Abstract
Soluble recombinant proteins that comprise the extracellular part of a surface expressed receptor attached to the Fc region of an IgG antibody have facilitated the determination of ligand specificity for an array of immune system receptors. Among such receptors is the family of killer cell immunoglobulin-like receptors (KIR) that recognize HLA class I ligands. These receptors, expressed on natural killer (NK) cells and T cells, play important roles in both immune defense and placental development in early pregnancy. Here we describe a method for the production of two domain KIR-Fc fusion proteins using baculovirus infected insect cells. This method is more scalable than traditional mammalian cell expression systems and produces efficiently folded proteins that carry posttranslational modifications found in native KIR. We also describe a multiplex binding assay using the Luminex platform that determines the avidity and specificity of two domain KIR-Fc for a panel of microbeads, each coated with one of 97 HLA class I allotypes. This assay is simple to perform, and represents a major improvement over the assays used previously, which were limited in the number of KIR and HLA class I combinations that could be assayed at any one time. The results obtained from this assay can be used to predict the response of NK cell and T cells when their KIR recognize HLA class I.
Keywords: Natural Killer Cells, MHC, Comparative Immunology/Evolution, Antigens/Peptides/Epitopes
1. Introduction
Killer-cell immunoglobulin like receptors (KIR) are a family of germ-line encoded cell surface receptors that regulate the activity of natural killer (NK) cells and T cells in immunity and reproduction through interaction with HLA class I molecules (Parham and Moffett 2013). Both KIR and HLA are encoded by polymorphic genes, which have numerous alleles encoding unique KIR and HLA class I proteins, which are known as allotypes. HLA class I proteins are expressed on the surface of most cell types and present a diverse repertoire of peptides that furnish ligands for KIR and other immune system receptors (Bjorkman et al. 1987; Colonna et al. 1992; Colonna and Samaridis 1995; Moretta et al. 1993). The HLA class I locus contains three highly polymorphic genes, called HLA-A, B and C. HLA-C is the most recently evolved and the only one for which all the variant forms are KIR ligands (Guethlein et al. 2007; Older Aguilar et al. 2010; Older Aguilar et al. 2011). Dimorphism at position 80 in HLA-C defines two epitopes, C1 (asparagine 80) and C2 (lysine 80), which are ligands for two different forms of two-domain KIR (Mandelboim et al. 1996; Winter and Long 1997). KIR2DL1 encodes methionine at position 44 and binds to C2 bearing HLA-C, KIR2DL2/3 encodes lysine at position 44 and binds to C1 bearing HLA-C allotypes.
Because the genes encoding KIR and HLA class I are on different chromosomes, their independent segregation during meiosis produces diversity in the number and type of KIR-HLA gene combinations inherited by individuals (Norman et al. 2013; Wilson et al. 2000). Further, NK cells can express more than one KIR at a time (Lanier 1997; Valiante et al. 1997). This inherent diversity has complicated the investigation of the specific KIR-HLA class I interactions that modulate immune response. Development of soluble KIR proteins for which the reactivity for single HLA class I molecules was determined by direct binding assay, facilitated understanding of how particular receptor-ligand combinations contributed to NK cell reactivity (Winter et al. 1998). These recombinant proteins were made in a mammalian cell expression system by fusing the extracellular domains of a two-domain KIR with two Fc domains of a human IgG1 to form a soluble homodimer (Winter and Long 2000).
We have adapted this method for the production of soluble KIR-Fc fusion proteins by using baculovirus-infected insect cells. The advantage of this approach is that insect cells are simple to culture. They have short doubling times that facilitate scaling and they are capable of higher protein yields than mammalian cell systems of expression. Because of these advantages, the baculovirus-insect cell system is now one of the most widely used methods for the production of recombinant proteins (Hitchman et al. 2009). Although not equivalent to higher eukaryotic cells, most post-translational modifications are made correctly in insect cells, and proteins unable to be expressed in E. coli have been successfully expressed in the insect cell system (Victor et al. 2010). The baculovirus family are species-specific double-stranded DNA viruses that infect insects as their natural host (Kost and Condreay 1999). Once inserted into the host nucleus, the baculovirus is packaged into flexible nucleocapsids, into which foreign DNA may readily be inserted. The target gene, in this case the KIR-Fc fusion construct, is inserted into a transfer vector and positioned between sequences that are homologous to ones in the baculoviral genome. When the viral genome and transfer vector are transfected into insect cells, recombination occurs, and produces intact viral genomes harbouring the target gene sequence. The target gene replaces the non-essential baculoviral polyhedrin gene. The strong promoter of the polyhedrin gene is co-opted for production of recombinant target protein.
We have also developed a multiplex assay that tests the binding of soluble KIR-Fc to 97 HLA class I allotypes. This assay uses the Luminex platform, in which the antigenic targets are microbeads, each coated with a defined HLA class I allotype. Such beads were developed originally for studying the specificity of human alloantibodies (Pei et al. 1998; Pei et al. 2003), but our group has successfully adapted this platform for use with recombinant two-domain KIR-Fc fusion proteins and monoclonal antibodies (Hilton and Parham 2013; Moesta et al. 2008). By adjusting the relative concentration of two fluorescent dyes, a set of 100 individually identifiable beads is generated. Each bead is then coated with a different HLA class I allotype, allowing the results of the immunoassay to be correlated with HLA class I specificity.
The goal of the KIR-Fc HLA-bead binding assay is to determine the strength and specificity of the interactions between HLA class I and KIR using defined purified proteins. The results can be used to predict the reactivity of KIR expressing NK cells and T cells when their KIR recognize cognate HLA class I ligands. This assay represents a major advance from the cell-based direct binding assay in which the reactivity of only a few KIR and HLA class I combinations could be determined at any one time (Winter and Long 2000). Moreover, the KIR-Fc HLA bead-binding assay is designed to inform cellular assays of lymphocyte function in which receptor deficient effector NK cells transfected with a specific KIR are incubated with ligand-deficient target cells transfected with a specific HLA class I molecule (Moesta et al. 2008).
