Abstract
Epithelial ovarian cancer consists of 5 major histotypes: high-grade serous carcinoma (HGSC), endometrioid carcinoma (EC), clear cell carcinoma (CCC), mucinous carcinoma (MC) and low-grade serous (LGSC). Each can have a broad spectrum of morphological appearances, and one histotype can closely mimic histopathological features more typical of another. Historically, there has been a relatively high frequency of mixed, defined by 2 or more distinct histotypes present based on routine histopathological assessment, histotype carcinoma diagnoses (3–11%), however recent immunohistochemical studies identifying histotype specific markers and allowing more refined histotype diagnoses suggests a much lower incidence. We reviewed hematoxylin and eosin stained slides from 871 cases of epithelial ovarian cancer and found the frequency of mixed carcinomas to be 1.7% when modern diagnostic criteria are applied. Through international collaboration, we established a cohort totaling 22 mixed epithelial ovarian cancers, consisting of 9 EC/CCC, 4 EC/LGSC, 3 HGSC/CCC, 2 CCC/MC and 4 other combinations. We interrogated the molecular differences between the different components of each case using immunohistochemistry, gene expression and hotspot sequencing analyses. Immunohistochemical data alone suggested 9 of the 22 cases were not mixed tumors as they presented a uniform immuno-phenotype throughout, and these cases most probably represent morphological mimicry and variation within tumors of a single histotype. Synthesis of molecular data further reduces the incidence of mixed carcinomas. Based on these results, true mixed carcinomas with both morphological and molecular support for the presence of more than one histotype within a given tumor represent less than 1% of epithelial ovarian cancers.
Keywords: Ovarian carcinoma, Ovarian cancer, Mixed Histology, Diagnosis, Immunohistochemistry, Heterogeneity
Introduction
Epithelial ovarian cancers (EOC) occur as five major distinct histotypes, i.e., high-grade serous carcinoma (HGSC), endometrioid [ovarian] carcinoma (EC), clear cell carcinoma (CCC), mucinous carcinoma (MC) and low-grade serous carcinoma (LGSC) (1–4). Each is distinct in presentation, outcome, and molecular characteristics. HGSC is the most common histotype (70% of EOC), typically presenting at advanced stage, and most commonly showing papillary architecture; TP53 mutation is near ubiquitous (~97%) and BRCA1/2 loss are frequent (30–45%, including germline and somatic alterations) (5, 6). Reflecting typical histology of the most common form of endometrial carcinomas, EC (10% of EOC) generally presents at low stage with glandular architecture and squamous differentiation (7). PTEN, CTNNB1, PIK3CA and ARID1A are commonly mutated in EC, and tumors frequently show immunohistochemical positivity for Progesterone Receptor (PR) and Trefoil Factor 3 (TFF3) (2, 8). Also frequently presenting at low stage, CCC (10% of EOC) is defined by an abundance of clear cells occurring in tubulocystic, papillary or solid architecture (7). These generally chemoresistant tumors express HNF-1β and frequently possess mutations in PIK3CA and ARID1A (9). MC is typically a low grade EOC characterised by goblet cells and intracellular mucin; this is rare histotype (2–4% of EOC) known to harbor Ras-pathway alterations (7, 10). Finally, LGSC (2% of EOC) frequently presents at advanced stage, and is defined by low-grade nuclear atypia and low mitotic activity (7). LGSC-associated molecular alterations include BRAF and KRAS mutations, and standard platinum-based chemotherapy is generally considered ineffective (7, 11, 12).
Historically, all EOC histotypes were thought to arise from the ovarian surface epithelium (OSE); however, contemporary hypotheses suggest distinct sites of origin and molecular events during oncogenesis for the different histotypes. HGSC have been shown to arise from serous tubal intraepithelial carcinomas of the fallopian tube in the majority of cases (1, 13–15). Endometriosis is associated with both EC and CCC and ectopic endometrial glandular epithelium is thought to be the tissue of origin (16–19). The origins of MC remain elusive (4, 20, 21). Lastly, LGSC are still believed to originate from the OSE, although this is the subject of some debate (3, 22–24). Many LGSC appear to arise from serous borderline tumors (SBT) and these are generally accepted to be a precursor of the invasive form. Despite histological architectural similarity between HGSC and LGSC/SBT they are accepted to arise through mutually exclusive pathways. Considering the apparently distinct origins of EOC histotypes, tumors with mixed cell types would be anticipated to be rare.
Interobserver agreement in EOC histotype diagnosis has dramatically improved in recent years, with concordance rates rising from less than 60% to 85–92% with the integration of biomarker-assisted diagnosis in equivocal cases (25, 26). Previously, reproducibility was particularly poor in diagnosing EOC of pure histology versus those with mixed features (25), and the reported frequency of mixed EOC ranged from 3–11% between 1975 and 2006 (27–33). With modern diagnostic criteria and the use of molecular markers it is now appreciated that ovarian carcinoma histotypes can show a spectrum of morphological features. A corollary of this observation is that papillary architecture does not equate to serous, glandular architecture does not equate to endometrioid, and the presence of clear cells is not pathognomonic of clear cell carcinoma. Recognition of the spectrum of architectural features associated with each histotype, especially HGSC, has resulted in fewer diagnoses of mixed cell type cancer and better interobserver concordance in histotype diagnosis (34–36).
In this study, we set out to catalogue the largely understudied group of mixed ovarian carcinomas, assessing whether there is a molecular basis for morphological heterogeneity and establishing a true frequency within a large population-based cohort. Disparate regions of mixed tumors were further interrogated using molecular analyses including immunohistochemistry, gene expression and hotspot sequencing. Herein, we report mixed-type ovarian carcinomas to occur at a frequency of 1.7% after morphological review. With further molecular analysis, this frequency drops to less than 1%, where the use of common IHC biomarkers generally suffices in identifying true mixed-type EOC.
