). Values below each lane indicate the Runx2/Gapdh protein ratio as determined by densitometry. PC3-luc cells were detected after 16 d by in vivo bioluminescence imaging (A, C). Mice were sacrificed at d 17. Tumors were photographed (D), weighed (E) and analyzed for Vegf (F), Mmp9 (G) or Spp-1 (H) mRNA normalized to 18S rRNA. Statistics: Panels C & E, aSignificantly different from LacZ control (p < 0.05), bSignificantly different from Runx2-WT (p < 0.01); Panels F-H, aSignificantly different from LacZ control (p < 0.05), bSignificantly different from Runx2-WT (p < 0.01), N = 6/group.
To further explore the basis for differences in tumor growth, tumors were stained for human Ki67, a cell proliferation marker, and Caspase 3, a marker of apoptosis. The Ki67 antibody uniformly stained control, Runx2-WT and Runx2-SA tumors, indicating that Runx2 did not significantly affect PC3 cell proliferation (Suppl. Fig. S5). In contrast, Caspase 3 staining was reduced by approximately 60 percent in wild type Runx2 tumors. Values for the Runx2-SA group were highly variable and not significantly different from controls (Suppl. Fig. S6). These results suggest that Runx2 phosphorylation increases tumor growth by preferentially suppressing apoptosis.
Association of Runx2 phosphorylation with prostate cancer in a patient population
A tissue microarray (TMA) of normal and malignant prostate tissue and metastases was prepared from biopsies from 129 patients and examined for the presence of P-S319-Runx2 by immunohistochemistry. Results were correlated with clinical tumor parameters as described in Materials and Methods. Figure 7 shows representative immunohistochemistry of normal prostate, benign prostate hyperplasia (BPH), prostate intraepithelial neoplasia (PIN), low and high Gleason score primary prostate cancer and a lymph node metastasis. Little or no staining was seen in normal tissue. Light nuclear staining was observed in a small number of cells in the BPH sample while intense, localized nuclear staining was seen in PIN, particularly in basal cells of crypts and luminal cells. Nuclear staining was more dispersed in advanced cancer and further elevated in high Gleason score tumors and lymph node metastases. Table 1 provides quantitation of P-Runx2 staining for all 129 biopsies, expressed as the percentage of cells showing positive staining in each sample. Overall, high P-Runx2 staining was strongly correlated with primary prostate cancer (PPC) when compared with BPH or prostatitis (p < 0.001), with very low staining observed in the latter two conditions. Within the PPC cohort, significant correlations were also noted between percent P-Runx2 staining and high Gleason score (p = 0.047), primary prostate cancers with lymph node metastases at the time of initial clinical staging (p = 0.033) and primary prostate cancer with prostatic intraepithelial neoplasia (PIN, p = 0.041). Although strong P-Runx2 staining was observed in lymph node metastases, levels were not significantly different from than those observed in the primary tumors. Based on these results, we conclude that Runx2 S319 phosphorylation is a common event in PCa that can be used to distinguish between BPH, prostatitis and neoplastic disease and is also correlated with disease severity and metastases.
Figure 7. Immunohistochemical staining of prostate tissue and metastases using anti-P-S319-Runx2 antibody.
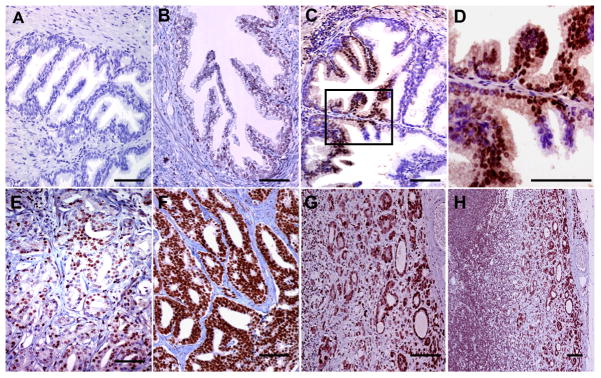
Representative samples from TMAs are shown. (A) normal prostate tissue, (B) benign prostate hyperplasia, (C) prostate intraepithelial neoplasia (PIN), (D) higher power view of boxed region in C showing strong nuclear staining of basal cells, (E) moderate staining in low Gleason score PCa, (F) very strong staining in high Gleason score PCa, (G) lymph node metastasis. (H) lower power view of lymph node metastasis showing strong staining in extra-capsular spread (ECS). Bars = 100 μm
Table 1.