The HLA class I specificity of several KIR allotypes has been investigated using various assays. Our initial study with the multiplex bead-binding assay showed that KIR2DL2*001-Fc recognized HLA-C2 allotypes with higher avidity than its allotypic variant KIR2DL3*001-Fc (Moesta et al. 2008). A cellular cytotoxicity assay subsequently showed that KIR2DL2*001, but not KIR2DL3*001 effectively inhibited lysis when incubated with HLA class I deficient 221 cells transfected with the HLA-C2 allotype, HLA-C*04:01 (Moesta et al. 2008). Another group investigated a second allotypic variant, KIR2DL3*005 (Frazier et al. 2013) using a similar multiplex assay. They showed that KIR2DL3*005-Fc bound HLA-C1 allotypes with higher avidity than KIR2DL3*001-Fc. This result was concordant with a cellular assay in which NK cells expressing either 2DL3*005 or 2DL3*001 respectively were incubated with 221 cells expressing the C1 bearing allotype HLA-C*03:04. Natural killer cells expressing 2DL3*005 exhibited a more potent inhibitory signal than those transfected with 2DL3*001. (Frazier et al. 2013). A third type of assay, surface plasmon resonance, confirmed that the 2DL3*005 variant bound with greater avidity than the 2DL3*001 variant to the HLA-C1 allotype HLA-C*03:04 (Frazier et al. 2013).
In summary, we have developed a simplified method for the production of KIR-Fc and designed an assay that tests their binding to 97 HLA class I allotypes simultaneously. The assay is easy to perform and correlates well with more complicated experimental techniques such as cellular cytotoxicity and surface plasmon resonance that have traditionally been used to determine the avidity and specificity of KIR for HLA class I ligands.
2. Materials and Methods
KIR-Fc fusion protein generation
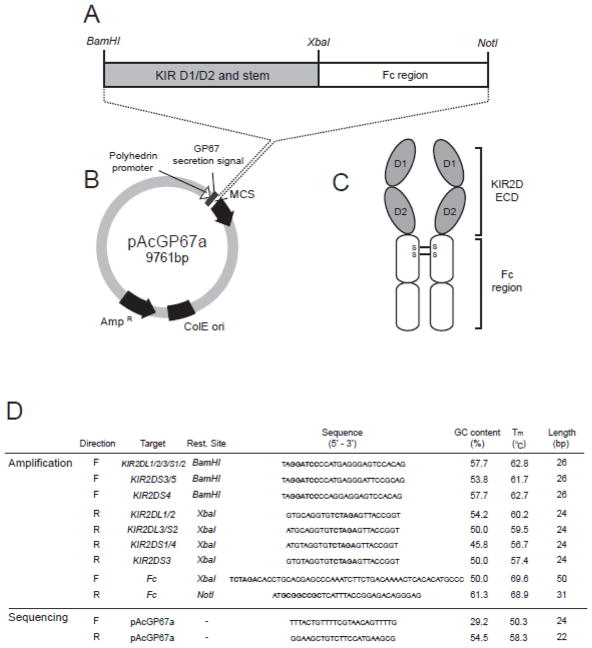
This section describes the generation of a DNA insert, flanked by restriction sites, that encodes the first 224 amino acids of the KIR2D of interest and the Fc region of human IgG1 (Figure 1A). This insert is first cloned into the pAcGP67a transfer vector (Figure 1B). Subsequently it is co-transfected with linearized baculovirus into insect cells.
Figure 1.
(A) Schematic diagram showing the configuration of a recombinant KIR-Fc fusion gene. The recombinant fusion gene consisting of the D1, D2 and stem domains of a KIR2D molecule (grey box) and the Fc region of a human IgG1 antibody (white box) is cloned using BamH1 and Not1 restriction sites into the multiple cloning site (MCS) of the pAcGP67a vector (B) in frame behind the GP67 secretion signal sequence. Transfection into insect cells produces a soluble recombinant KIR-Fc dimer (C) consisting of the D1, D2 and stem regions (extracellular domain – ECD) of the KIR molecule (grey ovals) and the Fc region (white ovals) of the IgG1 antibody. (S-S) shows the location of the disulphide bonds that lead to formation of a dimer.
(D) Table listing the primers for amplification and sequencing of KIR-Fc fusion genes representing inhibitory KIR2DL1, 2DL2/3 and activating KIR2DS1, 2DS2, 2DS3, 2DS4 and 2DS5. The properties of each primer, including the GC content, melting temperature (Tm) and length are listed to the right. Restriction sites are shown in bold when present in a primer sequence.
2.1 KIR-Fc fusion construct generation
Amplify the sequences encoding the D1, D2 and stem region of the selected KIR2D molecule by PCR (Primers from Figure 1D, upper panel). The forward primer annealing site is immediately downstream of the KIR signal sequence cleavage site and should contain a BamHI (New England BioLabs, Cat. #R0136) site. The reverse primer annealing site is immediately upstream of the transmembrane region and contains an XbaI (New England Biolabs, Cat. #R0145s) site.
Add a mix consisting of 20.2μl PCR grade sterile water, 2.5μl 10X cloned Pfu buffer, 0.5μl forward primer (10mM), 0.5μl reverse primer (10mM), 0.2μl dNTP mix (40mM) and 0.25μl HotStart TAQ polymerase (Kit from Qiagen, Cat. #203203) to 1μl of template DNA (10ng/μl) and perform PCR using the following conditions: (5min at 95°C) × 1, (30s at 94°C, 1min25s at 62°C, 40s at 72°C) × 35, (10min at 72°C) × 1, hold at 4°C. Digest the amplification products with BamHI and XbaI and ligate into the BamH1 and XbaI digested pAcGP67a vector (BD Pharmigen, Cat. #554758). Amplify the sequences encoding the hinge, CH2 and CH3 regions of human IgG1 using a forward primer containing an XbaI site and a reverse primer containing a NotI (New England Biolabs, Cat. #R0189) site (Primers from Figure 1D, upper panel). Digest the amplification products with XbaI and NotI and ligate into the XbaI and NotI digested pAcGP67a vector. Transform the ligation products into E.coli competent cells (Kit from Invitrogen, Cat. #K4510-20). Following overnight incubation select individual E.coli colonies, clonally amplify and prepare transfection quality DNA using Qiagen columns (Kit from Qiagen, Cat. #27104). Sequence each clone to confirm that the desired KIR-Fc fusion gene is inserted in frame behind the GP67 secretion signal sequence (primers from Figure 1D, lower panel).
