Abstract
Estrogen receptors, comprised of ERα and ERβ isoforms in mammals, act as ligand-modulated transcription factors and orchestrate a plethora of cellular functions from sexual development and reproduction to metabolic homeostasis. Herein, I revisit the structural basis of the binding of ERα to DNA and estradiol in light of the recent discoveries and emerging trends in the field of nuclear receptors. A particular emphasis of this review is on the chemical and structural diversity of an ever-increasing repertoire of physiological, environmental and synthetic ligands of estrogen receptors that ultimately modulate their interactions with cognate DNA located within the promoters of estrogen-responsive genes. In particular, modulation of estrogen receptors by small molecule ligands represents an important therapeutic goal toward the treatment of a wide variety of human pathologies including breast cancer, cardiovascular disease, osteoporosis and obesity. Collectively, this article provides an overview of a wide array of small organic and inorganic molecules that can fine-tune the physiological function of estrogen receptors, thereby bearing a direct impact on human health and disease.
Keywords: Estrogen receptors, Endoestrogens, Phytoestrogens, Xenoestrogens, SERMs, Metalloestrogens
1. Introduction
In mammals, estrogen receptor is expressed in two alternative isoforms designated ERα and ERβ (1, 2). Together, these mediate a plethora of cellular functions from sexual development and reproduction to metabolic homeostasis. ERα and ERβ are members of a family of ligand-modulated transcription factors that have come to be known as nuclear receptors (NRs) (3–6). As their name implies, the binding of ligand is a pre-requisite for the subsequent binding of NRs in a sequence-specific manner to their cognate DNA within the promoters of target genes. This mechanism of action is in sharp contrast to the binding of classical transcription factors to DNA, which are not dependent upon prior activation with a specific ligand.
Notably, all members of NR family share a core modular architecture comprised of a central DNA-binding (DB) domain flanked between an N-terminal trans-activation (TA) domain and a C-terminal ligand-binding (LB) domain (7–9). A typical scenario for the activation of nuclear receptors, as schematically illustrated for ERα in Figure 1, involves the secretion of lipophilic messengers such as hormones and vitamins by appropriate tissues. Upon their diffusion through the cell membrane, the binding of these ligands to the LB domain culminates in a series of events involving the translocation of nuclear receptors into the nucleus and subsequent modulation of expression of target genes (10–12). While the DB domain recognizes specific promoter elements, the LB domain additionally serves as a platform for the recruitment of a multitude of cellular proteins, such as transcription factors, co-activators and co-repressors, to the site of DNA transcription and thereby allowing nuclear receptors to exert their action at genomic level in a concerted fashion (13, 14). While the trans-activation function of the LB domain is ligand-dependent, the TA domain operates in an autonomous manner and it is believed to be responsive to growth factors acting through the MAPK signaling and may further synergize the action of various co-activators and co-repressors recruited by the LB domain at the site of DNA transcription (15, 16). In this manner, nuclear receptors orchestratea diverse array of cellular functions from embryonic development to metabolic homeostasis and their malfunction has been widely implicated in disease (7, 17–21).
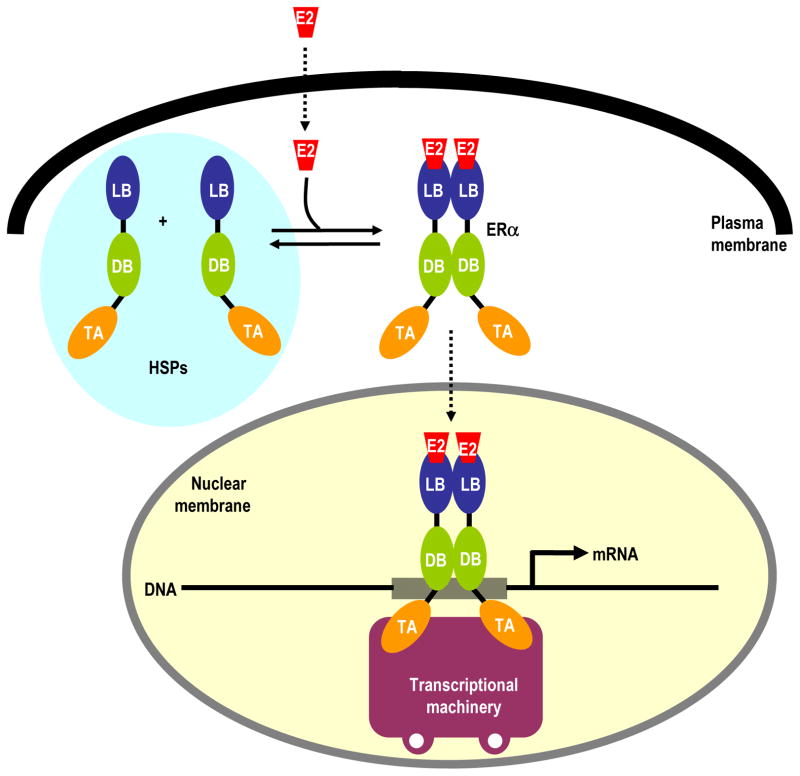
Figure 1.
A schematic illustrating the activation of estrogen receptor (ERα) by estradiol (E2). ERα is comprised of the core TA-DB-LB modular architecture that is also shared by other members of the nuclear receptor family, where DB is the central DNA-binding (DB) domain flanked between an N-terminal trans-activation (TA) domain and a C-terminal ligand-binding (LB) domain. Notably, ERα exists in dimer-monomer equilibrium in the cytoplasm in association with heat shock proteins (HSPs). Upon its diffusion through the plasma membrane, E2 binds to the LB domain and shifts the monomer-dimer equilibrium in favor of the latter allowing ERα to translocate into the nucleus where it binds to estrogen response element (ERE) element within the promoters of target genes via the DB domain, thereby leading to the recruitment of transcriptional machinery.
First discovered more than half a century ago, ERα and ERβ mediate the action of estrogens such as estradiol and their hyperactivation leads to the genesis of large fractions of breast cancer (22–28). In addition to breast cancer, estrogen receptors are also implicated in a plethora of other human pathologies including cardiovascular disease (29), osteoporosis (30), and obesity (31). While the physiological action of ERα is highly complex and involves multiple layers at both the genomic and non-genomic level, two major pathways by which ERα participates in the regulation of transcriptional machinery are the classical and the non-classical pathways. In the classical pathway, ERα binds to the promoters of estrogen-responsive genes containing the estrogen response element (ERE) through its DB domain in an estradiol-dependent manner. Remarkably, the ability of ERα to bind to the promoters of target genes in an estradiol-independent manner upon post-translational phosphorylation within the TA domain by kinases such as Cdk2 is also well-documented (32–34). Examples of ERα-responsive genes regulated by ERα through the classical pathway include Myc, Fos, cathepsin D and pS2 (27, 35–39). In the non-classical pathway, ERα regulates gene transcription without directly binding to DNA but in an estradiol-dependent manner. This is made possible by the fact ERα tethers to other transcription factors such as ATF2, Fos, Jun, Elk and SP1 already bound to their cognate elements within the promoters of their target genes and, in so doing, aids in the recruitment of a wide array of transcriptional co-regulators (27, 35, 40, 41). In this manner, ERα modulates expression of estrogen-responsive genes that do not contain an ERE motif within their promoters solely through protein-protein interactions. Examples of estrogen-responsive genes regulated by ERα through the non-classical pathway include Bcl2, ovalbumin, collagenase, cyclin D1 and IGF1 (42–46).
In this article, I revisit the structural basis of the binding of ERα to DNA and estradiol in light of the recent discoveries and emerging trends in the field of nuclear receptors. A particular emphasis of this review is on the chemical and structural diversity of an ever-increasing repertoire of physiological, environmental and synthetic ligands of estrogen receptors that ultimately modulate their interactions with cognate DNA located within the promoters of estrogen-responsive genes. The role of these ligand-mediated interactions on human health and disease is discussed.
