Abstract
The well-studied mechanism of ribonuclease A is believed to involve concerted general acid–base catalysis by two histidine residues, His12 and His119. The basic features of this mechanism are often cited to explain rate enhancement by both protein and RNA enzymes that catalyze RNA 2’-O-transphosphorylation. Recent kinetic isotope effect analyses and computational studies are providing a more chemically detailed description of the mechanism of RNase A and the rate limiting transition state. Overall, the results support an asynchronous mechanism for both solution and ribonuclease catalyzed reactions in which breakdown of a transient dianoinic phosphorane intermediate by 5’O-P bond cleavage is rate limiting. Relative to non-enzymatic reactions catalyzed by specific base, a smaller KIE on the 5’O leaving group and a less negative βLG are observed for RNase A catalysis. Quantum mechanical calculations consistent with these data support a model in which electrostatic and H-bonding interactions with the non-bridging oxygens and proton transfer from His119 render departure of the 5'O less advanced and stabilize charge buildup in the transition state. Both experiment and computation indicate advanced 2’O-P bond formation in the rate limiting transition state. However, this feature makes it difficult to resolve the chemical steps involved in 2’O activation. Thus, modeling the transition state for RNase A catalysis underscores those elements of its chemical mechanism that are well resolved, as well as highlighting those where ambiguity remains.
Graphical Abstract
1. Introduction
Ribonuclease A has long served as an experimental system for exploring the fundamental principles that describe how enzymes achieve catalysis[1]. Extensive structural and biochemical studies have resulted in a model for its mechanism that is widely cited as a foundational example of biological catalysis. In this model, RNase A catalysis of RNA 2’-O-transphosphorylation involves a general acid catalytic mode involving donation of a proton to the 5’O leaving group by a protonated His119. Activation of the 2’O nucleophile is attributed to His12, which is proposed to act in its neutral form as a general base (Fig. 1). Parallels to this concerted acid/base mechanism have been drawn to many different phosphoryl transfer enzymes including ribozymes[2–4].
Fig. 1.
The concerted general acid/base mechanism for RNase A catalysis. A scheme for active site interactions involving His12, His119 and Lys41 in the ground state, transition state (TSc) and product state are shown from left to right. Note that nucleophilic addition (2’O-P bond formation) and leaving group departure (5’O-P bond cleavage) occur in a single transition state. Proton transfer involving the nucleophile and leaving group occur concomitantly with changes in O-P bonding. General base catalysis involves transfer of the 2’O proton (shown in red) to the deprotonated form of His12. General acid catalysis occurs via transfer of a proton (shown in blue) from the protonated form of His119 to the departing 5’O.
The development of models for the catalytic mechanism of RNase A has advanced in parallel with increased understanding of the mechanism of solution RNA 2’-O-transphosphorylation[1, 5]. In solution, RNA 2’-O-transphosphorylation reactions can proceed by concerted or stepwise mechanisms with a range of potential transition state structures. Decades of research have shown that these reactions can occur by anionic or neutral species that are distinguished by the relative stability of a phosphorane intermediate, and the protonation state of the non-bridging oxygens during the reaction[6, 7]. Early controversies regarding the overall mechanism of RNase A have largely resolved toward an concerted anionic transition state[1, 8].
However, new questions are suggested by the classic model for RNase A catalysis that are universal for enzyme catalyzed phosphoryl transfer reactions: What is the structure of the rate limiting transition state? How do proposed active site interactions alter the transition state relative to solution reactions? The goal of this review is to integrate recent results from novel KIE studies and advanced computational multi-scale modeling methods into the extensive existing framework for this paradigmatic enzyme. Resolving enzyme mechanism at this level of detail is valuable because it can provide insight into general principles of biological catalysis and suggest strategies for engineering novel catalysts. Also, areas of ambiguity identified in the process help to identify limitations to our current understanding of enzyme mechanism.
RNase A is good system to address questions regarding transition state stabilization by enzymes because it is a potent catalyst, providing greater than 1010-fold rate enhancement over the uncatalyzed RNA 2’-O-transphosphorylation reaction at neutral pH[9]. Although spontaneous RNA 2’-O-transphosphorylation is slow, it can be catalyzed by both acid and base. These reactions proceed at experimentally accessible rate constants permitting detailed mechanistic analysis[6, 7, 10]. Comparison of solution and enzyme mechanisms has permitted a better understanding of the simultaneous application of multiple catalytic modes that defines biological catalysis[1, 6–8]. Protein ribonucleases like RNase A contain amino acids in their active sites, such as histidine, glutamine and lysine, capable of acid/base catalysis and electrostatic catalytic modes[8, 11]. Similarly, the active sites of small self-cleaving ribozymes contain nucleobases that are proposed to employ general acid/base catalytic modes analogous to His12 and His119[2, 3]. Thus, investigation of the details of RNase A and RNA 2’-O-transphosphorylation can facilitate the comparison of strategies for transition state stabilization across a wide range of biological catalysts.
2. Mechanisms and transition states of solution RNA 2’-O-transphosphorylation
RNA 2’-O-transphosphorylation reactions involve nucleophilic attack of the 2’O on the adjacent phosphoryl group to generate a 2’,3’ cyclic phosphate and a 5’ hydroxyl products (Fig. 2). This reaction is catalyzed by both acid (pathways 1 and 2) and base (pathway 3). At high pH (>9) the rate constant increases log-linear with increasing pH and displays an apparent pKa at ca. pH 13.4. The observed pH dependence reflects an underlying mechanism involving equilibrium deprotonation of the 2’OH to generate a 2’ oxyanion followed by nucleophilic attack[6, 10, 12]. The rate constant also increases with decreasing pH (pH < 5). The observed pH dependence is believed to reflect ground state protonation of one or more non-bridging oxygens, and reaction via either monoanionic or neutral stepwise reaction channels. Under acidic conditions isomerization products are also observed that result from pseudorotation of singly (Imonoanion) or a doubly protonated (Ineutral) phosphorane intermediate[6]. The observation that only transphosphorylation products are observed at high pH indicates that the base catalyzed reaction occurs by a concerted mechanism or an asynchronous one in which the dianionic (Idianion) phosphorane is too short lived to undergo pseudorotation.
Fig. 2.
Reaction pathways and transition states for RNA 2’-O-transphosphorylation in solution. Acid catalysis of the neutral diester (1) and monoanion (2) involve stepwise mechanisms via a phosphorane that may be neutral (Ineutral) or monoanionic (Imonoanion). Scheme 3 shows the potential stepwise (via a dianionic phosphorane, Idianion) and concerted (via a single transition state, TSc) mechanisms for base catalysis. Stepwise mechanisms are associative involving first a transition state for 2’O nucleophilic attack (TS1) and a second transition state for breakdown of the intermediate by departure of the 5’O leaving group.