TMA Analysis of P-S319-Runx2 in Prostatic Diseases, Primary Cancers and Metastases.
| GROUPS | n° | MEAN P-S319-Runx-2+ (percent) | Standard Deviation | Standard Error | Comparisons | ANOVA (P) | Student-Newman-Keuls P<0.05 |
|---|---|---|---|---|---|---|---|
| Primary Prostate Cancer (PPC) | 93 | 57.48 | 30.75 | 3.19 | PPC vs BPH | < 0.001 | YES |
| Benign prostatic hyperplasia (BPH) | 15 | 1.87 | 2.07 | 0.53 | |||
| Primary Prostate Cancer (PPC) | 93 | 57.48 | 30.75 | 3.19 | PPC vs LNM | 0.464 | NO |
| Lymph node metastases (LNM) | 13 | 64.23 | 33.16 | 9.20 | |||
| Primary Prostate Cancer (PPC) | 93 | 57.48 | 30.75 | 3.19 | PPC vs P | < 0.001 | YES |
| Prostatitis (P) | 8 | 4 | 0 | 0 | |||
| Low Gleason (LG-PPC) | 68 | 53.65 | 29.20 | 3.54 | LG-PPC vs HG-PPC | 0.047 | YES |
| High Gleason (HG-PPC) | 25 | 67.92 | 33.01 | 6.60 | |||
| Perineural permeation (PNP-PPC) | 75 | 58.43 | 30.39 | 3.51 | PNP-PPC vs NoPNP-PPC | 0.549 | NO |
| No perineural permeation (NoPNP-PPC) | 18 | 53.56 | 32.79 | 7.73 | |||
| Vascular permeation (VP-PPC) | 1 | - | - | - | VP-PPC vs NVP-PPC | N/A | |
| No vascular permeation (NVP-PPC) | 92 | 57.5 | 30.8 | 3.18 | |||
| PPC with lymph node metastasis (N+PPC) | 13 | 74.23 | 29.29 | 8.12 | N+PPC vs N0-PPC | 0.033 | YES |
| PPC without lymph node metastasis (N0-PPC) | 80 | 54.76 | 30.29 | 3.39 | |||
| PIN-PPC | 16 | 71.75 | 25.51 | 6.38 | PIN-PPC vs NoPIN-PPC | 0.041 | YES |
| NoPIN-PPC | 77 | 54.52 | 31.05 | 3.54 | |||
| Involved surgical margins (ISM-PPC) | 56 | 58.36 | 30.81 | 4.12 | ISM-PPC vs FSM-PPC | 0.738 | NO |
| Free surgical margins (FSM-PPC) | 37 | 56.16 | 31.03 | 5.10 | |||
| Extra prostatic extension (EPE-PPC) | 13 | 64.62 | 27.50 | 7.63 | EPS-PPC vs I-PPC | 0.370 | NO |
| Intraprostatic (I-PPC) | 80 | 56.33 | 31.25 | 3.49 |
Discussion
Here we establish the importance of Runx2 S301, S319 phosphorylation sites in PCa gene expression, migration, invasion and in vivo tumor growth and identify P-S319-Runx2 as a marker for more aggressive metastatic disease in a patient population. Based on these findings, we propose that the elevated MAPK activity in PCa cells phosphorylates Runx2 and stimulates transcription of genes necessary for increased tumor growth and metastasis. These studies provide a potential link between previously reported increases in MAPK signaling and Runx2 in PCa as well as a plausible mechanism to explain how Runx2 and MAPK cooperatively interact to regulate the expression of metastasis-related genes.