2.2 Site-directed mutagenesis of KIR-Fc constructs
Site-directed mutagenesis can be used to introduce non-synonymous mutations into the newly generated KIR-Fc fusion gene. This technique is useful for making comparisons of closely related KIR2D-Fc allotypes or to investigate the effect of alternative amino acids at a particular position on HLA class I recognition. Mutagenesis primers should have a melting temperature of at least 78°C and the desired mutation should be situated in the centre of the primer sequence. The website www.bioinformatics.org/primerx was used to design mutagenesis primers. Add a mix consisting of 40μl PCR grade sterile water, 5μl 10X cloned Pfu buffer (Agilent Technologies, Cat. #600153-82), 1μl forward primer (10mM), 1μl reverse primer (10mM), 1μl dNTP mix (40mM) and 1μl Turbo polymerase (Agilent Technologies, Cat. #600252-52) to 1μl of template DNA (50ng/μl) and perform PCR using the following conditions: (30s at 95°C) × 1, (30s at 95°C, 60s at 55°C, 15min at 68°C) × 20, (7min at 72°C) × 1, hold at 4°C. A 15 min extension time is recommended, because of the large size of the pAcGP67a vector (9761bp). Following the PCR, add 1μl DpnI (New England BioLabs, Cat. #R0167S) to the reaction mixture and incubate for 30min at 37°C. DpnI digests the methylated template DNA and effectively increases the efficiency of mutated DNA transformation into competent E.coli cells. Following overnight incubation select individual E.coli colonies and then clonally amplify and prepare transfection quality DNA using Qiagen columns (Kit from Qiagen, Cat. #27104). Sequence each clone to confirm that the desired KIR-Fc fusion gene is inserted in frame with the GP67 secretion signal (primers from Figure 1D, lower panel).
2.3 Co-transfection of KIR-Fc containing transfer vector and baculovirus into insect cells
Having constructed the KIR-Fc gene, cloned it into the pAcGP67a transfer vector and mutated it if required, the construct must now be combined with linearized baculovirus and transfected into insect cells. For the initial transfection, the insect cells derived from Spodoptera frugiperda (Sf9) (Invitrogen, Cat. #11496-015) are used. Both the Sf9 insect cells and baculovirus should be handled in a sterile laminar flow hood. The Sf9 cells should be cultured in Sf-900 II media (Invitrogen, Cat. #10902-096) supplemented with 10% heat inactivated fetal bovine serum (FBS), 1% Penicillin-Streptomycin and 1% L-Glutamine, shaking at 120 rpm at 27°C. The Sf9 cells should be maintained at between 4 × 105 and 2 × 106 cells/ml. When maintained at this density, Sf9 cells have an approximate doubling time of 24h; a slower doubling time may signify unhealthy cells (see section 2.11). For each KIR-Fc transfection, seed one well of a sterile six-well plate (BD Falcon, Cat. #353046) with 2ml Sf9 cells at a density of 1 × 106 cells/ml and incubate for 60 mins at 27°C to allow adherence of Sf9 cells. Concurrently, mix (gently using a pipette) 2μg of purified transfer vector DNA with KIR-Fc insert from step 2.1 with 0.5μg of linearized baculovirus DNA (Expression Systems, Cat. #91-002) in a sterile 2ml microcentrifuge tube and incubate for 5 mins at room temperature. This specific baculovirus has the chitinase and cathepsin loci deleted to remove a detrimental protease and reduce competition for resources for protein synthesis during late gene expression. Meanwhile, for each transfectant, mix 10μl Cellfectin II (Invitrogen, Cat. #10362-100) with 100μl un-supplemented Sf-900 II media (Invitrogen, Cat. #10902-096) and pipette to mix them together. Add this to the baculovirus/KIR-Fc construct mixture (immediately following the 5 min incubation), mix gently using a pipette and incubate for a further 30 mins at room temperature. Following the 30 min incubation, add 1.8ml un-supplemented Sf-900 II media to complete each KIR-Fc transfection mixture. In summary, each KIR-Fc transfection mixture should contain Sf9 cells, KIR-Fc fusion gene DNA in pAcGP67a transfer vector, linearized baculovirus DNA, Cellfectin and un-supplemented Sf-900 II media.
Remove the 6-well plate from incubation and replace the medium covering the adherent cells with the KIR-Fc transfection mixture. Incubate, covered but not shaking, for 3–5 hours at 27°C. Remove the supernatant from each well and replace with 3ml Sf-900 II media (warmed to 27°C and supplemented with 10% heat inactivated FBS, 1% Penicillin-Streptomycin and 1% L-Glutamine). Incubate, covered and sealed, with laboratory film (Pechiney, Cat. #PM996), for 7 days at 27°C. Remove the supernatant from each well and transfer to a sterile 15ml tube. Separate the cells by centrifugation (1000g for 10 mins), discard the pellet and transfer the supernatant to a sterile 15ml tube and store in the dark at 4°C. This first transfectant, termed P0, and each subsequent Sf9 amplification supernatant (P1–P3) may be placed without further modification at −80°C for long-term storage.
2.4 Amplification of baculovirus in Sf9 cells – P1-3 production
Successive rounds of Sf9 cell amplification are now used to amplify the baculovirus (now containing the KIR-Fc gene added in step 2.3). Add 30ml Sf9 cells at a density of 1 × 106 cells/ml to a 125ml vented sterile Erlenmeyer flask (Thermo-Scientific, Cat. #4116-0125) and leave shaking at 115 rpm overnight at 27°C. The following day, add 500μl of P0 virus stock to the flask and incubate for 4–5 days shaking at 120rpm at 27°C. Collect 14ml suspension from each flask and transfer to a sterile 15ml tube. Separate the cells by centrifugation (1000g for 5mins) and transfer each supernatant to a new sterile 15ml tube. Discard the pellet and store in the dark at 4°C. This forms the P1 viral stock. A second (P2) or third (P3) round of amplification in which each round is seeded with 50μl of the preceding amplified supernatant, may be required to produce Sf9 supernatant with a sufficiently high baculoviral titre for adequate protein production.