2.1 Linear free energy relationship analyses
Linear free energy relationship (LFER) analyses provide information on mechanism by analyzing the dependence of reaction rate constants on the pKa of the nucleophile and leaving group[13]. To relate these values to transition state bonding, the observed β(β = Δlogk/ΔpKa) is calibrated against the βEQ, which is the effect changing pKa on reaction equilibria. The parameter α = (βLG or βNUC)/βEQ is often used to express the fraction of total charge development in the transition state, which is related to the amount of bond breakage or formation[13]. Quantum chemical calculations provide a molecular level characterization of the structure and bonding in the transition state that can aid in the interpretation of experimental LFER analysis[32]. For reactions involving phosphoryl transfer reactions of ribose hydroxyls, the βEQ is not available, however a value of −1.35 is measured for equilibrium ionization of substituted phenols[14] and a value of −1.56 has been estimated[15]. A βLG of −1.28 is measured for the reaction of a series of uridine 3’ alkyl phosphodiesters indicating a late transition state with respect to bond cleavage (α = 0.71)[16]. For reaction of uridine 3’ aryl phosphates, however, a βLG of −0.56 was measured for hydroxypropyl aryl phosphate cyclization (α = 0.44)[17]. These results are consistent with less charge accumulation on the leaving group in the transition state and therefore an earlier transition state for diesters with better aryl leaving groups. A breakpoint in the relationship between leaving group pKa and logk obtained by linear extrapolation of the two data sets is observed at ca. 12.5, and this feature has been attributed to a change in mechanism from a concerted to stepwise mechanism[16]. A βNUC of 0.75 for solution RNA transphosphorylation has been measured using a series of 2’ substituted analogs[15]. For comparison, the βNUC values measured for oxygen and nitrogen nucleophiles attacking monoesters are in the range of 0.1–0.2[18]. This result suggests that the 2′O nucleophile has a relatively smaller effective charge in the rate limiting transition state for reactions of alkyl phosphate diesters.
2.2 Kinetic isotope effects
The substitution of a stable heavy isotope for a reacting atom can affect both the rate and equilibrium constants for the reaction. Kinetic isotope effects (KIEs) arise primarily due to differences in bond vibrational properties between the ground state and transition state[19]. Differences in the observed KIEs for transphosphorylation reactions with good (aryl) and poor (alkyl) leaving groups also indicate that there are significant differences in rate limiting transition states for the two classes of substrates. Computational studies reveal that these different transition states have distinct kinetic isotope signatures[33].
The transphosphorylation reaction of hydroxypropyl-p-nitrophenol phosphate shows a nucleophile KIE that is normal (1.0327) while the nucleophile KIE measured for RNA 2’-O-transphosphorylation is inverse (0.995)[12, 20]. The fact that these values are so different suggests immediately that there are significant differences in transition state structure, at least with respect to nucleophile bonding. However, in order to interpret these effects in terms of specific mechanistic differences it is important to understand that observed KIEs reflect O-P bonding as well as all other vibrational modes that change on going from the ground state to the transition state.
Oxygen isotope effects on transphosphorylation can reflect the fractionation due to deprotonation of the hydroxyl group and the kinetic isotope effect on nucleophilic attack or leaving group departure. Fractionation due to deprotonation or protonation can have a large contribution to the observed effect since the equilibrium deprotonation of the 2’O is estimated to be 1.024[21]. The full contribution is expected for a mechanism involving equilibrium deprotonation, while a smaller contribution would result from partial transfer in the rate limiting transition state. The former mechanism defines specific base nucleophilic activation while the later exemplifies general base catalysis. For example, the observed effect of 0.995 for specific base catalysis cited above changes to 0.984 above pH 13 due to loss of the O-H stretching mode in the ground state[12, 22]. The observation of a nucleophile KIE closer to unity at lower pH is interpreted as reflecting a large inverse contribution that is offset by the normal contribution due to the equilibrium isotope effect on 2’O deprotonation.
The contribution of nucleophilic attack to the observed KIE is itself composed of two factors, the temperature-independent factor (TIF) and the temperature-dependent factor (TDF)[23, 24]. The TDF is due to differences in bond vibrational modes in the transition state compared to the ground state and reflect differences in zero point vibrational energies proportional to, and in the direction of equilibrium isotope effects. Inverse nucleophile KIEs are are observed for reactions involving oxygen which hydrogen is replaced by a larger chemical group such as carbon introducing new vibrational modes[25, 26]. Formation of the O-P bond, therefore, favors the heavier isotope and results in an inverse contribution to the KIE. The TIF reflects the extent to which the labeled atom participates in reaction coordinate motion. While all other modes are at their minima in the transition state, this motion is uniquely and by definition at a maxima at the transition state. As a consequence the TIF will always favor 16O resulting in a normal contribution to the observed KIE proportional to the extent it its motion is involved in the reaction coordinate.
Thus, observed nucleophile KIEs are normal for early transition states and become less normal as O-P bonding in the transition state increases, eventually becoming inverse as nucleophile bonding becomes more complete[27–30]. In this light, the normal (1.023) nucleophoile KIE observed for the transphosphorylation reaction of hydroxypropyl-p-nitrophenol phosphate reflects an early transition state for a substrate with an aryl leaving group. Inverse KIEs reflect mechanisms in which the nucleophile participates very little in reaction coordinate motion, and the TDF dominates the observed KIE. The inverse KIE measured for the RNA cleavage reaction (0.984) therefore can be interpreted as representing advanced O-P bond formation and in a stepwise mechanism essentially becomes the EIE for formation of a dianionic phosphorane intermediate[21, 22].
The leaving group KIEs for transphosphorylation reactions with good (nitrophenol) and poor (ribose) leaving groups are also very different consistent with a change in rate limiting transition state. The leaving group KIE for transphosphorylation of hydroxypropyl-p-nitrophenol phosphate is small (1.005) while a much larger leaving group KIE (1.034) is measured for RNA strand cleavage. The EIE on deprotonation of the alcohol of malate is reported to be 1.032 and the calculated EIE between methanol and dimethyl ether is estimated at 1.022[25, 31]. Therefore, the large normal leaving group KIE of 1.034 observed for the RNA reaction represents advanced O-P bond cleavage. In contrast, the minimal value observed for an aryl leaving group is consistent with minimal O-P bond cleavage.
Thus, the KIE measurements to date and the LFER data discussed in the previous section are consistent in revealing very significant differences in transition state structure for transphosphorylation reactions with good and poor leaving groups. For good leaving groups the transition state is early with only a small degree of nucleophile bond formation and leaving group O-P bond cleavage. The RNA transition state however is distinct with advanced nucleophile bond formation and leaving group bond cleavage. The observation of an inverse nucleophile isotope suggests a stepwise mechanism is likely in which breakdown of a phosphorane intermediate is rate limiting.