2. Structural basis of ERα-DNA interactions
Upon the binding of estradiol to the LB domain, ERα translocates to the nucleus and binds as a homodimer with a two-fold axis of symmetry to the estrogen response element (ERE), containing the AGGTCAnnnTGACCT consensus sequence, located within the promoters of target genes (47, 48). DNA-binding is accomplished through a pair of tandem C4-type Zinc fingers located within the DB domain, with each finger containing a Zn2+ ion coordinated in a tetrahedral arrangement by four highly conserved cysteine residues to generate the Zn2+[Cys]4 metal-protein complex (49, 50) (Figure 2a). Importantly, while the first Zinc finger (ZF-I) within each monomer of DB domain recognizes the hexanucleotide sequence 5’-AGGTCA-3’ within the major groove at each end of the ERE duplex, the second Zinc finger (ZF-II) is responsible for the homodimerization of DB domain. It is noteworthy that environmental metals can exert dramatic effects on ERα-DNA interactions by virtue of their ability to replace the Zn2+ divalent ion within the zinc fingers of the DB domain (51–53). In particular, the DB domain of ERα reconstituted with divalent ions of zinc, cadmium, mercury and cobalt binds to DNA with affinities in the nanomolar range, while divalent ions of barium, copper, iron, lead, manganese, nickel and tin are unable to regenerate DB domain with DNA-binding potential though they can compete with zinc for coordinating the cysteine ligands within the zinc fingers.
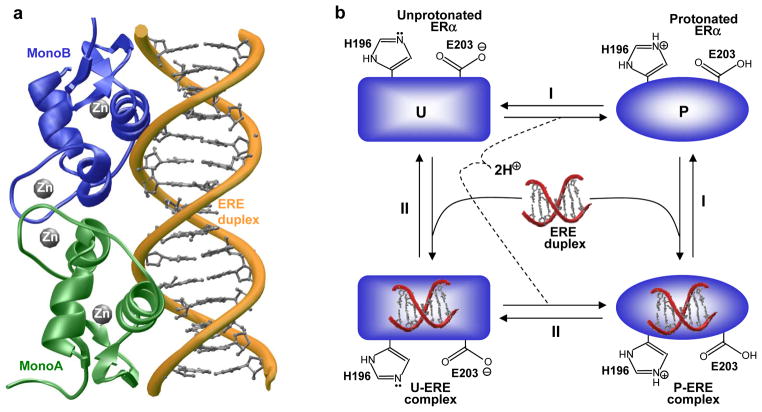
Figure 2.
Structural basis of DNA-binding to ERα. (a) Structural model of the DB domain of ERα as a homodimer in complex with ERE duplex containing the AGGTCAcagTGACCT consensus sequence. One monomer of the DB domain is colored green (MonoA) and the other in blue (MonoB). The Zn2+ divalent ions are depicted as gray spheres. The DNA backbone is shown in yellow and the bases are colored gray for clarity. The structural model was built using the MODELLER software based on homology modeling (140). The crystal structure of DB domain of ERα in complex with a dsDNA oligo containing the ERE motif but with varying flanking sequences, as determined by Rhodes and co-workers (49), was used as a template (PDBID 1HCQ). The structure was rendered using RIBBONS (141). (b) A thermodynamic cycle for the various equilibria linked to the binding of ERα to DNA: (I) ERα becomes protonated in the free form and the resulting protonated form (P) binds to DNA; (II) ERα binds to DNA in the unprotonated form (U) and the resulting liganded form becomes protonated.
Interestingly, recent work from our laboratory suggests that the binding of ERα to DNA is coupled to proton uptake by two ionizable residues, H196 and E203, located at the protein-DNA interface (54). Unsurprisingly, alanine substitution of these ionizable residues decouples protonation and hampers the binding of ERα to DNA by nearly an order of magnitude. This pH-dependent effect of ERα-DNA interaction is further illustrated via a thermodynamic cycle (Figure 2b). It is clearly evident from such a cycle that the binding of ERα to DNA may or may not be coupled to proton uptake depending on solution pH. Thus, under low pH values, ERα may become fully protonated prior to binding DNA. On the other hand, under high pH values, ERα may become fully unprotonated and the proton uptake may be decoupled for its binding to DNA, albeit with a much diminished affinity. Although the binding of DB domain of ERα to DNA appears to be coupled to proton uptake, it is not clear how such coupled equilibrium might dictate the physiological role of this important nuclear receptor. It is noteworthy that changes in intracellular pH regulate a multitude of cellular processes such as metabolic homeostasis and apoptosis (55). Furthermore, it is believed that ionizable residues within proteins sense such changes and activate a variety of proton pumps and ion transporters that in turn mediate extracellular transport of protons and anions to regulate intracellular pH (56–58). It is thus conceivable that changes in intracellular pH may also tightly regulate the transcriptional activity of ERα through direct modulation of two ionizable residues, H196 and E203, located at the protein-DNA interface. It should also be noted that H196 and E203 are predominantly conserved across ~50 members of the nuclear receptor family, implying that proton-coupled equilibrium may serve as a key regulatory switch for modulating protein-DNA interactions central to nuclear receptor function and regulation.
3. Structural basis of ERα-estradiol interactions
As noted above, the binding of estradiol to the LB domain represents a key priming step for the subsequent binding of ERα via its DB domain to gene promoters. Importantly, the LB domain of ERα binds to estradiol as a homodimer with a two-fold axis of symmetry, wherein each monomer is largely comprised of a triple-layered anti-parallel α-helical sandwich capped at one end by a small two-stranded anti-parallel β-sheet (20) (Figure 3). Within this α-helical sandwich, the central layer comprised of H5/H6/H9/H10 α-helices is flanked by one layer of H1/H2/H3/H4 α-helices on the left and one layer of H7/H8/H11 α-helices on the right as viewed from the direction of the sole β-sheet (S1/S2) or the C-terminal α-helix H12. Notably, the liganded and unliganded conformations of the LB monomers are very similar except for the spatial orientation and structural ordering of H12 helix relative to the rest of the helical-bundle. In the unliganded conformation (Figure 3a), the H12 helix flips outwards so as to act as a dynamic lid and make way for estradiol to enter into the deep interior hydrophobic pocket. Subsequent to estradiol entry, the repositioning of H12 helix within each monomer seals the pocket accommodating the ligand. Importantly, such ligand-induced trigger allows the entire LB domain to undergo structural re-arrangement to adopt a conformation that best “attracts” a diverse array of transcriptional co-activators (59–61). This model thus exquisitely explains how the binding of estradiol activates ERα for its role in transcriptional regulation.
Figure 3.
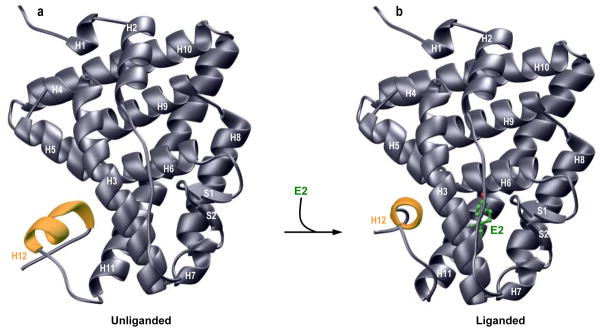
Structural models of unliganded (a) and liganded (b) LB domain of ERα in complex with estradiol (E2). In each case, only one monomer of LB domain is shown in gray with the C-terminal H12 helix colored yellow for contrast. E2 is depicted as a ball-and-stick model with various atom types and associated bonds shown in green (C) and red (O). In (a), the H12 helix is partially unstructured and displaced resulting in the disorganization of the ligand binding pocket in the unliganded conformation. In (b), subsequent binding of E2 induces the formation of ligand binding pocket, wherein the H12 helix winds up by an extra turn and undergoes ~180° rotation about an axis along the H11 helix. Such structural rearrangement allows H12 helix to block the mouth of the pocket accommodating E2 in what has come to be known as the “mouse trap” model. In both (a) and (b), the structural models were built using the MODELLER software based on homology modeling (140). For the unliganded conformation (a), the crystal structure of the LB domain of PPARγ nuclear receptor, as determined by Milburn and co-workers (142), was used as a template (PDBID 1PRG). It should be noted that the ligand-binding pockets within each monomer of PPARγ are structurally disordered. For the liganded conformation, the crystal structure of the LB domain of ERα in complex with E2, as determined by Carlquist and co-workers (20), was used as a template (PDBID 1ERE). It is noteworthy that a portion of the H9-H10 loop is missing in the crystal structure of ERα bound to E2. The structures were rendered using RIBBONS (141).