Quantification of baculovirus amplification in Sf9 supernatant
Before proceeding it is necessary to quantify the amount of baculovirus generated from the amplification of the Sf9 cells in Sections 2.3 and 2.4. The amount of baculovirus generated is critical, as insufficient baculoviral amplification will result in low or absent protein yield. An estimation of viral titre can be made in two ways: directly, using small-scale protein production (protein mini-prep) (see Section 2.5) or indirectly, by flow cytometry, assessing baculoviral-induced up-regulation of surface glycoprotein 64 (GP64) (see Section 2.6).
2.5 KIR-Fc fusion protein – mini-prep
This small-scale protein production tests the capacity of the baculoviral infected Sf9 supernatant from steps 2.3 and 2.4 to produce protein from Trichopulsia ni (Hi5) (Invitrogen, Cat. #B855-02) insect cells. Hi5 cells are similar to Sf9 cells but are optimized for recombinant protein production rather than baculoviral amplification. Hi5 cells should be cultured, shaking at 27°C in Express Five serum free media (Invitrogen, Cat. #10486-025) supplemented with 1% L-Glutamine. The cell density should be kept at between 4 × 105 and 3 × 106 cells/ml. Aggregation of cells, which is more likely to occur at densities higher than 3 × 106 cells/ml, is a sign of unhealthy cells and will reduce the possible protein yield from any preparation in which they are used (see Section 2.11). Seed each well of a sterile 6-well tissue culture plate with 2ml Hi5 cells at a density of 1 × 106 cells/ml. Add 100μl of the viral stock under test (P1, P2 or P3) to each well, seal the plate with laboratory film and incubate for 48 hours (covered, shaking at 120 rpm, at 27°C). To serve as a negative control, one well should contain Hi5 cells but no Sf9 supernatant. To serve as a positive control, one well should be transfected with an Sf9 supernatant previously used to produce KIR-Fc protein. Transfer the contents of each well to a 2ml microcentrifuge tube and separate the cells by centrifugation (1000g for 5 mins). Discard the pellet and transfer 1.5ml of the supernatant to a clean 2ml microcentrifuge tube, add 150μl 10x Hepes Buffered Saline (HBS), pH 7.2 and 10μl protein A Sepharose bead slurry (Invitrogen, Cat. #101142). Rotate the mixture overnight at 4°C.
Separate the beads by centrifugation (2500g, 15min, 4°C) and discard the supernatant. Add 20μl Laemmli sample buffer (Bio-Rad, Cat. #161-0737) with 5% β-mercaptoethanol (Bio-Rad, Cat. #161-0710) and incubate the sample at 95°C for 10 minutes. Load the sample onto a 12% SDS-PAGE gel (Bio-Rad, Cat. #456-1043) and run at 150v for 1hr. Stain with Coomassie reagent (Bio-Rad, Cat. #161-0786) to identify an appropriately sized band. Although each KIR-Fc exists as a 102Kda homodimer in native format (Figure 1C), they run as a band of 51Kda monomers (Figure 3A) on an SDS gel. The reducing conditions in the sample application buffer disrupt the disulphide bonds that link the two monomers. The bands produced from successive Sf9 amplifications of the same KIR-Fc construct should show increasing intensity. Amplification is considered sufficient when successive rounds of amplification produce bands of similar intensity. This indicates that the amplification of baculovirus from Sf9 cells has reached capacity on SDS-PAGE.
Figure 3.
(A) Shown is a reducing (SDS) gel stained with Coomasie Blue with a protein ladder in the left column (Precision plus Protein Kaleidescope Standards, Bio-Rad, Cat # 1610375) and a band corresponding to KIR2DL3-Fc at 50.7kDa in the right column. The gel was loaded with 15 μl of KIR2DL3-Fc protein (100μg/ml).
(B) KIR2D-Fc bound to IgG coated beads (Bangs Laboratories) are stained with KIR specific antibodies to assess their integrity. Representative flow cytometry plots showing staining of KIR2DL1 (left panel, red) and KIR2DL3-Fc (right panel, blue) fusion proteins with EB6 and DX27 antibodies respectively. (C) Titrations of the binding of KIR2D to beads coated with HLA class I molecules. Each of three KIR2DL1-Fc fusion proteins distinguished by substitutions at residue 70 bind to HLA-C*06:02 with different avidities. Both mutant and wild type KIR2DL1 show saturated binding at concentrations greater than 100μg/ml. KIR2DL3-Fc binds to three HLA class I allotypes (HLA-B*73:01, HLA-C*03:04 and HLA-C*16:01) with different avidities. The binding of KIR2DL3-Fc to each allotype becomes saturated at concentrations above 100μg/ml. (D) KIR2DL1-Fc binds to HLA-C2 bearing allotypes but not to HLA-C1, HLA-A or HLA-B allotypes. KIR2DL3-Fc binds to HLA-C1 allotypes and to HLA-C2 allotypes with lower avidity. KIR2DL3-Fc also binds to two HLA-B allotypes that encode the C1 epitope. (E) KIR-Fc fusion proteins are amenable to long-term storage at −80°C. The binding of KIR2DL1-Fc stored at −80°C for 12 months is compared to 2DL1-Fc stored at 4°C.