Materials and Methods
Case Selection
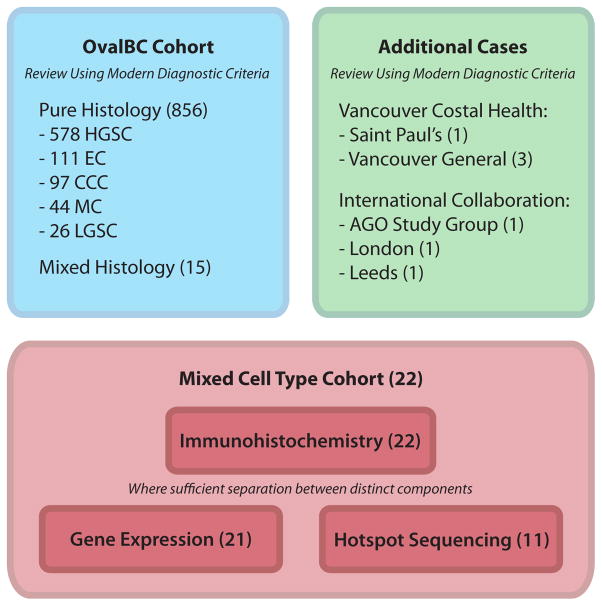
Review of the 871 case Ovarian Cancer in Alberta and British Columbia study (OVAL-BC) cohort of EOC was performed, and cases were diagnosed as mixed EOC when, based on routine H&E stains, there were two or more distinct histotypes present, and each comprised at least 10% of the total tumor volume. For the purpose of this study we accepted as “mixed carcinomas” any tumor in which one component was carcinoma and the other borderline, as long as they were different histotypes; there were only two such cases in the series and we acknowledge that some would not consider these to be true examples of mixed carcinoma, reserving that designation for cases in which both/all components of the tumor are carcinomatous. The frequency of mixed carcinoma was established using this cohort. Also included were seventeen cases, initially diagnosed as mixed-type, that were referred to us from the AGO (Arbeitsgemeinschaft Gynäkologische Onkologie) study group. Following review using modern criteria, a single case retained a diagnosis of mixed-type and was included in this study (See Supplemental Digital Content 1). Additional mixed-type cases from the hospitals of Vancouver Costal Health (British Columbia), The Leeds Teaching Hospital (Leeds), and Barts Health (London) were reviewed for inclusion on a case-by-case basis (Figure 1). Formalin-fixed paraffin embedded (FFPE) tissue was available for all cases.
Figure 1.
Overview of mixed cell cohort and experimental process including the 871-case fully reviewed Ovarian Cancer in Alberta and British Columbia study (OVAL-BC) cohort and a small collection of referred cases from collaborating institutions. Our experimental design began with morphological assessment, followed by immunohistochemical profiling using a standardized IHC panel, gene expression profiling on a limited set of histotype-specific genes, and mutational analysis of common cancer-associated genes.
Immunohistochemistry
We refined a previously described 10-marker algorithm termed COSPv2 (Calculator of ovarian carcinoma subtype prediction version 2) (37), to a 8 marker panel (p53, p16, PR, WT1, ARID1A and HNF1B, VIM and TFF3) that distinguish between OC histotypes with 90% accuracy. Immunohistochemistry (IHC) was performed on freshly cut full sections of FFPE tissue and scored independently for the distinct regions of each mixed-type case (8, 37). A binary scoring method was employed with a cutoff of 1% for PR, and WT1, 10% for ARID1A, 50% for HNF1B, TFF3, VIM and 90% for p16 (37). Expression of p53 was quantified using a 3-tiered scoring system (0=complete loss, 1=wild type/variable expression intensity in 1–70% of tumor cell nuclei, 2=overexpression/>70% of tumor cell nuclei) and binned accordingly (score of 1 = bin of 0/normal; score of 0 or 2 = bin of 1/abnormal) (37, 38). A histotype diagnosis was independently calculated from the immunostaining results for each component of the mixed EOC, as described previously (37, 38). Additional detail on antibodies, dilutions and protocol can be found in Supplemental Digital Content 2.
Macrodissection and Nucleic Acid Extraction
Nucleic acids were extracted from FFPE tissue blocks sectioned at 10μm when a single histotype was present within a given block. When different histotypes were present on the same block, distinct regions were separated by macrodissection using the H&E slide as a guide after having been marked by an expert pathologist (CBG). DNA and RNA were extracted using the Qiagen AllPrep FFPE kit, according to the manufacturers recommended protocols, with the following exceptions: deparaffinization was done using xylene, with digestion of tissue prior to RNA extraction for 30min, and use of RBC buffer (Qiagen RNeasy FFPE kit) rather than buffer FRN.
Gene Expression
Mixed-type cases with RNA extraction yields of sufficient quantity (>400ng) and purity (OD 260/280 1.7–2.5) were selected for expression analysis. Using a custom codeset synthesized by NanoString (Seattle, WA), the expression levels of 363 genes were quantified (39). Five endogenous housekeeping genes (ACTB, RPL19, SDHA, PGK1, POLR1B), plus spiked-in, non-human positive and negative controls were used to normalize and ensure assay success. RNA (400ng) was hybridized and run on the nCounter Analysis System as described previously (39, 40). Gene expression was analyzed using a pre-selected gene panel enriched for genes that were found to be differentially expressed between histological types of ovarian cancer. As this was not a genome-wide profile it partially restricted the scope of statistical analysis of these data. A qualitative analysis using Multi-dimensional scaling (MDS) was used as this provides a means of visualizing the degree of similarity between cases in our datasets. Generally speaking, the closer two points are, the more similar their gene expression profiles are. MDS plots were used to assess the extent of gene expression similarity between components of a mixed tumor, and whether these components fall within a highlighted region of the plot typically populated by EOC of corresponding ‘pure-type’. These ‘pure-type’ EOC were sourced from the OVCARE tumor bank and included 19 CCC, 12 EC, 95 HGSC, 12 LGSC, and 10 MC. All ‘pure-type’ tumors had high cellularity, an immuno-profile characteristic of their representative histotype (chosen from a subset of samples described in Kalloger et al. (8)), and, on review, did not exhibit morphological characteristics that suggested overlap with another histological type.
Mutation Analysis
Components from 11 mixed-type tumors were sequenced independently using the IonTorrent platform. Amplicon libraries were prepared and barcoded using the Cancer Hotspot Panel v2 primer pool and IonXpress barcode adapter kit. Libraries were quantified using Agilent High Sensitivity DNA chips, diluted to 20pM, and pooled together. Pooled libraries were clonally amplified onto IonSphere particles using the OneTouch2 system and loaded onto Ion 316 or 318 chips depending on the number of cases pooled together (2 cases - 316, 3 cases - 318). The Ion Torrent Variant Caller (version 3.6) was used to detect variants with hg19 as a reference genome.