Effects of MAPK on Runx2 transcriptional activity in PCa cells are similar to those previously reported in osteoblasts.(21, 22, 26) Specifically, transfection of PC3 cells with constitutively active MEK1 strongly stimulated Runx2 phosphorylation and activation of an Spp1 promoter-luciferase reporter. Furthermore, this response required intact S301 and S319 Runx2 phosphorylation sites. Similarly, endogenous metastasis-related genes such as Vegf, Mmp9 and Spp1, which all contain functional Runx2-specific enhancer sequences in their proximal promoter, were strongly induced by wild type Runx2 while a phosphorylation-resistant S301A/S319A Runx2 mutant was uniformly less active (Fig. 3).(8, 10, 31, 35)
Intact S301 and S319 phosphorylation sites were also shown to be required for two previously demonstrated Runx2 actions in PCa, stimulation of in vitro cell migration and invasion (Figs. 4, 5).(6, 8, 9) More importantly, this same requirement was seen when growth of PCa cell xenografts was measured in vivo (Fig 6). However, in this experiment wild type and phosphorylation-deficient Runx2 were transiently expressed using adenovirus vectors, which remain active for only 7 to 10 days.(36) For this reason, effects of Runx2 on longer-term responses including metastasis to peripheral tissues could not be measured. The basis for the stimulatory effects of Runx2 on tumor mass appears to be mainly through suppression of apoptosis since Caspase 3 immunostaining was reduced in tumors expressing wild type Runx2 (Suppl. Figs. S5 & S6). This is consistent with a previous report showing that Runx2 can suppress apoptosis via direct up-regulation of Bcl-2.(37) The in vivo finding of reduced tumor growth with expression of the less active, phosphorylation site mutant Runx2 is consistent with previous studies that demonstrated orthotopic injection of metastatic breast or prostate cell lines modified by nearly complete knockdown of Runx2, resulted in inhibited tumor growth and osteolytic bone disease).(6, 8)
In osteoblasts, differentiation is initiated by synthesis of a collagenous extracellular matrix that activates MAPK signaling via α1 integrins. This activation is accompanied by translocation of P-ERK from the cytoplasm to nucleus where it binds and phosphorylates Runx2 already associated with regulatory regions of target genes; MAPK activation does not affect Runx2 distribution, which remains exclusively nuclear in undifferentiated and differentiated cells.(25, 27) Nuclear Runx2 localization was also seen in the more highly metastatic PCa cell lines, PC3 and C4-2B. However, in non-metastatic LNCaP cells, considerable cytoplasmic staining was also observed (Fig. 2). Although Runx2 is generally considered to be a nuclear protein, cytosolic localization has previously been reported.(7, 38, 39) For example, one study proposed that Runx2 can shuttle between nucleus and cytoplasm where it binds to microtubules.(38) Also cytoplasmic Runx2 staining was previously reported in BPH samples.(39) Thus, the concept that Runx2 can exist in the cytoplasm under certain conditions remains an intriguing subject for further study.
Cellular distribution studies using anti-Runx2 antibody are to be contrasted with results obtained with antibody against S319-P-Runx2, which in all cases showed an exclusively nuclear localization. While most nuclei in the 3 PCa cell lines were positively stained with anti-Runx2 antibody, the percentage of nuclei staining positive with anti-S319-P-Runx2 antibody progressively increased when LNCaP, C4-2B and PC3 cells were compared and this correlated with the level of MAPK activation and metastatic potential (Fig. 1).