2.6 Identification of baculovirus-infected Hi5 cells by flow cytometry
GP64 is a baculovirus encoded glycoprotein that is expressed on insect cells upon infection with baculovirus (Blissard and Rohrmann 1989). Because the titre of the transfecting supernatant corresponds to the degree of cell-surface expression of GP64, this can be used as a proxy to assess viral amplification in Sf9 supernatant. Seed an appropriate number of wells of a sterile six-well tissue culture plate with 2ml Hi5 cells at a density of 1 × 106 cells/ml. Add 100μl of P1, P2 or P3 viral stock to each well, seal the plate with laboratory film and incubate for 24 hours (covered, shaking at 120 rpm, at 27°C). To serve as a negative control, one well should contain Hi5 cells but no Sf9 supernatant. To serve as a positive control for the ability of the Hi5 cells to express GP64, one well should be transfected with a high baculoviral titre Sf9 supernatant. Mix the cells and add 250μl of the contents of the well to a 2ml microcentrifuge tube and separate the cells via centrifugation (50g for 2 mins). In the following steps, all wash solutions should be at 4°C. Resuspend the cells in flow cytometry buffer (FCB) (1x phosphate buffered saline [PBS - Cellgro, Cat. #21-031-CV] supplemented with 0.5% EDTA and 1% BSA) and wash twice. Following the second wash, resuspend the cells in 25μl FCB and stain the cells with 1μl anti-baculovirus GP64 antibody (eBioscience, Cat. #14-6995-81). Incubate for 30 mins at 4°C. Wash the cells a further three times in FCB, resuspending in 1ml FCB after the final wash. Flow cytometry is then used to detect the presence of the PE-conjugated antibody bound to cell surface GP64. Hi5 cells transfected with low viral titre P0 or P1 viral stocks show low or absent levels of surface GP64. Successive Sf9 amplifications of the same KIR-Fc construct should show increasing levels of cell-surface GP64 (Figure 2). Amplification is considered complete when successive rounds of amplification result in similar levels of cell-surface GP64. This indicates that the amplification of baculovirus from Sf9 cells has reached capacity and typically occurs at the P2 stage.
Figure 2.

(A) GP64 is up-regulated on the surface of Hi5 cells following transfection with baculovirus infected Sf9 supernatant. Shown is the gating strategy and representative flow cytometry plots from Hi5 cells transfected with Sf9 supernatant from P0, P1, P2 and P3 viral stocks and stained with PE conjugated anti-GP64 antibody. P0 viral stocks do not induce up-regulation of GP64 on the surface of Hi5 cells. Transfection of Hi5 cells with P1 viral stocks induces partial up-regulation of GP64 whereas transfection of Hi5 cells with P2 or P3 viral stocks maximally up-regulates GP64.
2.7 Harvest and purification of soluble KIR-Fc fusion proteins from baculovirus infected Hi5 cells
Once a suitable baculoviral Sf9 stock has been produced and assessed for viral titre, a full-scale experiment to produce recombinant KIR-Fc protein can be performed. Grow a 1L Hi5 cell culture to a density of 2 × 106 cells/ml. Add 1ml of high-titre P2 or P3 viral stock and incubate for 60 hours (covered, shaking at 120 rpm, 27°C). Separate cells by centrifugation (2500g for 15 mins, 4°C). Discard the pellet and pass the supernatant through a 0.2μm filter (Nalgene, Cat. #450-0020) into a sterile container. Add 100ml 10x HBS, pH 7.2 per 900ml filtered Hi5 supernatant. Add 1ml protein A Sepharose bead slurry and rotate bottles slowly overnight at 4°C.
In the following steps, all wash solutions should be at 4°C. Separate the protein A Sepharose beads from the Hi5 supernatant by filtration using gravity flow through a Buchner funnel with fritted glass disc (Pyrex, Cat. #36060). Care should be taken to avoid the Sepharose beads running dry. Wash the beads with approximately 500ml of 1x PBS and transfer to an empty 13ml PD-10 column with a 20–85μm frit filter (GE Healthcare, Cat. #17-0435-01) using a serological pipette. The beads should collect on the frit filter as the wash solution passes through by gravity flow.
Elute the KIR-Fc fusion protein from the collected beads in the PD-10 column using 100mM glycine, pH 2.7 in 8 fractions of 800μl each, neutralizing by elution into 8 separate microcentrifuge tubes, each containing 200μl 1M Tris, pH 9.0. The protein content of each elution fraction should be determined by Bradford assay (Kit from Bio-Rad, Cat. #500-0001).
Wash a Sephadex G-25 desalting column (GE Healthcare, Cat. #17-0851-01) with 25ml 1x PBS. Load 2.5ml of the fractions having the highest protein concentration onto the desalting column. Add 3.5ml 1x PBS to the desalting column in 0.5ml aliquots and collect seven 0.5ml fractions in separate microcentrifuge tubes. Determine the protein content of each fraction by Bradford assay.
At this stage, it is best to estimate the purity of each eluted protein fraction by gel electrophoresis. Add 10μl of each KIR-Fc eluent to an equal volume of Laemmli sample buffer with 5% β-mercaptoethanol and incubate the sample at 95°C for 10 minutes. Transfer the sample onto a 15% SDS-PAGE gel and electrophorese at 150–200v for 30–60 minutes. Stain with Coomassie reagent to identify the appropriately sized band (Figure 3A).
2.8 Assessment of KIR-Fc integrity by flow cytometry
Having produced recombinant KIR-Fc protein from the full-scale prep as described in Section 2.7, it is now necessary to determine the integrity (ie. correct protein folding) of the protein. This is achieved by flow-cytometry, using monoclonal antibodies specific for KIR. In the following steps, all wash solutions should be kept at 4°C. Add 20μl of anti-human IgG-coated beads (Bangs Laboratories, Cat. #BM562) to 500μl KIR-Fc fusion protein (diluted in 1x PBS to 100μg/ml). Ensure that the beads are vortexed gently to resuspend them prior to their use. Incubate, shaking gently for 30 min at 4°C. Collect the beads by centrifugation (50g for 2 mins) and wash twice with FCB. Following the second wash, resuspend the beads in 25μl FCB and add 2μl PE-conjugated mouse anti-human KIR antibody (KIR2DL1: Beckman Coulter, Cat # EB6-PE; KIR2DL2/3, Beckman Coulter, Cat # DX27-PE; Lineage III KIR: AbD Serotec, Cat # NKVFS1). Incubate, shaking gently for 30 min at 4°C. Collect the beads by centrifugation (50g for 2 mins) and wash a further two times with FCB. Resuspend the beads in 150μl FCB. Flow cytometry is then used to detect the presence of the PE conjugated anti-KIR antibody bound to individual IgG coated beads (Figure 3B).