2.3 A general framework for solution transphosphorylation reactions
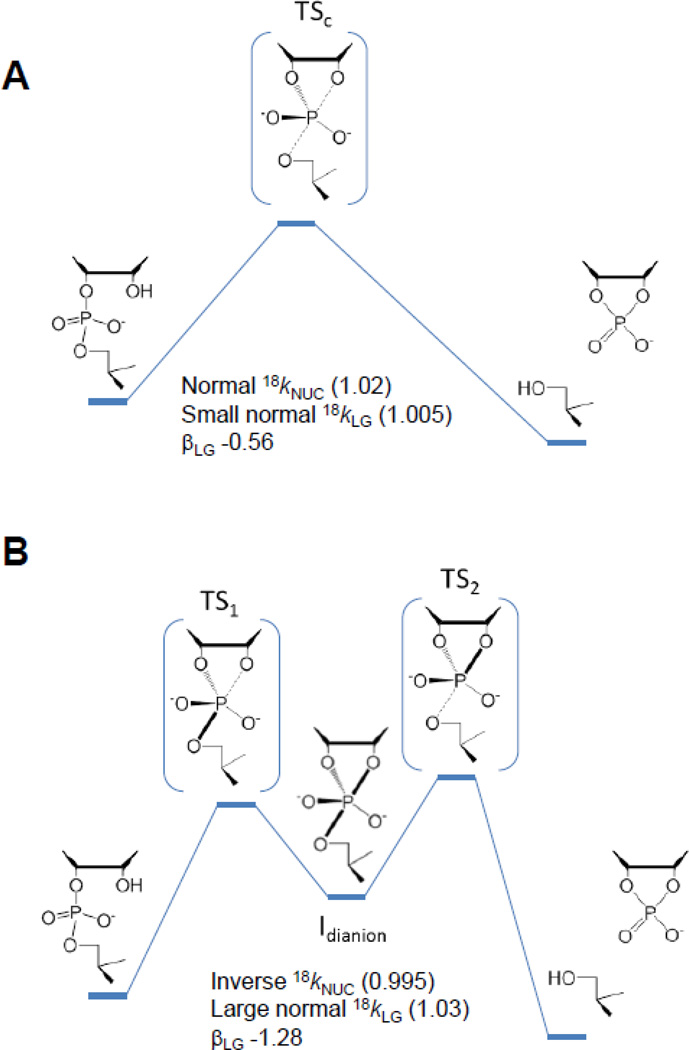
Computational studies of nucleophilic attack on diesters including cyclization reactions has consistently illustrated the potential for stepwise mechanism for RNA transphosphorylation with a transient monoanionic phosphorane intermediate[32–36]. Recently, density-functional calculations[33] were used to analyze a series of transphosphorylation models with different leaving groups to provide a direct connection between observed Brønsted coefficients and KIEs with the structure and bonding in the transition state (Fig. 3).
Fig. 3.
Associative mechanisms for intramolecular phosphodiester transesterification reactions. A concerted mechanism (A) that passes through a single transition state and a stepwise mechanism (B) that proceeds via two transition states separated by an intermediate are shown. Concerted mechanisms can be classified as synchronous (having similar degrees of bonding to the nucleophile and leaving group in the transition state) or asynchronous (having differing degrees of bonding to the nucleophile and leaving group in the transition state). For both stepwise mechanisms and concerted asynchronous mechanisms, the transition states can be further designated as either “early” or “late”, depending on the location of the transition state along a reaction coordinate ξ that involves the difference in the leaving group (R2) and nucleophile (R1) distances with the reactive phosphorus.
In this framework, diesters with sufficiently reactive leaving groups react via a concerted mechanism that proceeds through a single early transition state, and LFER analysis predicts a βLG value with a small magnitude in correspondence with experimental results. This transition state is characterized by normal nucleophile KIE and a small leaving group KIE as reported for base catalysis of hydroxypropyl-m-nitrobenzyl phosphate. For poor leaving groups like the 5’O of RNA, the computationally observed mechanism is stepwise as reported previously. For this class of reactions the rate-controlling transition state is late, leading to a large negative βLG value[33]. The KIEs for this mechanism are in line with the inverse nucleophile and large normal leaving group KIEs observed for specific base catalyzed RNA transphosphorylation described, above. The resulting computational framework can assist in identifying and predicting KIEs that provide insight into mechanism and provide a benchmark? for interpreting the effects of enzymes such as RNase A.
3. Evidence for acid/base catalytic modes in the active site of RNase A
3.1 Roles of key active site residues
Structures of complexes of RNase A together with cyclic nucleotides, substrate analogs and a transition state analog have played a large role in defining potential active site interactions important for catalysis. His12 and His119 are proximal to the scissile phosphoryl group in enzyme-transition state analog uridine vanadate complexes[37, 38] and complexes with inactive substrate oligonucleotides, e.g. [39–41]. His119 is consistently near the 5’O leaving group, or a non-bridging phosphoryl oxygen in product complexes. The interactions of His12 are more variable and it interacts with the 2’ position in the 2’ fluoro substrate and vanidate complexes. However, it interacts with a non-bridging oxygen in the 2’-deoxy substrate analogs. In structures of RNase A with product or deoxy modified substrates, Lys41 is positioned to interact with either the 2’O or a non-bridging oxygen.
Substitution of either His12 or His119 with alanine decreases the value of kcat/KM for cleavage of a UpA dinucleotide substrate by more than 104-fold[42]. The general acid/base mechanism is predicted to underlie the observed bell-shaped pH-kcat/Km profile. Association of polynucleotide substrates is rate limiting for kcat/Km and these kinetics distort the observed kinetic pKa values from their intrinsic values[43]. However, for cyclic nucleotide and dinucleotide substrates for which the first irreversible step is not binding, but catalysis or product release, the observed reaction pKa values are near 6[44–46] consistent with the microscopic pKa values of the active-site histidine residues determined by NMR spectroscopy[47, 48]. An RNase A variant containing a 4-fluorohistidine, which has a pKa of 4.5, at both His12 and His119 retains pH dependence that is still bell-shaped, but shifted to lower pKa consistent with both 4-fluorohistidine residues participating in catalysis[49]. However, there is a relatively small decrease in kcat for the modified enzyme, which suggested that the proton transfer steps may not be rate limiting when the enzyme is in the correct protonation state. Subsequent detailed analyses of RNase A kinetics and structural dynamics shows that product release coupled with protein loop motion is rate limiting for kcat [50, 51].