Phosphorylation of Runx2 by the RAS/MAPK pathway was previously analyzed using a combination of in vivo and in vitro kinase assays, mass spectroscopy and mutational analysis.(21) In addition to the serine 301 and 319 sites discussed here, phosphorylation sites were identified at serine 43, 282 and 510. The role of these additional sites in osteoblast differentiation was considered to be minor because S301A/S319A mutations eliminated most MAPK-dependent transcriptional activity.(21) However, S43, S282 and S510 may have greater roles in other cell types including PCa cells. Notably, in the present study, S301A, S319A mutant Runx2, while always being considerably less active than wild type protein, still had appreciable ability to induce gene expression and stimulate migration/invasion. In this regard, studies on Runx1, a runt domain transcription factor required for hematopoiesis and associated with acute myelogenous leukemia, may be relevant. Runx1 is also phosphorylated by the RAS/MAPK pathway on serine residues homologous to S301, 319, and 510 in Runx2 as well as additional sites homologous to T326 and S329. While mutation of the first two sites reduced the transforming activity of Runx1, it did not block hematopoietic activity, which required mutation of three additional sites. (40, 41)
Runt domain transcription factors are also substrates for other kinases including Src, Pim-1, Cdc2, GSK3β, PKA and AKT.(42–47) Of these, the AKT pathway is of particular interest due to its role in PCa.(19) Inactivation of PTEN, a negative regulator of AKT, is frequently observed in metastatic PCa.(48) In fact, prostate-selective knockout of PTEN is a widely-used mouse model for PCa.(49) Activation of the RAS/MAPK pathway in PTEN-null mice dramatically accelerates PCa progression suggesting possible cooperativity between RAS/MAPK and PI3K/AKT pathways.(17, 50) Interestingly, Runx2 is elevated in the prostates of PTEN-null mice.(51) Consistent with these findings, AKT phosphorylation of Runx2 stimulates transcriptional activity and in vitro invasion by breast carcinoma cells.(47) The putative AKT phosphorylation sites (S203, T205, T207) are all in the runt domain of Runx2 and distinct from MAPK sites located mainly in the C-terminal region. Like results obtained with MAPK sites, AKT site mutation reduced, but did not abolish, all Runx2 activity. It is, therefore, possible that both RAS/MAPK and PI3K/AKT pathways phosphorylate Runx2 in cancer cells and that both types of phosphorylation contribute to overall Runx2 activity.
An anti-Runx2-S319-P antibody previously developed in the project laboratory was found to be an excellent reagent for discriminating between PCa and benign prostatic disease.(26) This antibody strongly stains nuclei of primary prostate tumors at both early (PIN) and later stages of disease progression, but minimally stains BPH or prostatitis samples. Furthermore, the amount of staining in primary tumors correlated with Gleason score and presence of lymph node metastases. Previous studies using commercially available anti-Runx2 antibodies gave variable results with noncancerous and early-stage PCa tissues. Some studies reported diffuse cytoplasmic and intense nuclear staining for total Runx2 in normal and cancerous prostate tissue while others reported staining only in more advanced high Gleason score cancers and not in early PIN.(6, 7) In contrast, anti-Runx2-S319-P antibody strongly stained basal and luminal cell nuclei in PIN (Fig 7 and Table 1). This may indicate both that this antibody is more sensitive and that early Runx2 phosphorylation is a hallmark of PCa that becomes more intense with disease progression. As noted above, anti-Runx2-S319-P antibody staining is exclusively nuclear and associated with transcriptional activation.(25, 27) Thus, detection of P-S319-Runx2 in patient samples may indicate that Runx2 is in an active form capable of stimulating the genetic program necessary for invasion and metastasis. Detection of P-S319-Runx2 in primary tumors may also have predictive value in discriminating between tumors that will remain localized and those that will progress to metastasis.
Materials and Methods
Antibodies
Antibodies used in this study were obtained from the following sources: Phospho-ERK and total-ERK antibodies from Cell Signaling, total Runx2 antibodies from MBL (cat. no. D130-3, mouse monoclonal antibody) or Abcam (cat. no. ab76956, mouse monoclonal antibody), anti-human Ki67 monoclonal antibody from Dako (cat. no. F7268) and Caspase-3 polyclonal antibody from Cell Signaling (cat. no. 9661). A rabbit polyclonal antibody specifically detecting Runx2-S319-P was previously described.(26)
Cell culture and measurement of metastasis-associated mRNAs
LNCaP, C4-2B, PC3 and RWPE1 cells were obtained from the ATCC. LNCaP, PC3 and C42B cells were maintained in RPMI 1640, and 10% FBS while RWPE1 cells were cultured in Keratinocyte SFM (Gibco Life Technologies). RNA was isolated from cells and tumors using TriZol reagent (Invitrogen) and mRNAs were measured by quantitative real-time RT/PCR as described previously.(25)
Western blot analysis and immunofluorescence
Western blot analysis of cell and tissue extracts was conducted as previously described.(26) All primary antibodies were used at a 1:500 dilution. Sheep anti-mouse IgG or donkey anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase was used at a 1:10000 dilution and HRP activity was detected by ECL (Amersham). Films were scanned for densitometric analysis using ImageJ 1.46 image analysis software. For immunofluorescence, cells were grown on glass cover slips, fixed and permeabilized with 0.25% Triton X-100 as previously described.(25) Total Runx2 (Abcam antibody) and S319-P-Runx2 antibodies were used at a 1:500 dilution. Fluorescence was detected using an Olympus FluoView 500 Laser Scanning Confocal Microscope.