In the liganded conformation (Figure 3b), estradiol is trapped between the three α-helical layers such that the S1-S2 β-sheet serves as the floor and H12 α-helix as the lid in what is often described as a “mouse trap” model of ligand binding to nuclear receptors (62). The recognition of estradiol is in part achieved through intermolecular hydrogen bonding involving the 3-OH of the A-ring in estradiol with carboxylate moiety of E353 and the guanidinium group of R394 as well as the 17β-OH of the D-ring with imidazole sidechain of H524. Additionally, van der Waals contacts are established as a result of sandwiching of the A/B-ring interface ofestradiol with A350 and L387 on one side and F404 on the other side as well as sandwiching of the D-ring with I424 on one side and L525 on the opposite side. In short, the liganded conformation of LB domain harbors no route for estradiol entry into the deep hydrophobic pocket within the interior of the protein constituted by various α-helices and capped by the sole S1-S2 β-sheet at one end and H12 α-helix at the other. To overcome this physical constraint, the binding of estradiol is coupled to conformational changes in an allosteric manner so as to allow ligand entry and its subsequent entrapment within the deep pocket completely shielded from the external sea of water.
4. Diversity of estrogen receptor ligands
In addition to naturally produced estrogens within the body, a diverse array of small organic and inorganic molecules serve as estrogen receptor (ER) ligands by virtue of their ability to recognize the LB domain in a specific manner. While majority of these ligands display higher selectivity toward ERα, a number of ERβ-selective compounds have also recently been discovered in spite of the rather high similarity between the ligand binding pockets of the two isoforms. It is important to note that ERα and ERβ usually act in an antagonistic manner. For example, while the mitogenic action of ERα in breast tissue is responsible for cellular proliferation leading to the development of breast carcinoma, activation of ERβ has an anti-proliferative effect in breast (63). In a similar manner, ERβ is also believed to exert anti-proliferative effect on other tissues such as prostate, colon, lung, and brain (2, 64–68). Accordingly, the development of small molecule ligands that can selectively activate ERβ hold promise for the treatment of indications such as cancers and neurological disorders (69–78). Below, a brief overview of the five major classes of ER ligands is provided.
4.1 Endoestrogens
Endoestrogens are physiological estrogens endogenously produced within the body. Examples include estradiol, estriol and estrone (Figure 4). Endoestrogens are steroidal compounds produced from cholesterol by the sex glands such as ovaries in females and testes in males among other organs such as the liver, brain and adrenal glands. While estradiol is the predominant form in both females and males, estriol is primarily produced during pregnancy and estrone during menopause. In addition to its role in sexual development and reproductive functions in females, estradiol is also involved in a plethora of other physiological functions including lipid metabolism, cardiovascular function and bone maintenance in both males and females (79–83). Endoestrogens are also constituents of oral contraceptives and hormone replacement therapy.
Figure 4.
Chemical structures of endoestrogens estrone (E1), 17β-estradiol (E2) and estriol (E3).
4.2 Phytoestrogens
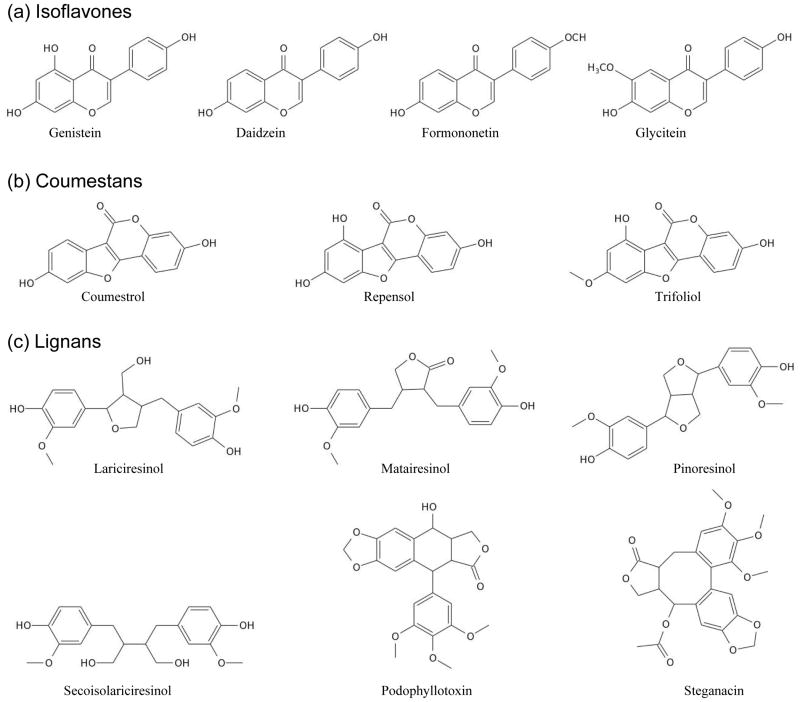
Phytoestrogens are non-steroidal polyphenolic compounds naturally produced by plants. Three major groups are isoflavones, coumestans and lignans (Figure 5). Because they share chemical and structural similarities with estradiol, phytoestrogens can exert estrogenic or antiestrogenic effects by virtue of their ability to bind to the LB domain of estrogen receptors and block the binding of estradiol in a competitive manner (84). In particular, genistein, coumestrol and liquiritigenin appear to be more selective toward ERβ by virtue of their ability to bind to its LB domain with much higher affinity compared to that of ERα (75, 85–87). Dietary sources rich in phytoestrogens include nuts, oilseeds, soy products, cereals, breads, legumes, vegetables and fruits (88). However, phytoestrogens are not considered dietary supplements as their lack in diet does not lead to any deficiency syndrome nor they play any essential biological function. Finally, the role of phytoestrogens in reducing the risk of diseases such as cancer, heart disease and osteoporosis remains controversial (89–94). On the contrary, it isbelieved that some phytoestrogens may be harmful to health, particularly if their intake is excessive.
Figure 5.
Chemical structures of three major groups of phytoestrogens. (a) Isoflavones include genistein, daidzein, formononetin and glycitein. (b) Coumestans include coumestrol, repensol andtrifoliol. (c) Lignans include lariciresinol, matairesinol, pinoresinol, secoisolariciresinol, podophyllotoxin and steganacin.
4.3 Xenoestrogens
Xenoestrogens are comprised of a wide variety of non-natural synthetic chemical compounds with estrogenic effects. These can be broadly divided into five major categories: medicinal drugs, food additives, body cosmetics, environmental pesticides and industrial chemicals (Figure 6). Although medicinal drugs such as diethylstilbestrol (DES) and ethinyl estradiol were specifically developed to possess estrogenic activity, other xenoestrogens are also believed to mimic the action of endoestrogens in humans and other animals and, as such, they can also interfere with cellular processes leading to serious implications on health (95–101). In particular, a large body of data suggests that there is a positive correlation between the uptake of xenoestrogens and the development of breast cancer in humans (102–107). There is also growing evidence that xenoestrogens may impact human male reproductive health (108–110). Because of their structural resemblance to endoestrogens, xenoestrogens are believed to exert their estrogenic effects by virtue of their ability to bind to the LB domain of estrogen receptors. It is noteworthy that synthetic non-steroidal compounds with preferential selectivity toward ERβ have also been developed (2, 74, 75, 111).
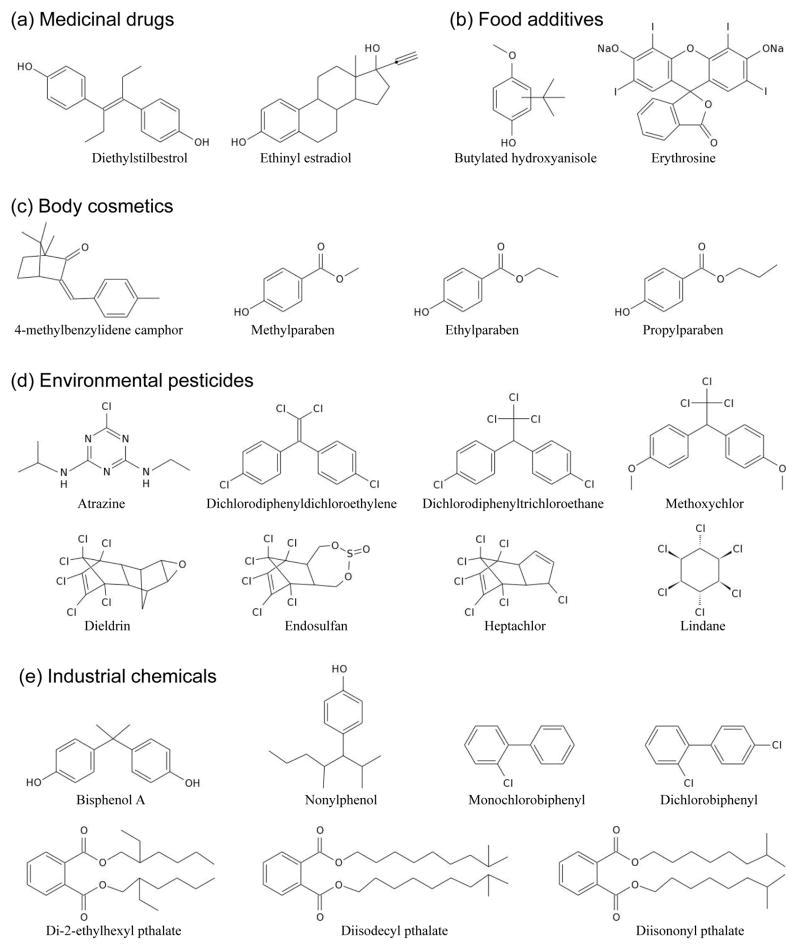
Figure 6.
