Abstract
Background & Aims
Pigment epithelium-derived factor (PEDF) is a secretory protein that inhibits multiple tumor types. PEDF inhibits the Wnt coreceptor, low-density lipoprotein receptor-related protein 6 (LRP6), in the eye, but whether the tumor-suppressive properties of PEDF occur in organs such as the liver is unknown.
Methods
Wnt-dependent regulation of PEDF was assessed in the absence and presence of the Wnt coreceptor LRP6. Whole genome expression analysis was performed on PEDF knockout (KO) and control livers (7 months). Interrogation of Wnt/β-catenin signaling was performed in whole livers and human hepatocellular carcinoma (HCC) cell lines after RNA interference of PEDF and restoration of a PEDF-derived peptide. Western diet feeding for 6 to 8 months was used to evaluate whether the absence of PEDF was permissive for HCC formation (n = 12/group).
Results
PEDF levels increased in response to canonical Wnt3a in an LRP6-dependent manner but were suppressed by noncanonical Wnt5a protein in an LRP6-independent manner. Gene set enrichment analysis (GSEA) of PEDF KO livers revealed induction of pathways associated with experimental and human HCC and a transcriptional profile characterized by Wnt/β-catenin activation. Enhanced Wnt/β-catenin signaling occurred in KO livers, and PEDF delivery in vivo reduced LRP6 activation. In human HCC cells, RNA interference of PEDF led to increased levels of activated LRP6 and β-catenin, and a PEDF 34-mer peptide decreased LRP6 activation and β-catenin signaling, and reduced Wnt target genes. PEDF KO mice fed a Western diet developed sporadic well-differentiated HCC. Human HCC specimens demonstrated decreased PEDF staining compared with hepatocytes.
Conclusions
PEDF is an endogenous inhibitor of Wnt/β-catenin signaling in the liver.
Keywords: Extracellular Matrix, PEDF, Wnt/β-Catenin
Abbreviations used in this paper: BABB, benzyl alcohol/benzyl benzoate; CM, conditioned medium; ECM, extracellular matrix; ERK, extracellular signal-regulated kinase; FDR, false-discovery rate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GO, Gene Ontology; GSEA, gene set enrichment analysis; GSK, glycogen synthase kinase; HCC, hepatocellular carcinoma; KO, knockout; LRP6, low-density lipoprotein receptor-related protein 6; PCR, polymerase chain reaction; PEDF, pigment epithelium-derived factor; SHG, second harmonic generation; siRNA, small interfering RNA; WT, wild type
Summary.
The absence of pigment epithelium-derived factor (PEDF) in hepatocellular carcinoma (HCC) enhances Wnt/β-catenin signaling. Genomic profiling of PEDF knockout livers correlates with gene expression signatures of human HCC associated with aberrant Wnt/β-catenin signaling. PEDF is an endogenous inhibitor of Wnt/β-catenin signaling.
Hepatocellular carcinoma (HCC) is a major cause of cancer-related deaths worldwide.1 Genomic profiling has classified HCC based on molecular “signatures” that correlate with biological characteristics and clinical outcomes.2, 3 One finding from these studies is the role of the extracellular matrix (ECM) in determining tumor behavior.4, 5, 6 For instance, modulators of the ECM can activate developmental pathways such as Wnt/β-catenin signaling, thereby connecting liver fibrosis to a signaling pathway that drives hepatocarcinogenesis.3
Pigment epithelium-derived factor (PEDF) is a circulating 50-kDa protein with ECM binding domains and broad tumor suppressive properties.7, 8, 9, 10 In PEDF knockout (KO) mice, stromal abnormalities occur in multiple organs including the prostate, pancreas, and liver.11, 12, 13, 14, 15 Endogenous liver levels of PEDF decline in experimental and human cirrhosis, and PEDF delivery ameliorates experimental liver fibrosis.14, 16 PEDF null mice crossed with the KrasG12D mice resulted in marked stromal changes in the pancreas and an invasive malignant phenotype not seen in the KrasG12D mutant mice alone.15 These results indicate that PEDF regulates tissue matrix quiescence and its absence is permissive for malignant transformation.
The antitumor properties of PEDF are typically attributed to an antiangiogenic effect.10, 17 PEDF, however, inhibits tumor cells in culture, indicating other mechanisms.17, 18 Park et al19 identified PEDF’s ability to inhibit Wnt/β-catenin signaling in the eye with avid binding to the Wnt coreceptor, low-density lipoprotein receptor-related protein 6 (LRP6). Whether PEDF has systemic effects beyond the eye and inhibits tumor development through an inhibitory effect on Wnt/β-catenin signaling is unclear. Because PEDF is most highly expressed by the liver, a finding recently confirmed in the Human Protein Atlas,20, 21 and modulates Wnt/β-catenin signaling,19, 22 we asked whether PEDF functions as an LRP6 antagonist in the liver.
We establish that canonical Wnt3a ligand directly regulates PEDF levels. PEDF, in turn, inhibits Wnt/β-catenin signaling. Consistent with this, livers from PEDF KO mice have a transcriptional profile closely aligned with murine models of hepatocarcinogenesis and human HCC characterized by aberrant Wnt/β-catenin signaling. Knockout and knock-in experiments demonstrate that PEDF inhibits Wnt/β-catenin signaling in murine livers and human HCC cells through its ability to inhibit LRP6 and β-catenin activity. Finally, a chronic Western diet elicited sporadic HCC formation in PEDF KO mice, while the human HCC specimens demonstrated diminished PEDF staining.
Materials and Methods
Human Hepatocellular Carcinoma, Animals, and Liver Tumor Induction
Archival human HCC tissues and their corresponding adjacent livers from 14 patients were obtained from the VA Connecticut Healthcare System according to an approved institutional review board protocol. The PEDF KO mice were bred with age-matched wild-type (WT) littermates on the C57BL/6J background to generate heterozygous breeding pairs, and then PEDF KO and WT offspring were backcrossed for more than 10 generations.11 The mice were genotyped using a commercially available polymerase chain reaction (PCR) kit (Sigma-Aldrich, St. Louis, MO). All procedures were approved by the Institutional Animal Care and Use Committee of VA CT Healthcare System. A commercial Western diet—TestDiet 4342 (TestDiet, St. Louis, MO): energy (% kcal) from fat (40%), carbohydrate (44%), protein (16%)—or standard chow was given for 26 to 32 weeks to PEDF KO and age-matched controls (n = 12/group) starting at 8 to 12 weeks of age.
RNA Extraction and Gene Arrays
Frozen whole liver tissue from five PEDF KO animals and WT controls were maintained in liquid nitrogen until total RNA extraction using the TRIzol method (Invitrogen, Carlsbad, CA). TRIzol-extracted RNA was further purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA), yielding high-quality RNA suitable for microarray analyses (RNA integrity number >9). The RNA quality was verified using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), and the RNA was quantified by NanoDrop (NanoDrop Technologies, Wilmington, DE). For gene expression analysis, 500 ng of total RNA was used to generate biotin-labeled cRNA using the Illumina Total RNA amplification and labeling kit (Ambion, Austin, TX) according to the manufacturer’s instructions. The biotinylated cRNA was labeled with fluorescent dye at the Yale Keck Genomic Core Facility (West Haven, CT), hybridized onto a MouseRef-8 v2.0 Expression BeadChip expression array bead chip (Illumina, San Diego, CA) and scanned.
Expression data were analyzed by Genespring GX12 software (Agilent Technologies) after normalization by 75th percentile shift. Only genes with a present signal (signal above background noise) in more than 50% of samples were included in the analysis. Group samples with gene expression correlation coefficients ≤0.95 were excluded (one KO sample). For the statistical analysis, replicate samples were averaged. Differences in gene expression were determined using a moderated t test, and multiple hypothesis testing adjustment was made using Benjamini–Hochberg method at a false-discovery rate (FDR) ≤ .05 and by adding a fold expression cutoff of 1.3. Genes differentially expressed in KO mice versus WT were subjected to Gene Ontology (GO) (http://www.geneontology.org) and WikiPathways (http://www.wikipathways.org) enrichment analysis using the hypergeometric test corrected by Benjamini–Yekutieli method at FDR q ≤0.05.
To further extend the analysis, gene set enrichment analysis (GSEA) was used (http://www.broadinstitute.org/gsea). GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant differences between two phenotypes.23 To identify the gene sets that were statistically significantly enriched, we created a rank-order list by gene expression differences between KO and WT sets. Gene Ontology, KEGG pathways (http://www.genome.jp), Reactome (http://www.reactome.org), Biocarta (http://www.biocarta.org), Pathway interaction database (http://pid.nci.nih.gov), and curated gene sets reflecting changes induced by various chemical and genetic perturbances were used to interpret results. FDR q value was used to rank the results. Gene sets enriched at FDR q value ≤ .05 and nominal P < .05 were considered statistically significant. Gene array data were deposited at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63643.
PEDF and PEDF Peptide Restoration
Human full-length PEDF was generated in human embryonic kidney cells as described elsewhere, and its purity confirmed using Coomassie and silver staining (Invitrogen).12 PEDF was administered (25 μg/kg bwt) by intraperitoneal injection on alternate days for a period of 4 weeks.24 A 34-mer of human PEDF corresponding to amino acids 44–77 has been previously shown to inhibit neovascularization and inhibit tumor growth, but its role in Wnt signaling is unclear.17, 25 We interrogated Wnt signaling with a 34-mer that was commercially obtained (NeoBiolab, Cambridge, MA) and used at a concentration of 100μM to evaluate Wnt/β-catenin signaling in vitro.
Cell Culture
The human HCC cell lines HepG2 and Huh7 were obtained from the American Type Culture Collection (Manassas, VA), propagated, and kept at the Yale Liver Center (P30DK034989). To obtain conditioned medium (CM), the cells were grown to 80% confluence, washed twice with serum-free media, and then incubated with serum-free media overnight. The CM was obtained after 18–20 hours and was concentrated approximately 40-fold using Amicon Ultra centrifugal filters (Millipore, Billerica, MA) with a 10-kDa cutoff. For PEDF peptide experiments, the medium was removed, washed three times with serum medium, and PEDF 34-mer was added for 2 hours before the lysates were obtained. For the lysates, the cells were scraped in radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors, incubated on ice, and centrifuged at 10,000g for 10 minutes.
Silencing of PEDF and LRP6 With RNAi in Hepatocellular Carcinoma Cells
To reduce PEDF levels in human HCC cells, commercial small interfering RNA (siRNA) constructs targeting PEDF (cat. no. 4392420, 4390771) or scrambled (cat. no. 4390843) sequences (Ambion) were transfected according to the manufacturer’s instructions. After 6 hours, the transfection medium was replaced with fresh medium lacking siRNA. After an additional 48 hours, the medium was changed to serum-free medium for 24 hours. CM and cell lysates were obtained as described earlier. The HepG2 cells stably transfected with small-hairpin RNA constructs targeting LRP6 were a gift of Dr. Arya Mani (Yale University School of Medicine). The integrity of PEDF and LRP6 KO was assessed in conditioned medium and in lysates. Measurement of PEDF levels in culture was performed with by a commercial enzyme-linked immunosorbent assay kit (BioProducts, Frederick, MD).
RNA Analysis and Quantitative Polymerase Chain Reaction
The RNA was isolated using the RNAEasy mini kit (Qiagen). The primer probe sets were obtained from a commercial source (Applied Biosystems, Foster City, CA), and quantitative reverse-transcription PCR was performed on a TaqMan ABI 7500 system (Applied Biosystems) as described elsewhere.13 Target gene expression was normalized against β-actin.
Immunoblotting
Immunoblotting was performed as described elsewhere.12 Protein content was determined by Bradford assay. Lysates (20–30 μg total protein) were separated under denaturing conditions on a gradient gel (Bio-Rad Laboratories, Hercules, CA), and transferred to polyvinylidene fluoride membranes. After they were blocked in a 5% milk solution, the membranes were incubated overnight with antibodies. Primary antibodies used were PEDF from Chemicon (Temecula, CA); transforming growth factor-β1 (TGF-β1; 3711S), phospho-LRP6 (2568), total LRP6 (2560), nonphosphorylated (active) β-catenin and total β-catenin, phospho-glycogen synthase kinase-3β (p-GSK3β), total GSK3β, phospho-extracellular-signal-regulated kinase (p-ERK), total ERK (4370), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5174S) from Cell Signaling Technology (Beverly, MA); collagen I (ab6308) from Abcam (Cambridge, MA); collagen III (15946) from Novus Biologicals (Oakville, ON, Canada); and β-actin from Sigma-Aldrich.
Collagen I blots were run under reducing and nonreducing conditions. After washing in Tris-buffered saline and 0.05% Tween, the primary antibody was labeled using a peroxidase-conjugated secondary antibody specific for the primary antibody species. Samples were resolved on a gradient gel and transferred to nitrocellulose membranes. Equivalence of loading was confirmed using β-actin or GAPDH for lysates, or Coomassie stains for CM. Densitometry was performed using the National Institutes of Health ImageJ software (http://imagej.nih.gov/ij/).
Hydroxyproline Assays
Hydroxyproline assays were performed using a commercial kit (BioVision Research, Mountain View, CA). Measurements were performed on four separate occasions using three different sets (n = 3–4/group) of age-matched PEDF KO and control livers.
Second Harmonic Generation Imaging
Second harmonic generation (SHG) imaging preferentially detects type I, and to a lesser extent type III, fibrillar collagen.26 Multiphoton stimulation combined with tissue clearing was used to visualize fibrillar collagen deposition in volume sections of both WT and KO liver specimens measuring approximately 5 × 5 × 1 mm. Tissue clearing was performed on formalin-fixed organs using benzyl alcohol/benzyl benzoate (BABB) in 2:1 ratio as previously described elsewhere.27 Briefly, tissue specimens were then dehydrated by graded methanol incubations in 30-minute intervals and then incubated overnight with BABB. SHG was measured on a TriM Scope II multiphoton microscope (LaVision BioTec, Bielefeld, Germany) with 780 nm excitation and 390 nm band pass emission filter using a 0.95 NA, 25× objective (Leica Microsystems GmbH, Wetzlar, Germany) designed specifically for BABB immersion. Tissue volume was determined using intrinsic fluorescence with 960 nm excitation and 600–50 nm band pass filter detection. The SHG signal was collected in reflection: the specimen was placed on a deep-well slide, and a mirror was placed underneath to improve collection efficiency. The imaging parameters were kept constant among the specimens, including laser power and scanning speed as well as detector distance from the specimen. Data were collected in 16-bit depth, and contrast was adjusted using identical intensity thresholds for all images, allowing for direct intensity comparison.