Transient transfections, luciferase reporter assays and virus transduction
Transient transfection of PC3 cells was accomplished using lipofectamine reagent (Invitrogen), 0.5 μg pOPN-Luc reporter plasmid containing a -1454/+5 bp region of the proximal murine Spp1 promoter, 0.05 μg of pRL-SV40 plasmid encoding renilla reformis luciferase with or without 0.5 μg Runx2 and/or MEK1 expression plasmids.(32, 52, 53) Cells were harvested 48 hours after transfection and assayed using a dual luciferase kit (Promega). Adenovirus expressing β-galactosidase (AdLacZ), wild type Runx2 (AdRunx2-WT) or phosphorylation-deficient S301A, S319A mutant Runx2 (AdRunx2-SA) were previously described.(21) Viral titers were adjusted to give equivalent expression of Runx2 protein.
Cell migration assay
For PC3 cells, migration was measured using a RadiusTM 24 Well Cell Migration Assay Kit according to the manufacturer’s instructions (Cell Biolabs). Cells were transduced with adenovirus vectors and, after 48 h, were trypsinized and plated in migration wells at a density of 50,000 cells/cm2. After 24 h, the RadiusTM biocompatible gel layer was removed and cells were allowed to migrate to the cell-free area. For RWPE1 cells, migration was measured by plating cells in 12 well tissue culture plates and, after 24 h, removing a consistent area of cells by scraping with a Pasteur pipette. In both cases, pre-migration and post-migration images were captured with an inverted microscope at various time points and migration of cells into the initially cell-free area was analyzed using ImageJ 1.46 image analysis software.
Cell invasion assay
Cell invasion was measured using a CytoselectTM 24 Well Cell Invasion Assay Kit according to the manufacturer’s instructions (Cell Biolabs Inc.). Cells were trypsinized, suspended in serum-free medium and plated at 3 × 105 cells/well in the upper well containing MatrigelTM and allowed to invade toward the lower chamber, which contained 10% FBS and 100 ηg/mL SDF-1. After 24 h, invasion of cells to the bottom side of the insert membrane was measured by staining with Coomasie Blue and counting cells in 5 separate fields per insert.
Subcutaneous tumor growth and immunohistochemistry
All procedures were approved by the University Committee on the Use and Care of Animals. PC3 cells stably expressing firefly luciferase(54) were transduced with AdLacZ, AdRunx2-WT or AdRunx-SA vectors. After 24 h, cells were trypsinized and 1 × 106 cells were mixed with MatrigelTM and subcutaneously injected into 16 week-old immunodeficient mice (Foxn1nu/Foxn1nu; 2 injection sites/mouse). Tumor growth was monitored over a 16-day period using bioluminescence imaging. For analysis, 50 uL of 40 mg/mL D-luciferin substrate in sterile PBS was injected into each mouse and the luminescence signal was captured on an IVIS Spectrum instrument (Perkin-Elmer, Waltham, MA, USA) in the University of Michigan Small Animal Imaging Core. Image analysis was performed using Living Image software version 4.0 (Perkin-Elmer). Mice were then sacrificed and tumors were dissected free of connective tissue beofe analysis.
For immunohistochemistry, tumors were fixed in 4 percent paraformaldehyde before paraffin embedding, sectioning and immunohistochemical staining. Sections were stained with anti-human Ki67 or Caspase 3 primary antibody followed by HRP-anti-mouse or rabbit IgG. Samples were counterstained with H&E.