Chemical structures of five major groups of xenoestrogens. (a) Medicinal drugs include diethylstilbestrol and ethinyl estradiol. (b) Food additives include butylated hydroxyanisole and erythrosine. (c) Body cosmetics include camphors such as 4-methylbenzylidene camphor and parabens such as methylparaben, ethylparaben and propylparaben. (d) Environmental pesticides include atrazine, dichlorodiphenyldichloroethylene, dichlorodiphenyltrichloroethane, methoxychlor, dieldrin, endosulfan, heptachlor and lindane. (e) Industrial chemicals include phenols such as bisphenol A and nonylphenol, biphenyls such as monochlorobiphenyl and dichlorobiphenyl, and phthalates such as di-2-ethylhexyl phthalate, diisodecyl phthalate and diisononyl phthalate.
4.4 SERMs
In the so-called ER-positive breast cancer, which accounts for nearly three quarters of all breast cancers, the estrogen receptors are overexpressed and the cancer heavily relies on continuous supplies of endoestrogens to develop. Among the first lines of defense against ER-positive breast cancer and other ER-related syndromes are a group of non-steriodal compounds that have come to be known as the selective estrogen receptor modulators (SERMs) (112). Examples include tamoxifen, raloxifene, clomifene, ormeloxifene andtoremifene (Figure 7). What distinguishes SERMs from xenoestrogens is that they harbor functional duality in their ability to act both as agonists and antagonists of ERα in different tissues (113–115). For example, tamoxifen acts as an antagonist in breast but as an agonist in uterus. Thus, while tamoxifen is often the preferred choice of treatment for ER-positive breast cancer, it can also induce endometrial cell growth resulting in uterine cancer. On the other hand, raloxifene acts as an antagonist in both breast and uterus coupled with its agonistic action in bone, making it a preferred choice of therapy in postmenopausal women with osteoporosis. While SERMs are largely selective for ERα, synthetic steroidal analogs that can preferentially modulate ERβ have also been reported (2, 74, 75, 116–119).
Figure 7.
Chemical structures of SERMs. Examples include tamoxifen, raloxifene, clomifene, ormeloxifene and toremifene.
Importantly, while SERMs exert their antagonistic action by virtue of their ability to compete with estradiol (E2) for binding to a deep interior hydrophobic pocket within the LB domain of ERα, they do so via a distinct mechanism (120, 121) (Figure 8). Binding of E2 agonist induces a conformational change in the LB domain that results in the repositioning of H12 helix so as to enable it to seal the ligand binding pocket (Figure 8a). Additionally, such structural rearrangement ensures that the subsequent binding of transcriptional co-activators, such as TIF2, to the canonical LXXLL motif located within a binding cleft right above H12 helix is not sterically hindered. By sharp contrast, owing to their bulkier groups, the binding of SERMS such as 4-hydroxytamoxifen (OHT)—the active form of tamoxifen prodrug after liver metabolism—to the LB domain results in a slightly differential conformational change compared to that observed upon the binding of E2 (Figure 8b). This in part involves unwinding of H3 and H11 helices by about one turn upon the binding of OHT, thereby relieving the tension on H12 helix and allowing it to occupy the LXXLL binding cleft in lieu of acting as a “lid” on the ligand binding pocket. Accordingly, while the agonist-bound conformation of H12 helix promotes the binding of transcriptional co-activators (Figure 8a), the antagonist-bound conformation hampers the binding of co-activators in a competitive manner (Figure 8b).
Figure 8.
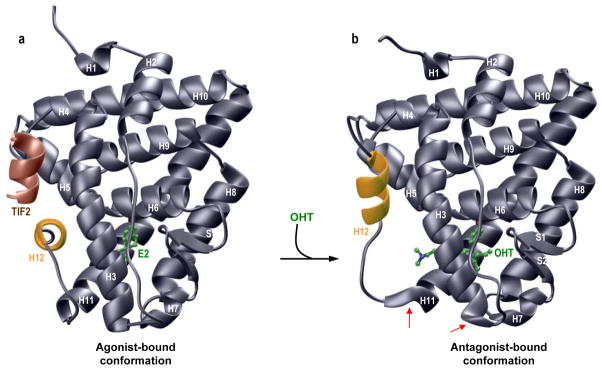
Crystal structures of LB domain of ERα in agonist-bound (a) and antagonist-bound (b) conformations. In each case, only one monomer of LB domain is shown in gray with the C-terminal H12 helix colored yellow for contrast. In (a), LB domain is bound to estradiol (E2) and a short helical peptide (colored brown), whichis derived from the TIF2 transcriptional co-activator and harbors the canonical LXXLL motif. E2 is depicted as a ball-and-stick model with various atom types and associated bonds shown in green (C) and red (O). In (b), LB domain is bound to 4-hydroxytamoxifen (OHT), depicted as a ball-and-stick model with various atom types and associated bonds shown in green (C), red (O), and blue (N). Note the similarity in the mode of docking of TIF2 peptide to LB domain in response to E2 agonist (a), and that adopted by H12 helix in response to OHT antagonist (b). In (b), the red arrows indicate the unwinding of H3 and H11 helices by about one turn upon the binding of OHT. The structures shown in (a) and (b) were respectively rendered from atomic coordinates provided by PDB codes 1GWR and 3ERT using RIBBONS (141).
How does this explain the antagonistic action of OHT in breast but its agonistic role in uterus? The ability of H12 helix to occupy two distinct positions within the LB domain in response to agonist versus antagonist binding is governed by dynamic equilibrium. In other words, the H12 helix bears the propensity to transiently resonate between these two distinct positions depending on the biological context. In the absence of co-activators, the H12 helix would predominantly occupy the LXXLL site upon OHT binding. As the cellular concentration of transcriptional co-activators rises, the H12 helix would become competitively displaced from the LXXLL site and re-engage at the agonist site, thereby enabling transcriptional activation of LB domain in response to OHT binding. It thus goes without saying that the ability of SERMs to act as agonists or antagonists arises from the differential ratios of transcriptional co-activators to co-repressors in different tissues (27, 122–124).
From a therapeutic perspective, the design of ER agonists should be such that their binding favors the orientation of H12 helix to occupy the agonist site within the LB domain in a manner akin to that induced by the binding of E2. This mode of binding is indeed shared by synthetic agonists such as diethylstilbestrol (DES) (121). On the other hand, the design of ER antagonists should be such that their binding reorients H12 helix to occupy the LXXLL site with high affinity within the LB domain so that it cannot be competitively displaced by rising cellular levels of transcriptional co-activators. As discussed above, this latter phenomenon is indeed widely adopted by SERMS such as tamoxifen and raloxifene (20, 121). Equally importantly, it should also be important to design or screen for new small molecules that could directly bind with high affinity to the LXXLL site in lieu of competing with E2 for binding to the interior hydrophobic pocket within the LB domain. Such novel antagonists may serve as more effective drugs for the treatment of ailments such as breast cancer. Regardless of their precise mechanism of action, a whole new plethora of SERMs is currently under development to modulate the action of ERα in numerous cellular activities. Unsurprisingly, however, clinical trials on many of these promising candidates, such as ERB-041 and ORG37663, have failed to live up to their therapeutic promise (125, 126).