Mutations of Lys41 affect the reaction rates for RNA, cyclic nucleotide and uridine-3’-p-nitrophenol substrates equivalently, arguing for a common role in stabilizing the transition states for these substrates. Although the solution TS for aryl substrates is different than RNA and cyclic nucleotides, each proceeds by an anionic transition state. The location of Lys41 adjacent to the reactive phosphoryl group is consistent with a role in electrostatic stabilization. Based on mutagenesis and modification data, however, the contribution of Lys41 has been argued to donate a hydrogen bond to the transition state during catalysis rather than simply stabilize negative charge [52].
3.2. Mechanistic information from linear free energy relationship analysis
In order to fully appreciate the roles of Lys41, His12 and His119 in transition state stabilization it is necessary to identify the rate limiting transition state for RNase A catalysis and understand its structure. Davis et al. analyzed the dependence of kcat/Km for RNase A catalyzed reactions of uridine-3’-arylphosphate esters on leaving group pKa and obtained a βLG value of ca. −0.2 [53]. As indicated, above, LFER analysis of uridine-3’-arylphosphate reactions catalyzed by specific base has a more negative βLG value (−0.52) indicating less development of negative charge on the leaving group in the reaction catalyzed by RNase A.
Mutation of His119 has a relatively small effect on the rate constant for transphosphorylation of the aryl diester uridine-3’-p-nitrophenol, while it decreases the rate constant for cleavage of RNA substrates by 104-fold. Because the pKa of the nitrophenol is much lower than ribose (ca. 7 versus 14), it is argued that for the uridine-3’-p-nitrophenol substrate, the function of His119 as a general acid is no longer required[8, 9] and therefore its mutation has little effect. As detailed above, aryl phosphate reactions appear to proceed by a different mechanism relative to RNA in solution, proceeding by an early transition state with little P-5’O bond cleavage. If His119 does not interact with this transition state, then it follows that the observation of a lower βLG value for RNase A reflects an overall change in transition state structure, rather than the influence of general acid catalysis. A much larger βLG value of −1.28 is observed for non-enzymatic reactions of alkyl diesters such as RNA as described, above [54]. However, the βLG for RNase A reaction for substrates in this pKa range has not been reported, but such analyses could help resolve the extent to which His119 influences the accumulation of negative charge on the 5’O leaving group.
Corresponding analyses of βNUC for RNase A and the potential role of His12 in altering this parameter relative to solution reactions are not available. A 2’-deoxy-2’-thio-UpA substrate was reported to bind the active site of RNase A, but are apparently is not a substrate[8]. The rate of thiolate attack on the adjacent phosphodiester bond is estimated to be 107-fold slower than that of the corresponding alkoxide, although at neutral pH there is a much higher concentration of the thiolate, due to its lower pKa,(~8 versus ~13) resulting in only a 25-fold slower observed rate constant for the solution reaction[55]. The inactivity of RNase A toward this substrate is not understood, but could be an issue related to reaction geometry, or due to stable non-catalytic interactions that form due to the presence of a full negative charge at the 2’ position in the ground state. A βNUC of 0.75 for solution RNA transphosphorylation has been measured using a series of 2’ substituted analogs[15]. This result suggests that the 2′O nucleophile has an effective charge of only −0.25 in the rate limiting transition state for the spontaneous reaction. A comparative determination of βNUC for RNase A is not available, but may be feasible if the requisite analogs do not otherwise interfere with enzyme function.
3.3 Transition state by analysis using heavy atom (18O) isotope effects
In principle, measurement of heavy atom KIEs on the RNase A catalyzed RNA 2’-O-transphosphorylation can provide an greater extent of mechanistic clarity. This optimism is based in part on the fact that KIEs involve the smallest possible perturbation of substrate structure that can be used to determine reaction mechanism. Moreover, the work of W. W. Cleland and colleagues on KIEs of phosphoryl transfer reactions including RNase A catalysis provides a firm foundation for interpreting mechanistic detail[56–59]. These classic studies involved application of a remote label method in which the heavy oxygen isotope was monitored indirectly using a 15N label in the nitrophenol leaving group. The required isotope ratio measurements were made to high precision using an isotope ratio mass spectrometer. They further measured RNase A catalyzed transphosphorylation of uridine-m-nitrobenzyl phosphate (UmNB) substrate by the same remote label method. The pKa of the m-nitrobenzyl alcohol leaving group is close to that of a natural substrate nucleotide ribose 5’O leaving group and is expected to react with a similar transition state[59].
The results obtained for RNase A transphosphorylation of uridine-m-nitrobenzyl phosphate (UmNB) by Cleland and colleagues[59] are shown in Fig. 4. These values are compared to KIEs determined for RNA 2’-O-transphosphorylation reactions using whole molecule mass spectrometry by Gu et al., 2013. In this alternative approach the isotope ratio in a 5’-UpG-3’ substrate was measured using tandem quadrupole/time of flight mass spectrometry[60]. The substrate dinucleotide RNA molecule is selected in the first round of mass spectrometry, and the fragments of this parent ion resulting from inert gas collision are analyzed by a second round of mass spectrometry. The enhanced signal to noise allows the 16O/18O ratio to be measured with sufficient precision to allow mechanistic interpretations[12, 34, 36, 60]. The tandem MS/MS analysis of whole RNA molecules permits the KIE on the 2’O nucleophile to be measured, which is more difficult to obtain using the remote label approach. However, the application of isotope ratio MS by Cleland provided superior precision as can be seen in the values compared in Fig. 4. The correspondence between the values measured by these two different methods for the leaving group and non-bridging oxygens is excellent, and provides confidence in the accuracy of the results.
Fig. 4.