Tissue microarray construction and analysis
Tissue samples from a total of 129 caucasian patients with prostate disease were used to construct TMAs. Among these were 106 tumor cases represented by 93 primary prostate cancers (PPC) treated by total prostatectomy with curative intent and 13 lymph node metastases (LNM) from biopsies of patients with inoperable primary prostate adenocarcinomas, 15 cases of benign prostate hyperplasia (BPH) and 8 cases of prostatitis (P) (4 p. granulomatosa and 4 p. xantogranulomatosa). Paraffin blocks were retrieved from the archives of University of Foggia, Italy. All tissues were obtained with patient informed consent. Tissue procurement and analysis was approved by the University of Foggia Ethical Committee. For TMA construction, areas rich in non-necrotic tumor cells were identified on corresponding haematoxylin and eosin-stained sections and marked on the source paraffin block. Cores (3 mm) were obtained from the source block and transferred to the recipient master block. H&E staining of a 4-μm TMA section was used to verify all samples. Immunohistochemical analysis was performed using Ventana Benchmark® XT autostainer and/or manual standard linked streptavidin-biotin horseradish peroxidase technique (LSAB-HRP). Primary anti-Runx2-S319-P antibody was diluted 1:200 with PBS and incubated with TMA sections for 30 min. Negative control slides without primary antibody were included for each staining. Two pathologists (PB, GP) independently evaluated immunostaining results. The study was double-blinded and investigators did not have real-time access to the clinical data. For quantitative analysis, a score based on the percentage of P-Runx2-stained cells was calculated by evaluating 10 high-power representative fields (400x) on complete tumour sections. P-Runx2 staining in PPC samples was compared with benign prostatic hyperplasia (BPH) and prostatitis as well as lymph node metastases. Within the PPC group, P-Runx2 staining was related to Gleason score (Low Gleason 3+3, 3+4, 4+3; High Gleason 3+5, 4+4, 4+5, 5+3, 5+4), perineural permeation, presence or absence of lymph node metastases at initial clinical staging, presence or absence of Prostatic Intraepithelial Neoplasia (PIN); positive or negative surgical margins; and intraprostatic or extraprostatic extension.
Statistical analysis
For cell culture experiments, statistical analyses were performed using SPSS 16.0 Software. Unless indicated otherwise, each reported value is the mean ± S.D. of triplicate independent samples. Statistical significance was assessed using a one-way analysis of variance. For TMAs, analysis was conducted using Stanton Glantz statistical software 3 (MS-DOS) and GraphPad Prism software version 4.00 for Windows (Graph Pad software San Diego California, USA; www.graphpad.com). P-values <0.05 were considered significant. The one-way analysis of variance (ANOVA) and the Student-Newman-Keuls tests were used to study the differences between groups.
Supplementary Material
Acknowledgments
This work was supported by award number W81XWH-11-1-0317 from the Department of Defense (to RTF), and P01 CA093900 from the National Institutes of Health (EK) and I.R.C.C.S Centro di Riferimento Oncologico Della Basilicata Rionero in Vulture (PB). The pOPN-Luc plasmid was a generous gift from Dr. Martha Somerman (NIDCR).
Non-standard abbreviations used
- PCa
prostate cancer
- TMA
tissue microarray
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest to disclose.