Post-sequencing quality control steps were implemented to account for procedural and nucleic acid integrity limitations involved in sequencing DNA extracted from FFPE tissue. Initial screening limited reported variants to those observed at a minimum 5% allelic frequency and 10x coverage. Normal tissue was unavailable for sequencing, so our analysis was restricted to include only nonsynonymous alterations occurring in hotspot regions that have been reported in a mutation database (Catalogue of somatic mutations in cancer, COSMIC (41)). Insertions or deletions resulting in a truncated protein were also included in our dataset.
Results
Mixed cell type carcinomas in a population based series
After review of the 871 case OVAL-BC cohort of EOC, we identified 856 cases of so-called pure type EOC: 578 (66%) HGSC, 111 (13%) EC, 97 (11%) CCC, 44 (5%) MUC, 26 (3%) LGSC, and 15 with mixed histology. As OVAL-BC is a population-based cohort with representative proportions of the major histotypes, the overall frequency of mixed-type EOC can be approximated at 1.7% when diagnosed based solely on morphological criteria. Amongst these mixed EOC, EC/CCC mixes were most common (6 cases), followed by EC/LGSC (4 cases) and CCC/MC (2 cases), with other mixes comprising the remainder of the observed mixed tumors (1 case each of MC/SBT, MC/borderline Brenner, and MC/HGSC).
To expand our series for molecular analysis, additional mixed EOC cases were acquired: seventeen from the AGO study group (see Table, Supplemental Digital Content 1 for initial and review diagnosis of AGO study group cases) and six cases from collaborating centres (4 from hospitals of Vancouver Costal Health (VCH), 1 from Barts Health NHS Trust and 1 from The Leeds Teaching Hospital NHS Trust). After review of H&E stained slides, we retained all cases from VCH, Barts Health and The Leeds Teaching Hospital and one case from the AGO study group (Figure 1). Our final cohort of mixed-type carcinomas comprised 9 EC/CCC, 4 EC/LGSC, 3 HGSC/CCC, 2 MUC/CCC and 4 additional cases that were grouped as ‘other’ mixes (Table 1).
Table 1.
Case identifier, mixed histology type and cohort grouping (VCH – Hospitals of Vancouver Coastal Health) of the 28 cases of mixed-type EOC studied herein.
| Case ID | Mixed histologies | Cohort | |
|---|---|---|---|
| MX2 | EC/CCC | OVAL-BC | Endometrioid and Clear Cell |
| MX3 | EC/CCC | OVAL-BC | |
| MX9 | EC/CCC | OVAL-BC | |
| MX11 | EC/CCC | OVAL-BC | |
| MX15 | EC/CCC | OVAL-BC | |
| MX26 | EC/CCC | OVAL-BC | |
| MX12 | EC/CCC | VCH | |
| MX13 | EC/CCC | VCH | |
| MX14 | EC/CCC | VCH | |
|
| |||
| MX4 | EC/LGSC | OVAL-BC | Endometrioid and Low Grade Serous |
| MX5 | EC/LGSC | OVAL-BC | |
| MX1 | EC/SBT | OVAL-BC | |
| MX6 | EC/SBT | OVAL-BC | |
|
| |||
| MX21 | CCC/HGSC | AGO | Clear Cell and High Grade Serous |
| MX27 | CCC/HGSC | Barts | |
| MX28 | CCC/HGSC | Leeds | |
|
| |||
| MX8 | MBOT/CCC | OVAL-BC | Mucinous and Clear Cell |
| MX7 | MC/CCC | OVAL-BC | |
|
| |||
| MX16 | MC/HGSC | OVAL-BC | Other Mixes |
| MX17 | MC/SBT | OVAL-BC | |
| MX25 | MC/Borderline Brenner | OVAL-BC | |
| MX10 | EC/CCC/Seromucinous | VCH | |
Immunohistochemical analysis
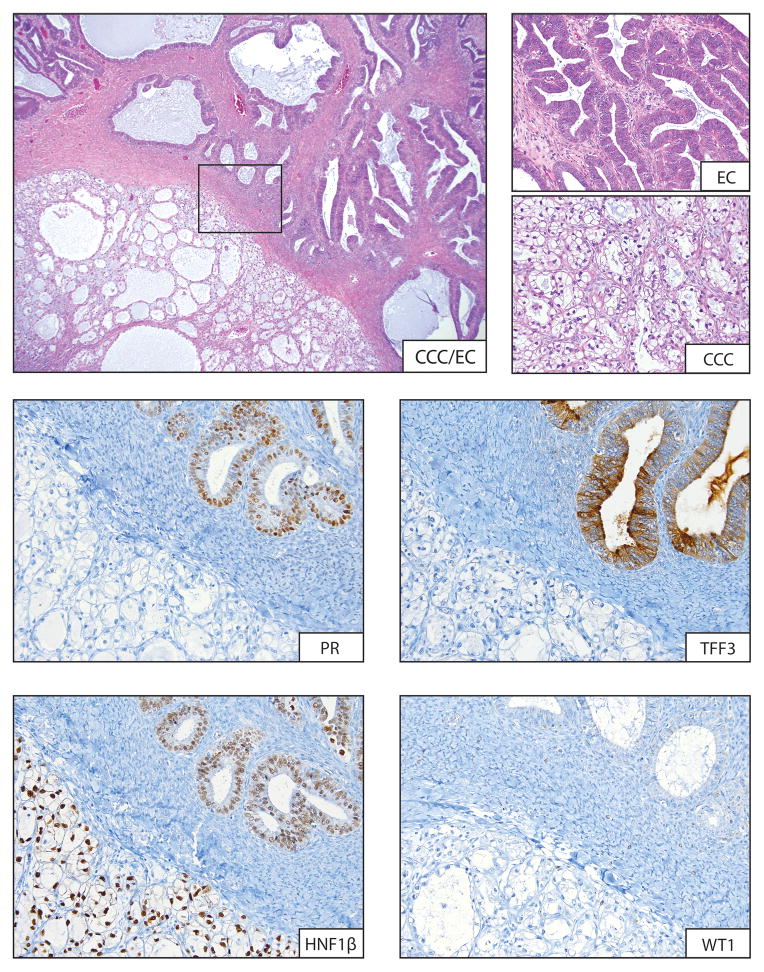
Immunohistochemical profiling of mixed-type tumors identified nine cases where components were predicted to be the same histotype, i.e., the immunoprofile of morphologically distinct components in these “mixed” tumors was indistinguishable (Table 2). For the 13 remaining cases, the immunoprofiles of the different components were distinct, and for 23 of 26 (88%) individual elements, the predicted histotype, based on the immunoprofile, corresponded to what was predicted based on H&E morphological examinations, a level of agreement with morphological assessment similar to what has been observed previously (42). Figure 2 shows a selection of IHC stains for MX11, a CCC and EC admixed case with distinct immunoprofiles. The EC/LGSC mixed case, MX4, was predicted to be a mix of EC and CCC based on immunohistochemical profiling. MX25, an admixture of MC and borderline Brenner tumor, presented a unique case as Brenner tumors are not included at all in the IHC prediction algorithm. While the immuno-phenotypes differed between components this was solely the result of expression of TFF3, with no additional molecular information available.