4.5 Metalloestrogens
Unlike all of the ligands of estrogen receptors discussed above, small inorganic compounds in the form of heavy metal ions is rapidly emerging as a new class of ER ligands with estrogenic activity. Several lines of evidence suggest that heavy metal ions adsorbed from various environmental sources mimic the action of endoestrogens and can potentiate the transcriptional activity of estrogen receptors within the body (127–129). High levels of many of these heavy metal ions have indeed been detected in various tissues, including the breast, and there appears to be a direct correlation between their accumulation and the subsequent development and progression of cancer (104, 130, 131). Because of their estrogenic activity, these heavy metal ions have come to be known as metalloestrogens. Examples include a wide variety of cations such as those of aluminum (Al3+), antimony (Sb3+), barium (Ba2+), cadmium (Cd2+), chromium (Cr2+), cobalt (Co2+), copper (Cu2+), lead (Pb2+), mercury (Hg2+) and nickel (Ni2+), and anions such as arsenite (AsO33−), selenite (SeO32−) and vanadate (VO43−). Given the environmental contamination of air and water, food and drinks, cigarettes and medicines, and a plethora of cosmetic products with such heavy metal ions coupled with the knowledge that the hyperactivation of estrogen receptors is linked to the genesis of large fractions of breast cancer (22–27, 104, 130), efforts are being undertaken to decipher the molecular basis of how metalloestrogens activate estrogen receptors. Several studies indicate that the metalloestrogens exert their effects by virtue of their ability to coordinate to specific amino acid residues within the LB domain of estrogen receptors so as to block the binding of estradiol in a non-competitive manner (129, 132–134). It is believed that metalloestrogens may employ various geometries to coordinate with specific residues in the LB domain in a manner akin to their coordination with other proteins (135).
5. Concluding Remarks
In addition to naturally produced estrogens within the body, the library of small organic and inorganic molecules identified as potential ligands of estrogen receptors over the past half century or so runs into hundreds and it is likely that there are thousands of other potential ER ligands that are yet to be discovered from a plethora of environmental and synthetic sources. On the basis of structure activity relationships (136, 137), it has been argued that despite chemical and structural diversity, all organic ligands of ER bind to the LB domain with a remarkably similar mechanism as follows: (1) Hydroxyl groups or similar moieties engage in hydrogen bonding with the LB domain in a manner akin to 3-OH and 17β-OH moieties of estradiol; (2) The hydroxyl groups, if present, are usually separated by about 12 , the distance between 3-OH and 17β-OH moieties of estradiol; and (3) A rigid hydrophobic planar ring structure mimicking estradiol is usually present. Additionally, a set of hierarchical rules for the identification of novel potential organic ligands of ER have also been developed as summarized here: (1) If a chemical contains no ring structure or a non-aromatic ring structure that does not contain O, S, N, or other heteroatoms for H-bonding, then it is unlikely to be an ER ligand; (2) If a chemical has a non-OH aromatic structure, then it could serve as an ER ligand provided other structural requirements are met; and (3) If a chemical contains a phenolic ring, then it tends to be an ER ligand if it contains any additional structural features similar to estradiol. However, given that a wide spectrum of heavy metals ions can also serve as potential ER ligands, the above set of rules may be of little use in identifying novel ligands of estrogen receptors.
In sum, this review has revisited the structural basis of the binding of ERα to DNA and estradiol in light of the recent discoveries and emerging trends in the field of nuclear receptors. Additionally, I have also provided herein an overview of the chemical diversity of ER ligands and how they employ distinct mechanisms to recognize estrogen receptors. Modulation of estrogen receptors by small molecule ligands represents an important step toward thetreatment of a wide variety of human pathologies including breast cancer (22–28), cardiovascular disease (29), osteoporosis (30), and obesity (31). In particular, development of drugs that can selectively activate ERβ over ERα would be highly desirable for combating estrogen-related disorders. Toward this goal, clinical trials on MF101 for the treatment of hot flushes in menopausal women show a great deal of promise for the future development of ERβ-selective therapy (138, 139). Further research is thus clearly warranted to not only identify highly selective small molecule ligands of ERα and ERβ but also to advance our molecular understanding of how they modulate estrogen receptors.
Acknowledgments
I am grateful to Brian Deegan for his help with the preparation of some of the figures used here and for many thoughtful discussions. This work was supported by the National Institutes of Health Grant R01-GM083897 and funds from the Sylvester Comprehensive Cancer Center to AF.
Abbreviations
- DB
DNA-binding
- ER
Estrogen receptor
- ERα
Estrogen receptor alpha
- ERβ
Estrogen receptor beta
- ERE
Estrogen response element
- LB
Ligand-binding
- MAPK
Mitogen-activated protein kinase
- NR
Nuclear receptor
- SERM
Selective estrogen receptor modulator
- TA
Trans-activation
- ZF
inc finger
References
- 1.Planey SL, Kumar R, Arnott JA. Estrogen receptors (ERalpha versus ERbeta): friends or foes in human biology? J Recept Signal Transduct Res. 2014;34:1–5. doi: 10.3109/10799893.2013.853188. [DOI] [PubMed] [Google Scholar]
- 2.Paterni I, Granchi C, Katzenellenbogen JA, Minutolo F. Estrogen receptors alpha (ERalpha) and beta (ERbeta): subtype-selective ligands and clinical potential. Steroids. 2014;90:13–29. doi: 10.1016/j.steroids.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- 5.Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, Steffen DL, Tsai MJ, Tsai SY, Yu R, Margolis RN, Evans RM, O'Malley BW. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- 8.Barnett P, Tabak HF, Hettema EH. Nuclear receptors arose from pre-existing protein modules during evolution. Trends Biochem Sci. 2000;25:227–228. doi: 10.1016/s0968-0004(00)01579-6. [DOI] [PubMed] [Google Scholar]
- 9.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64:310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 10.Egea PF, Klaholz BP, Moras D. Ligand-protein interactions in nuclear receptors of hormones. FEBS Lett. 2000;476:62–67. doi: 10.1016/s0014-5793(00)01672-0. [DOI] [PubMed] [Google Scholar]
- 11.Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- 12.Green S, Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988;4:309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- 13.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ham J, Parker MG. Regulation of gene expression by nuclear hormone receptors. Curr Opin Cell Biol. 1989;1:503–511. doi: 10.1016/0955-0674(89)90012-4. [DOI] [PubMed] [Google Scholar]
- 15.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 16.Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- 17.Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46:13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb B, Beitel LK, Wu J, Elhaji YA, Trifiro M. Nuclear receptors and disease: androgen receptor. Essays Biochem. 2004;40:121–136. doi: 10.1042/bse0400121. [DOI] [PubMed] [Google Scholar]
- 19.Gurnell M, Chatterjee VK. Nuclear receptors in disease: thyroid receptor beta, peroxisome-proliferator-activated receptor gamma and orphan receptors. Essays Biochem. 2004;40:169–189. doi: 10.1042/bse0400169. [DOI] [PubMed] [Google Scholar]
- 20.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 21.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen EV, Jacobson H. Basic guides to the mechanism of estrogen action. Recent Prog Horm Res. 1962;18:318–414. [Google Scholar]
- 23.Jensen EV. On the mechanism of estrogen action. Perspect Biol Med. 1962;6:47–59. doi: 10.1353/pbm.1963.0005. [DOI] [PubMed] [Google Scholar]