Supplementary information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA: Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Logothetis CJ, Lin SH. Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer. 2005;5(1):21–8. doi: 10.1038/nrc1528. [DOI] [PubMed] [Google Scholar]
- 3.Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, et al. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25(4):589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 4.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 5.Chua CW, Chiu YT, Yuen HF, Chan KW, Man K, Wang X, et al. Suppression of androgen-independent prostate cancer cell aggressiveness by FTY720: validating Runx2 as a potential antimetastatic drug screening platform. Clin Cancer Res. 2009;15(13):4322–35. doi: 10.1158/1078-0432.CCR-08-3157. [DOI] [PubMed] [Google Scholar]
- 6.Akech J, Wixted J, Bedard K, Van der Bedard K, Hussain S, Guise T, et al. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2009;2009:1–11. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Cao W, Chellaiah MA. Integrin alphavbeta3 and CD44 pathways in metastatic prostate cancer cells support osteoclastogenesis via a Runx2/Smad 5/receptor activator of NF-kappaB ligand signaling axis. Mol Cancer. 2012;11:66. doi: 10.1186/1476-4598-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, et al. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Deen M, Akech J, Wang T, FitzGerald TJ, Altieri DC, Languino LR, et al. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010;109(4):828–37. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zelzer E, Glotzer DJ, Hartmann C, Thomas D, Fukai N, Soker S, et al. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech Dev. 2001;106(1–2):97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 11.Stewart M, Mackay N, Hanlon L, Blyth K, Scobie L, Cameron E, et al. Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res. 2007;67(11):5126–33. doi: 10.1158/0008-5472.CAN-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5(5):376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 13.Blyth K, Vaillant F, Jenkins A, McDonald L, Pringle MA, Huser C, et al. Runx2 in normal tissues and cancer cells: A developing story. Blood Cells Mol Dis. 2010;45(2):117–23. doi: 10.1016/j.bcmd.2010.05.007. Epub 2010/06/29. [DOI] [PubMed] [Google Scholar]
- 14.Cho NY, Choi M, Kim BH, Cho YM, Moon KC, Kang GH. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006;119(8):1858–62. doi: 10.1002/ijc.22071. [DOI] [PubMed] [Google Scholar]
- 15.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Jr, Weber MJ. Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res. 1999;59(2):279–84. [PubMed] [Google Scholar]
- 17.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong JH, Wang Z, Guimaraes AS, Ouyang X, Figueiredo JL, Ding Z, et al. BRAF activation initiates but does not maintain invasive prostate adenocarcinoma. PLoS ONE. 2008;3(12):e3949. doi: 10.1371/journal.pone.0003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin L, Teitell MA, Lawson DA, Kwon A, Mellinghoff IK, Witte ON. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc Natl Acad Sci USA. 2006;103(20):7789–94. doi: 10.1073/pnas.0602567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang YM, Kung HJ, Evans CP. Nonreceptor tyrosine kinases in prostate cancer. Neoplasia. 2007;9(2):90–100. doi: 10.1593/neo.06694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, et al. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J Biol Chem. 2009;284(47):32533–43. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275(6):4453–9. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 23.Ge C, Xiao G, Jiang D, Franceschi RT. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol. 2007;176(5):709–18. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120(7):2457–73. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Ge C, Franceschi RT. Differentiation-dependent association of phosphorylated extracellular signal-regulated kinase with the chromatin of osteoblast-related genes. J Bone Miner Res. 2010;25(1):154–63. doi: 10.1359/jbmr.090705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge C, Yang Q, Zhao G, Yu H, Kirkwood KL, Franceschi RT. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J Bone Miner Res. 2012;27(3):538–51. doi: 10.1002/jbmr.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Ge C, Long JP, Begun DL, Rodriguez JA, Goldstein SA, et al. Biomechanical stimulation of osteoblast gene expression requires phosphorylation of the RUNX2 transcription factor. J Bone Miner Res. 2012;27:1263–74. doi: 10.1002/jbmr.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ware JL, Paulson DF, Mickey GH, Webb KS. Spontaneous metastasis of cells of the human prostate carcinoma cell line PC-3 in athymic nude mice. J Urol. 1982;128(5):1064–7. doi: 10.1016/s0022-5347(17)53345-5. [DOI] [PubMed] [Google Scholar]