2.9 Multiplex assay to detect binding of soluble KIR proteins to HLA class I single antigen beads
The purified, functional KIR-Fc fusion proteins are now ready to be tested for their capacity to recognize HLA class I allotypes. Each KIR-Fc protein is first incubated with approximately 10,000 individual beads, each coated with one of 97 HLA class I allotypes; the goal being to test the binding of the KIR-Fc to each HLA class I allotype approximately 100 times. In the second step of the assay, a secondary antibody is added that binds to the Fc portion of the KIR-Fc. The Luminex reader is able to simultaneously detect the identity of the bead (which correlates with a specific HLA class I) and the fluorescence of the antibody, which indicates the amount of KIR-Fc bound to the bead.
For each KIR-Fc protein to be tested, pre-wet one well of a 96-well 0.65μm filter plate (Millipore, Cat. #MSDVN6510) with 200μl 1x PBS. Remove the PBS from the wells by vacuum aspiration (manifold from Qiagen, Cat. #19504). Add 50μl of soluble KIR protein (100μg/ml) and 3μl LABscreen microbeads (One Lambda, Cat. #LS1A04) to each pre-wetted well. Ensure that the beads are vortexed gently to resuspend them prior to aliquoting into each well. For each assay, 50μl W6/32 antibody (Biolegend, Cat. #311402) (50μg/ml) should be added to one well to control for antigen density on individual beads (see Section 2.10). Incubate the KIR-Fc proteins and W6/32 with the beads for 60 mins, shaking gently and covered at 4°C.
In the following steps, all wash and resuspension solutions should be at 4°C. Wash the beads in each well four times with 1x Luminex wash buffer (One Lambda, Cat. #LS1A04). For each wash, add 200μL of wash buffer to each well and gently pipette up and down several times. If more than one protein is being tested use a multi-channel pipette to ensure even washing of each KIR-Fc protein being tested. Care should be taken to avoid introducing air bubbles into the wells. Following each wash, aspirate the wash solution using a vacuum manifold, ensuring that the well does not become dry. The vacuum pressure should not exceed 100mmHg.
Following the first complete wash cycle (4 individual washes), resuspend the KIR-Fc fusion proteins in 100μl PBS with 1% PE-conjugated goat anti-human IgG-Fc antibody (One Lambda, Cat. #LS-AB2). Suspend the beads incubated with W6/32 in 100μl PBS with 1% PE-conjugated goat anti-mouse IgG-Fc antibody (BD Pharmigen, Cat. #550589). Incubate both KIR-Fc and W6/32 for a further 60 mins, shaking gently and covered at 4°C. Following the second incubation, wash the beads a further four times with 1x Luminex wash buffer, resuspend in 100μl 1x PBS and transfer into a 96-well, 250μl ‘V’ bottom microplate (Whatman, Cat. #7701-3250). The test plate should be transferred immediately into the Luminex reader (with pre-warmed lasers and beadset template entered to avoid delay in starting the assay).
2.10 Calculating fluorescence relative to HLA class I antigen density
To correct for differences in the absolute amount of HLA class I annealed to each microbead, the binding of KIR-Fc fusion proteins to a specific HLA class I should be calculated relative to the amount of HLA class I as determined by binding of W6/32, an antibody that recognizes an epitope common to all HLA class I allotypes. The relative fluorescence ratio of a given KIR-Fc is calculated using the formula (KIR-Fc binding – negative control bead binding)/(W6/32 binding – negative control bead binding).
2.11 Common problems and their solutions
The most common problem encountered in the protocol is failure of amplification of baculovirus in Sf9 cell preps and failure of protein production from Hi5 cell preps. We have introduced several steps to test baculoviral amplification, the goal being to identify insufficient baculoviral amplification early and take steps to correct it to minimize lost time. A common cause of amplification failure is unhealthy Sf9 or Hi5 cells. The following steps can be used to ensure healthy cells and detect unhealthy cells should they occur.
Sf9 cells should have a doubling time of between 24 and 30h. Hi5 cells should have a doubling time of between 18 and 24h. Slow doubling times usually indicate that Sf9 or Hi5 cell cultures are unhealthy. Unhealthy cells will not amplify baculovirus successfully or produce adequate recombinant protein. Cell viability for both Sf9 and Hi5 cells, as determined by trypan blue staining, should be greater than 95% at all times. Both bacterial and fungal infections in the insect cell culture will reduce cell viability and doubling times. These can be prevented with good cell culture technique and the addition of antibiotics (1% Penicillin-streptomycin) and/or anti-fungals (0.25μg/ml Amphotericin B) to the culture medium. Cultures should be discarded immediately if there is evidence of microorganism contamination. A second reason for slow doubling is oxygen restriction. If the cell culture does not have sufficient surface area exposed to the air, cell growth will be retarded. This can be prevented by ensuring that flasks are filled no more than one-third (by volume) with culture medium. Ensuring that the flasks are shaken at between 120–150 rpm also ensures adequate oxygenation. Shaking at a higher rpm leads to cell damaging shearing stress and should be avoided. Further information on the culture of insect cells is available (Shrestha et al. 2008).
For the formation of intact baculovirus it is essential that transfer vector and linearized baculoviral DNA recombine during initial co-transfection of Sf9 cells. We have obtained the best results with freshly isolated transfer vector and baculoviral DNA that has not been stored at 4°C for more than two weeks. Additionally, freeze/thaw cycles of the transfer vector should be avoided where possible. Because the linearized baculovirus is a large DNA fragment (~130Kb), it is particularly susceptible to shear stress; over-zealous pipetting during transfection should therefore be avoided to minimize DNA and maximize transfection success.
The most common problems associated with the Luminex assay are high background and inter-assay variability. We have found that the following precautions minimize these problems. In this context we define high background readings as those in which the negative control bead binding is greater than 1% of the highest positive reading obtained. During the wash and incubation phases of the protocol, care should be taken to minimize any warming of the reagents. All wash reagents should be chilled to 4°C and wash steps should be completed as quickly as possible to avoid unnecessary warming of the samples. This may mean reducing the number of KIR-Fc proteins under test in any one assay to expedite this phase.