Histology
Immunohistochemical analysis was performed as described on 14 sequentially obtained human HCC specimens. Sections were deparaffinized, treated to inhibit endogenous peroxidase, and subjected to antigen retrieval. After incubation with primary antibody, sections were washed and then incubated with biotinylated anti-mouse antiserum. Streptavidin complexed with horseradish peroxidase was added, and labeling was detected using diaminobenzidine. Semiquantitative scoring of the immunohistochemical labeling was evaluated by a pathologist (S.E.C.) using a numerical grading score (1, no staining; 2, focal positivity; 3, moderate; 4, diffuse, strong immunostaining) on 10 nonoverlapping fields per case with normal hepatocytes distant to the tumor margin assessed as “Nl liver.”
Statistical Analysis
The P values were calculated, assuming equal sample variance, using a two-tailed Student t test on Prism software. P < .05 was considered statistically significant. Values were stated as mean ± standard deviation (SD) or standard error of the mean.
Results
PEDF Secretion Is Wnt3a-Responsive and Depends on the Wnt Coreceptor LRP6
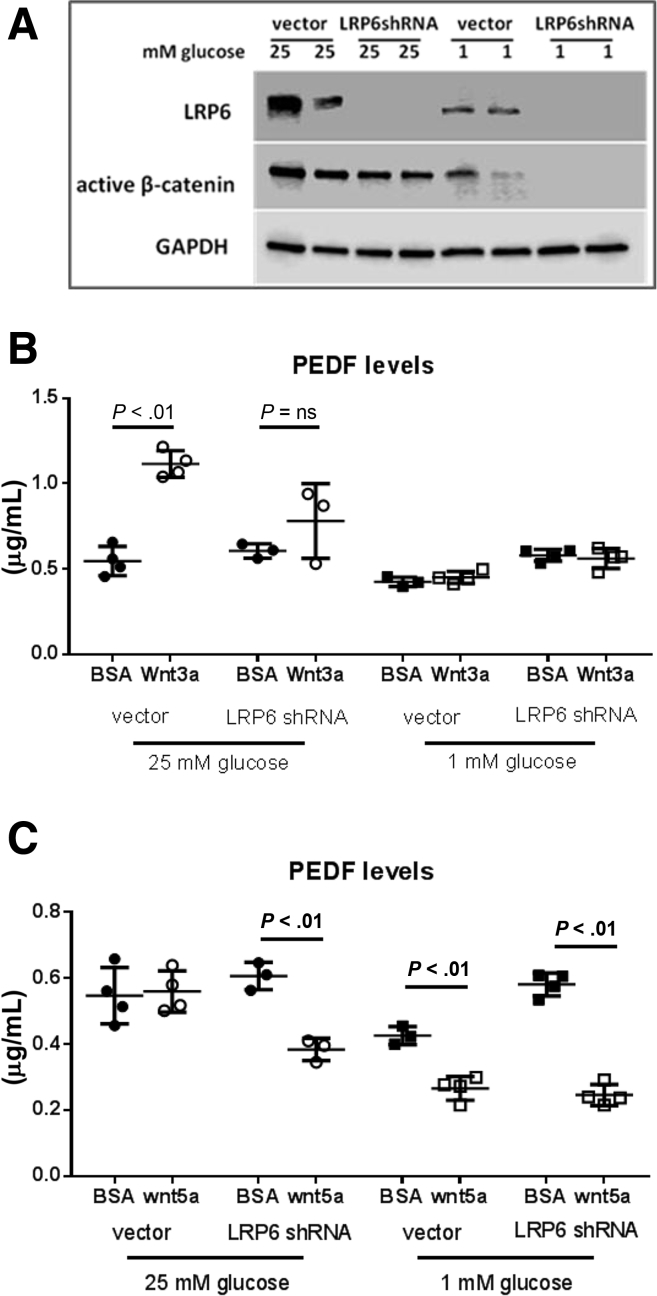
We evaluated PEDF regulation by Wnt ligands and dependence upon LRP6. The integrity of the LRP6 KO and the stimulatory effects of high (25 mM) versus low (1 mM) glucose on LRP6 and its effector active (nonphosphorylated) β-catenin were shown (Figure 1A). Canonical Wnt3a (50 ng/mL) led to a greater than twofold increase in PEDF levels that was LRP6 dependent (Figure 1B, P < .01). In the absence of the LRP6, Wnt3a had no effect on PEDF levels. Similarly, Wnt3a had no effect on PEDF levels under 1 mM glucose conditions, likely reflecting markedly suppressed LRP6 levels seen in this condition. Thus, Wnt3a-stimulated induction of PEDF levels are LRP6 dependent.
Figure 1.
PEDF secretion is regulated by Wnt ligands in an LRP6-dependent manner. (A) Integrity of the LRP6 small-hairpin RNA-mediated knockdown in HepG2 cells was demonstrated under high (25 mM) and low (1 mM) glucose conditions. (B) Canonical Wnt3a ligand significantly induces PEDF levels in the presence of the Wnt coreceptor LRP6 (P < .01). Genetic knockdown of LRP6 or its functional depletion with 1 mM glucose abrogates this effect (not statistically significant). (C) The noncanonical Wnt5a suppresses PEDF levels when LRP6 is genetically deleted under 25 mM glucose or reduced by low glucose (P < .01). Experiments were conducted in duplicate with n = 3–4/group. Data are presented as mean ± SD.
The noncanonical Wnt pathway includes the Wnt5a ligand and its orphan receptor, ROR2 (receptor tyrosine kinase-like orphan receptor 2), and counters the effects of the canonical pathway.28 To determine whether PEDF could be modulated by the noncanonical pathway, Wnt5a was added to HepG2 cells with and without LRP6. Wnt5a did not alter PEDF levels under high-glucose conditions in the presence of the LRP6 receptor. When the canonical receptor LRP6 was deleted, Wnt5a significantly suppressed PEDF protein levels (Figure 1C, P < .01). Thus, deletion of LRP6 favors the noncanonical pathway and lowers PEDF under high-glucose conditions.
Similarly, the 1 mM glucose condition leads to a functional depletion of the LRP6 receptor (Figure 1A) without genetic manipulation. Here, the Wnt5a ligand significantly decreased PEDF under scrambled and LRP6 KO conditions indicating that the noncanonical Wnt ligands can decrease PEDF in the setting of diminished LRP6 levels (Figure 1C, P < .01 for low glucose with and without LRP6). Thus, canonical Wnt3a and the noncanonical Wnt5a differentially regulate PEDF levels.
PEDF Knockout Livers Resemble Experimental and Human Hepatocellular Carcinoma Marked by Wnt/β-Catenin Signaling
To explore PEDF’s role in the liver, gene expression profiling was done in KO versus WT livers. There were 1113 gene entities differentially expressed between WT and KO animals at FDR ≤ .05 and 1.3-fold expression cutoffs. Out of 1113 genes 344 were up-regulated in KOs, and 769 were down-regulated (Supplementary Table 1). Grouping these genes by GO categories using hypergeometric model showed that most up-regulated GO categories were related to extracellular matrix function, lipid metabolism, immune response, DNA replication, phase I and II enzymes (FDR ≤ .05). Most down-regulated GO categories were related to ribosomal and mitochondrial function and numerous primary metabolic processes such as nitrogen compound metabolism, glutamine family amino acid metabolic process, urea cycle, and carboxylic acid metabolism, and peptidase inhibitory activity (FDR < .05).
To further characterize the gene expression changes in KO mice, GSEA using curated pathways as well as GO categories were performed. Consistent with analysis by moderated t test, the GSEA showed that most up-regulated pathways were related to cell proliferation, inflammatory responses, collagen expression, extracellular matrix function, and phase I and phase II enzymatic activity (Supplementary Table 2). Subsequently, another GSEA was performed to test for similarities between gene expression profiles in PEDF KO mouse livers and curated gene sets representing expression signatures of genetic and chemical perturbation. This analysis showed that the most significantly enriched gene sets represented rodent models and human samples of HCC tissues and various inflammatory liver conditions, suggesting that loss of PEDF leads to gene expression changes similar to those found in HCC (Table 1, Supplementary Table 3). In fact, eight out of top 10 enriched gene sets represented rodent models of HCC (Table 1).
Table 1.
Top 10 Enriched Chemical and Genetic Perturbation Gene Sets Corresponding to PEDF Null Livers
| Gene Set Name | FDR q Valuea | Gene Set Description |
|---|---|---|
| LEE_LIVER_CANCER_ACOX1_UP | <.001 | Genes up-regulated in HCC of ACOX1 knockout mice |
| LEE_LIVER_CANCER_E2F1_UP | <.001 | Genes up-regulated in HCC induced by overexpression of E2F1 |
| LEE_LIVER_CANCER_MYC_E2F1_UP | <.001 | Genes up-regulated in HCC from MYC and E2F1 double transgenic mice |
| LEE_LIVER_CANCER_MYC_TGFA_UP | <.001 | Genes up-regulated in HCC tissue of MYC and TGFA double transgenic mice |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_UP | <.001 | Hepatic graft versus host disease day 35: genes up-regulated in allogeneic vs syngeneic bone marrow transplant |
| KHETCHOUMIAN_TRIM24_TARGETS_UP | <.001 | Retinoic acid-responsive genes up-regulated in HCC samples of TRIM24 knockout mice |
| LEE_LIVER_CANCER_CIPROFIBRATE_UP | <.001 | Genes up-regulated in HCC induced by ciprofibrate |
| LEE_LIVER_CANCER_DENA_UP | <.001 | Genes up-regulated in HCC induced by diethylnitrosamine |
| WIELAND_UP_BY_HBV_INFECTION | <.001 | Genes induced in the liver during hepatitis B viral clearance in chimpanzees |
| BORLAK_LIVER_CANCER_EGF_UP | <.001 | Genes up-regulated in HCC developed by transgenic mice overexpressing a secreted form of epidermal growth factor in liver |
Note: Gene set enrichment analysis showed that expression signatures in PEDF knockout mouse livers resembled those found in genetic and chemical models of HCC. Of the top 10 enriched chemical and genetic perturbation gene sets, eight represented rodent models of HCC, and two (Ichiba and Wieland) sets are related to inflammatory liver conditions. HCC, hepatocellular carcinoma.
FDR (false-discovery rate): adjusted P value (FDR q value).
PEDF Knockout Livers Display a Genomic Signature Resembling Hepatocellular Carcinoma Categorized by Wnt/β-Catenin Signaling
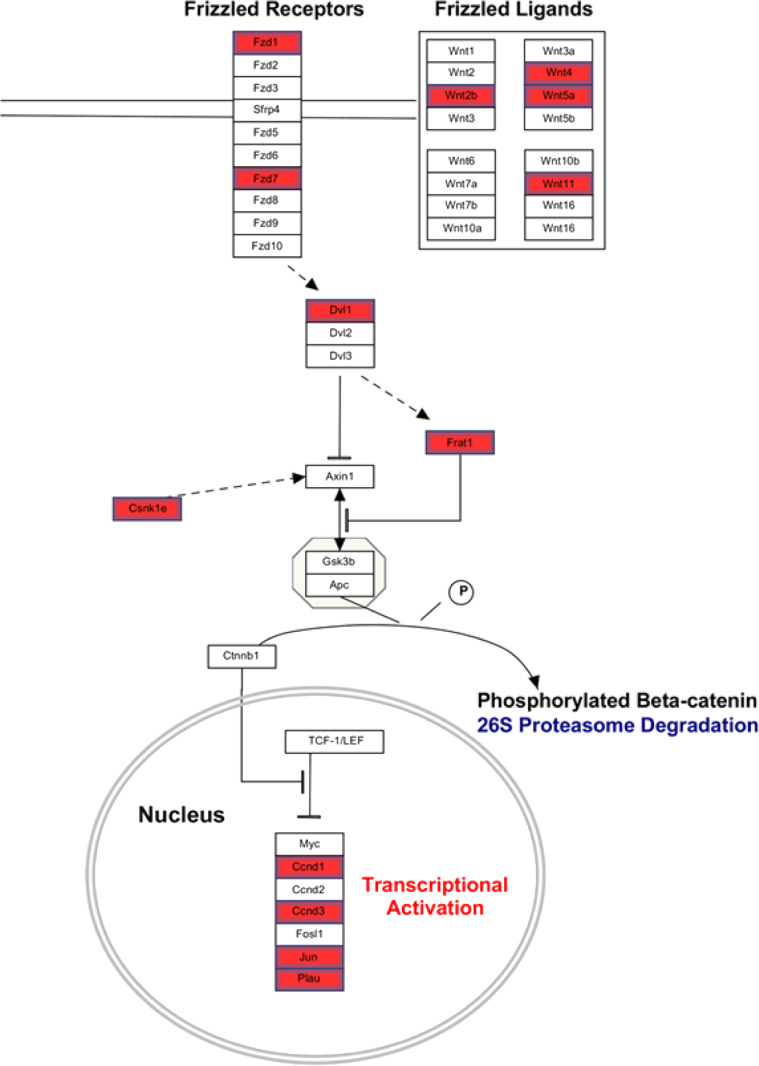
Comparison of liver-specific gene expression signatures of genetic and chemical perturbation to PEDF KO livers showed a striking resemblance to various human HCC subsets marked by overactive Wnt/β-catenin signaling (Table 2).3, 29, 30, 31 Furthermore, PEDF KO liver expression profiles also correlated with the gene expression patterns of nonliver tissue experimental models where constitutively active mutant β-catenin was overexpressed (Table 2).32 Additionally, we observed overexpression of both Fzd 1 and 7, Wnt coreceptors that have been reported to be induced in human HCC specimens and cell lines (Figure 2).33 Downstream targets of Wnt/β-catenin signaling, such as Ccnd1, Ccnd3, and c-Jun, were also found to be up-regulated in PEDF KO livers.
Table 2.