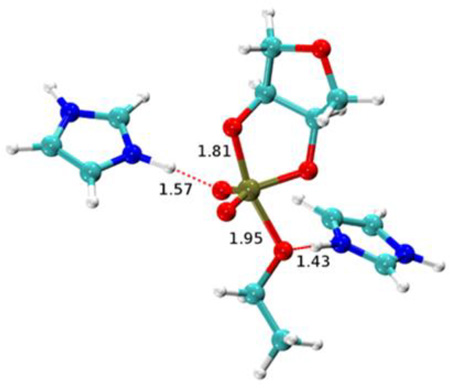
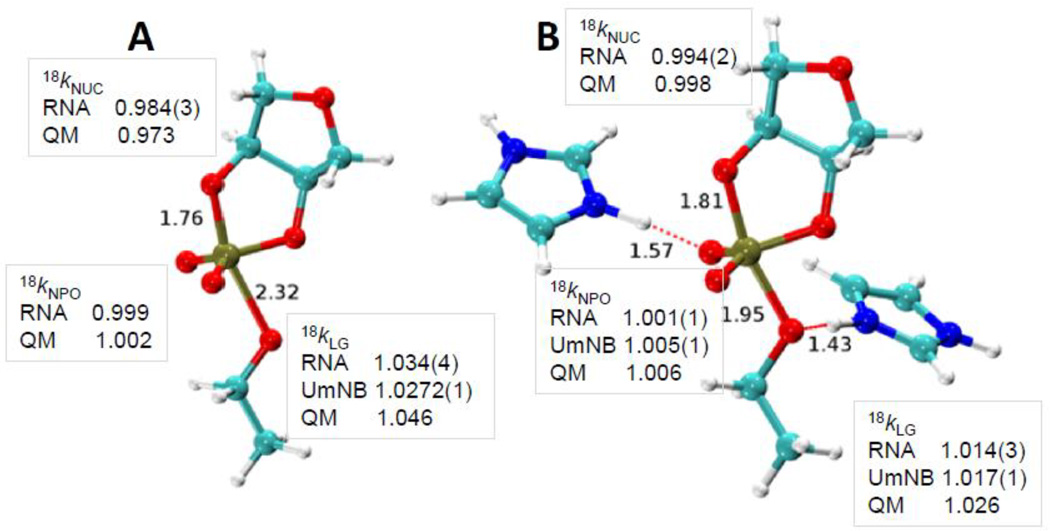
Transition state structures for nonenzymatic specific base catalysis (A) and for the enzymatic transition state model based on hydrogen bonding in the active site of the RNase A (B). The 2’O-P and 5’O-P bond lengths are indicated. For the RNase A transition state model the hydrogen bonds involving His119 and the 5’O, and His12 and a non-bridging oxygen are also indicated. The observed and calculated KIEs for the 2’O nucleophile (18kNUC), 5’O leaving group (18kLG) and non-bridging phosphoryl oxygens (18kNPO) are shown adjacent to the corresponding atoms in the transition state models. KIE values obtained for RNA and for uridine-m-nitrobenzyphosphate (UmNB) are labeled accordingly. Note that the computational results were obtained using a simplified ribose 3’-ethylphosphate model.
The KIE values for RNase A are overall very similar to the values measured for the specific base reaction. The nucleophile KIE is inverse and its value is similar to the solution value consistent with advanced 2’O-P bond formation in both transition states (0.994 for specific base catalysis and 0.995 for RNase A). Although as described, above, the contributions to observed nucleophile isotope effects can be complex. Both reactions have non-bridging oxygen effects of essentially unity, indicating that the bonding environment at these positions is similar in the ground state and transition state. This result is consistent with a product-like transition state since the product of the transphosphorylation reactions is also a diester. Although, taken alone it must be noted that this result is also consistent with a reactant like transition state. The 5’O leaving group KIE is normal for both reactions, however, the value for RNase A is significantly smaller (1.015 versus 1.034). This large difference could arise from less advanced O-P bond cleavage, the presence of a new O-H vibrational mode due to general acid catalysis, or a combination of both.
3.4 Integration of theory and experiment to develop a model for the RNase A transition state
Theoretical calculations provide powerful tools to aid in the interpretation of KIE measurements (for a detailed discussion in the context of RNase A, see the work of Chen et al. [61] in this same special issue). Alternative mechanistic models involving protonation of a non-bridging phosphoryl oxygen or the 5’O alone and 5’O protonation in the presence of H-bonding to an non-bridging oxygen were evaluated using QM calculations of model transition states[33, 60]. The calculated values obtained using a ribose-3’-ethylphosphate model and measured KIE values agree best for a model involving H-bonding between the active site and non-bridging oxygen atoms along with proton transfer from His119 to the 5’O as in the classic mechanistic model[59] [18kNUC (0.998/0.994), 18kLG (1.026/1.014), and 18kNPO (1.006/1.001) (calculated/measured)] (Fig. 4). The calculated 5’O isotope effect for the base-catalyzed reaction simulation (1.048) is larger than the value for the enzymatic model (1.026), consistent with the observed experimental trend (1.034 vs. 1.017). In the simulations, the P-5’O bond length is considerably shorter for the RNase A transition state than that for the base-catalyzed reaction (1.95 Å vs. 2.3 Å) and it retains a higher degree of covalent bond character. Moreover, proton transfer from the general acid (His119) further creates a stiffer bonding environment for stabilizing the leaving group.
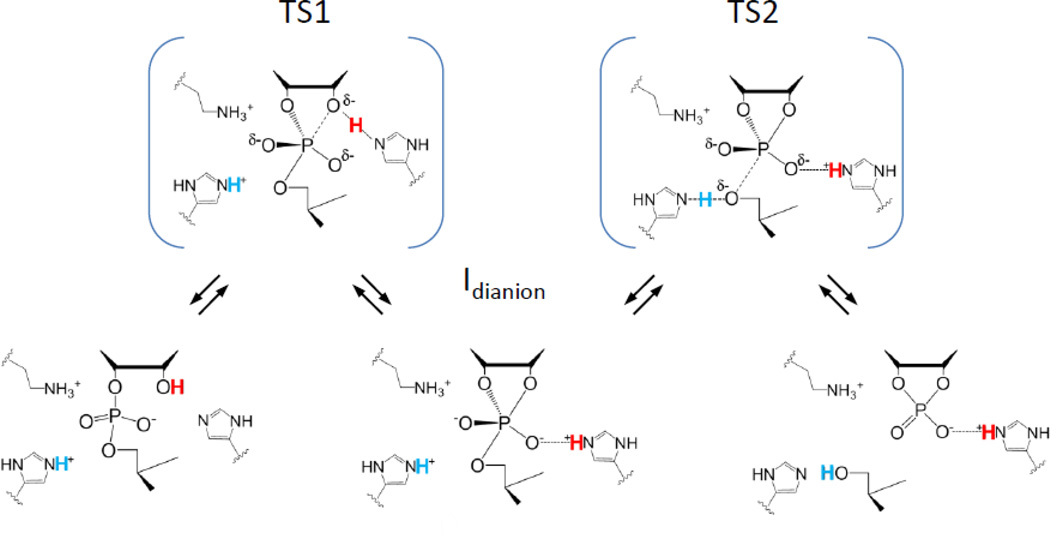
Systematic and detailed QM/MM studies of RNase A mechanism have also been reported by Weare and colleagues[62]. This investigation also supports a stepwise mechanism although with two key features that differ from the model described, above (Fig. 5). In the alternative mechanism, the transition state for nucleophilic addition (TS1) is observed to be rate limiting. However, the observation of inverse nucleophile and normal leaving group KIEs similar to solution reactions unambiguously supports a late, product-like transition state dominated by leaving group departure (TS2). The second difference regards the structure of the phosphorane intermediate. In the alternative mechanism, the proton initially coordinating the O2′ migrates first to His12 and then to the nonbridging O1P resulting in a triester mechanism with a monoanionic intermediate. However, simple interpretation of weak thio effects on RNase A and the observation of a small nonbridging oxygen isotope effect is that such triester mechanism is unlikely[63].