- 29.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, et al. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54(10):2577–81. [PubMed] [Google Scholar]
- 30.Wai PY, Kuo PC. Osteopontin: regulation in tumor metastasis. Cancer Metastasis Rev. 2008;27(1):103–18. doi: 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- 31.Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, et al. Transcriptional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene. 1998;17(12):1517–25. doi: 10.1038/sj.onc.1202064. [DOI] [PubMed] [Google Scholar]
- 32.Zheng CF, Guan KL. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J Biol Chem. 1993;268(32):23933–9. [PubMed] [Google Scholar]
- 33.Zhang H, Pan Y, Zheng L, Choe C, Lindgren B, Jensen ED, et al. FOXO1 Inhibits Runx2 Transcriptional Activity and Prostate Cancer Cell Migration and Invasion. Cancer Res. 2011;71(9):3257–67. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18(6):1215–23. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 35.Kwon TG, Zhao X, Yang Q, Li Y, Ge C, Zhao G, et al. Physical and functional interactions between Runx2 and HIF-1alpha induce vascular endothelial growth factor gene expression. J Cell Biochem. 2011;112(12):3582–93. doi: 10.1002/jcb.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M, Zhao Z, Koh JT, Jin T, Franceschi RT. Combinatorial gene therapy for bone regeneration: cooperative interactions between adenovirus vectors expressing bone morphogenetic proteins 2, 4, and 7. J Cell Biochem. 2005;95(1):1–16. doi: 10.1002/jcb.20411. [DOI] [PubMed] [Google Scholar]
- 37.Browne G, Nesbitt H, Ming L, Stein GS, Lian JB, McKeown SR, et al. Bicalutamide-induced hypoxia potentiates RUNX2-mediated Bcl-2 expression resulting in apoptosis resistance. Br J Cancer. 2012;107(10):1714–21. doi: 10.1038/bjc.2012.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, et al. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J Cell Physiol. 2006;206(2):354–62. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- 39.Yun SJ, Yoon HY, Bae SC, Lee OJ, Choi YH, Moon SK, et al. Transcriptional repression of RUNX2 is associated with aggressive clinicopathological outcomes, whereas nuclear location of the protein is related to metastasis in prostate cancer. Prostate Cancer and Prostatic Disease. 2012;15(4):369–73. doi: 10.1038/pcan.2012.31. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16(7):3967–79. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimi M, Goyama S, Kawazu M, Nakagawa M, Ichikawa M, Imai Y, et al. Multiple phosphorylation sites are important for RUNX1 activity in early hematopoiesis and T-cell differentiation. Eur J Immunol. 2012;42(4):1044–50. doi: 10.1002/eji.201040746. [DOI] [PubMed] [Google Scholar]
- 42.Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW, et al. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J Biol Chem. 2010;285(13):10122–9. doi: 10.1074/jbc.M109.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HR, Oh BC, Choi JK, Bae SC. Pim-1 kinase phosphorylates and stabilizes RUNX3 and alters its subcellular localization. J Cell Biochem. 2008;105(4):1048–58. doi: 10.1002/jcb.21906. [DOI] [PubMed] [Google Scholar]
- 44.Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281(11):7118–28. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura N, Kugimiya F, Oshima Y, Ohba S, Ikeda T, Saito T, et al. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS ONE. 2007;2(10):e1058. doi: 10.1371/journal.pone.0001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selvamurugan N, Pulumati MR, Tyson DR, Partridge NC. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor alpha1. J Biol Chem. 2000;275(7):5037–42. doi: 10.1074/jbc.275.7.5037. [DOI] [PubMed] [Google Scholar]
- 47.Pande S, Browne G, Padmanabhan S, Zaidi SK, Lian JB, van Wijnen AJ, et al. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J Cell Physiol. 2013;228(8):1784–92. doi: 10.1002/jcp.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22(8):1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 50.Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci U S A. 2013;110(37):E3506–15. doi: 10.1073/pnas.1303558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim M, Zhong C, Yang S, Bell AM, Cohen MB, Roy-Burman P. Runx2 regulates survivin expression in prostate cancer cells. Lab Invest. 2010;90(2):222–33. doi: 10.1038/labinvest.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fatherazi S, Matsa-Dunn D, Foster BL, Rutherford RB, Somerman MJ, Presland RB. Phosphate regulates osteopontin gene transcription. J Dent Res. 2009;88(1):39–44. doi: 10.1177/0022034508328072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18(7):4197–208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Havens AM, Pedersen EA, Shiozawa Y, Ying C, Jung Y, Sun Y, et al. An in vivo mouse model for human prostate cancer metastasis. Neoplasia. 2008;10(4):371–80. doi: 10.1593/neo.08154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.