Up-Regulated Gene Sets From PEDF KO Livers Matching Gene Expression Signatures Associated With Aberrant Wnt/β-Catenin Signaling
| Name | FDRa | Description, Web Link, and PubMed ID |
|---|---|---|
| HOSHIDA LIVER CANCER SUBCLASS S1 | <.001 | Gene signature from HCC subset with aberrant Wnt activation http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SUBCLASS_S1 PUBMED ID: 19723656 |
| KENNY CTNNB1 TARGETS UP | .002 | Genes up-regulated in mammary epithelial cells with constitutively active mutant β-catenin gene http://www.broadinstitute.org/gsea/msigdb/cards/KENNY_CTNNB1_TARGETS_UP.html PUBMED ID: 15642117 |
| CAVARD LIVER CANCER MALIGNANT VS BENIGN | .003 | Genes identified by subtractive hybridization to compare gene expression between malignant and benign components of a human HCC occurring from pre-existing adenoma with activated β-catenin http://www.broadinstitute.org/gsea/msigdb/cards/CAVARD_LIVER_CANCER_MALIGNANT_VS_BENIGN.html PUBMED ID: 16314847 |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | .031 | Genes up-regulated in the subclass of HCC characterized by activated β-catenin (CTNNB1) gene http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_CTNNB1_UP.html PUBMED ID: 18701503 |
| CAIRO HEPATOBLASTOMA UP | .050 | Gene signature from human hepatoblastoma characterized by Wnt/β-catenin activation http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_UP.html PUBMED ID: 19061838 |
FDR (false-discovery rate): adjusted P value (FDR q value).
Figure 2.
Expression profiling of PEDF knockout (KO) livers demonstrates up-regulation of genes involved in Wnt/β-catenin signaling. Items in red represent genes that were up-regulated <1.1-fold in PEDF KO compared with wild-type (WT) livers. In particular, Frizzled ligands known to play a role in hepatocarcinogenesis were up-regulated. Induction of multiple downstream targets of Wnt/β-catenin (Ccnd1, Ccnd3, Jun, and Plau) suggests transcriptional activation of Wnt/β-catenin signaling.
PEDF Inhibits Activation of the Wnt Coreceptor LRP6 In Vivo
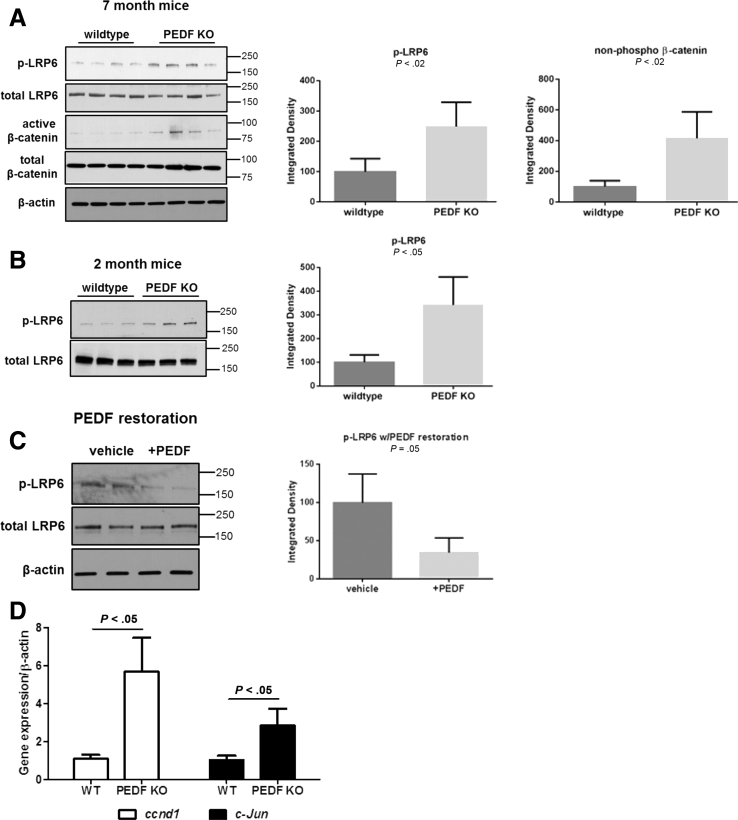
To evaluate concordance with the genomic analysis, we interrogated components of the Wnt/β-catenin signaling pathway in PEDF KO livers before and after PEDF reconstitution. PEDF KO livers showed enhanced phospho-LRP6 levels and active β-catenin compared with WT controls in 7-month-old mice (Figure 3A, P < .02). A similar activation of LRP6 was seen in 2-month-old mice (Figure 3B, P < .05). Restoration of PEDF in KO mice resulted in decreased LRP6 phosphorylation without affecting total LRP6 levels (Figure 3C, P = .05). Moreover, gene expression of downstream canonical Wnt signaling pathway targets Ccnd1 and c-Jun was increased in PEDF KO livers versus controls (Figure 3D, P < .05). These results indicate that PEDF functions as an antagonist of hepatic LRP6 activation in vivo and that exogenous PEDF can inhibit LRP6 activation in vivo.
Figure 3.
PEDF inhibits LRP6 phosphorylation in murine livers. (A) Increased phospho-LRP6 and nonphosphorylated (active) β-catenin in 7-month-old PEDF knockout (KO) mice and corresponding quantification of immunoblots (P < .02). (B) Younger 2 month-old) PEDF KO mice also show increased phosphorylation of LRP6 (P < .05). (C) PEDF restoration in vivo reduces LRP6 activation (P = .05). (D) Gene expression of Ccnd1 and c-Jun in murine control and PEDF KO livers. Representative data from duplicate experiments conducted with n = 3–4/group for immunoblots. Quantitative reverse-transcription polymerase chain reaction data, n = 6/group. Data are presented as mean ± SD.
PEDF Loss Is Associated With Increased Fibrogenic Markers and Enhanced Cellular Proliferation
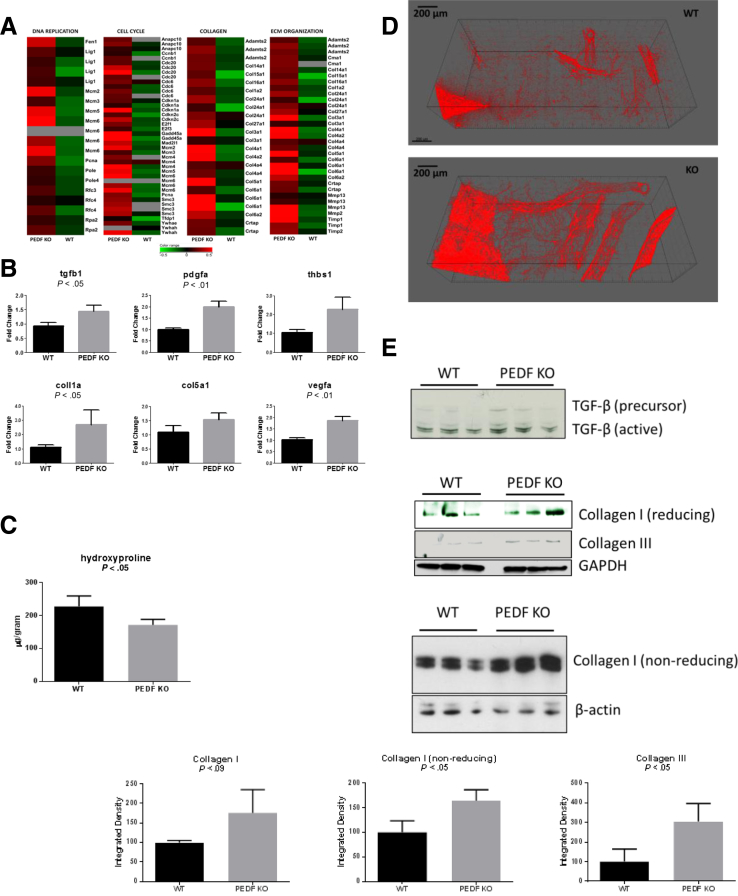
PEDF expression is reduced in human cirrhosis, and its restoration in two different models of experimental liver cirrhosis mitigates fibrotic changes.14, 16 Consistent with this finding, the GSEA revealed an induction of pathways related to extracellular matrix deposition in PEDF KO liver tissue (Figure 4A, Supplementary Table 1). Biochemical assessment of collagen content and specific collagen subtypes, however, revealed a more complex picture of the matricellular changes in the absence of PEDF.
Figure 4.
Absence of PEDF is permissive for induction of fibrogenic markers. (A) Gene expression heat maps show up-regulation of DNA replication, collagen, and extracellular matrix organization pathways. Heat maps represent graphic gene expressions of the genes contributing most to statistically significant enrichment score in gene set enrichment analysis (core enriched genes). The log transformed color expression scale is shown at the bottom of the figure. (B) PEDF KO livers demonstrate enhanced expression of profibrotic cytokines (tgfb1, P < .05; pdgfa, P < .01, vegfa, P < .01) and fibrillar collagen. (C) Decreased hydroxyproline in PEDF KO livers. (D) Second harmonic generation (SHG) imaging demonstrates enhanced fibrillar collagen deposition adjacent to vessels in PEDF KO livers. (E) Transforming growth factor-β (TGF-β) and fibrillar type I and III collagens were increased in PEDF KO livers under reducing and nonreducing conditions. Quantitative reverse-transcription polymerase chain reaction data, n = 5–6/group; data are presented as mean ± S.E.M. Representative SHG images taken from n = 3/group. Representative hydroxyproline data from n = 4 separate experiments from three different sets of age-matched livers; data are presented as mean ± SD. Immunoblots are from n = 3 livers/group from three separate experiments; data are presented as mean ± SD.
Confirmation of fibrogenic cytokines with quantitative PCR showed that tgfb1 and pdgfa were significantly increased, and thbs1, an activator of transforming growth factor-β, showed a trend toward increased expression (Figure 4B). Angiogenic factors play a role in promoting fibrogenesis and can be regulated by Wnt pathway activation.34 Enhanced expression of vegfa was present in PEDF KO livers (Figure 4B). Similarly, expression of col1a was increased but not that of other fibrillar collagen types such as col5a1. Surprisingly, the total hydroxyproline content of PEDF KO livers was 75% of the control livers (Figure 4C), indicating that overall the collagen content was decreased. However, SHG imaging revealed visual evidence of increased fibrillar collagen in PEDF KO livers (Figure 4D). Consistent with the SHG imaging, the fibrillar collagen types I and III levels in PEDF KO livers were higher than in the controls (Figure 4E). Thus, a preferential induction of fibrillar collagen occurs in PEDF KO livers, but it is accompanied by an overall decrease in other collagen or structural proteins that contain hydroxyproline residues.
PEDF Is a Secreted Antagonist of Wnt/β-Catenin Signaling in Hepatocellular Carcinoma Cells
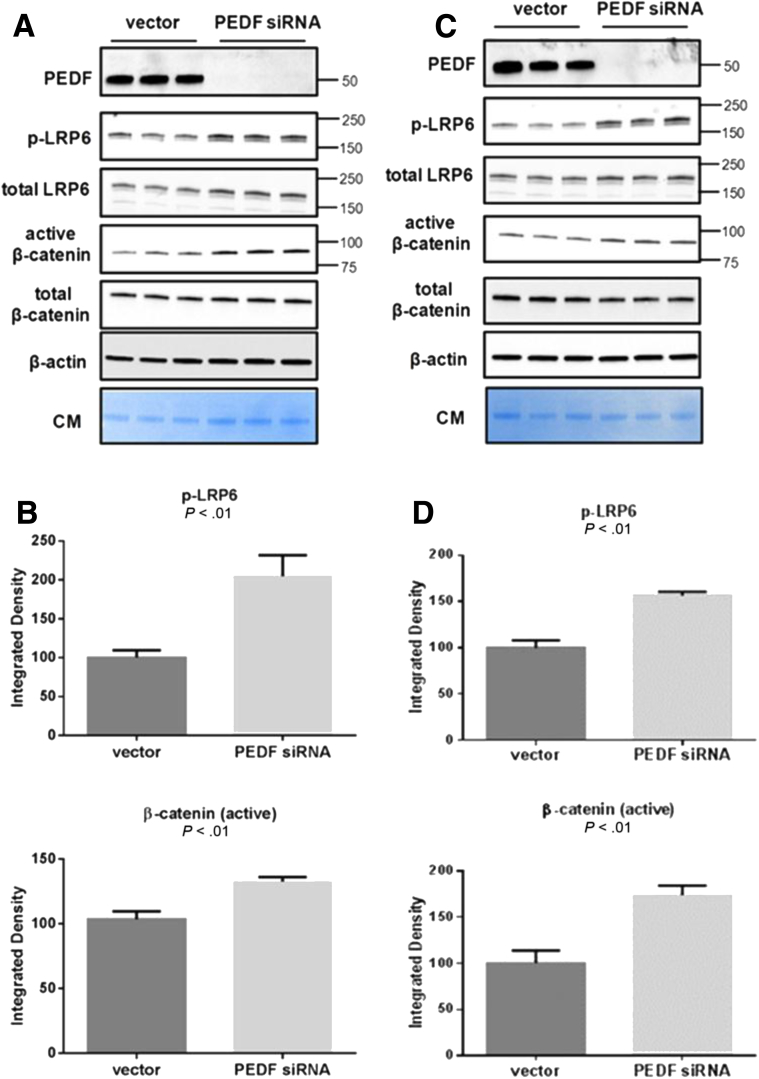
Findings in murine livers were extended to human HCC cells to determine whether PEDF functions as a Wnt antagonist. Both HepG2 and Huh7 cells secreted PEDF into the CM (Figure 5A and C). In HepG2 cells, siRNA-mediated PEDF knockdown led to increased phospho-LRP6 and active β-catenin levels (Figure 5A and B, P < .01). Similar results were observed in Huh-7 cells after PEDF knockdown (Figure 5C and D, P < .01).
Figure 5.