Fig. 5.
Stepwise model for RNase A catalysis. An alternative to the concerted general acid/base model consistent with experimental data and indicated by computation involves transfer of the 2’O proton (red) to His12 followed by formation of the 2’O-P bond. The resulting intermediate is shown as a dianion in which an H-bond between His12 and the substrate phosphoryl group is maintained. However, computational studies have also suggested the potential for formation of a protonated phosphorane in which this proton resides on the phosphoryl oxygen[62]. The rate limiting transition state involves departure of the 5’O leaving group and transfer of a proton from His119 (blue).
4. Conclusions and future directions
Experiment and computation together indicate that RNase A stabilizes a product-like transition state that is very similar to the anionic solution mechanism catalyzed by specific base in solution[12, 34, 36, 60]. Nonetheless, the enzyme significantly alters leaving group bonding attributable to general acid catalysis from His119. The overall asynchrony of the mechanism that is inherent in both the computational and experimental perspectives, however, complicates experimentally evaluating modes of nucleophilic activation. Similarly, the specific catalytic interactions between the active site and the non-bridging oxygens remain difficult to resolve with precision.
Because the interpretation of LFER and KIE are necessarily grounded in transition state theory, assumptions or interpretations of available mechanistic data must be made in order to frame the results in terms of overall transition state structure[64]. The current model framework for RNA transphosphorylation predicts the solution mechanism to be stepwise and that the transition state for leaving group departure (TS2) is rate limiting. It is worth noting that Cleland and Cook demonstrated using adenosine and cytosine deaminases that if proton transfer and changes in 18O bonding occur in the same step, then the 18O KIEs will be the same in both H2O and D2O[65]. For a mechanism in which rate constants for both formation and breakdown of an intermediate are kinetically significant, then the observed heavy atom KIE will be decreased in proportion to the change in proton fractionation. For phosphate groups the equilibrium deuterium isotope effect on protonation is large (3–5)[26], and thus differences in the observed nucleophile and leaving group KIEs may be detectable such an experiment could provide an additional incisive test of concerted/stepwise mechanism.
As described above, the asynchronous mechanism for solution and RNase A reactions makes it difficult to experimentally interrogate the chemical details of nucleophilic activation because this step is not rate limiting. An attractive strategy, given the emerging framework for solution reactions, would be to adjust the reactivity of the leaving group to make nucleophilic attack rate-limiting. In such a mechanism a normal nucleophile KIE would be predicted, but its value would be reduced by the degree of loss of the 2’O-H bond. In general, the normal contribution to the nucleophile KIE from deprotonation is diminished by the degree of O-P bonding in the transition state. Therefore, in a general base mechanism in which deprotonation and nucleophilic attack occur in the same step, a more inverse KIE is expected. Complete loss of the 2’O-H stretching mode prior to nucleophilic attack in a specific base mechanismresults in a large normal contribution to the observed nucleophile KIE of ca. 1.027 based on observed effects on phenols and quantum calculations. Mutation of His12 would be predicted result in a change to a specific base mechanism with a concomitant increase in the observed nucleophile KIE.
In sum, KIE analyses of RNA transphosphorylation have significant power to distinguish between alternative mechanisms, however, they cannot always be interpreted simply. Thus, additional benchmarks must be sought and integration of theory and experiment continued.
Highlights.
Kinetic isotope effects and computational studies provide a chemically detailed description of the RNase A transition state.
2’O-P bond formation is advanced in the rate limiting transition state is advanced.
Proton transfer from His119 renders departure of the 5'O less advanced than solution reactions.
Modeling the RNase A transition state highlights mechanistic features where ambiguity remains.
Acknowledgements
The authors are grateful for the support and encouragement of Dr. W. W. Cleland especially during the initial development of our work on isotope effects. We thank Andrew Knappenberger and Ben Weisman for comments on the manuscript. This work was supported by GM096000 to MEH, AI081987 to JAP, and GM062248 to DMY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cuchillo CM, Nogues MV, Raines RT. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Charged nucleobases and their potential for RNA catalysis. Acc Chem Res. 2011;44:1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lilley DM. Catalysis by the nucleolytic ribozymes. Biochem Soc Trans. 2011;39:641–646. doi: 10.1042/BST0390641. [DOI] [PubMed] [Google Scholar]
- 4.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 5.Findlay D, Herries DG, Mathias AP, Rabin BR, Ross CA. The active site and mechanism of action of bovine pancreatic ribonuclease. 7. The catalytic mechanism. Biochemical Journal. 1962;85:152–153. doi: 10.1042/bj0850152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oivanen M, Kuusela S, Lonnberg H. Kinetics and mechanism for the cleavage and isomerization of phosphodiester bonds of RNA by Bronsted acids and bases. Chemical reviews. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 7.Perreault DM, Anslyn EV. Unifying the Current Data on the Mechanism of Cleavage–Transesterification of RNA. Angewandte Chemie International Edition in English. 1997;36:432–450. [Google Scholar]
- 8.Raines RT. Ribonuclease A. Chemical reviews. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 9.Thompson JE, Kutateladze TG, Schuster MC, Venegas FD, Messmore JM, Raines RT. Limits to Catalysis by Ribonuclease A. Bioorganic chemistry. 1995;23:471–481. doi: 10.1006/bioo.1995.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Breaker RR. Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2'-Hydroxyl Group. J Am Chem Soc. 1999;121:5364–5372. [Google Scholar]