Although not always the case, some KIR-Fc are not stable when stored at 4°C and their use in the assay can lead to high background readings. To control for this issue, divide each batch of protein into 100 μl aliquots and store them at −80°C. Similarly, HLA class I beads that have been thawed and stored at 4°C are not typically stable for more than 3 months. The beads should also be divided into 10μl aliquots and frozen at −80°C if they are unlikely to be used within this time frame. Each of these precautions also helps to reduce both high background and inter assay variability. Additionally, we include well characterized HLA-C1 and HLA-C2 receptors (e.g KIR2DL3*001-Fc and KIR2DL1*003-Fc respectively) in every assay as positive controls. Changes in the avidity and specificity of the KIR-Fc under test can then be related not only to W6/32, but also to these KIR-Fc controls.
In addition to the failure of baculoviral amplification described above, a further limitation of this system is that we have, as yet, been unable to produce functional three domain KIR using the Fc fusion system. Although the KIR3D protein is expressed by insect cells, it does not fold into a tertiary structure that binds anti-KIR antibody or to HLA class I. As such, the HLA class I reactivity of these receptors remains under investigation.
3. Results and Discussion
The purpose of this protocol is to provide a simplified method with which to produce and test the reactivity of soluble two-domain KIR-Fc fusion proteins. We chose to use an insect cell expression system because most post-translational modifications are made correctly in insect cells and the system is scalable, allowing production of large quantities of soluble recombinant protein in a comparatively short time. We have sought to reduce variability in final protein yield by implementing a series of quality controls at points during the baculoviral amplification phase. These methodological improvements allow us to better track baculoviral amplification in Sf9 cells and prevent the Hi5 preparations having insufficient protein yield.
We have also designed a novel assay that tests the binding of KIR-Fc to HLA class I allotypes. This assay simultaneously detects the binding of KIR to 97 HLA class I allotypes. As a result it holds a clear advantage over previous cell-based binding assays that allowed examination of only a few KIR-HLA class I interactions at any one time. The assay is sensitive over a two-log range, permitting both strong and weak reactions to be interpreted with confidence and correlated with structural polymorphisms in both KIR and HLA class I (Frazier et al. 2013; Gendzekhadze et al. 2009; Graef et al. 2009; Hilton et al. 2012; Moesta et al. 2008).
The KIR-Fc HLA-bead binding assay has also been used to explore the binding characteristics of KIR in simian primates. The HLA class I specificity of primate KIR has been difficult to determine because of a comparative lack of cellular reagents. As such, the combination of methods described in this paper has led to a number of critical discoveries on the immunologic function and co-evolution of KIR and MHC class I. In Old World Monkeys, the lineage III KIR (precursor to the MHC-C receptors of higher primates) is represented by a single gene (Sambrook et al. 2005) while lineage II KIR genes have expanded and diversified. To identify their MHC epitope-specificity and avidity, a panel of rhesus macaque lineage II KIR-Fc was assayed using the methods present here (Older Aguilar et al. 2011). Although MHC-C is not present in macaques, their KIR recognize HLA-C epitopes more effectively than they recognize HLA-A and HLA-B, suggesting that MHC-C evolved to become a stronger ligand for KIR than HLA-A and -B. The emergence of MHC-C in the orangutan was accompanied by an expansion of lineage III KIR and their evolution as MHC-C receptors (Guethlein et al. 2007). All orangutan MHC-C allotypes have asparagine at position 80 and display the C1 epitope. Correspondingly, results from the KIR-Fc HLA bead binding assay showed that the orangutan has C1-specific KIR but no C2-specific KIR (Older Aguilar et al. 2010). In chimpanzees the MHC-C gene became fixed and the C2 epitope emerged. As a consequence, out of nine chimpanzee lineage III KIR genes (Abi-Rached et al. 2010), eight encode receptors with high avidity for HLA-C, comprising three C1-specific receptors and 5 C2-specific receptors (Moesta et al. 2009). Thus, results from the multiplex binding assay show that changes in the character of the KIR locus correlate with change in the MHC class I genes, suggesting co-evolution between these receptors and ligands and uncovering a progression in which the complexity of the KIR locus gets increasingly sophisticated across higher primate species.
3.1 Flow cytometry can be used to confirm baculoviral transfection
Surface glycoprotein GP64 is a baculovirus encoded glycoprotein that can be used as a marker for successful transfection and amplification of baculovirus in Hi5 insect cells. (Blissard and Rohrmann 1989; Volkman and Goldsmith 1988). Figure 2 shows that GP64 expression is not detected on un-transfected Hi5 cells but is expressed following transfection with high viral titre Sf9 supernatant. P0 viral stock was not sufficient to induce GP64 surface expression whereas surface expression was typically detected after transfection with P1 viral stock and with each subsequent amplified viral stock (P2 and P3) (Figure 2). Surface expression of GP64 was sensitive to the baculoviral titre with P2 transfected Hi5 cells showing a 40% increase in surface expression of GP64 as compared to Hi5 cells transfected with P1 viral stocks. That P3 stocks induced only marginally greater GP64 surface expression than P2 viral stocks suggests that GP64 is either maximally up-regulated by a given viral titre or Sf9 cells reach maximal viral amplification between the second and third amplification rounds.
3.2 Confirmation of KIR-Fc integrity by flow cytometry
Both EB6 antibody, which recognizes KIR2DL1, and DX27 antibody, which recognizes KIR2DL2/3, bound to KIR-Fc immobilized on anti-human IgG flow cytometry beads (Figure 3B). This test provides a cost-effective way to test the integrity of both the KIR region and Fc region of the final fusion protein, as both are required to have folded correctly to produce a positive result in the Luminex binding assay.
3.3 Titration of KIR2D-Fc fusion proteins against HLA class I shows that 100μg/ml is an appropriate concentration to use in the binding assay
The binding of KIR-Fc fusion proteins to HLA class I increases with an increasing concentration of KIR-Fc fusion protein until binding becomes saturated at approximately 100μg/ml. Use of KIR-Fc at greater concentrations (100–400μg/ml) does not increase binding to HLA class I and avidity differences between individual allotypes are found to be consistent at this concentration. Additionally, this saturation point applies to between different HLA class I allotypes and for different naturally occurring and mutant KIR2D (Figure 3C).