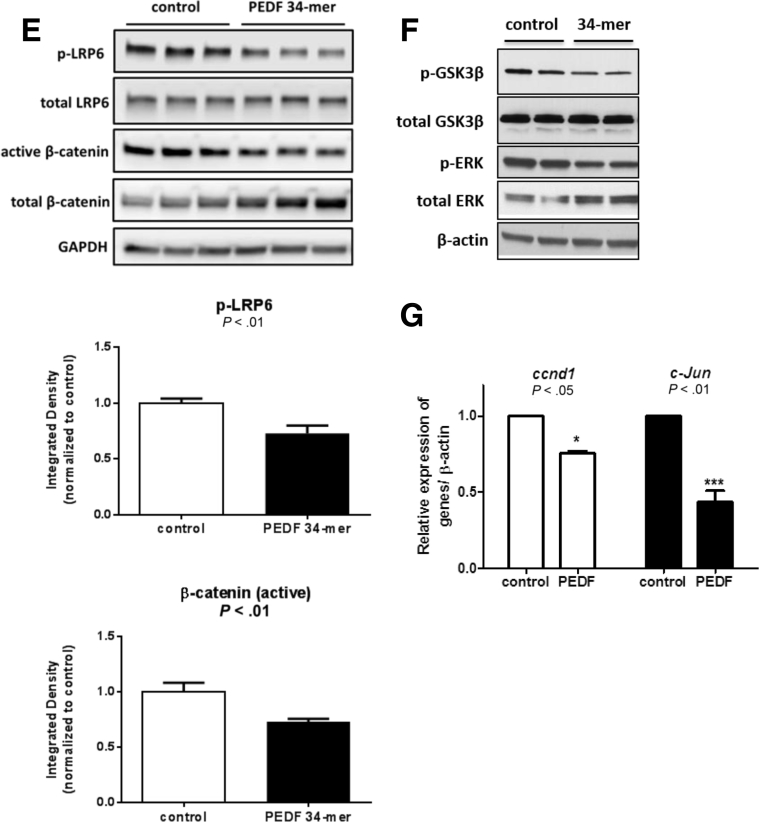
PEDF inhibits canonical Wnt/β-catenin signaling in human hepatocellular carcinoma (HCC) cells. (A) PEDF knockdown in HepG2 cells results in increased LRP6 phosphorylation and increased active β-catenin. (B) Corresponding quantification of phospho-LRP6 and active β-catenin after RNA interference of PEDF in HepG2 cells (P < .01). (C) Huh-7 cells display increased LRP6 phosphorylation and active β-catenin after depletion of endogenous PEDF. (D) Quantification of phospho-LRP6 and active β-catenin after RNA interference of PEDF in Huh-7 cells (P < .01). (E) A PEDF 34-mer peptide decreased LRP6 phosphorylation and active β-catenin levels in Huh-7 cells (P < .01). (F) Changes in the levels of downstream targets of canonical Wnt signaling such as phospho-GSK3β/total GSK3β and phospho-ERK/total ERK reflect inhibition of Wnt signaling with the PEDF 34-mer. (G) Gene targets of the Wnt pathway, ccnd1 and c-Jun, were significantly suppressed with PEDF 34-mer (P < .05 and P < .01, respectively). Representative data are shown from three separate experiments conducted with n = 3/group for siRNA experiments. Data from 34-mer peptide experiments were performed in duplicate and n = 3/group. Data are presented as mean ± SD.
A 34-mer sequence within PEDF mediates its well-documented antiangiogenic effects.25 Because angiogenesis requires Wnt signaling, we surmised that the PEDF 34-mer is responsible for its effects on Wnt/β-catenin signaling. Adding the PEDF 34-mer decreased the levels of active phospho-LRP6 and active β-catenin (Figure 5E, P < .01). Downstream regulators and targets of Wnt signaling such as GSK3β and phospho-ERK levels corresponded to the effects of Wnt blockade with PEDF 34-mer (Figure 5F). Levels of phospho-GSK3β (inactive form) were diminished consistent with increased intracellular active GSK3β and enhanced degradation of β-catenin seen with Wnt blockade. The downstream targets of β-catenin such as phospho-ERK were decreased. Moreover, transcriptional targets of canonical Wnt signaling such as ccnd1 and c-Jun were suppressed with the 34-mer (Figure 5G). These results demonstrate that PEDF antagonizes Wnt/β-catenin signaling in human HCC cells and point to a 34-amino-acid peptide fragment derived from PEDF that mediates LRP6 blockade.
Induction of Liver Fibrosis and Sporadic Hepatocellular Carcinoma in PEDF Knockout Mice After Western Diet Feeding
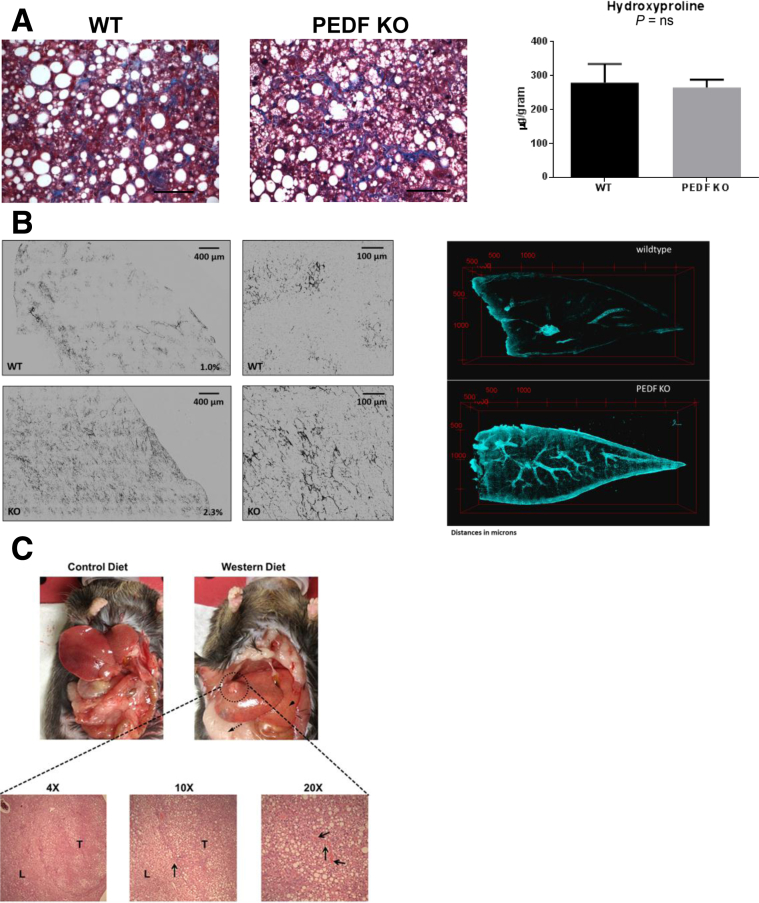
Genomic profiling of PEDF KO livers corresponded to various human HCC subsets marked by overactive Wnt/β-catenin, but spontaneous HCC did not develop in PEDF KO mice up to 1 year of age (data not shown). To test whether diet-induced obesity could induce HCC formation in the absence of PEDF, a Western diet (40% fat, 44% carbohydrate, 16% protein) was given to PEDF KO and WT mice for 6 to 8 months. A Western diet increased fibrosis in WT and PEDF KO mice as shown by trichrome staining and hydroxyproline measurements (Figure 6A).
Figure 6.
A Western diet induces liver fibrosis and sporadic hepatocellular carcinoma (HCC) in PEDF knockout (KO) mice. (A) Six months of Western diet feeding induced liver fibrosis in wild-type (WT) and PEDF KO mice as demonstrated by trichrome staining (magnification 20×; size bars: 100 μM) and measured by hydroxyproline content. (B) Second harmonic generation (SHG) imaging shows increased fibrillar type I/III collagen deposition in PEDF KO mice livers (bottom panels) compared with WT (top panels) mice fed a Western diet. Magnification: left 4×; right 20×. Three-dimensional reconstruction of serial SHG images reveals prominence of fibrillar collagen around blood vessels in PEDF KO livers. (C) PEDF KO mice showing macroscopic tumor in mice fed the Western diet versus control diet. Bottom panel shows histology of a well-differentiated HCC arising in KO mouse fed a Western diet. L, liver; T, tumor; magnification 10×, arrow at demarcation between liver and HCC; 20×, arrows highlighting unpaired blood vessels in HCC.
Increased fibrillar collagen deposition as seen with SHG imaging was more apparent in PEDF KO than WT livers (Figure 6B). Three-dimensional reconstructed images from SHG imaging revealed an increase in fibrillar collagen adjacent to vessels, outlining their structures (Figure 6B). A subset of PEDF KO mice (3 of 12) developed macroscopic tumor formation compared with none (0 of 12) in the control mice (Figure 6C) after chronic Western diet feeding. Histologic examination showed features consistent with a well-differentiated HCC with the increased presence of unpaired blood vessels (Figure 6C, arrows). In contrast to the diet-induced HCC, a one-time diethylnitrosamine injection did not result in HCC formation in either the WT or KO mice at 6 months (data not shown). Thus, PEDF deficiency combined with a chronic Western diet led to sporadic HCC formation.
PEDF Expression Is Reduced in Human Hepatocellular Carcinoma Specimens
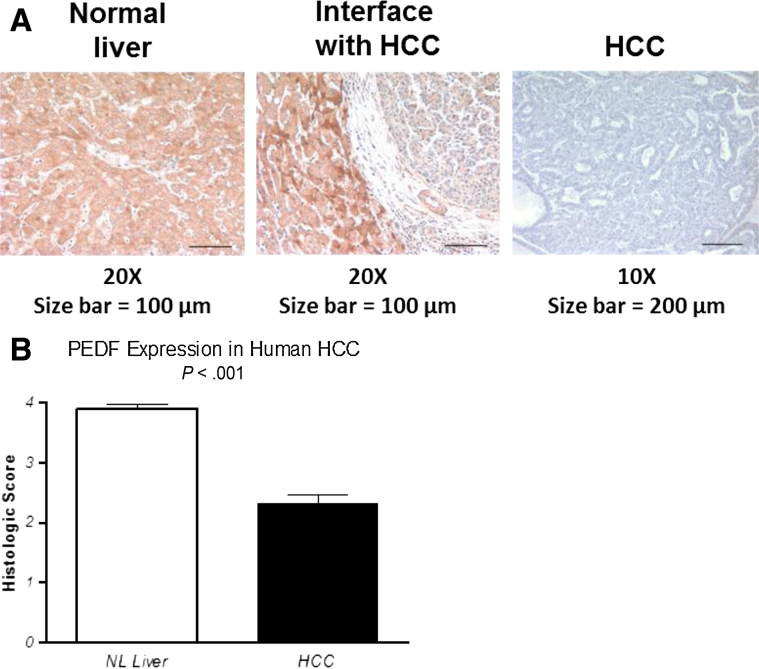
A previous study of embryonic and adult human tissue sites demonstrated that the liver has the highest expression levels of the PEDF gene, and the recent tissue-based map of the human proteome confirmed this finding.20, 21 Relative to the high endogenous levels in the normal liver, we asked whether PEDF levels in HCC specimens were diminished. Staining of PEDF showed diffuse and strong immunoreactivity for PEDF in normal liver tissue (Figure 7A, left). In contrast, PEDF immunolabeling was statistically significantly reduced in HCC compared with the adjacent liver (Figure 7A, middle and right, and B; P < .01). Thus, human HCC specimens demonstrated decreased PEDF expression compared with the adjacent nontransformed hepatocytes.
Figure 7.
PEDF expression is reduced in human hepatocellular carcinoma (HCC). (A) Immunostaining for PEDF in human livers (top) and human HCC specimens (bottom). (B) Semiquantitative scoring of PEDF staining demonstrates increased labeling in normal liver compared with HCC specimens (P < .01; n = 14). NL, normal.
Discussion
Aberrant Wnt/β-catenin signaling underlies a number of malignancies, including HCC.3, 35 Our study has identified PEDF as an endogenous inhibitor of LRP6 activation that is secreted in response to canonical Wnt ligands. Enhanced LRP6 and β-catenin activation was seen in the livers of PEDF KO mice and in two human HCC cell lines where PEDF was depleted. Further, adding a PEDF 34-mer inhibited LRP6, active β-catenin, and downstream targets of Wnt signaling, thereby identifying the region on PEDF that mediates Wnt inhibitory effects. These data support the idea that PEDF functions as a part of a negative feedback loop to modulate Wnt signaling. Gene enrichment data supported this interaction. Further, biochemical analyses of PEDF KO murine livers before and after PEDF reconstitution in vivo confirmed that PEDF can block Wnt signaling in the liver. PEDF knockdown in two human HCC cell lines led to increased Wnt/β-catenin signal transduction with a specific 34-amino-acid region mediating these effects. Thus, PEDF is regulated by and inhibits the canonical Wnt/β-catenin pathway in the murine liver and in two human HCC cell lines.
The genomic analysis in this study correlated with genetic profiles of murine hepatocarcinogenesis and human HCC subsets marked by overactive Wnt/β-catenin signaling, but PEDF deficiency alone did not result in HCC formation. A prolonged nutritional challenge induced only a fraction of animals to develop a well-differentiated HCC. These results are consistent with models of hepatic overexpression of normal and mutant β-catenin that do not result in spontaneous HCC.35 Paradoxically, deletion of β-catenin from the liver is permissive for HCC formation after injection with diethylnitrosamine.36 This surprising effect of β-catenin deletion conferring an increased rate of HCC development in murine models, rather than its overexpression, reflects the importance of this pathway for liver tissue homeostasis. In its absence, the liver is prone to injury from oxidative stress and enhanced fibrosis.36 Thus, findings from β-catenin transgenic mice are at odds with those from genomic and immunohistochemical studies in human HCC, which point to Wnt/β-catenin signaling as a significant driver in a subset of HCC.3 The absence of HCC found in transgenic models of β-catenin overexpression and the occurrence of HCC with β-catenin deletion highlights the limitations of constitutively active or deletion of β-catenin, where temporal and context-specific activity of β-catenin may more accurately capture its role in human disease.
Absence of PEDF led to complex changes to the ECM of the liver. Despite lower total hydroxyproline levels, type I/III collagen content and SHG imaging demonstrated increased deposition of fibrillar collagen in PEDF KO livers. In experimental and human cirrhosis specimens, PEDF levels are also depleted.14 Restoration of PEDF in experimental models of CCl4 [chemokine (C-C motif) ligand 4] and bile-duct ligated cirrhosis ameliorates tissue fibrosis, suggesting an important role for endogenous PEDF in maintaining quiescence of the liver ECM.14, 16 These findings are consistent with studies that demonstrate Wnt/β-catenin signaling as a regulator of the fibrotic response in diverse organs.37, 38, 39 Further, examination of the PEDF null state in humans, osteogenesis imperfecta type VI, points to abnormalities in the extracellular matrix.24, 40 These findings suggest that PEDF may regulate matricellular content in multiple organ sites.