- 11.Dyer K, Rosenberg H. The RNase a superfamily: Generation of diversity and innate host defense. Mol Divers. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 12.Harris ME, Dai Q, Gu H, Kellerman DL, Piccirilli JA, Anderson VE. Kinetic isotope effects for RNA cleavage by 2'-O-transphosphorylation: nucleophilic activation by specific base. J Am Chem Soc. 2010;132:11613–11621. doi: 10.1021/ja103550e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leffler JE. Parameters for the Description of Transition States. Science. 1953;117:340–341. doi: 10.1126/science.117.3039.340. [DOI] [PubMed] [Google Scholar]
- 14.Bourne N, Williams A. Effective charge on oxygen in phosphoryl (-PO32-) group transfer from an oxygen donor. The Journal of Organic Chemistry. 1984;49:1200–1204. [Google Scholar]
- 15.Ye JD, Li NS, Dai Q, Piccirilli JA. The mechanism of RNA strand scission: an experimental measure of the Bronsted coefficient, beta nuc. Angew Chem Int Ed Engl. 2007;46:3714–3717. doi: 10.1002/anie.200605124. [DOI] [PubMed] [Google Scholar]
- 16.Lonnberg H, Stromberg R, Williams A. Compelling evidence for a stepwise mechanism of the alkaline cyclisation of uridine 3'-phosphate esters. Org Biomol Chem. 2004;2:2165–2167. doi: 10.1039/b406926a. [DOI] [PubMed] [Google Scholar]
- 17.Davis AM, Hall AD, Williams A. Charge description of base-catalyzed alcoholysis of aryl phosphodiesters: A ribonuclease model. J Am Chem Soc. 1988;110:5105–5108. [Google Scholar]
- 18.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleland WW. The use of isotope effects to determine enzyme mechanisms. Arch Biochem Biophys. 2005;433:2–12. doi: 10.1016/j.abb.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Humphry T, Iyer S, Iranzo O, Morrow JR, Richard JP, Paneth P, Hengge AC. Altered transition state for the reaction of an RNA model catalyzed by a dinuclear zinc(II) catalyst. J Am Chem Soc. 2008;130:17858–17866. doi: 10.1021/ja8059864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphry T, Forconi M, Williams NH, Hengge AC. Altered mechanisms of reactions of phosphate esters bridging a dinuclear metal center. J Am Chem Soc. 2004;126:11864–11869. doi: 10.1021/ja047110g. [DOI] [PubMed] [Google Scholar]
- 22.Humphry T, Forconi M, Williams NH, Hengge AC. An altered mechanism of hydrolysis for a metal-complexed phosphate diester. J Am Chem Soc. 2002;124:14860–14861. doi: 10.1021/ja027671c. [DOI] [PubMed] [Google Scholar]
- 23.Melander L, Saunders WH. Reaction Rates of Isotopic Moleculaes. Wiley, Place Published; 1980. [Google Scholar]
- 24.Westaway KC, Fang Y-r, Persson J, Matsson O. Using 11C/14C Incoming Group and Secondary α-Deuterium KIEs To Determine How a Change in Leaving Group Alters the Structure of the Transition State of the SN2 Reactions between m-Chlorobenzyl para-Substituted Benzenesulfonates and Cyanide Ion. Journal of the American Chemical Society. 1998;120:3340–3344. [Google Scholar]
- 25.Blanchard JS, Cleland WW. Use of isotope effects to deduce the chemical mechanism of fumarase. Biochemistry. 1980;19:4506–4513. doi: 10.1021/bi00560a019. [DOI] [PubMed] [Google Scholar]
- 26.Rishavy MA, Cleland WW. 13C, 15N and 18O equilibrium isotope effects and fractionation factors. Can. J. Chem. 1999;77:967–977. [Google Scholar]
- 27.Marlier JF, Dopke NC, Johnstone KR, Wirdzig TJ. A Heavy-Atom Isotope Effect Study of the Hydrolysis of Formamide. J. Am. Chem. Soc. 1999;121:4356–4363. [Google Scholar]
- 28.Marlier JF. Heavy-atom isotope effects on the alkaline hydrolysis of methyl formate: The role of hydroxide ion in ester hydrolysis. J Am Chem Soc. 1993;115:5953–5956. [Google Scholar]
- 29.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–10559. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 30.Cassano AG, Anderson VE, Harris ME. Evidence for direct attack by hydroxide in phosphodiester hydrolysis. J Am Chem Soc. 2002;124:10964–10965. doi: 10.1021/ja020823j. [DOI] [PubMed] [Google Scholar]
- 31.Cleland WW. Measurement of isotope effects by the equilibrium perturbation technique. Methods Enzymol. 1980;64:104–125. doi: 10.1016/s0076-6879(80)64007-5. [DOI] [PubMed] [Google Scholar]
- 32.Uchimaru T, Storer JW, Tanabe K, Uebayasi M, Nishikawa S, Taira K. RNA hydrolysis via an oxyphosphorane intermediate. Biochemical and Biophysical Research Communications. 1992;187:1523–1528. doi: 10.1016/0006-291x(92)90475-z. [DOI] [PubMed] [Google Scholar]
- 33.Huang M, York DM. Linear free energy relationships in RNA transesterification: theoretical models to aid experimental interpretations. Physical Chemistry Chemical Physics. 2014;16:15846–15855. doi: 10.1039/c4cp01050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Giese TJ, Huang M, Wong KY, Harris ME, York DM. Mechanistic insights into RNA transphosphorylation from kinetic isotope effects and linear free energy relationships of model reactions. Chemistry-A European Journal. 2014 doi: 10.1002/chem.201403862. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez X, Dejaegere A, Leclerc F, York DM, Karplus M. Nucleophilic attack on phosphate diesters: a density functional study of in-line reactivity in dianionic, monoanionic, and neutral systems. J Phys Chem B. 2006;110:11525–11539. doi: 10.1021/jp0603942. [DOI] [PubMed] [Google Scholar]
- 36.Wong KY, Gu H, Zhang S, Piccirilli JA, Harris ME, York DM. Characterization of the reaction path and transition States for RNA transphosphorylation models from theory and experiment. Angew Chem Int Ed Engl. 2012;51:647–651. doi: 10.1002/anie.201104147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladner JE, Wladkowski BD, Svensson LA, Sjolin L, Gilliland GL. X-ray structure of a ribonuclease A-uridine vanadate complex at 1.3 A resolution. Acta Crystallogr D Biol Crystallogr. 1997;53:290–301. doi: 10.1107/S090744499601582X. [DOI] [PubMed] [Google Scholar]
- 38.Wlodawer A, Miller M, Sjolin L. Active site of RNase: neutron diffraction study of a complex with uridine vanadate, a transition-state analog. Proc Natl Acad Sci U S A. 1983;80:3628–3631. doi: 10.1073/pnas.80.12.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zegers I, Maes D, Dao-Thi MH, Poortmans F, Palmer R, Wyns L. The structures of RNase A complexed with 3'-CMP and d(CpA): active site conformation and conserved water molecules. Protein Sci. 1994;3:2322–2339. doi: 10.1002/pro.5560031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilar CF, Thomas PJ, Moss DS, Mills A, Palmer RA. Novel non-productively bound ribonuclease inhibitor complexes--high resolution X-ray refinement studies on the binding of RNase-A to cytidylyl-2',5'-guanosine (2',5'CpG) and deoxycytidylyl-3',5'-guanosine (3',5'dCpdG) Biochim Biophys Acta. 1991;1118:6–20. doi: 10.1016/0167-4838(91)90435-3. [DOI] [PubMed] [Google Scholar]