Table 2.
Immunohistochemical scores and corresponding histotype predictions for distinct regions of mixed-type EOC.
| Case ID | Morphological Histotype | IHC Markers | Histotype Prediction* (%) | Immunophenotypeτ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT1 | p53 | p16 | HNF1B | TFF3 | VIM | ARID1A | PR | ||||
| MX5 | EC | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | HGSC (89) | Pure (HGSC) |
| LGSC | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | HGSC (100) | ||
| MX6 | EC | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | EC (100) | Pure (EC) |
| SBT | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | EC (100) | ||
| MX7 | CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | Pure (CCC) |
| MC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | ||
| MX3 | EC | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | CCC (88) | Pure (CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | ||
| MX9 | EC | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | EC (99) | Pure (EC) |
| CCC | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | EC (100) | ||
| MX26 | EC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | Pure (CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | CCC (78) | ||
| MX21 | HGSC | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | HGSC (100) | Pure (HGSC) |
| CCC | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | HGSC (100) | ||
| MX17 | MC | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | EC (99) | Pure (EC) |
| SBT | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | EC (99) | ||
| MX10 | CCC/seromucinous | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | EC (100) | Pure (EC) |
| EC | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | EC (96) | ||
| MX4 | EC | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | CCC (78) | Mixed (CCC/EC) |
| LGSC | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | EC (76) | ||
| MX1 | EC | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | EC (99) | Mixed (EC/LGSC) |
| SBT | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | LGSC (63) | ||
| MX8 | CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | Mixed (CCC/MC) |
| MBT | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | MC (100) | ||
| MX2 | EC | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | EC (100) | Mixed (EC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | CCC (100) | ||
| MX11 | EC | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | EC (100) | Mixed (EC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | ||
| MX12 | EC | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | EC (99) | Mixed (EC/CCC) |
| CCC | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (80) | ||
| MX13 | EC | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | EC (100) | Mixed (EC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | CCC (92) | ||
| MX14 | EC | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | EC (99) | Mixed (EC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | CCC (100) | ||
| MX15 | EC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | EC (93) | Mixed (EC/CCC) |
| CCC | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | CCC (100) | ||
| MX27 | HGSC | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | HGSC (100) | Mixed (HGSC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | CCC (100) | ||
| MX28 | HGSC | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | HGSC (100) | Mixed (HGSC/CCC) |
| CCC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | ||
| MX25 | MC | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | MC (100) | Mixed (MC/CCC) |
| Borderline Brenner | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | CCC (95) | ||
| MX16 | MC | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | MC (100) | Mixed (MC/HGSC) |
| HGSC | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | HGSC (100) | ||
Histotype predictions are given as the percent probability that a tumor with the indicated IHC profile is a tumor of the corresponding histotype.
For our purposes, immunoprofiles of serous borderline tumors (SBT) and LGSC, as well as mucinous borderline tumors (MBOT) and MC were considered to be equivalent in the context of distinguishing them from other histologies.
Figure 2.
A typical example of immuno-profile used to assess immuno-profiles in each mixed-cell type pair. In this case the CCC and EC admixed case MX11 is shown at low power (4x) and at higher power (20x) in regions of CCC and EC. IHC for PR and TFF3 commonly differentiate the distinct regions, staining positively in the EC and negative in the CCC regions. Conversely HNF1β and WT1 showed uniform positive and negative staining in both regions, respectively.
Gene expression
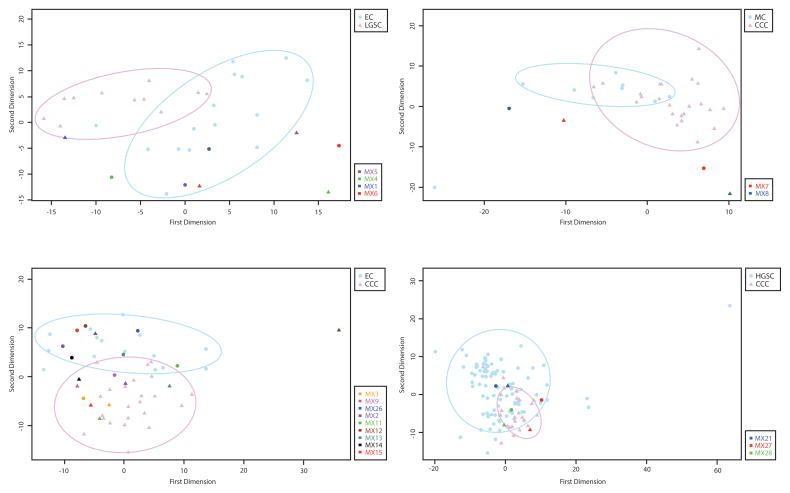
Gene expression results were obtained for 21/22 mixed EOC cases (all but MX25; insufficient RNA was extracted). Similar expression patterns, represented by co-segregation of mixed components within a cloud, or region, of ‘pure-type’ EOC on the multidimensional scaling (MDS) plot were observable in all 9 cases where components had indistinguishable immunoprofiles (Figure 3; see Figures, Supplemental Digital Content 3, 4 for additional MDS plots). Five of six EC and CCC admixed cases with distinct immunoprofiles for the two components had gene expression results in agreement with the immunoprofile, i.e. those components showing CCC morphology fell within their corresponding cloud of ‘pure’ CCC type, with the same holding true for the EC component. The sixth case (MX12) was also concordant in that the components separated, however the CCC component also showed some distance from a pure CCC type (Figure 3C). Similar trends were observed for two HGSC/CCC cases (MX27 and MX28), and one EC/LGSC mixed EOC (MX1). Although distinct components of MX8 (MC/CCC) did not lie within either cluster of pure type EOC (Figure 3B), there is a large amount of separation between the components, indicating dissimilarity in the expression profiles of the two regions. Thus the results of gene expression analysis were concordant with immunophenotypic characterization. When considering both immunohistochemical and gene expression data, 9/22 (41%) of cases did not show molecular evidence for the distinct morphological appearance and are considered tumors of pure histotype with a spectrum of morphological heterogeneity (Table 2).