This study provides further evidence to support the role of PEDF in Wnt/β-catenin signaling. The discovery through exome sequencing that null mutations in PEDF cause osteogenesis imperfecta type VI implicated PEDF’s role in modulating Wnt/β-catenin signaling in human disease.22, 40, 41 We and others have shown that PEDF could induce differentiation of progenitor cells and that these effects were LRP6 dependent.22, 42 In the eye, PEDF inhibited Wnt3a-mediated β-catenin nuclear translocation, and recent studies showed that PEDF directly suppressed other Wnt modulators such as sclerostin.19, 41 Exogenous PEDF protein and a peptide derived from PEDF demonstrate inhibitory effects on Wnt signaling in the liver and in two HCC cell lines, thereby pointing to its role in attenuating Wnt signaling in a negative feedback loop.
Interestingly, PEDF appears to promote Wnt/β-catenin signaling in stem cell populations but inhibits Wnt signaling in differentiated cells.22, 41 Differential effects are also seen in Wnt ligands and Wnt-related proteins such as Wnt5a and Dickkopf2, and stem from selective expression patterns of Wnt coreceptors.28, 43, 44 Future studies detailing the expression patterns of different Fzd species should allow identification of the receptor combination that directs PEDF’s different functional outcomes as they pertain to Wnt signaling.
In summary, PEDF functions as an endogenous inhibitor of Wnt/β-catenin signaling in the liver and in human HCC cells. These findings provide a framework for understanding the antitumor properties of PEDF in other cancer types.
Acknowledgements
The authors thank Kathy Harry (Yale Liver Center) and Edward J. Edmonds (VA Connecticut Healthcare) for expert technical assistance.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by Digestive Diseases Research Core Centers–5P30DK034989 (to C.C.); Yale Core Center for Musculoskeletal DisordersP30-AR46032; Veterans Affairs Merit Grant (to C.C.).
Appendix
Supplementary Table 1.
List of Statistically Significant Differentially Expressed Gene Entities in PEDF Knockout Animals Versus Wild-Type (FDR < .05 and Expression Cutoff of 1.3-Fold)
| Symbol | Fold Up | Symbol | Fold Down |
|---|---|---|---|
| Cyp2b9 | 6.730 | Serpinf1 (KO GENE) | −143.796 |
| Gsta1 | 4.853 | Serpina1e | −8.049 |
| Cyp2b23 | 4.285 | Hsd3b5 | −5.549 |
| Ly6d | 4.166 | Lpin1 | −3.815 |
| Gsta2 | 3.822 | Serpina4-ps1 | −3.642 |
| Tubb2b | 3.390 | Nnmt | −3.193 |
| Lcn2 | 3.346 | Nnmt | −3.105 |
| Anxa2 | 2.943 | Egfr | −3.080 |
| Mod1 | 2.733 | C6 | −3.023 |
| Cidec | 2.654 | Egfr | −2.957 |
| S100a11 | 2.579 | Aatk | −2.871 |
| Bcl6 | 2.524 | C8b | −2.749 |
| Orm2 | 2.467 | Egfr | −2.578 |
| Wfdc2 | 2.254 | C6 | −2.577 |
| Aqp8 | 2.222 | Cyp7b1 | −2.553 |
| Insig2 | 2.192 | Cyp4a12b | −2.465 |
| Tceal8 | 2.169 | Clca3 | −2.437 |
| Lgals3 | 2.167 | Sds | −2.380 |
| Spon2 | 2.165 | Slc38a2 | −2.378 |
| Aqp8 | 2.127 | Ela1 | −2.333 |
| Ubd | 2.079 | 2200001I15Rik | −2.305 |
| H2-Ab1 | 2.062 | Cyp7a1 | −2.302 |
| Apoa4 | 2.056 | Fbxo31 | −2.246 |
| Cbr3 | 2.053 | EG13909 | −2.223 |
| Hsd17b6 | 1.980 | Selenbp2 | −2.181 |
| Lpl | 1.978 | Socs2 | −2.177 |
| Cd74 | 1.972 | Upp2 | −2.167 |
| Raet1b | 1.967 | Fbxo31 | −2.165 |
| Egr1 | 1.965 | Tsc22d3 | −2.161 |
| Pdk4 | 1.927 | C8a | −2.142 |
| Ttc39a | 1.900 | Cyp4f14 | −2.136 |
| Slc17a4 | 1.878 | 2810439F02Rik | −2.120 |
| Gstm2 | 1.850 | Slc29a1 | −2.110 |
| Sqle | 1.829 | F11 | −2.106 |
| Spp1 | 1.793 | Siat9 | −2.095 |
| Ccnd1 | 1.788 | Mup4 | −2.095 |
| H2-Aa | 1.786 | LOC100047762 | −2.070 |
| Sepp1 | 1.785 | Cish | −2.065 |
| H2-Ab1 | 1.780 | Ptpre | −2.059 |
| Insig2 | 1.735 | Por | −2.055 |
| Cd74 | 1.734 | Ccrn4l | −2.022 |
| Insig2 | 1.734 | Hpd | −2.013 |
| Aqp4 | 1.724 | Upp2 | −1.984 |
| Vldlr | 1.724 | Prei4 | −1.980 |
| Srxn1 | 1.722 | F11 | −1.978 |
| Elovl6 | 1.711 | Prodh | −1.961 |
| Slpi | 1.707 | Zap70 | −1.960 |
| Lgals1 | 1.706 | Por | −1.960 |
| Gdf15 | 1.696 | Zxda | −1.954 |
| Ptp4a2 | 1.690 | Gne | −1.950 |
| LOC100047046 | 1.689 | Kcnk5 | −1.918 |
| LOC641240 | 1.684 | Pptc7 | −1.906 |
| Limk1 | 1.683 | Eif4ebp3 | −1.894 |
| Ntrk2 | 1.680 | 2810439F02Rik | −1.890 |
| H2-Ab1 | 1.679 | Tk1 | −1.879 |
| Gstm2 | 1.679 | Zap70 | −1.875 |
| Fam129b | 1.676 | Tef | −1.871 |
| Col6a1 | 1.673 | Angptl4 | −1.851 |
| Ctps | 1.672 | Chac1 | −1.838 |
| Nudt18 | 1.669 | Gm129 | −1.836 |
| Anxa5 | 1.668 | Agxt | −1.835 |
| Zfp36l1 | 1.664 | Lpin2 | −1.835 |
| Ankrd56 | 1.658 | EG13909 | −1.834 |
| H2-Eb1 | 1.652 | Asl | −1.824 |
| S100a10 | 1.648 | Afmid | −1.821 |
| Ccnd1 | 1.642 | Tle1 | −1.820 |
| Srxn1 | 1.641 | Afmid | −1.820 |
| Lyzs | 1.639 | Spata2L | −1.817 |
| Cd63 | 1.637 | Upp2 | −1.814 |
| Idh2 | 1.634 | St3gal5 | −1.802 |
| Uhrf1 | 1.633 | Chka | −1.802 |
| Uap1l1 | 1.632 | rp9 | −1.797 |
| Dusp6 | 1.625 | Smarcd2 | −1.796 |
| Aldh1a1 | 1.623 | Gpt1 | −1.792 |
| Mfge8 | 1.619 | Cyp4f13 | −1.784 |
| Acnat2 | 1.614 | Nrbp2 | −1.784 |
| Cxcl9 | 1.614 | Cps1 | −1.778 |
| Acly | 1.609 | Cebpb | −1.773 |
| Tsc22d1 | 1.609 | Serpinf2 | −1.762 |
| Rtn4rl1 | 1.597 | Hs6st1 | −1.757 |
| Plekha1 | 1.596 | Gls2 | −1.750 |
| Cyp4a14 | 1.595 | Susd4 | −1.750 |
| Usp18 | 1.594 | Apom | −1.744 |
| Cd9 | 1.591 | Klhl21 | −1.744 |
| Smpd3 | 1.591 | Fkbp5 | −1.743 |
| Hist1h2ao | 1.590 | Prhoxnb | −1.743 |
| Col4a1 | 1.587 | Cbs | −1.741 |
| Lypla1 | 1.583 | Lgals4 | −1.732 |
| Gstm2 | 1.583 | Ccbl1 | −1.727 |
| Rcan1 | 1.582 | Upp2 | −1.726 |
| Tlr2 | 1.571 | Foxa3 | −1.724 |
| Prune | 1.569 | Sardh | −1.711 |
| LOC100043671 | 1.567 | Clec2d | −1.710 |
| Lgals3bp | 1.567 | B230342M21Rik | −1.708 |
| Ccbl2 | 1.567 | Afmid | −1.708 |
| Hrsp12 | 1.566 | Bach1 | −1.708 |
| Elovl5 | 1.562 | Tmem50a | −1.707 |
| Ccnd1 | 1.560 | Mlxipl | −1.706 |
| Gadd45a | 1.559 | Gcgr | −1.703 |
| Samd9l | 1.556 | Agpat6 | −1.697 |
| Gas6 | 1.555 | Cyp4f15 | −1.693 |
| Esd | 1.549 | Plg | −1.692 |
| Cyp3a11 | 1.548 | Cyp1a2 | −1.687 |
| Sparc | 1.547 | Serpina3k | −1.676 |
| LOC100047934 | 1.542 | D4Bwg0951e | −1.675 |
| Ctsa | 1.538 | Nfic | −1.673 |
| Ppic | 1.536 | Hhex | −1.671 |
| Nipsnap3a | 1.536 | Ush2a | −1.670 |
| Mcm6 | 1.533 | Ang | −1.668 |
| Axl | 1.531 | Hyal1 | −1.666 |
| Tmem43 | 1.530 | Pgls | −1.663 |
| Plscr1 | 1.528 | Itih3 | −1.662 |
| Lum | 1.526 | Rnase4 | −1.659 |
| 2410004L22Rik | 1.526 | Ttc36 | −1.655 |
| Cdkn1a | 1.523 | 1700019G17Rik | −1.651 |
| Arl8a | 1.521 | Cps1 | −1.650 |
| Acot4 | 1.519 | Rps5 | −1.646 |
| Laptm5 | 1.515 | 9530058B02Rik | −1.643 |
| St5 | 1.515 | 2310076L09Rik | −1.642 |
| Gbp2 | 1.515 | Il6ra | −1.638 |
| Sirpa | 1.513 | Mbd1 | −1.638 |
| Ifi27 | 1.512 | Atp5sl | −1.635 |
| Sqle | 1.511 | Kcnk5 | −1.635 |
| Acot3 | 1.509 | Gnat1 | −1.634 |
| Spc25 | 1.505 | Abcg8 | −1.629 |
| B930041F14Rik | 1.504 | Tmem160 | −1.622 |
| Tgfbr2 | 1.502 | Hist1h2bm | −1.621 |
| Tgm2 | 1.502 | Hes6 | −1.619 |
| Cbr3 | 1.500 | Asl | −1.618 |
| Aldh1a7 | 1.500 | Acaa2 | −1.617 |
| Hprt1 | 1.497 | Zfp259 | −1.617 |
| Entpd5 | 1.497 | Klf13 | −1.612 |
| Cyba | 1.497 | Hist2h2aa1 | −1.607 |
| Tpm1 | 1.496 | Ccbl1 | −1.593 |
| Acox1 | 1.495 | Npr2 | −1.593 |
| Cyp4a31 | 1.493 | Map1lc3a | −1.592 |
| Atp5a1 | 1.492 | Ephx2 | −1.590 |
| Col6a1 | 1.489 | Tmem183a | −1.587 |
| Dusp6 | 1.488 | Scnn1a | −1.586 |
| Tmem77 | 1.486 | Afmid | −1.586 |
| Fos | 1.483 | Igfals | −1.582 |
| LOC100048346 | 1.483 | Fgf1 | −1.582 |
| Ifit3 | 1.482 | Cnpy2 | −1.582 |
| Ctsc | 1.480 | 0610012D14Rik | −1.582 |
| Serpina7 | 1.480 | Tspan31 | −1.581 |
| Pex11a | 1.478 | Rbm5 | −1.580 |
| Ccdc80 | 1.475 | Ugt2b1 | −1.580 |
| Rnf125 | 1.475 | Med25 | −1.578 |
| Tm4sf4 | 1.475 | Eif4ebp1 | −1.578 |
| St5 | 1.474 | Serpina11 | −1.577 |
| Col5a1 | 1.474 | Ass1 | −1.577 |
| Cyp2a5 | 1.474 | Nfkbia | −1.574 |
| Enc1 | 1.473 | Lcat | −1.573 |
| Plscr1 | 1.472 | Rps10 | −1.572 |
| Aldh1b1 | 1.470 | Atad3a | −1.571 |
| Lamb3 | 1.469 | Hist1h2bn | −1.570 |
| Cdc20 | 1.468 | Upb1 | −1.568 |
| Cxcl10 | 1.468 | Cpsf4l | −1.563 |
| H2-DMb1 | 1.467 | Fn3k | −1.563 |
| Vps29 | 1.466 | Klf9 | −1.561 |
| Igf2bp2 | 1.466 | Ivd | −1.558 |
| Csf1r | 1.464 | Prodh2 | −1.558 |
| Ear2 | 1.463 | Mcm10 | −1.558 |
| Cyp2c29 | 1.460 | Mrpl55 | −1.557 |
| Dsp | 1.460 | Prodh2 | −1.556 |