- 24.Toft D, Gorski J. A receptor molecule for estrogens: isolation from the rat uterus and preliminary characterization. Proc Natl Acad Sci U S A. 1966;55:1574–1581. doi: 10.1073/pnas.55.6.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toft D, Shyamala G, Gorski J. A receptor molecule for estrogens: studies using a cell-free system. Proc Natl Acad Sci U S A. 1967;57:1740–1743. doi: 10.1073/pnas.57.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- 27.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 29.Murphy E. Estrogen signaling and cardiovascular disease. Circ Res. 2011;109:687–696. doi: 10.1161/CIRCRESAHA.110.236687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson S, Gustafsson JA. Estrogen receptors: therapies targeted to receptor subtypes. Clin Pharmacol Ther. 2011;89:44–55. doi: 10.1038/clpt.2010.226. [DOI] [PubMed] [Google Scholar]
- 31.Mauvais-Jarvis F, Clegg DJ, Hevener AL. The role of estrogens in control of energy balance and glucose homeostasis. Endocr Rev. 2013;34:309–338. doi: 10.1210/er.2012-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogatsky I, Trowbridge JM, Garabedian MJ. Potentiation of human estrogen receptor alpha transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A-CDK2 complex. J Biol Chem. 1999;274:22296–22302. doi: 10.1074/jbc.274.32.22296. [DOI] [PubMed] [Google Scholar]
- 33.Ali S, Metzger D, Bornert JM, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Goff P, Montano MM, Schodin DJ, Katzenellenbogen BS. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269:4458–4466. [PubMed] [Google Scholar]
- 35.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 36.Coleman KM, Smith CL. Intracellular signaling pathways: nongenomic actions of estrogens and ligand-independent activation of estrogen receptors. Front Biosci. 2001;6:D1379–1391. doi: 10.2741/coleman. [DOI] [PubMed] [Google Scholar]
- 37.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor alpha. Mol Cell Endocrinol. 2001;174:151–166. doi: 10.1016/s0303-7207(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 39.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 40.Gottlicher M, Heck S, Herrlich P. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J Mol Med (Berl) 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- 41.Wang MM, Traystman RJ, Hurn PD, Liu T. Non-classical regulation of estrogen receptor-alpha by ICI182,780. J Steroid Biochem Mol Biol. 2004;92:51–62. doi: 10.1016/j.jsbmb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Gaub MP, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;63:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- 43.Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J Biol Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- 44.Webb P, Lopez GN, Uht RM, Kushner PJ. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- 45.Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc Natl Acad Sci U S A. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Jr, Pestell RG, Kushner PJ. Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- 47.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deegan BJ, Bhat V, Seldeen KL, McDonald CB, Farooq A. Genetic variations within the ERE motif modulate plasticity and energetics of binding of DNA to the ERalpha nuclear receptor. Arch Biochem Biophys. 2011;507:262–270. doi: 10.1016/j.abb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 50.Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- 51.Predki PF, Sarkar B. Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions. J Biol Chem. 1992;267:5842–5846. [PubMed] [Google Scholar]
- 52.Predki PF, Sarkar B. Metal replacement in “zinc finger” and its effect on DNA binding. Environ Health Perspect. 1994;102(Suppl 3):195–198. doi: 10.1289/ehp.94102s3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deegan BJ, Bona AM, Bhat V, Mikles DC, McDonald CB, Seldeen KL, Farooq A. Structural and thermodynamic consequences of the replacement of zinc with environmental metals on estrogen receptor alpha-DNA interactions. J Mol Recognit. 2011;24:1007–1017. doi: 10.1002/jmr.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deegan BJ, Seldeen KL, McDonald CB, Bhat V, Farooq A. Binding of the ERalpha nuclear receptor to DNA is coupled to proton uptake. Biochemistry. 2010;49:5978–5988. doi: 10.1021/bi1004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boron WF. Regulation of intracellular pH. Adv Physiol Educ. 2004;28:160–179. doi: 10.1152/advan.00045.2004. [DOI] [PubMed] [Google Scholar]
- 56.Khaled AR, Moor AN, Li A, Kim K, Ferris DK, Muegge K, Fisher RJ, Fliegel L, Durum SK. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol Cell Biol. 2001;21:7545–7557. doi: 10.1128/MCB.21.22.7545-7557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khaled AR, Reynolds DA, Young HA, Thompson CB, Muegge K, Durum SK. Interleukin-3 withdrawal induces an early increase in mitochondrial membrane potential unrelated to the Bcl-2 family. Roles of intracellular pH, ADP transport, and F(0)F(1)-ATPase. J Biol Chem. 2001;276:6453–6462. doi: 10.1074/jbc.M006391200. [DOI] [PubMed] [Google Scholar]
- 58.Puceat M, Roche S, Vassort G. Src family tyrosine kinase regulates intracellular pH in cardiomyocytes. J Cell Biol. 1998;141:1637–1646. doi: 10.1083/jcb.141.7.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beato M, Sanchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- 60.Cavailles V, Dauvois S, Danielian PS, Parker MG. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci U S A. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.L'Horset F, Dauvois S, Heery DM, Cavailles V, Parker MG. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 63.Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology. 2006;147:4831–4842. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- 64.McPherson SJ, Hussain S, Balanathan P, Hedwards SL, Niranjan B, Grant M, Chandrasiri UP, Toivanen R, Wang Y, Taylor RA, Risbridger GP. Estrogen receptor-beta activated apoptosis in benign hyperplasia and cancer of the prostate is androgen independent and TNFalpha mediated. Proc Natl Acad Sci U S A. 2010;107:3123–3128. doi: 10.1073/pnas.0905524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 2010;17:319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pinton G, Thomas W, Bellini P, Manente AG, Favoni RE, Harvey BJ, Mutti L, Moro L. Estrogen receptor beta exerts tumor repressive functions in human malignant pleural mesothelioma via EGFR inactivation and affects response to gefitinib. PLoS One. 2010;5:e14110. doi: 10.1371/journal.pone.0014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu CP, Ho JY, Huang YT, Cha TL, Sun GH, Yu DS, Chang FW, Chen SP, Hsu RJ. Estrogen inhibits renal cell carcinoma cell progression through estrogen receptor-beta activation. PLoS One. 2013;8:e56667. doi: 10.1371/journal.pone.0056667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rudolph A, Toth C, Hoffmeister M, Roth W, Herpel E, Jansen L, Marx A, Brenner H, Chang-Claude J. Expression of oestrogen receptor beta and prognosis of colorectal cancer. Br J Cancer. 2012;107:831–839. doi: 10.1038/bjc.2012.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris HA. Estrogen receptor-beta: recent lessons from in vivo studies. Mol Endocrinol. 2007;21:1–13. doi: 10.1210/me.2005-0459. [DOI] [PubMed] [Google Scholar]
- 70.Harris HA, Albert LM, Leathurby Y, Malamas MS, Mewshaw RE, Miller CP, Kharode YP, Marzolf J, Komm BS, Winneker RC, Frail DE, Henderson RA, Zhu Y, Keith JC., Jr Evaluation of an estrogen receptor-beta agonist in animal models of human disease. Endocrinology. 2003;144:4241–4249. doi: 10.1210/en.2003-0550. [DOI] [PubMed] [Google Scholar]
- 71.Manas ES, Unwalla RJ, Xu ZB, Malamas MS, Miller CP, Harris HA, Hsiao C, Akopian T, Hum WT, Malakian K, Wolfrom S, Bapat A, Bhat RA, Stahl ML, Somers WS, Alvarez JC. Structure-based design of estrogen receptor-beta selective ligands. J Am Chem Soc. 2004;126:15106–15119. doi: 10.1021/ja047633o. [DOI] [PubMed] [Google Scholar]