Figure 3.
Multi-dimensional scaling plots of NanoString-based gene expression analysis comparing components of mixed-type ovarian carcinoma with a background of pure-type EOCs. Panel A shows endometrioid (EC) and low grade serous (LGSC) admixed cases, panel B shows mucinous (MC) and clear cell (CCC) mixed-type EOC, panel C shows EC/CCC admixed cases, and panel D shows high grade serous (HGSC) and CCC admixed EOC. Each point on the two dimensional plot represents gene expression data from 361 genes that are scaled and projected on two dimensions, permitting qualitative similarity analysis. Ellipsoids highlight regions representative of pure-type EOCs in each case.
Mutation analysis
Finally, cancer hotspot sequencing analysis was performed for 11 available cases of mixed EOC (Table 3). The 11 cases were selected such that at least one representative of each of the major groups (Outlined in Table 1) was sequenced. In three cases, sequencing was concordant with IHC and gene expression results of molecularly homogenous tumors with morphological heterogeneity showing the characteristic TP53 (HGSC; MX5, MX21) and PI3KCA (CCC; MX7) mutations at moderately equal proportions throughout the tumors.
Table 3.
Ion Torrent hotspot sequencing data for select mixed-type tumors.
| Case ID | Gene | DNA Alteration | Protein change | Histology | Frequency (%) | Difference | Coverage | COSMIC ID | Mix Type |
|---|---|---|---|---|---|---|---|---|---|
| MX11 | PIK3CA | c.1624G>A | p.E542K | EC | 87.7 | 65.6 | 16201 | COSM760 | Endometrioid and Clear Cell |
| CCC | 22.1 | 444 | |||||||
| MX2 | CTNNB1 | c.98C>G | p.S33C | EC | 30.8 | 30.8 | 2599 | COSM5677 | |
| CCC | 0 | 2520 | |||||||
| MX12 | PTEN | c.522_523delTG | p.Y176Lfs*4 | EC | 30.2 | 5 | 1450 | N/A | |
| CCC | 25.2 | 4745 | |||||||
| TP53 | c.524G>A | p.R175H | EC | 56.3 | 36.9 | 1593 | COSM10648 | ||
| CCC | 93.2 | 2627 | |||||||
| MX3 | TP53 | c.481G>A | p.A161T | EC | 20.8 | 20.8 | 2264 | COSM10739 | |
| CCC | 0 | 4350 | |||||||
| MX27 | TP53 | c.524G>A | p.R175H | HGSC | 83.7 | 83.7 | 4846 | COSM10648 | High Grade Serous and Clear Cell |
| CCC | 0 | 4410 | |||||||
| MX21 | TP53 | c.404G>A | p.C135Y | HGSC | 33.6 | 1.4 | 9799 | COSM10801 | |
| CCC | 35 | 9808 | |||||||
| IDH1 | c.395G>A | p.R132H | HGSC | 1 | 6.1 | 3238 | COSM28746 | ||
| CCC | 7.1 | 4251 | |||||||
| MX28 | MET | c.1124A>G | p.N375S | HGSC | 46.8 | 8.7 | 2964 | COSM710 | |
| CCC | 38.1 | 3121 | |||||||
| TP53 | c.380C>T | p.S127F | HGSC | 44.1 | 5.1 | 4686 | COSM44226 | ||
| CCC | 39 | 8064 | |||||||
| MX7 | PIK3CA | c.3140A>T | p.H1047L | CCC | 50.7 | 29.5 | 1495 | COSM776 | Mucinous and Clear Cell |
| MBOT | 21.2 | 811 | |||||||
| MX8 | KRAS | c.35G>A | p.G12D | CCC | 82.9 | 60.4 | 5006 | COSM521 | |
| MC | 22.5 | 7139 | |||||||
| MX4 | KRAS | c.35G>T | p.G12V | EC | 55.6 | 7 | 6023 | COSM520 | Endometrioid and Low Grade Serous |
| LGSC | 62.6 | 6890 | |||||||
| CTNNB1 | c.94G>A | p.D32N | EC | 37 | 37 | 2836 | COSM5672 | ||
| LGSC | 3.3 | 3240 | |||||||
| MX5 | TP53 | c.528C>G | p.C176W | EC | 71.2 | 33.2 | 2108 | COSM11114 | |
| LGSC | 38 | 3285 |
Identical mutations were observed within the distinct regions of two EC and CCC admixed cases: MX11 contains a PIK3CA p.E542K, while MX12 harbors PTEN (p.Y176Lfs*4) and TP53 (p.R175H) mutations. Found exclusively within the EC region of the tumor, CTNNB1 p.S33C and TP53 p.A161T mutations were found in MX2 and MX3, respectively. TP53 mutations were also found in all three mixed HGSC/CCC cases, presenting in both regions of MX28; however, only the HGSC region of MX27 had the observed TP53 p.R175H alteration. Both sequenced cases of MC/CCC mixed EOC harboured identical mutations in their distinct components: PIK3CA p.H1047L in MX7 and KRAS p.G12D in MX8.
Discussion
In summary, 9 of 22 (41%) tumors which were mixed based on morphological assessment were pure histotypes based on molecular studies, while 12 of 22 (55%) were mixed based on both morphological and molecular (immunohistochemical and genomic) analysis (Table 4) one case was inconclusive (MX25; Brenner/MC). Of the 12 ‘true’ mixed tumors, molecular results confirmed the morphological impression of histotype in 92% (11/12) of cases. In the 1 discordant case and the 1 ambiguous case, morphological assessment suggested LGSC and Brenner tumor components respectively, both of which were interpreted as CCC based on molecular analysis. In the latter cases the discordant component was an uncommon histotype, for which the molecular characteristics are not well defined, and based on review neither showed characteristic morphological features of CCC. Overall this suggests that these are best considered examples of molecular misclassification and, in the context of our study, were more accurately classified based on morphological assessment.
Table 4.