| Tubb6 | 1.459 | Sri | −1.555 |
| Ctsc | 1.459 | Eef2 | −1.552 |
| Tnfrsf22 | 1.458 | Pla1a | −1.551 |
| Gadd45a | 1.458 | Cisd1 | −1.551 |
| Mmp2 | 1.453 | Serpinf2 | −1.549 |
| Gstm1 | 1.451 | P2ry1 | −1.547 |
| Cd52 | 1.448 | 4933426M11Rik | −1.547 |
| Vldlr | 1.445 | Alas1 | −1.546 |
| Aadac | 1.444 | Dedd2 | −1.540 |
| Glul | 1.444 | Pipox | −1.536 |
| Bcap31 | 1.443 | Fam152b | −1.535 |
| Ly6a | 1.442 | 1300017J02Rik | −1.530 |
| Abcb11 | 1.441 | Abcc6 | −1.529 |
| Loxl1 | 1.441 | Sema4g | −1.529 |
| Col4a2 | 1.436 | BC031353 | −1.529 |
| Mvd | 1.436 | Brap | −1.528 |
| Il1b | 1.434 | Lsm4 | −1.527 |
| Mcm4 | 1.433 | Slc38a3 | −1.527 |
| Slc13a3 | 1.432 | Pex6 | −1.526 |
| Net1 | 1.432 | Nfix | −1.521 |
| Hist1h2ak | 1.430 | Als2 | −1.520 |
| Ugt2b35 | 1.430 | Errfi1 | −1.519 |
| Apoc2 | 1.428 | Ldb1 | −1.518 |
| Mcm5 | 1.426 | Hc | −1.516 |
| Rnd2 | 1.426 | Ptms | −1.516 |
| Gclc | 1.426 | Agt | −1.516 |
| Pparg | 1.425 | Igf1 | −1.515 |
| Gpam | 1.425 | Akr7a5 | −1.515 |
| Osbpl3 | 1.425 | Ddx6 | −1.513 |
| Slc41a2 | 1.422 | Hpn | −1.513 |
| Tnfaip2 | 1.422 | Stard10 | −1.513 |
| Tmsb4x | 1.421 | Mafg | −1.511 |
| Mcm6 | 1.420 | Tsku | −1.510 |
| Khk | 1.417 | Cyp2c37 | −1.508 |
| Fam110a | 1.416 | Rpl34 | −1.508 |
| Mme | 1.416 | Rbm4b | −1.507 |
| LOC677317 | 1.415 | Bmp1 | −1.506 |
| 4931406C07Rik | 1.414 | Wdr45l | −1.506 |
| Klf6 | 1.412 | Afmid | −1.505 |
| Ywhah | 1.411 | Ppp1r10 | −1.505 |
| LOC100047963 | 1.409 | Afmid | −1.504 |
| Nampt | 1.409 | Pop5 | −1.504 |
| Hist1h2af | 1.408 | 4833421E05Rik | −1.502 |
| Emr1 | 1.406 | OTTMUSG00000000231 | −1.502 |
| Dapk2 | 1.405 | Pscd1 | −1.500 |
| S100a8 | 1.404 | Vgll4 | −1.499 |
| Hprt1 | 1.403 | F7 | −1.499 |
| 1810023F06Rik | 1.401 | Tmem42 | −1.499 |
| Ndufa5 | 1.401 | Gm129 | −1.497 |
| Bmp4 | 1.399 | Saps3 | −1.496 |
| Akr1c14 | 1.397 | Cyp2c67 | −1.496 |
| Cyp2c39 | 1.395 | Oat | −1.495 |
| Vldlr | 1.395 | 1110001J03Rik | −1.494 |
| Nqo1 | 1.394 | Glyctk | −1.494 |
| Jun | 1.394 | Srrm2 | −1.492 |
| LOC100048733 | 1.394 | Tst | −1.492 |
| Tm4sf4 | 1.393 | Sdsl | −1.492 |
| Rtn4 | 1.391 | Fgg | −1.491 |
| Iqgap1 | 1.391 | Hist1h2bk | −1.490 |
| Arhgdib | 1.391 | Klf1 | −1.490 |
| Rcan2 | 1.389 | F2 | −1.490 |
| Palmd | 1.388 | Elovl3 | −1.487 |
| Hist1h2an | 1.387 | Ctdsp2 | −1.485 |
| Rcan2 | 1.386 | Cbs | −1.485 |
| Tmem49 | 1.385 | Ppm1k | −1.483 |
| Entpd5 | 1.384 | Cyp1a2 | −1.483 |
| Idi1 | 1.382 | Hsp105 | −1.482 |
| Nsdhl | 1.381 | 1110032A13Rik | −1.481 |
| Slamf9 | 1.381 | H2afy | −1.480 |
| Trim2 | 1.380 | Dnajb6 | −1.480 |
| Lip1 | 1.377 | Keg1 | −1.480 |
| 6330409N04Rik | 1.376 | Slc35b2 | −1.480 |
| 9030625A04Rik | 1.376 | Tmem19 | −1.479 |
| Cxadr | 1.376 | Fam125a | −1.478 |
| Pltp | 1.375 | Gde1 | −1.478 |
| Agpat9 | 1.375 | Gpr182 | −1.477 |
| Zfp608 | 1.375 | D9Wsu20e | −1.477 |
| Gale | 1.375 | Gpr108 | −1.475 |
| Rasl11b | 1.374 | Rps8 | −1.475 |
| Tpm4 | 1.374 | Lman1 | −1.475 |
| Saa2 | 1.374 | Rpl23 | −1.474 |
| BC005537 | 1.372 | Zfp91-cntf | −1.474 |
| Mcm2 | 1.372 | Upf1 | −1.473 |
| Lhfp | 1.370 | F10 | −1.473 |
| Slc23a2 | 1.369 | Acot1 | −1.472 |
| Usp18 | 1.368 | Slc1a2 | −1.472 |
| Ppp1r3c | 1.367 | Prodh2 | −1.468 |
| Cpxm1 | 1.366 | Ap3m1 | −1.468 |
| LOC100046254 | 1.365 | Fbxo33 | −1.467 |
| Junb | 1.364 | Slc25a42 | −1.467 |
| 2010311D03Rik | 1.363 | Qdpr | −1.466 |
| Krt8 | 1.363 | Hbs1l | −1.466 |
| Angptl3 | 1.360 | Pcsk4 | −1.465 |
| Orm1 | 1.360 | Tut1 | −1.465 |
| Emp1 | 1.359 | Sec14l4 | −1.465 |
| Gca | 1.359 | Slc7a2 | −1.464 |
| Nid1 | 1.358 | Gstp1 | −1.464 |
| Ddx3x | 1.357 | Glt25d1 | −1.464 |
| Ebpl | 1.354 | Tmem160 | −1.464 |
| Slc13a3 | 1.353 | Smarca2 | −1.464 |
| Btg1 | 1.353 | Pxmp2 | −1.463 |
| Tnip1 | 1.352 | Zfp276 | −1.463 |
| Tmem43 | 1.352 | Nosip | −1.463 |
| Pparg | 1.352 | Cml1 | −1.463 |
| Serinc2 | 1.351 | Tmprss6 | −1.462 |
| Col15a1 | 1.350 | Scara5 | −1.462 |
| LOC433801 | 1.350 | Mup5 | −1.460 |
| Cyp3a25 | 1.349 | Cadps2 | −1.459 |
| Gale | 1.347 | Rsn | −1.459 |
| Id2 | 1.347 | EG665378 | −1.458 |
| LOC668837 | 1.346 | Pop5 | −1.458 |
| Slc6a8 | 1.345 | Igfbp4 | −1.456 |
| Vim | 1.345 | Mbl1 | −1.455 |
| Cd274 | 1.345 | Rps25 | −1.455 |
| Mfge8 | 1.342 | Crcp | −1.455 |
| Tnfrsf19 | 1.342 | Plxna1 | −1.454 |
| Rnf125 | 1.341 | Zfp771 | −1.454 |
| Mlkl | 1.341 | Serpina1a | −1.454 |
| EG277333 | 1.340 | Trfr2 | −1.453 |
| Adam9 | 1.339 | Foxo1 | −1.453 |
| Pgrmc1 | 1.338 | Dnajc3 | −1.452 |
| Mat2a | 1.337 | LOC622404 | −1.452 |
| Ccbl2 | 1.336 | Sec63 | −1.451 |
| Tnfaip2 | 1.336 | Tmem150 | −1.451 |
| Pip4k2a | 1.335 | Sepx1 | −1.451 |
| Mad2l1 | 1.335 | Slc6a12 | −1.451 |
| Adra2b | 1.334 | Hist1h2bj | −1.451 |
| Snx7 | 1.333 | Xpa | −1.449 |
| Chmp5 | 1.332 | Syt1 | −1.448 |
| Gsta4 | 1.331 | Trap1 | −1.447 |
| Pigp | 1.330 | Tnrc6a | −1.446 |
| Mreg | 1.329 | Ercc5 | −1.446 |
| Rnd3 | 1.329 | Sil1 | −1.445 |
| Nit2 | 1.328 | 9430029K10Rik | −1.445 |
| Cxadr | 1.328 | Gltpd2 | −1.445 |
| Cotl1 | 1.323 | Cxxc1 | −1.444 |
| 2900064A13Rik | 1.323 | Trp53inp2 | −1.443 |
| Lrrc39 | 1.323 | Serinc3 | −1.443 |
| Dld | 1.322 | Trak1 | −1.443 |
| Pmpcb | 1.321 | Arfgap2 | −1.443 |
| Rab34 | 1.320 | Iyd | −1.442 |
| Fas | 1.320 | Tnrc6c | −1.442 |
| Hist1h2ah | 1.320 | Hint2 | −1.442 |
| Fen1 | 1.320 | 0610012G03Rik | −1.442 |
| Hsd17b11 | 1.320 | Coq5 | −1.441 |
| Tnxb | 1.319 | Gls2 | −1.441 |
| Saa1 | 1.319 | Rpain | −1.439 |
| Tnfrsf12a | 1.317 | Surf1 | −1.438 |
| Acot2 | 1.317 | Ube3b | −1.437 |
| Cd53 | 1.317 | Mrps21 | −1.437 |
| Entpd2 | 1.316 | Eif4g1 | −1.436 |
| Ermp1 | 1.315 | Hamp | −1.435 |
| Cd86 | 1.315 | Os9 | −1.435 |
| Tapbp | 1.315 | Ganab | −1.434 |
| Cyp2c55 | 1.315 | Mcm10 | −1.432 |
| 2610305D13Rik | 1.314 | Rab43 | −1.432 |
| Ccl4 | 1.314 | Rshl2a | −1.431 |
| 1700047I17Rik1 | 1.314 | Spg20 | −1.431 |
| Snx3 | 1.313 | Josd2 | −1.430 |
| Mcm6 | 1.312 | Cyp2c70 | −1.430 |
| Ccdc120 | 1.310 | Aldh16a1 | −1.428 |
| Slc16a6 | 1.310 | Vkorc1 | −1.428 |
| Nipa1 | 1.308 | Gorasp1 | −1.428 |
| Arl2bp | 1.308 | Dap | −1.427 |
| 1190002N15Rik | 1.308 | Pim3 | −1.426 |
| Cryl1 | 1.308 | Aox3 | −1.425 |
| Litaf | 1.307 | Rps15 | −1.425 |
| Jak1 | 1.306 | Cyp27a1 | −1.425 |
| Cdkn2c | 1.306 | 2310007F21Rik | −1.425 |
| Rhod | 1.306 | Acy1 | −1.424 |
| Bcl2l13 | 1.306 | Mug2 | −1.424 |
| Acot10 | 1.305 | Stk11 | −1.424 |
| Aifm1 | 1.303 | Yif1b | −1.424 |
| Phca | 1.303 | Irf3 | −1.423 |
| Arcn1 | 1.303 | Fbxl10 | −1.423 |
| Esr1 | 1.303 | Rapgef4 | −1.423 |
| Palld | 1.301 | Tm2d2 | −1.422 |
| Ldlr | 1.301 | Serpinf2 | −1.422 |
| Rab8b | 1.300 | Ceacam1 | −1.422 |
| Csnk1g2 | −1.422 | ||
| Hnrpc | −1.421 | ||
| Gpld1 | −1.421 | ||
| Hist1h2bh | −1.421 | ||
| Ssr4 | −1.421 | ||
| Bst2 | −1.421 | ||
| Acox2 | −1.421 | ||
| Sra1 | −1.420 | ||
| Cyp2c37 | −1.420 | ||
| Eif4ebp2 | −1.420 | ||
| Atp13a1 | −1.420 | ||
| Abat | −1.420 | ||
| Per2 | −1.419 | ||
| Polr2f | −1.419 | ||
| Slc1a2 | −1.419 | ||
| Bckdhb | −1.418 | ||
| Itih1 | −1.418 | ||
| Pbld | −1.418 | ||
| Fam134a | −1.417 | ||
| Lgals4 | −1.416 | ||
| LOC100047856 | −1.416 | ||
| LOC100044324 | −1.416 | ||
| 2900010M23Rik | −1.415 | ||
| Rnase4 | −1.415 | ||
| Vtn | −1.415 | ||
| Mrpl17 | −1.414 | ||
| Stat3 | −1.414 | ||
| Ankzf1 | −1.414 | ||
| 5133401N09Rik | −1.414 | ||
| Prpf8 | −1.414 | ||
| Bckdha | −1.413 | ||
| Sirt7 | −1.413 | ||
| C1rl | −1.413 | ||
| Ndufb10 | −1.413 | ||
| EG13909 | −1.413 | ||
| Mug4 | −1.412 | ||
| Gnmt | −1.412 | ||
| Bloc1s1 | −1.411 | ||
| Cuta | −1.411 | ||
| Vrk3 | −1.411 | ||
| Fetub | −1.410 | ||
| Lims2 | −1.409 | ||
| Tm7sf2 | −1.407 | ||
| Gltpd2 | −1.407 | ||
| Ppap2b | −1.407 | ||
| Prei4 | −1.407 | ||
| Arl3 | −1.407 | ||
| A430005L14Rik | −1.406 | ||
| Rpl36a | −1.406 | ||
| Dnajc7 | −1.406 | ||
| Map2k2 | −1.405 | ||
| Dym | −1.405 | ||
| Wdr45l | −1.404 | ||
| Plekhg3 | −1.404 | ||
| Rps21 | −1.404 | ||
| Ghr | −1.403 | ||
| Bmp1 | −1.403 | ||
| Tle1 | −1.403 | ||
| Ppargc1b | −1.402 | ||
| Acad10 | −1.402 | ||
| Rpl12 | −1.402 | ||
| Pnpo | −1.401 | ||
| Ddx3y | −1.401 | ||
| Galt | −1.401 | ||
| Smoc1 | −1.401 | ||
| Cyp27a1 | −1.399 | ||
| Clmn | −1.399 | ||
| 3110056O03Rik | −1.399 | ||
| Tex264 | −1.399 | ||
| Nat6 | −1.398 | ||
| Pla2g12a | −1.397 | ||
| Srm | −1.396 | ||