3.4 Multiplex Luminex assay comparing the binding of W6/32, KIR2DL1-Fc and KIR2DL3-Fc to HLA class I coated microbeads
KIR2DL1-Fc binds specifically and with high avidity to HLA-C2 but not HLA-C1 or any HLA-A or HLA-B allotypes (Figure 3D). HLA-C2 allotypes display a range of avidity for KIR2DL1 with HLA-C*15:02 and HLA-C*04:01 having the highest and lowest avidities respectively (Figure 3D). KIR2DL3-Fc binds specifically to HLA-C1 but does not bind to HLA-C2 or any HLA-A allotype. Unlike KIR2DL1, KIR2DL3 does bind to two unusual HLA-B allotypes (HLA-B*46:01 and HLA-B*73:01) that have the C1 epitope (Figure 3D). HLA-C1 allotypes have a range of avidity for KIR2DL3 with HLA-C*03:04 and HLA-C*16:01 having the highest and lowest avidities, respectively. Both HLA-B*46:01 and HLA*73:01 bind with high avidity to KIR2DL3.
3.5 KIR-Fc fusion proteins remain functional following long-term storage at −80°C
Each 1 litre prep of Hi5 cells yields between 2 and 4 mg of KIR-Fc protein. Given that the KIR-Fc are used at a dilution of 100ug/ml in the multiplex binding assay, there is typically an excess of KIR-Fc reagent. KIR-Fc remain stable at 4°C for 1–6 months, however, to allow repetition of a particular experiment over longer time periods and to provide stable positive controls, we investigated the storage of KIR-Fc aliquots at −80°C (Figure 3E). KIR-Fc show similar binding in the multiplex binding assay up to 12 months following initial freezing. Freezing the KIR-Fc is assumed to prevent the protein degradation, aggregation and misfolding that occurs at an unpredictable rate when stored at 4°C.
4. Conclusions
We have described the production of KIR-Fc fusion proteins in an insect cell expression system and their interaction in a multiplex binding assay with a panel of 97 HLA class I allotypes. KIR-Fc production in insect cells is relatively simple allowing production of large amounts of recombinant protein in around 20 days. The assay is sensitive enough to discriminate between single amino acid substitutions in the extracellular domains of the KIR molecule and has, as a result, greatly facilitated investigation of even closely related KIR allotypes. Furthermore, the results of this direct binding assay appear to correlate well with the results obtained in the limited cellular assays that were used to discover the KIR and first investigate their specificity for HLA class I.
Highlights.
KIR and HLA class I ligand interactions modulate NK cell reactivity
Soluble recombinant KIR-Fc fusion proteins are made efficiently in insect cells
Production method described is simple and scalable
KIR-Fc used in multiplex HLA class I binding assay to characterize ligand specificity
Results inform functional and evolutionary studies of NK cell immunity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abi-Rached L, et al. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010;6(11):e1001192. doi: 10.1371/journal.pgen.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman PJ, et al. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329(6139):512–8. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Blissard GW, Rohrmann GF. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170(2):537–55. doi: 10.1016/0042-6822(89)90445-5. [DOI] [PubMed] [Google Scholar]
- Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268(5209):405–8. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- Colonna M, et al. Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc Natl Acad Sci U S A. 1992;89(17):7983–5. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier WR, et al. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. 2013;190(12):6198–208. doi: 10.4049/jimmunol.1300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendzekhadze K, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106(44):18692–7. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef T, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206(11):2557–72. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein LA, et al. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179(1):491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- Hilton HG, Parham P. Direct binding to antigen-coated beads refines the specificity and cross-reactivity of four monoclonal antibodies that recognize polymorphic epitopes of HLA class I molecules. Tissue Antigens. 2013;81(4):212–20. doi: 10.1111/tan.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton HG, et al. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 2012;189(3):1418–30. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchman RB, Possee RD, King LA. Baculovirus expression systems for recombinant protein production in insect cells. Recent Pat Biotechnol. 2009;3(1):46–54. doi: 10.2174/187220809787172669. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Curr Opin Biotechnol. 1999;10(5):428–33. doi: 10.1016/s0958-1669(99)00005-1. [DOI] [PubMed] [Google Scholar]
- Lanier LL. Natural killer cells: from no receptors to too many. Immunity. 1997;6(4):371–8. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- Mandelboim O, et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184(3):913–22. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, et al. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182(6):3628–37. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesta AK, et al. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180(6):3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Moretta A, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178 (2):597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman PJ, et al. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9(10):e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Older Aguilar AM, et al. Rhesus macaque KIR bind human MHC class I with broad specificity and recognize HLA-C more effectively than HLA-A and HLA-B. Immunogenetics. 2011;63(9):577–85. doi: 10.1007/s00251-011-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Older Aguilar AM, et al. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185(7):4238–51. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13(2):133–44. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R, et al. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation. 2003;75(1):43–9. doi: 10.1097/00007890-200301150-00008. [DOI] [PubMed] [Google Scholar]
- Pei R, et al. Simultaneous HLA Class I and Class II antibodies screening with flow cytometry. Hum Immunol. 1998;59(5):313–22. doi: 10.1016/s0198-8859(98)00020-2. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15(1):25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Smee C, Gileadi O. Baculovirus expression vector system: an emerging host for high-throughput eukaryotic protein expression. Methods Mol Biol. 2008;439:269–89. doi: 10.1007/978-1-59745-188-8_19. [DOI] [PubMed] [Google Scholar]
- Valiante NM, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- Victor ME, et al. Insect cells are superior to Escherichia coli in producing malaria proteins inducing IgG targeting PfEMP1 on infected erythrocytes. Malar J. 2010;9:325. doi: 10.1186/1475-2875-9-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA. Resistance of the 64K protein of budded Autographa californica nuclear polyhedrosis virus to functional inactivation by proteolysis. Virology. 1988;166(1):285–9. doi: 10.1016/0042-6822(88)90176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158(9):4026–8. [PubMed] [Google Scholar]
- Winter CC, Long EO. Binding of soluble KIR-Fc fusion proteins to HLA class I. Methods Mol Biol. 2000;121:239–50. doi: 10.1385/1-59259-044-6:239. [DOI] [PubMed] [Google Scholar]
- Winter CC, et al. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161(2):571–7. [PubMed] [Google Scholar]