Summary of molecular histotype results for 22 cases of mixed epithelial ovarian cancer.
| Histotypes based on morphological review* | Pure histotype based on molecular studies (# of cases) | Type of pure EOC | Mixed based on molecular studies (# of cases) | Histotypes in mixed EOC by molecular studies |
|---|---|---|---|---|
| EC/CCC (n=9) | 3 | CCC (n=2) | 6 | EC/CCC (n=6) |
| EC(n=1) | ||||
| EC/LGSC/SBT (n=4) | 2 | HGSC(n=1) | 2 | EC/CCC (n=1) |
| EC(n=1) | EC/LGSC (n=1) | |||
| HGSC/CC (n=3) | 1 | HGSC (n=1) | 2 | HGSC/CCC (n=2) |
| CCC/MC (n=2) | 1 | CCC (n=1) | 1 | CCC/MC (n=1) |
| MC/SBT (n=1) | 1 | EC(n=1) | 0 | - |
| CCC/SMBT/EC (n=1) | 1 | EC (n=1) | 0 | - |
| MC/Brenner BT (n=1) | 0 | - | 1 | CCC/MC (n=1) |
| MC/HGSC (n=1) | 0 | - | 1 | MC/HGSC (n=1) |
| Total: 22 cases | Total: 9 (41%) | Total mixed based on molecular studies: 13 (59%) | Morphology concordant with molecular results in 11/13 mixed cases (85%) |
SMBT: seromucinous tumor; Brenner BT: brenner borderline tumor
Historically, mixed-type EOC accounted for up to 11% of cases (27–33). Even as recently as 2010 it was stated that “Careful study of [ovarian] tumors often reveals two or even three or more cell types” (43); however, the criteria for diagnosis of EOC histotypes have evolved (44) and recent studies suggest a much lower frequency. Currently, fewer mixed EOC diagnoses are being made, however no concrete figure exists for the frequency of mixed-type EOC based on current diagnostic criteria, and few studies examining the molecular alterations of these admixtures have been published. To address this, we conducted a population-based review, establishing a frequency of 1.7% for EOC of apparent mixed histology based on routine histopathological assessment, and suggest an even lower figure (8/871 OVAL-BC Cohort; 0.9%) based on molecular analysis.
Generally, the cases of EOC with mixed morphological features studied herein fall within one of two main groupings: 1, tumors with uniform molecular features throughout, consistent with their being a single histotype, but displaying morphological plasticity within distinct regions, and 2, tumors where the observed heterogeneity represents a more complete divergence of molecular and morphological features stemming from a common ancestral clone. Examples from the former category can be identified based on similar immunohistochemical and gene expression profiles in the morphologically distinct regions of these tumors, and include cases MX3, MX5 and MX21. Identical mutations occurring within distinct regions of MX21 and MX5 further support this conclusion. Interestingly, a TP53 p.A161T mutation was observed to occur solely in the EC region of MX3. This is presumably a new mutation acquired during tumor progression, occurring within a subpopulation of the tumor and potentially driving the divergence of morphology.
Microscopic heterogeneity observed in the morphology of mixed EOC falling within the second of the two categories extends to the molecular level, where distinct immunohistochemical and gene expression profiles exist for the disparate regions. However, a unifying mutation exists in nearly every case: e.g. a KRAS p.G12V mutation in both EC and LGSC regions of MX4, a TP53 mutation in the CCC and HGSC regions of MX28, and identical KRAS p.G12D mutations in the CCC and MC regions of MX8. Additionally, most of the cases that fall within this category are admixed CCC and EC, two histotypes that are largely accepted to arise from the same cell of origin, i.e. glandular epithelial cells of endometriosis (18, 19, 45–48). The mutational data, where informative, indicate that in such cases, the morphologically and molecularly distinct regions have arisen from the same initiation event. The sequencing panel employed in this study is limited to hotspot regions of 50 genes and, in cases such as MX2 with only a single CTNNB1 mutation in EC component and no detectable mutations in the CCC, many ancestral mutations, such as ARID1A (48) may have been missed.
Previous studies identified the presence of cells with clear cytoplasm within serous carcinoma as representing regions of morphological plasticity within a pure serous tumor (26, 34). One such example exists in our cohort (MX21); however, the remaining two HGSC/CCC cases (MX27 and MX28) have remarkable disparity in immunohistochemical staining patterns. The distinct regions of MX27 exhibit classical immunohistochemical profiles of pure type HGSC and CCC, and a TP53 mutation was found exclusively in the HGSC region (see Figure, Supplemental Digital Content 5, which shows distinct immunohistochemical staining for MX27). From our analysis, it is not possible to determine whether this case has arisen as the result of a distant common precursor that has undergone drastic subclonal divergence, or whether it is a true mixed, or ‘collision’ tumor. In MX28, however, a TP53 mutation was found within both regions suggesting that this unusual tumor is likely to have arisen from a common precursor.
Molecular analysis of admixed EC and LGSC shows different results for each case examined. Based on the molecular analyses, both MX6 and MX5 can be considered tumors of uniform histotype that display morphological plasticity within distinct regions. Immunohistochemical and gene expression results suggest MX1 and MX4 are true mixed tumors.
Epithelial ovarian tumors with mixed-type histology are uncommon, and bona fide mixed EOC where there is both morphological and molecular evidence of two, or more, histotypes account for approximately 1% of cases. CCC/EC admixtures are most commonly encountered, and this is likely in part related to their common ancestral lineage. In clinical practice, selective use of IHC can distinguish between true mixed EOC, and pure EOC with morphological variation, mimicking different histotypes. Although one immunomarker may be sufficient to determine the relatedness between two regions of a mixed EOC, use of an IHC panel is ideal in these cases. Some of the immunostaining in this study was done using polyclonal purified antibodies (specifically those for HNF1B and ARID1A). Although the same reagent lot was used throughout this study, and no variation in staining was observed across our controls, the potential for batch-to-batch variability of such preparation may be problematic for routine clinical use. Finally, the clinical significance of true mixed histotypes is unclear, and more cases with follow-up will be needed to know if their natural history is comparable to that of EOC of pure histotype.
Supplementary Material
Original and review diagnosis of OVAR11 mixed-type carcinomas (TCC – Transitional Cell Carcinoma).
Detailed methods of antibodies and immunohistochemical staining
MDS analysis of MX5 gene expression profile compared to ‘pure-type’ EC, LGSC and HGSC. MCT-09 and MCT-10 were diagnosed with LGSC and EC histology, respectively.
MDS analysis of MX4 gene expression profile compared to ‘pure-type’ EC and CCC. MCT-07 and MCT-08 were diagnosed with EC and LGSC histology, respectively.