| LOC100048020 | −1.396 | ||
| Bat3 | −1.396 | ||
| Tsc22d3 | −1.396 | ||
| Mupcdh | −1.396 | ||
| Acat1 | −1.396 | ||
| Cib1 | −1.396 | ||
| Exosc5 | −1.396 | ||
| 1300007L22Rik | −1.396 | ||
| Sort1 | −1.394 | ||
| LOC545056 | −1.394 | ||
| Gtf3c1 | −1.392 | ||
| Myo18a | −1.392 | ||
| LOC100048105 | −1.392 | ||
| Csnk2a2 | −1.391 | ||
| Csnk1g3 | −1.391 | ||
| Serpinc1 | −1.391 | ||
| Mrps28 | −1.391 | ||
| Aamp | −1.391 | ||
| Tha1 | −1.391 | ||
| Aars | −1.390 | ||
| Cope | −1.390 | ||
| Bri3 | −1.390 | ||
| Nme3 | −1.389 | ||
| Ppp1r3b | −1.389 | ||
| Ccdc84 | −1.389 | ||
| Sirt3 | −1.388 | ||
| 1500032D16Rik | −1.388 | ||
| Mrps26 | −1.388 | ||
| Ict1 | −1.387 | ||
| Tpst1 | −1.387 | ||
| Prpf38b | −1.387 | ||
| Als2 | −1.387 | ||
| Klkb1 | −1.387 | ||
| MGC18837 | −1.386 | ||
| Dcxr | −1.386 | ||
| 1700029P11Rik | −1.386 | ||
| Gaa | −1.385 | ||
| 1700012H05Rik | −1.385 | ||
| Gnl3 | −1.385 | ||
| Hdgf | −1.385 | ||
| Aifm1 | −1.385 | ||
| Tcf25 | −1.384 | ||
| Sdc2 | −1.384 | ||
| Mtss1 | −1.384 | ||
| Atf2 | −1.384 | ||
| Cyp2c67 | −1.383 | ||
| Eef2 | −1.383 | ||
| Mrpl2 | −1.383 | ||
| Usp2 | −1.382 | ||
| Timm10 | −1.382 | ||
| Fkbp8 | −1.382 | ||
| 0610012D14Rik | −1.382 | ||
| 3300001P08Rik | −1.382 | ||
| F12 | −1.381 | ||
| 2010100O12Rik | −1.381 | ||
| Slc26a1 | −1.381 | ||
| Paox | −1.380 | ||
| Afmid | −1.380 | ||
| Dpp3 | −1.380 | ||
| Dpm2 | −1.379 | ||
| St3gal3 | −1.378 | ||
| Serpina1a | −1.378 | ||
| 2810428I15Rik | −1.377 | ||
| Akr7a5 | −1.377 | ||
| 6430527G18Rik | −1.377 | ||
| D19Wsu162e | −1.376 | ||
| Phb2 | −1.376 | ||
| Trabd | −1.376 | ||
| Txnl4a | −1.376 | ||
| Macrod1 | −1.376 | ||
| Gamt | −1.375 | ||
| Lgsn | −1.374 | ||
| Atp5g2 | −1.374 | ||
| Jmjd6 | −1.373 | ||
| Cyp27a1 | −1.373 | ||
| Cno | −1.373 | ||
| Naprt1 | −1.372 | ||
| Hpn | −1.372 | ||
| Il1rap | −1.371 | ||
| Rnf6 | −1.370 | ||
| Atp1a1 | −1.370 | ||
| Yeats4 | −1.370 | ||
| Lmf1 | −1.370 | ||
| Bcas3 | −1.370 | ||
| Echdc2 | −1.370 | ||
| Acot12 | −1.370 | ||
| Kng1 | −1.369 | ||
| Hsd17b10 | −1.369 | ||
| Upb1 | −1.369 | ||
| D17Wsu92e | −1.369 | ||
| Taf10 | −1.369 | ||
| Keap1 | −1.368 | ||
| Pdcd5 | −1.368 | ||
| Plekhb1 | −1.368 | ||
| Mthfd1 | −1.368 | ||
| Nr1h4 | −1.367 | ||
| BC031181 | −1.366 | ||
| Fpgs | −1.366 | ||
| Gphn | −1.366 | ||
| Ccar1 | −1.366 | ||
| Stard5 | −1.366 | ||
| Slc25a38 | −1.365 | ||
| Ccdc21 | −1.365 | ||
| Psmc5 | −1.364 | ||
| C130074G19Rik | −1.364 | ||
| 0610007P22Rik | −1.364 | ||
| Dalrd3 | −1.364 | ||
| Mib2 | −1.363 | ||
| Tsc2 | −1.363 | ||
| Sec63 | −1.363 | ||
| Myo6 | −1.362 | ||
| Abtb1 | −1.362 | ||
| 1110008F13Rik | −1.362 | ||
| Tspan33 | −1.362 | ||
| Mettl7b | −1.361 | ||
| LOC100048445 | −1.361 | ||
| BC021381 | −1.361 | ||
| H13 | −1.361 | ||
| Zfp91 | −1.361 | ||
| Arfl4 | −1.360 | ||
| 1810008A18Rik | −1.359 | ||
| Tlcd2 | −1.359 | ||
| Ube2l3 | −1.359 | ||
| 6430706D22Rik | −1.359 | ||
| Prpf6 | −1.359 | ||
| Cebpa | −1.358 | ||
| Tsta3 | −1.358 | ||
| Aspscr1 | −1.358 | ||
| Gphn | −1.357 | ||
| Ccnt1 | −1.357 | ||
| Prox1 | −1.357 | ||
| Dph2 | −1.356 | ||
| Nr1h2 | −1.356 | ||
| Dcxr | −1.355 | ||
| Arg1 | −1.355 | ||
| Per1 | −1.355 | ||
| Cox4i1 | −1.355 | ||
| 1700021F05Rik | −1.354 | ||
| Masp2 | −1.354 | ||
| 9530058B02Rik | −1.354 | ||
| Sf3b5 | −1.353 | ||
| Ctdsp1 | −1.353 | ||
| Akap8l | −1.352 | ||
| Slc37a4 | −1.351 | ||
| Rab18 | −1.351 | ||
| Mrps34 | −1.351 | ||
| Mfsd2 | −1.350 | ||
| Ext2 | −1.350 | ||
| Ttyh2 | −1.350 | ||
| Dnajb2 | −1.350 | ||
| Lsm12 | −1.349 | ||
| Ddx24 | −1.349 | ||
| Tmem201 | −1.349 | ||
| Fh1 | −1.348 | ||
| Cpn1 | −1.348 | ||
| Cxxc1 | −1.348 | ||
| Isy1 | −1.347 | ||
| Srm | −1.347 | ||
| Ythdf1 | −1.347 | ||
| Derl2 | −1.346 | ||
| Csrp2 | −1.346 | ||
| Gnmt | −1.346 | ||
| Mfn1 | −1.346 | ||
| Igfbp4 | −1.345 | ||
| Rnf166 | −1.345 | ||
| LOC100048105 | −1.345 | ||
| 2700038C09Rik | −1.345 | ||
| Herpud1 | −1.345 | ||
| Trfr2 | −1.344 | ||
| BC056474 | −1.343 | ||
| Mon1a | −1.343 | ||
| Itih4 | −1.343 | ||
| Upf1 | −1.342 | ||
| Rpl19 | −1.342 | ||
| Gdi1 | −1.342 | ||
| Echdc2 | −1.341 | ||
| 5730453I16Rik | −1.340 | ||
| Eif3g | −1.340 | ||
| Dgcr2 | −1.340 | ||
| Fbxo34 | −1.340 | ||
| Mett11d1 | −1.340 | ||
| Ngef | −1.340 | ||
| Fastk | −1.340 | ||
| Pex6 | −1.340 | ||
| Dexi | −1.340 | ||
| Bclaf1 | −1.339 | ||
| Use1 | −1.339 | ||
| Zfp607 | −1.338 | ||
| EG545056 | −1.338 | ||
| Ugt2a3 | −1.338 | ||
| Uspl1 | −1.337 | ||
| Cope | −1.337 | ||
| Arrdc2 | −1.337 | ||
| C1rl | −1.336 | ||
| Rabac1 | −1.336 | ||
| Anp32a | −1.336 | ||
| Rilp | −1.336 | ||
| Prr14 | −1.336 | ||
| 620807 | −1.336 | ||
| Limd1 | −1.335 | ||
| Ctsf | −1.335 | ||
| Lemd2 | −1.335 | ||
| Lamp2 | −1.335 | ||
| Cldn3 | −1.335 | ||
| Nol5 | −1.335 | ||
| Man2c1 | −1.334 | ||
| Scarb2 | −1.333 | ||
| Igf1 | −1.333 | ||
| S100a13 | −1.333 | ||
| LOC100047937 | −1.333 | ||
| Zbtb7a | −1.332 | ||
| Ogfod2 | −1.332 | ||
| B3gnt1 | −1.332 | ||
| Zbtb22 | −1.331 | ||
| Atp6v0a1 | −1.331 | ||
| Pnpla2 | −1.331 | ||
| Plg | −1.331 | ||
| Sdhb | −1.330 | ||
| Cdo1 | −1.329 | ||
| Ilvbl | −1.329 | ||
| 6720456B07Rik | −1.329 | ||
| Map1lc3b | −1.329 | ||
| Smarca2 | −1.328 | ||
| Fars2 | −1.328 | ||
| Whdc1 | −1.328 | ||
| 1110032A13Rik | −1.327 | ||
| Dmwd | −1.327 | ||
| Morc3 | −1.327 | ||
| Myg1 | −1.327 | ||
| Scap | −1.327 | ||
| Itfg3 | −1.327 | ||
| 1110007A13Rik | −1.326 | ||
| Cmtm8 | −1.326 | ||
| Wipi2 | −1.326 | ||
| 1110007L15Rik | −1.326 | ||
| Vkorc1 | −1.326 | ||
| Eif3eip | −1.325 | ||
| 1810020D17Rik | −1.325 | ||
| Dexi | −1.325 | ||
| Rpl28 | −1.325 | ||
| Slc6a9 | −1.324 | ||
| Jmjd3 | −1.324 | ||
| 1300001I01Rik | −1.324 | ||
| Cog8 | −1.324 | ||
| Irf3 | −1.324 | ||
| Chmp2a | −1.324 | ||
| D19Bwg1357e | −1.323 | ||
| Itpr2 | −1.323 | ||
| LOC100047935 | −1.323 | ||
| H2-Ke6 | −1.323 | ||
| Mrpl3 | −1.322 | ||
| Mrpl34 | −1.322 | ||
| Slc25a39 | −1.322 | ||
| Spcs3 | −1.321 | ||
| Dhrs4 | −1.321 | ||
| Ppp1r9a | −1.321 | ||
| Nags | −1.321 | ||
| Keap1 | −1.321 | ||
| Cox7a2l | −1.320 | ||
| Mocs1 | −1.320 | ||
| Sap30l | −1.320 | ||
| C630028N24Rik | −1.319 | ||
| Zfand2b | −1.319 | ||
| LOC100045697 | −1.319 | ||
| Pdcd2 | −1.318 | ||
| Yipf3 | −1.318 | ||
| Ctdsp1 | −1.318 | ||
| Mrps9 | −1.318 | ||
| Plg | −1.317 | ||
| Upb1 | −1.317 | ||
| B020018G12Rik | −1.317 | ||
| Il1rap | −1.317 | ||
| Gchfr | −1.316 | ||
| Rab3gap1 | −1.316 | ||
| Slc35e3 | −1.316 | ||
| Rufy3 | −1.316 | ||
| Tmem63b | −1.316 | ||
| Ndufv2 | −1.316 | ||
| Pde4dip | −1.315 | ||
| Avpr1a | −1.315 | ||
| Ogfr | −1.315 | ||
| Tec | −1.314 | ||
| Golga2 | −1.314 | ||
| Acads | −1.314 | ||
| Tnrc6a | −1.314 | ||
| Sbf1 | −1.314 | ||
| Faah | −1.314 | ||
| 1810026J23Rik | −1.314 | ||
| Arl3 | −1.313 | ||
| Tmem14c | −1.313 | ||
| Brms1 | −1.313 | ||
| Qprt | −1.313 | ||
| Atp5d | −1.312 | ||
| Slc2a9 | −1.312 | ||
| Sdc4 | −1.312 | ||
| Eif1b | −1.311 | ||
| Prdx4 | −1.311 | ||
| Dmtf1 | −1.311 | ||
| Il6st | −1.311 | ||
| Tmem204 | −1.311 | ||
| Rnaseh2c | −1.311 | ||
| Aldh1l1 | −1.310 | ||
| Fis1 | −1.310 | ||
| Clcn2 | −1.310 | ||
| Impdh2 | −1.310 | ||
| Cdk8 | −1.309 | ||
| Wdr45 | −1.309 | ||
| Creb3l3 | −1.308 | ||
| Aes | −1.308 | ||
| Riok3 | −1.308 | ||
| Mta2 | −1.308 | ||
| Slc12a2 | −1.308 | ||
| Morf4l1 | −1.307 | ||
| Trpc4ap | −1.307 | ||
| Tmem53 | −1.307 | ||
| 2310044H10Rik | −1.307 | ||
| Snrpd2 | −1.307 | ||
| Cxcl12 | −1.306 | ||
| Lcat | −1.306 | ||
| Depdc6 | −1.306 | ||
| Imp3 | −1.306 | ||
| 2610003J06Rik | −1.306 | ||
| Proc | −1.306 | ||
| Fbxo34 | −1.305 | ||
| Dbp | −1.305 | ||
| Etfb | −1.305 | ||
| Mrpl27 | −1.305 | ||
| Bola2 | −1.305 | ||
| Elof1 | −1.305 | ||
| Cmtm8 | −1.305 | ||
| Enpp1 | −1.305 | ||
| 2410015M20Rik | −1.304 | ||
| Polr1a | −1.304 | ||
| Pih1d1 | −1.304 | ||
| Xrcc6 | −1.304 | ||
| Mbl2 | −1.304 | ||
| Naca | −1.304 | ||
| F12 | −1.304 | ||
| 2310003H01Rik | −1.303 | ||
| Fxyd1 | −1.303 | ||
| Tacc1 | −1.303 | ||
| Gemin4 | −1.303 | ||
| Slc1a2 | −1.303 | ||
| Txn2 | −1.302 | ||
| Gpt2 | −1.302 | ||
| LOC100045782 | −1.302 | ||
| Slc9a3r1 | −1.302 | ||
| Elavl1 | −1.302 | ||
| AA415398 | −1.301 | ||
| Sox5 | −1.301 | ||
| Tmem143 | −1.300 | ||
| Rab8a | −1.300 | ||
| Atg2a | −1.300 |
Supplementary Table 2.