- 41.Pavlovsky AG, Borisova SN, Borisov VV, Antonov IV, Karpeisky MY. The structure of the complex of ribonuclease S with fluoride analogue of UpA at 2.5 A resolution. FEBS Lett. 1978;92:258–262. doi: 10.1016/0014-5793(78)80766-2. [DOI] [PubMed] [Google Scholar]
- 42.Park C, Schultz LW, Raines RT. Contribution of the active site histidine residues of ribonuclease A to nucleic acid binding. Biochemistry. 2001;40:4949–4956. doi: 10.1021/bi0100182. [DOI] [PubMed] [Google Scholar]
- 43.Park C, Raines RT. Catalysis by ribonuclease A is limited by the rate of substrate association. Biochemistry. 2003;42:3509–3518. doi: 10.1021/bi026076k. [DOI] [PubMed] [Google Scholar]
- 44.Findlay D, Herries DG, Mathias AP, Rabin BR, Ross CA. The active site and mechanism of action of bovine pancreatic ribonuclease. Nature. 1961;190:781–784. doi: 10.1038/190781a0. [DOI] [PubMed] [Google Scholar]
- 45.Herries DG, Mathias AP, Rabin BR. The active site and mechanism of action of bovine pancreatic ribonuclease. 3. The pH–dependence of the kinetic parameters for the hydrolysis of cytidine 2′,3′-phosphate. Biochemical Journal. 1962;85:127–134. doi: 10.1042/bj0850127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Rosario EJ, Hammes GG. Kinetic and equilibrium studies of the ribonuclease-catalyzed hydrolysis of uridine 2',3'-cyclic phosphate. Biochemistry. 1969;8:1884–1889. doi: 10.1021/bi00833a017. [DOI] [PubMed] [Google Scholar]
- 47.Eftink MR, Biltonen RL. Energetics of ribonuclease A catalysis. 1. pH, ionic strength, and solvent isotope dependence of the hydrolysis of cytidine cyclic 2',3'-phosphate. Biochemistry. 1983;22:5123–5134. doi: 10.1021/bi00291a011. [DOI] [PubMed] [Google Scholar]
- 48.Markley JL. Correlation proton magnetic resonance studies at 250 MHz of bovine pancreatic ribonuclease. I. Reinvestigation of the histidine peak assignments. Biochemistry. 1975;14:3546–3554. doi: 10.1021/bi00687a006. [DOI] [PubMed] [Google Scholar]
- 49.Jackson DY, Burnier J, Quan C, Stanley M, Tom J, Wells JA. A Designed Peptide Ligase for Total Synthesis of Ribonuclease A with Unnatural Catalytic Residues. Science. 1994;266:243–247. doi: 10.1126/science.7939659. [DOI] [PubMed] [Google Scholar]
- 50.Doucet N, Watt ED, Loria JP. The flexibility of a distant loop modulates active site motion and product release in ribonuclease A. Biochemistry. 2009;48:7160–7168. doi: 10.1021/bi900830g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci U S A. 2007;104:11981–11986. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messmore JM, Fuchs DN, Raines RT. Ribonuclease a: revealing structure-function relationships with semisynthesis. J Am Chem Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis AM, Regan AC, Williams A. Experimental charge measurement at leaving oxygen in the bovine ribonuclease A catalyzed cyclization of uridine 3'-phosphate aryl esters. Biochemistry. 1988;27:9042–9047. doi: 10.1021/bi00425a024. [DOI] [PubMed] [Google Scholar]
- 54.Kosonen M, Youseti-Salakdeh E, Stromberg R, Lonnberg H. Mutual isomerization of uridine 2[prime or minute]- and 3[prime or minute]-alkylphosphates and cleavage to a 2[prime or minute],3[prime or minute]-cyclic phosphate: the effect of the alkyl group on the hydronium- and hydroxide-ion-catalyzed reactions. Journal of the Chemical Society, Perkin Transactions. 1997;2:2661–2666. [Google Scholar]
- 55.Dantzman CL, Kiessling LL. Reactivity of a 2‘-Thio Nucleotide Analog. Journal of the American Chemical Society. 1996;118:11715–11719. [Google Scholar]
- 56.Hengge AC, Aleksandra TE, Cleland WW. Studies of transition-state structures of phosphoryl transfer reactions of phosphodiesters of p-nitrophenol. J Am Chem Soc. 1995;117:5919–5926. [Google Scholar]
- 57.Hengge AC, Bruzik KS, Tobin AE, Cleland WW, Tsai MD. Kinetic Isotope Effects and Stereochemical Studies on a Ribonuclease Model: Hydrolysis Reactions of Uridine 3'-Nitrophenyl Phosphate. Bio-Organic Chemistry. 2000;28:119–133. doi: 10.1006/bioo.2000.1170. [DOI] [PubMed] [Google Scholar]
- 58.Hengge AC, Cleland WW. Phosphoryl-transfer reactions of phosphodiesters: characterization of transition states by heavy-atom isotope effects. Journal of the American Chemical Society. 1991:5835–5841. [Google Scholar]
- 59.Sowa GA, Hengge AC, Cleland WW. 18O isotope effects support a concerted mechanism for ribonuclease A. J Am Chem Soc. 1997;119:2319–2320. [Google Scholar]
- 60.Gu H, Zhang S, Wong KY, Radak BK, Dissanayake T, Kellerman DL, Dai Q, Miyagi M, Anderson VE, York DM, Piccirilli JA, Harris ME. Experimental and computational analysis of the transition state for ribonuclease A-catalyzed RNA 2'-O-transphosphorylation. Proc Natl Acad Sci U S A. 2013;110:13002–13007. doi: 10.1073/pnas.1215086110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Piccirilli JA, Harris ME, York DM. Effect of Zn2+ binding and active site on the transition state for RNA 2'-O-transphosphorylation interpreted through kinetic isotope effects. Biochemica et Biophysica Acta: Proteins and Proteomics. 2015 doi: 10.1016/j.bbapap.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elsasser B, Valiev M, Weare JH. A dianionic phosphorane intermediate and transition states in an associative A(N)+D(N) mechanism for the ribonucleaseA hydrolysis reaction. J Am Chem Soc. 2009;131:3869–3871. doi: 10.1021/ja807940y. [DOI] [PubMed] [Google Scholar]
- 63.Herschlag D. Ribonuclease Revisited: Catalysis via the Classical General Acid-Base Mechanism or a Triester-like Mechanism? Journal of the American Chemical Society. 1994;116:11631–11635. [Google Scholar]
- 64.Åqvist J, Kolmodin K, Florian J, Warshel A. Mechanistic alternatives in phosphate monoester hydrolysis: What conclusions can be drawn from available experimental data? Chemistry & Biology. 1999;6:R71–R80. doi: 10.1016/S1074-5521(99)89003-6. [DOI] [PubMed] [Google Scholar]
- 65.Weiss PM, Cook PF, Hermes JD, Cleland WW. Evidence from nitrogen-15 and solvent deuterium isotope effects on the chemical mechanism of adenosine deaminase. Biochemistry. 1987;26:7378–7384. doi: 10.1021/bi00397a027. [DOI] [PubMed] [Google Scholar]