- 72.Mewshaw RE, Edsall RJ, Jr, Yang C, Manas ES, Xu ZB, Henderson RA, Keith JC, Jr, Harris HA. ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity. J Med Chem. 2005;48:3953–3979. doi: 10.1021/jm058173s. [DOI] [PubMed] [Google Scholar]
- 73.Carswell HV, Macrae IM, Gallagher L, Harrop E, Horsburgh KJ. Neuroprotection by a selective estrogen receptor beta agonist in a mouse model of global ischemia. Am J Physiol Heart Circ Physiol. 2004;287:H1501–1504. doi: 10.1152/ajpheart.00227.2004. [DOI] [PubMed] [Google Scholar]
- 74.Bertini S, De Cupertinis A, Granchi C, Bargagli B, Tuccinardi T, Martinelli A, Macchia M, Gunther JR, Carlson KE, Katzenellenbogen JA, Minutolo F. Selective and potent agonists for estrogen receptor beta derived from molecular refinements of salicylaldoximes. Eur J Med Chem. 2011;46:2453–2462. doi: 10.1016/j.ejmech.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Minutolo F, Macchia M, Katzenellenbogen BS, Katzenellenbogen JA. Estrogen receptor beta ligands: recent advances and biomedical applications. Med Res Rev. 2011;31:364–442. doi: 10.1002/med.20186. [DOI] [PubMed] [Google Scholar]
- 76.Nilsson S, Kuiper G, Gustafsson JA. ERbeta: a novel estrogen receptor offers the potential for new drug development. Trends Endocrinol Metab. 1998;9:387–395. doi: 10.1016/s1043-2760(98)00096-4. [DOI] [PubMed] [Google Scholar]
- 77.Kuiper GG, Shughrue PJ, Merchenthaler I, Gustafsson JA. The estrogen receptor beta subtype: a novel mediator of estrogen action in neuroendocrine systems. Front Neuroendocrinol. 1998;19:253–286. doi: 10.1006/frne.1998.0170. [DOI] [PubMed] [Google Scholar]
- 78.Stettner M, Kaulfuss S, Burfeind P, Schweyer S, Strauss A, Ringert RH, Thelen P. The relevance of estrogen receptor-beta expression to the antiproliferative effects observed with histone deacetylase inhibitors and phytoestrogens in prostate cancer treatment. Mol Cancer Ther. 2007;6:2626–2633. doi: 10.1158/1535-7163.MCT-07-0197. [DOI] [PubMed] [Google Scholar]
- 79.Vandenput L, Ohlsson C. Estrogens as regulators of bone health in men. Nat Rev Endocrinol. 2009;5:437–443. doi: 10.1038/nrendo.2009.112. [DOI] [PubMed] [Google Scholar]
- 80.Ohlsson C, Vandenput L. The role of estrogens for male bone health. Eur J Endocrinol. 2009;160:883–889. doi: 10.1530/EJE-09-0118. [DOI] [PubMed] [Google Scholar]
- 81.Komesaroff PA, Sudhir K, Esler MD. Effects of estrogen on stress responses in women. J Clin Endocrinol Metab. 1999;84:4292–4293. doi: 10.1210/jc.84.11.4292-a. [DOI] [PubMed] [Google Scholar]
- 82.Komesaroff PA, Esler MD, Sudhir K. Estrogen supplementation attenuates glucocorticoid and catecholamine responses to mental stress in perimenopausal women. J Clin Endocrinol Metab. 1999;84:606–610. doi: 10.1210/jcem.84.2.5447. [DOI] [PubMed] [Google Scholar]
- 83.Sudhir K, Komesaroff PA. Clinical review 110: Cardiovascular actions of estrogens in men. J Clin Endocrinol Metab. 1999;84:3411–3415. doi: 10.1210/jcem.84.10.5954. [DOI] [PubMed] [Google Scholar]
- 84.Turner JV, Agatonovic-Kustrin S, Glass BD. Molecular aspects of phytoestrogen selective binding at estrogen receptors. J Pharm Sci. 2007;96:1879–1885. doi: 10.1002/jps.20987. [DOI] [PubMed] [Google Scholar]
- 85.Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson JA. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- 86.Manas ES, Xu ZB, Unwalla RJ, Somers WS. Understanding the selectivity of genistein for human estrogen receptor-beta using X-ray crystallography and computational methods. Structure. 2004;12:2197–2207. doi: 10.1016/j.str.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 87.Mersereau JE, Levy N, Staub RE, Baggett S, Zogovic T, Chow S, Ricke WA, Tagliaferri M, Cohen I, Bjeldanes LF, Leitman DC. Liquiritigenin is a plant-derived highly selective estrogen receptor beta agonist. Mol Cell Endocrinol. 2008;283:49–57. doi: 10.1016/j.mce.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson LU, Boucher BA, Liu Z, Cotterchio M, Kreiger N. Phytoestrogen content of foods consumed in Canada, including isoflavones, lignans, and coumestan. Nutr Cancer. 2006;54:184–201. doi: 10.1207/s15327914nc5402_5. [DOI] [PubMed] [Google Scholar]
- 89.Adlercreutz H. Phyto-oestrogens and cancer. Lancet Oncol. 2002;3:364–373. doi: 10.1016/s1470-2045(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 90.Adlercreutz H. Phytoestrogens and breast cancer. J Steroid Biochem Mol Biol. 2002;83:113–118. doi: 10.1016/s0960-0760(02)00273-x. [DOI] [PubMed] [Google Scholar]
- 91.de Lemos ML. Effects of soy phytoestrogens genistein and daidzein on breast cancer growth. Ann Pharmacother. 2001;35:1118–1121. doi: 10.1345/aph.10257. [DOI] [PubMed] [Google Scholar]
- 92.Messina M, McCaskill-Stevens W, Lampe JW. Addressing the soy and breast cancer relationship: review, commentary, and workshop proceedings. J Natl Cancer Inst. 2006;98:1275–1284. doi: 10.1093/jnci/djj356. [DOI] [PubMed] [Google Scholar]
- 93.Messina M, Kucuk O, Lampe JW. An overview of the health effects of isoflavones with an emphasis on prostate cancer risk and prostate-specific antigen levels. J AOAC Int. 2006;89:1121–1134. [PubMed] [Google Scholar]
- 94.Messina M, Redmond G. Effects of soy protein and soybean isoflavones on thyroid function in healthy adults and hypothyroid patients: a review of the relevant literature. Thyroid. 2006;16:249–258. doi: 10.1089/thy.2006.16.249. [DOI] [PubMed] [Google Scholar]
- 95.Vajda AM, Barber LB, Gray JL, Lopez EM, Woodling JD, Norris DO. Reproductive disruption in fish downstream from an estrogenic wastewater effluent. Environ Sci Technol. 2008;42:3407–3414. doi: 10.1021/es0720661. [DOI] [PubMed] [Google Scholar]
- 96.Christin MS, Menard L, Gendron AD, Ruby S, Cyr D, Marcogliese DJ, Rollins-Smith L, Fournier M. Effects of agricultural pesticides on the immune system of Xenopus laevis and Rana pipiens. Aquat Toxicol. 2004;67:33–43. doi: 10.1016/j.aquatox.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 97.Aravindakshan J, Gregory M, Marcogliese DJ, Fournier M, Cyr DG. Consumption of xenoestrogen-contaminated fish during lactation alters adult male reproductive function. Toxicol Sci. 2004;81:179–189. doi: 10.1093/toxsci/kfh174. [DOI] [PubMed] [Google Scholar]
- 98.Aravindakshan J, Paquet V, Gregory M, Dufresne J, Fournier M, Marcogliese DJ, Cyr DG. Consequences of xenoestrogen exposure on male reproductive function in spottail shiners (Notropis hudsonius) Toxicol Sci. 2004;78:156–165. doi: 10.1093/toxsci/kfh042. [DOI] [PubMed] [Google Scholar]
- 99.Williams DE, Lech JJ, Buhler DR. Xenobiotics and xenoestrogens in fish: modulation of cytochrome P450 and carcinogenesis. Mutat Res. 1998;399:179–192. doi: 10.1016/s0027-5107(97)00255-8. [DOI] [PubMed] [Google Scholar]
- 100.Arukwe A, Celius T, Walther BT, Goksoyr A. Effects of xenoestrogen treatment on zona radiata protein and vitellogenin expression in Atlantic salmon (Salmo salar) Aquat Toxicol. 2000;49:159–170. doi: 10.1016/s0166-445x(99)00083-1. [DOI] [PubMed] [Google Scholar]
- 101.Golden RJ, Noller KL, Titus-Ernstoff L, Kaufman RH, Mittendorf R, Stillman R, Reese EA. Environmental endocrine modulators and human health: an assessment of the biological evidence. Crit Rev Toxicol. 1998;28:109–227. doi: 10.1080/10408449891344191. [DOI] [PubMed] [Google Scholar]
- 102.Pugazhendhi D, Sadler AJ, Darbre PD. Comparison of the global gene expression profiles produced by methylparaben, n-butylparaben and 17beta-oestradiol in MCF7 human breast cancer cells. J Appl Toxicol. 2007;27:67–77. doi: 10.1002/jat.1200. [DOI] [PubMed] [Google Scholar]
- 103.Buterin T, Koch C, Naegeli H. Convergent transcriptional profiles induced by endogenous estrogen and distinct xenoestrogens in breast cancer cells. Carcinogenesis. 2006;27:1567–1578. doi: 10.1093/carcin/bgi339. [DOI] [PubMed] [Google Scholar]
- 104.Darbre PD. Environmental oestrogens, cosmetics and breast cancer. Best Pract Res Clin Endocrinol Metab. 2006;20:121–143. doi: 10.1016/j.beem.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 105.Harvey PW, Darbre P. Endocrine disrupters and human health: could oestrogenic chemicals in body care cosmetics adversely affect breast cancer incidence in women? J Appl Toxicol. 2004;24:167–176. doi: 10.1002/jat.978. [DOI] [PubMed] [Google Scholar]