Immunoprofiling of MX27, a CCC and HGSC admixed (A–B) case shows striking disparity in staining between tumor regions. HGSC positivity and CCC negativity was seen for WT1 (C–D), ARID1A, (E–F), p16 (G–H), and p53 (I–J), while HGSC negativity and CCC positivity was seen for HNF1β (K–L).
Acknowledgments
Funding Declaration: This work was funded by the Carraresi Foundation OVCARE Research Grant, the BC Cancer Foundation, the VGH+UBC Hospitals foundation, The Canadian Institutes for Health Research, and WorkSafe BC
This work was funded by a Carraresi Foundation OVCARE Research Grant (to MSA), supported by the Carraresi Foundation, BC Cancer Foundation and the VGH+UBC Hospital Foundation. OVAL-BC study was supported by grants (to authors LSC and NL) from The National Institutes of Health/National Cancer Institute (R01 CA160669-01A1), The Canadian Institutes for Health Research and WorkSafe BC. Funding bodies have no influence on research. No writing assistance was utilized in the production of this manuscript.
List of Abbreviations
- CCC
Clear cell carcinoma
- COSMIC
Catalogue of somatic mutations in cancer
- EC
Endometrioid ovarian carcinoma
- EOC
Epithelial ovarian cancer
- FFPE
Formalin fixed paraffin embedded
- HGSC
High grade serous carcinoma
- IHC
Immunohistochemistry
- LGSC
Low grade serous carcinoma
- MC
Mucinous carcinoma
Footnotes
Competing Interests: Authors have no significant competing interests to declare, financial or otherwise.
Author contributions
Mixed cell type cases included in this study were obtained by the following authors: DC, NL, LSC and CBG (OVAL-BC); SK, FK and JP (AGO study group); NW (Leeds Teaching Hospital); JM and NS (Barts Health). SE and CBG conducted case review. IHC was performed by SL and CC, and was scored by RM with instruction from MK and CBG. MK interpreted IHC profiles and assigned histotype predictions. Extraction of nucleic acids, gene expression analysis and hotspot sequencing were performed by RM. Processing and analysis of NanoString data was done by AT. All data were interpreted by RM, MSA and CBG. Study design and oversight was provided by DGH, SK, CBG and MSA. RM, MSA and CBG wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Auersperg N. The origin of ovarian carcinomas: a unifying hypothesis. Int J Gynecol Pathol. 2011;30:12–21. doi: 10.1097/PGP.0b013e3181f45f3e. [DOI] [PubMed] [Google Scholar]
- 2.Kobel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurman RJ, Shih Ie M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer--shifting the paradigm. Hum Pathol. 2011;42:918–931. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soslow RA. Histologic subtypes of ovarian carcinoma: an overview. Int J Gynecol Pathol. 2008;27:161–174. doi: 10.1097/PGP.0b013e31815ea812. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. The Journal of pathology. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Press JZ, De Luca A, Boyd N, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurung A, Hung T, Morin J, et al. Molecular abnormalities in ovarian carcinoma: clinical, morphological and therapeutic correlates. Histopathology. 2013;62:59–70. doi: 10.1111/his.12033. [DOI] [PubMed] [Google Scholar]
- 8.Kalloger SE, Kobel M, Leung S, et al. Calculator for ovarian carcinoma subtype prediction. Mod Pathol. 2011;24:512–521. doi: 10.1038/modpathol.2010.215. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya A, Sakamoto M, Yasuda J, et al. Expression profiling in ovarian clear cell carcinoma: identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. The American journal of pathology. 2003;163:2503–2512. doi: 10.1016/s0002-9440(10)63605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anglesio MS, Kommoss S, Tolcher MC, et al. Molecular characterization of mucinous ovarian tumours supports a stratified treatment approach with HER2 targeting in 19% of carcinomas. The Journal of pathology. 2013;229:111–120. doi: 10.1002/path.4088. [DOI] [PubMed] [Google Scholar]
- 11.Gershenson DM, Sun CC, Bodurka D, et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecologic oncology. 2009;114:48–52. doi: 10.1016/j.ygyno.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Tsang YT, Deavers MT, Sun CC, et al. KRAS (but not BRAF) mutations in ovarian serous borderline tumor are associated with recurrent low-grade serous carcinoma. The Journal of pathology. 2013 doi: 10.1002/path.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson JW, Jarboe EA, Kindelberger D, et al. Serous tubal intraepithelial carcinoma: diagnostic reproducibility and its implications. Int J Gynecol Pathol. 2010;29:310–314. doi: 10.1097/PGP.0b013e3181c713a8. [DOI] [PubMed] [Google Scholar]
- 14.Cuff J, Longacre TA. Endometriosis does not confer improved prognosis in ovarian carcinoma of uniform cell type. The American journal of surgical pathology. 2012;36:688–695. doi: 10.1097/PAS.0b013e31824b6eed. [DOI] [PubMed] [Google Scholar]
- 15.Levanon K, Ng V, Piao HY, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29:1103–1113. doi: 10.1038/onc.2009.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anglesio MS, Carey MS, Kobel M, et al. Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecologic oncology. 2011;121:407–415. doi: 10.1016/j.ygyno.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38:87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 18.Prowse AH, Manek S, Varma R, et al. Molecular genetic evidence that endometriosis is a precursor of ovarian cancer. International journal of cancer. 2006;119:556–562. doi: 10.1002/ijc.21845. [DOI] [PubMed] [Google Scholar]
- 19.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. The New England journal of medicine. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess V, A’Hern R, Nasiri N, et al. Mucinous epithelial ovarian cancer: a separate entity requiring specific treatment. J Clin Oncol. 2004;22:1040–1044. doi: 10.1200/JCO.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 21.Schiavone MB, Herzog TJ, Lewin SN, et al. Natural history and outcome of mucinous carcinoma of the ovary. American journal of obstetrics and gynecology. 2011;205:480.e481–488. doi: 10.1016/j.ajog.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Laury AR, Ning G, Quick CM, et al. Fallopian tube correlates of ovarian serous borderline tumors. The American journal of surgical pathology. 2011;35:1759–1765. doi: 10.1097/PAS.0b013e318233b0f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Abushahin N, Pang S, et al. Tubal origin of ‘ovarian’ low-grade serous carcinoma. Mod Pathol. 2011;24:1488–1499. doi: 10.1038/modpathol.2011.106. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Fadare O, Xiang L, et al. Ovarian serous carcinoma: recent concepts on its origin and carcinogenesis. Journal of hematology & oncology. 2012;5:8. doi: 10.1186/1756-8722-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobel M, Kalloger SE, Baker PM, et al. Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study. The American journal of surgical pathology. 2010;34:984–993. doi: 10.1097/PAS.0b013e3181e1a3bb. [DOI] [PubMed] [Google Scholar]