- 106.Darbre PD, Aljarrah A, Miller WR, Coldham NG, Sauer MJ, Pope GS. Concentrations of parabens in human breast tumours. J Appl Toxicol. 2004;24:5–13. doi: 10.1002/jat.958. [DOI] [PubMed] [Google Scholar]
- 107.Darbre PD. Underarm cosmetics and breast cancer. Eur J Cancer Prev. 2004;13:153. doi: 10.1097/00008469-200404000-00013. [DOI] [PubMed] [Google Scholar]
- 108.Sharpe RM, Skakkebaek NE. Are oestrogens involved in falling sperm counts and disorders of the male reproductive tract? Lancet. 1993;341:1392–1395. doi: 10.1016/0140-6736(93)90953-e. [DOI] [PubMed] [Google Scholar]
- 109.Sharpe RM. The 'oestrogen hypothesis'- where do we stand now? Int J Androl. 2003;26:2–15. doi: 10.1046/j.1365-2605.2003.00367.x. [DOI] [PubMed] [Google Scholar]
- 110.Vidaeff AC, Sever LE. In utero exposure to environmental estrogens and male reproductive health: a systematic review of biological and epidemiologic evidence. Reprod Toxicol. 2005;20:5–20. doi: 10.1016/j.reprotox.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 111.Schopfer U, Schoeffter P, Bischoff SF, Nozulak J, Feuerbach D, Floersheim P. Toward selective ERbeta agonists for central nervous system disorders: synthesis and characterization of aryl benzthiophenes. J Med Chem. 2002;45:1399–1401. doi: 10.1021/jm015577l. [DOI] [PubMed] [Google Scholar]
- 112.Riggs BL, Hartmann LC. Selective estrogen-receptor modulators -- mechanisms of action and application to clinical practice. N Engl J Med. 2003;348:618–629. doi: 10.1056/NEJMra022219. [DOI] [PubMed] [Google Scholar]
- 113.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 114.Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 115.Martinkovich S, Shah D, Planey SL, Arnott JA. Selective estrogen receptor modulators: tissue specificity and clinical utility. Clin Interv Aging. 2014;9:1437–1452. doi: 10.2147/CIA.S66690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blizzard TA, Gude C, Morgan JD, 2nd, Chan W, Birzin ET, Mojena M, Tudela C, Chen F, Knecht K, Su Q, Kraker B, Mosley RT, Holmes MA, Sharma N, Fitzgerald PM, Rohrer SP, Hammond ML. Androstenediol analogs as ER-beta-selective SERMs. Bioorg Med Chem Lett. 2006;16:834–838. doi: 10.1016/j.bmcl.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 117.Blizzard TA, Gude C, Chan W, Birzin ET, Mojena M, Tudela C, Chen F, Knecht K, Su Q, Kraker B, Holmes MA, Rohrer SP, Hammond ML. Bridged androstenediol analogs as ER-beta selective SERMs. Bioorg Med Chem Lett. 2007;17:2944–2948. doi: 10.1016/j.bmcl.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 118.Blizzard TA, Gude C, Morgan JD, 2nd, Chan W, Birzin ET, Mojena M, Tudela C, Chen F, Knecht K, Su Q, Kraker B, Mosley RT, Holmes MA, Rohrer SP, Hammond ML. Androstene-3,5-dienes as ER-beta selective SERMs. Bioorg Med Chem Lett. 2007;17:6295–6298. doi: 10.1016/j.bmcl.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 119.Papapetropoulos A. A ginseng-derived oestrogen receptor beta (ERbeta) agonist, Rb1 ginsenoside, attenuates capillary morphogenesis. Br J Pharmacol. 2007;152:172–174. doi: 10.1038/sj.bjp.0707360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Warnmark A, Treuter E, Gustafsson JA, Hubbard RE, Brzozowski AM, Pike AC. Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor alpha. J Biol Chem. 2002;277:21862–21868. doi: 10.1074/jbc.M200764200. [DOI] [PubMed] [Google Scholar]
- 121.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 122.Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 123.Heldring N, Pawson T, McDonnell D, Treuter E, Gustafsson JA, Pike AC. Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J Biol Chem. 2007;282:10449–10455. doi: 10.1074/jbc.M611424200. [DOI] [PubMed] [Google Scholar]
- 124.Hubbard RE, Pike AC, Brzozowski AM, Walton J, Bonn T, Gustafsson JA, Carlquist M. Structural insights into the mechanisms of agonism and antagonism in oestrogen receptor isoforms. Eur J Cancer. 2000;36(Suppl 4):S17–18. doi: 10.1016/s0959-8049(00)00207-0. [DOI] [PubMed] [Google Scholar]
- 125.Roman-Blas JA, Castaneda S, Cutolo M, Herrero-Beaumont G. Efficacy and safety of a selective estrogen receptor beta agonist, ERB-041, in patients with rheumatoid arthritis: a 12-week, randomized, placebo-controlled, phase II study. Arthritis Care Res (Hoboken) 2010;62:1588–1593. doi: 10.1002/acr.20275. [DOI] [PubMed] [Google Scholar]
- 126.van Vollenhoven RF, Houbiers JG, Buttgereit F, In 't Hout J, Boers M, Leij S, Kvien TK, Dijkmans BA, Szczepanski L, Szombati I, Sierakowski S, Miltenburg AM. The selective estrogen receptor alpha agonist Org 37663 induces estrogenic effects but lacks antirheumatic activity: a phase IIa trial investigating efficacy and safety of Org 37663 in postmenopausal female rheumatoid arthritis patients receiving stable background methotrexate or sulfasalazine. Arthritis Rheum. 2010;62:351–358. doi: 10.1002/art.27196. [DOI] [PubMed] [Google Scholar]
- 127.McLachlan JA. Environmental signaling: what embryos and evolution teach us about endocrine disrupting chemicals. Endocr Rev. 2001;22:319–341. doi: 10.1210/edrv.22.3.0432. [DOI] [PubMed] [Google Scholar]
- 128.Colborn T, vom Saal FS, Soto AM. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ Health Perspect. 1993;101:378–384. doi: 10.1289/ehp.93101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martin MB, Reiter R, Pham T, Avellanet YR, Camara J, Lahm M, Pentecost E, Pratap K, Gilmore BA, Divekar S, Dagata RS, Bull JL, Stoica A. Estrogen-like activity of metals in MCF-7 breast cancer cells. Endocrinology. 2003;144:2425–2436. doi: 10.1210/en.2002-221054. [DOI] [PubMed] [Google Scholar]
- 130.Darbre PD. Metalloestrogens: an emerging class of inorganic xenoestrogens with potential to add to the oestrogenic burden of the human breast. J Appl Toxicol. 2006;26:191–197. doi: 10.1002/jat.1135. [DOI] [PubMed] [Google Scholar]
- 131.Antila E, Mussalo-Rauhamaa H, Kantola M, Atroshi F, Westermarck T. Association of cadmium with human breast cancer. Sci Total Environ. 1996;186:251–256. doi: 10.1016/0048-9697(96)05119-4. [DOI] [PubMed] [Google Scholar]
- 132.Stoica A, Katzenellenbogen BS, Martin MB. Activation of estrogen receptor-alpha by the heavy metal cadmium. Mol Endocrinol. 2000;14:545–553. doi: 10.1210/mend.14.4.0441. [DOI] [PubMed] [Google Scholar]
- 133.Stoica A, Pentecost E, Martin MB. Effects of selenite on estrogen receptor-alpha expression and activity in MCF-7 breast cancer cells. J Cell Biochem. 2000;79:282–292. doi: 10.1002/1097-4644(20001101)79:2<282::aid-jcb110>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 134.Stoica A, Pentecost E, Martin MB. Effects of arsenite on estrogen receptor-alpha expression and activity in MCF-7 breast cancer cells. Endocrinology. 2000;141:3595–3602. doi: 10.1210/endo.141.10.7704. [DOI] [PubMed] [Google Scholar]
- 135.Rulisek L, Vondrasek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 136.Shi LM, Fang H, Tong W, Wu J, Perkins R, Blair RM, Branham WS, Dial SL, Moland CL, Sheehan DM. QSAR models using a large diverse set of estrogens. J Chem Inf Comput Sci. 2001;41:186–195. doi: 10.1021/ci000066d. [DOI] [PubMed] [Google Scholar]
- 137.Fang H, Tong W, Shi LM, Blair R, Perkins R, Branham W, Hass BS, Xie Q, Dial SL, Moland CL, Sheehan DM. Structure-activity relationships for a large diverse set of natural, synthetic, and environmental estrogens. Chem Res Toxicol. 2001;14:280–294. doi: 10.1021/tx000208y. [DOI] [PubMed] [Google Scholar]
- 138.Grady D, Sawaya GF, Johnson KC, Koltun W, Hess R, Vittinghoff E, Kristof M, Tagliaferri M, Cohen I, Ensrud KE. MF101, a selective estrogen receptor beta modulator for the treatment of menopausal hot flushes: a phase II clinical trial. Menopause. 2009;16:458–465. doi: 10.1097/gme.0b013e31818e64dd. [DOI] [PubMed] [Google Scholar]
- 139.Stovall DW, Pinkerton JV. MF-101, an estrogen receptor beta agonist for the treatment of vasomotor symptoms in peri- and postmenopausal women. Curr Opin Investig Drugs. 2009;10:365–371. [PubMed] [Google Scholar]
- 140.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 141.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 142.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]