- 26.Han G, Gilks CB, Leung S, et al. Mixed ovarian epithelial carcinomas with clear cell and serous components are variants of high-grade serous carcinoma: an interobserver correlative and immunohistochemical study of 32 cases. The American journal of surgical pathology. 2008;32:955–964. doi: 10.1097/PAS.0b013e318164edf7. [DOI] [PubMed] [Google Scholar]
- 27.Fruscio R, Garbi A, Parma G, et al. Randomized phase III clinical trial evaluating weekly cisplatin for advanced epithelial ovarian cancer. Journal of the National Cancer Institute. 2011;103:347–351. doi: 10.1093/jnci/djq530. [DOI] [PubMed] [Google Scholar]
- 28.Hoskins P, Vergote I, Cervantes A, et al. Advanced ovarian cancer: phase III randomized study of sequential cisplatin-topotecan and carboplatin-paclitaxel vs carboplatin-paclitaxel. Journal of the National Cancer Institute. 2010;102:1547–1556. doi: 10.1093/jnci/djq362. [DOI] [PubMed] [Google Scholar]
- 29.Le T, Adolph A, Krepart GV, et al. The benefits of comprehensive surgical staging in the management of early-stage epithelial ovarian carcinoma. Gynecologic oncology. 2002;85:351–355. doi: 10.1006/gyno.2002.6636. [DOI] [PubMed] [Google Scholar]
- 30.Mannel RS, Brady MF, Kohn EC, et al. A randomized phase III trial of IV carboplatin and paclitaxel x 3 courses followed by observation versus weekly maintenance low-dose paclitaxel in patients with early-stage ovarian carcinoma: a Gynecologic Oncology Group Study. Gynecologic oncology. 2011;122:89–94. doi: 10.1016/j.ygyno.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markman M, Bundy BN, Alberts DS, et al. Phase III trial of standard-dose intravenous cisplatin plus paclitaxel versus moderately high-dose carboplatin followed by intravenous paclitaxel and intraperitoneal cisplatin in small-volume stage III ovarian carcinoma: an intergroup study of the Gynecologic Oncology Group, Southwestern Oncology Group, and Eastern Cooperative Oncology Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:1001–1007. doi: 10.1200/JCO.2001.19.4.1001. [DOI] [PubMed] [Google Scholar]
- 32.Seidman JD, Horkayne-Szakaly I, Haiba M, et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 33.Young RC, Brady MF, Nieberg RK, et al. Adjuvant treatment for early ovarian cancer: a randomized phase III trial of intraperitoneal 32P or intravenous cyclophosphamide and cisplatin--a gynecologic oncology group study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:4350–4355. doi: 10.1200/JCO.2003.02.154. [DOI] [PubMed] [Google Scholar]
- 34.DeLair D, Han G, Irving JA, et al. HNF-1beta in ovarian carcinomas with serous and clear cell change. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2013;32:541–546. doi: 10.1097/PGP.0b013e318273fd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilks CB, Ionescu DN, Kalloger SE, et al. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Human pathology. 2008;39:1239–1251. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 37.Kobel M, Kalloger SE, Lee S, et al. Biomarker-based ovarian carcinoma typing: a histologic investigation in the ovarian tumor tissue analysis consortium. Cancer Epidemiol Biomarkers Prev. 2013;22:1677–1686. doi: 10.1158/1055-9965.EPI-13-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobel M, Bak J, Bertelsen BI, et al. Ovarian carcinoma histotype determination is highly reproducible, and is improved through the use of immunohistochemistry. Histopathology. 2013 doi: 10.1111/his.12349. [DOI] [PubMed] [Google Scholar]
- 39.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen T, Wallden B, Schaper C, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobel M, Bak J, Bertelsen BI, et al. Ovarian carcinoma histotype determination is highly reproducible, and is improved through the use of immunohistochemistry. Histopathology. 2014;64:1004–1013. doi: 10.1111/his.12349. [DOI] [PubMed] [Google Scholar]
- 43.Mills S, Carter D, Greenson J, et al. Sternberg’s Diagnostic Surgical Pathology. 5. 2010. [Google Scholar]
- 44.Kurman RJ, Carcangiu ML, Herrington CS, et al. WHO Classification of Tumours of Female Reproductive Organs. WHO Classification of Tumours. (4) 2014;6 [Google Scholar]
- 45.Jiang X, Hitchcock A, Bryan EJ, et al. Microsatellite analysis of endometriosis reveals loss of heterozygosity at candidate ovarian tumor suppressor gene loci. Cancer research. 1996;56:3534–3539. [PubMed] [Google Scholar]
- 46.Munksgaard PS, Blaakaer J. The association between endometriosis and ovarian cancer: a review of histological, genetic and molecular alterations. Gynecologic oncology. 2012;124:164–169. doi: 10.1016/j.ygyno.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Sampson JA. Metastatic or Embolic Endometriosis, due to the Menstrual Dissemination of Endometrial Tissue into the Venous Circulation. The American journal of pathology. 1927;3:93–110. 143. [PMC free article] [PubMed] [Google Scholar]
- 48.Anglesio MS, Bashashati A, Wang Y, et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. The Journal of pathology. 2015 doi: 10.1002/path.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original and review diagnosis of OVAR11 mixed-type carcinomas (TCC – Transitional Cell Carcinoma).
Detailed methods of antibodies and immunohistochemical staining
MDS analysis of MX5 gene expression profile compared to ‘pure-type’ EC, LGSC and HGSC. MCT-09 and MCT-10 were diagnosed with LGSC and EC histology, respectively.
MDS analysis of MX4 gene expression profile compared to ‘pure-type’ EC and CCC. MCT-07 and MCT-08 were diagnosed with EC and LGSC histology, respectively.
Immunoprofiling of MX27, a CCC and HGSC admixed (A–B) case shows striking disparity in staining between tumor regions. HGSC positivity and CCC negativity was seen for WT1 (C–D), ARID1A, (E–F), p16 (G–H), and p53 (I–J), while HGSC negativity and CCC positivity was seen for HNF1β (K–L).