Full List of Significantly Enriched Canonical Pathways and Gene Ontology Categories Modulated in PEDF Knockout Mice Livers
| Up-Regulated Canonical Pathways [DATABASE_PATHWAY NAME] |
||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CP) | ||
| Name | NES | FDR q Value |
| KEGG_GLUTATHIONE_METABOLISM | 2.329 | <.001 |
| PID_INTEGRIN1_PATHWAY | 2.221 | .003 |
| REACTOME_COLLAGEN_FORMATION | 2.168 | .006 |
| REACTOME_GLUTATHIONE_CONJUGATION | 2.136 | .008 |
| REACTOME_NCAM1_INTERACTIONS | 2.089 | .013 |
| PID_SYNDECAN_1_PATHWAY | 2.080 | .013 |
| PID_FOXM1PATHWAY | 2.099 | .014 |
| REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | 2.057 | .016 |
| KEGG_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 | 2.010 | .020 |
| KEGG_ECM_RECEPTOR_INTERACTION | 2.029 | .021 |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 2.013 | .022 |
| PID_AVB3_INTEGRIN_PATHWAY | 1.986 | .024 |
| PID_NFAT_TFPATHWAY | 1.979 | .024 |
| REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | 1.966 | .024 |
| KEGG_DRUG_METABOLISM_CYTOCHROME_P450 | 1.971 | .025 |
| KEGG_CELL_CYCLE | 1.915 | .032 |
| KEGG_DNA_REPLICATION | 1.918 | .033 |
| REACTOME_DNA_STRAND_ELONGATION | 1.923 | .033 |
| PID_TOLL_ENDOGENOUS_PATHWAY | 1.928 | .034 |
| PID_FRA_PATHWAY | 1.879 | .046 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 1.864 | .049 |
| Down-Regulated Canonical Pathways [DATABASE_PATHWAY NAME] |
||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CP) | ||
| Name | NES | FDR q Value |
| REACTOME_PEPTIDE_CHAIN_ELONGATION | −2.878 | <.001 |
| REACTOME_TRANSLATION | −2.873 | <.001 |
| REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | −2.840 | <.001 |
| REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION | −2.827 | <.001 |
| KEGG_RIBOSOME | −2.821 | <.001 |
| REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCT_COMPLEX | −2.797 | <.001 |
| REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | −2.718 | <.001 |
| REACTOME_INFLUENZA_LIFE_CYCLE | −2.571 | <.001 |
| REACTOME_METABOLISM_OF_MRNA | −2.565 | <.001 |
| REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | −2.433 | .005 |
| REACTOME_METABOLISM_OF_RNA | −2.434 | .005 |
| REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX | −2.416 | .005 |
| REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX | −2.419 | .005 |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM | −2.232 | .049 |
| Up-Regulated Gene Ontology Categories |
||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=C5) | ||
| Name | NES | FDR q Value |
| GLUTATHIONE_TRANSFERASE_ACTIVITY | 2.041 | .039 |
| COLLAGEN | 2.049 | .071 |
| CYTOKINE_ACTIVITY | 1.907 | .109 |
| Down-Regulated Gene Ontology Categories |
||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=C5) | ||
| Name | NES | FDR q Value |
| STRUCTURAL_CONSTITUENT_OF_RIBOSOME | −2.790 | <.001 |
| MITOCHONDRIAL_PART | −2.287 | .048 |
| AMINO_ACID_AND_DERIVATIVE_METABOLIC_PROCESS | −2.185 | .050 |
| MITOCHONDRIAL_MATRIX | −2.173 | .051 |
| PROTEASE_INHIBITOR_ACTIVITY | −2.162 | .051 |
| CARBOXYLIC_ACID_METABOLIC_PROCESS | −2.244 | .052 |
| REGULATION_OF_ANGIOGENESIS | −2.198 | .053 |
| NITROGEN_COMPOUND_CATABOLIC_PROCESS | −2.186 | .053 |
| MITOCHONDRION | −2.292 | .054 |
| NITROGEN_COMPOUND_METABOLIC_PROCESS | −2.215 | .054 |
| MITOCHONDRIAL_LUMEN | −2.200 | .056 |
| SERINE_TYPE_ENDOPEPTIDASE_INHIBITOR_ACTIVITY | −2.224 | .056 |
| ANATOMICAL_STRUCTURE_FORMATION | −2.248 | .056 |
| MITOCHONDRIAL_ENVELOPE | −2.023 | .057 |
| ORGANELLE_INNER_MEMBRANE | −2.026 | .059 |
| AMINO_ACID_CATABOLIC_PROCESS | −2.005 | .059 |
| RIBOSOME | −2.031 | .060 |
| MITOCHONDRIAL_RIBOSOME | −2.047 | .060 |
| RIBOSOMAL_SUBUNIT | −2.250 | .062 |
| SPLICEOSOME | −1.977 | .062 |
| DNA_DIRECTED_RNA_POLYMERASEII_HOLOENZYME | −1.939 | .062 |
| MITOCHONDRIAL_MEMBRANE | −2.048 | .063 |
Note: FDR (false-discovery rate), FDR q value; NES, normalized enrichment score.
Supplementary Table 3.
Full List of Chemical and Genetic Perturbations That Were Significantly Enriched in PEDF KO Mice Livers
| Up-Regulated Chemical and Genetic Perturbation Datasets Up-Regulated List Truncated at FDR < .005 | ||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CGP) | ||
| Name | NES | FDR q Value |
| LEE_LIVER_CANCER_ACOX1_UP | 3.117 | .000 |
| LEE_LIVER_CANCER_E2F1_UP | 3.017 | .000 |
| LEE_LIVER_CANCER_MYC_E2F1_UP | 2.984 | .000 |
| LEE_LIVER_CANCER_MYC_TGFA_UP | 2.911 | .000 |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_UP | 2.892 | .000 |
| KHETCHOUMIAN_TRIM24_TARGETS_UP | 2.891 | .000 |
| LEE_LIVER_CANCER_CIPROFIBRATE_UP | 2.858 | .000 |
| LEE_LIVER_CANCER_DENA_UP | 2.780 | .000 |
| WIELAND_UP_BY_HBV_INFECTION | 2.772 | .000 |
| BORLAK_LIVER_CANCER_EGF_UP | 2.742 | .000 |
| SERVITJA_LIVER_HNF1A_TARGETS_UP | 2.742 | .000 |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM3 | 2.697 | .000 |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM2 | 2.635 | .000 |
| HESS_TARGETS_OF_HOXA9_AND_MEIS1_DN | 2.622 | .000 |
| DEMAGALHAES_AGING_UP | 2.605 | .000 |
| POOLA_INVASIVE_BREAST_CANCER_UP | 2.580 | .000 |
| HECKER_IFNB1_TARGETS | 2.516 | .000 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G5_DN | 2.507 | .000 |
| MCLACHLAN_DENTAL_CARIES_DN | 2.476 | .000 |
| LE_EGR2_TARGETS_UP | 2.445 | .000 |
| HOSHIDA_LIVER_CANCER_SUBCLASS_S1 | 2.403 | .000 |
| KIM_GLIS2_TARGETS_UP | 2.390 | .000 |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_D7_UP | 2.385 | .000 |
| ALTEMEIER_RESPONSE_TO_LPS_WITH_MECHANICAL_VENTILATION | 2.374 | .000 |
| JOHANSSON_GLIOMAGENESIS_BY_PDGFB_UP | 2.373 | .000 |
| MCBRYAN_PUBERTAL_TGFB1_TARGETS_DN | 2.366 | .000 |
| STEARMAN_TUMOR_FIELD_EFFECT_UP | 2.361 | .000 |
| BURTON_ADIPOGENESIS_3 | 2.357 | .000 |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_UP | 2.355 | .000 |
| MCBRYAN_PUBERTAL_BREAST_4_5WK_UP | 2.340 | .000 |
| ONDER_CDH1_TARGETS_2_DN | 2.328 | .000 |
| MCBRYAN_PUBERTAL_BREAST_6_7WK_DN | 2.322 | .000 |
| ISHIDA_E2F_TARGETS | 2.297 | .000 |
| KORKOLA_TERATOMA | 2.292 | .000 |
| LIU_VAV3_PROSTATE_CARCINOGENESIS_UP | 2.290 | .000 |
| PASINI_SUZ12_TARGETS_DN | 2.262 | .001 |
| MIKKELSEN_NPC_HCP_WITH_H3K27ME3 | 2.253 | .001 |
| CHANG_CYCLING_GENES | 2.253 | .001 |
| MOSERLE_IFNA_RESPONSE | 2.252 | .001 |
| ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_24HR | 2.250 | .001 |
| BOYLAN_MULTIPLE_MYELOMA_C_D_DN | 2.244 | .001 |
| PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_UP | 2.233 | .001 |
| WENG_POR_TARGETS_LIVER_UP | 2.233 | .001 |
| KANG_DOXORUBICIN_RESISTANCE_UP | 2.232 | .001 |
| ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_UP | 2.231 | .001 |
| MCLACHLAN_DENTAL_CARIES_UP | 2.229 | .001 |
| LENAOUR_DENDRITIC_CELL_MATURATION_UP | 2.226 | .001 |
| BROWN_MYELOID_CELL_DEVELOPMENT_UP | 2.222 | .001 |
| YAGI_AML_FAB_MARKERS | 2.216 | .001 |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_UP | 2.214 | .001 |
| JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP | 2.212 | .001 |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ERYTHROCYTE_UP | 2.209 | .001 |
| YAMASHITA_METHYLATED_IN_PROSTATE_CANCER | 2.195 | .001 |
| STEARMAN_LUNG_CANCER_EARLY_VS_LATE_DN | 2.193 | .001 |
| GAL_LEUKEMIC_STEM_CELL_DN | 2.193 | .001 |
| ODONNELL_TARGETS_OF_MYC_AND_TFRC_DN | 2.188 | .001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_UP | 2.186 | .001 |
| CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP | 2.163 | .001 |
| WALLACE_PROSTATE_CANCER_RACE_UP | 2.163 | .001 |
| WHITFIELD_CELL_CYCLE_LITERATURE | 2.162 | .001 |
| MORI_IMMATURE_B_LYMPHOCYTE_UP | 2.159 | .001 |
| SMID_BREAST_CANCER_LUMINAL_B_DN | 2.159 | .001 |
| CROONQUIST_NRAS_SIGNALING_DN | 2.155 | .001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_DN | 2.154 | .001 |
| CROONQUIST_IL6_DEPRIVATION_DN | 2.151 | .001 |
| DELYS_THYROID_CANCER_UP | 2.149 | .001 |
| MURATA_VIRULENCE_OF_H_PILORI | 2.148 | .001 |
| LI_INDUCED_T_TO_NATURAL_KILLER_UP | 2.144 | .001 |
| SERVITJA_ISLET_HNF1A_TARGETS_UP | 2.131 | .001 |
| HAN_JNK_SINGALING_DN | 2.129 | .001 |
| WIEDERSCHAIN_TARGETS_OF_BMI1_AND_PCGF2 | 2.122 | .001 |
| BERENJENO_ROCK_SIGNALING_NOT_VIA_RHOA_DN | 2.119 | .001 |
| GOLDRATH_ANTIGEN_RESPONSE | 2.119 | .001 |
| ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_6HR | 2.115 | .001 |
| YU_MYC_TARGETS_UP | 2.114 | .001 |
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_UP | 2.113 | .001 |
| RODWELL_AGING_KIDNEY_NO_BLOOD_UP | 2.107 | .001 |
| LIAN_LIPA_TARGETS_3M | 2.097 | .002 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_16D_UP | 2.097 | .002 |
| SWEET_KRAS_TARGETS_UP | 2.096 | .002 |
| TSAI_RESPONSE_TO_RADIATION_THERAPY | 2.094 | .002 |
| MIKKELSEN_MCV6_HCP_WITH_H3K27ME3 | 2.094 | .002 |
| MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_DN | 2.093 | .002 |
| RHODES_UNDIFFERENTIATED_CANCER | 2.092 | .002 |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_CDC25_DN | 2.090 | .002 |
| HAN_JNK_SINGALING_UP | 2.089 | .002 |
| AMIT_SERUM_RESPONSE_40_MCF10A | 2.088 | .002 |
| WORSCHECH_TUMOR_EVASION_AND_TOLEROGENICITY_UP | 2.087 | .002 |
| LIAN_LIPA_TARGETS_6M | 2.086 | .002 |
| AKL_HTLV1_INFECTION_UP | 2.081 | .002 |
| OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_UP | 2.081 | .002 |
| LEE_EARLY_T_LYMPHOCYTE_UP | 2.080 | .002 |
| LABBE_TARGETS_OF_TGFB1_AND_WNT3A_DN | 2.080 | .002 |
| KATSANOU_ELAVL1_TARGETS_UP | 2.079 | .002 |
| VANHARANTA_UTERINE_FIBROID_UP | 2.074 | .002 |
| CHICAS_RB1_TARGETS_GROWING | 2.073 | .002 |
| RODWELL_AGING_KIDNEY_UP | 2.072 | .002 |
| VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_UP | 2.067 | .002 |
| ABRAHAM_ALPC_VS_MULTIPLE_MYELOMA_UP | 2.067 | .002 |
| LIM_MAMMARY_LUMINAL_MATURE_DN | 2.066 | .002 |
| KENNY_CTNNB1_ßTARGETS_UP | 2.050 | .002 |
| BASAKI_YBX1_TARGETS_UP | 2.046 | .002 |
| LIANG_SILENCED_BY_METHYLATION_UP | 2.045 | .002 |
| CAVARD_LIVER_CANCER_MALIGNANT_VS_BENIGN | 2.038 | .003 |
| KEEN_RESPONSE_TO_ROSIGLITAZONE_DN | 2.036 | .003 |
| DAUER_STAT3_TARGETS_DN | 2.035 | .003 |
| KAMMINGA_EZH2_TARGETS | 2.032 | .003 |
| CHANG_IMMORTALIZED_BY_HPV31_DN | 2.031 | .003 |
| KOBAYASHI_EGFR_SIGNALING_24HR_DN | 2.027 | .003 |
| JEON_SMAD6_TARGETS_UP | 2.022 | .003 |
| IGLESIAS_E2F_TARGETS_UP | 2.017 | .004 |
| DAZARD_UV_RESPONSE_CLUSTER_G24 | 2.016 | .004 |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_UP | 2.008 | .004 |
| ROSS_AML_WITH_CBFB_MYH11_FUSION | 2.007 | .004 |
| URS_ADIPOCYTE_DIFFERENTIATION_DN | 2.006 | .004 |
| DAZARD_RESPONSE_TO_UV_SCC_UP | 2.006 | .004 |
| VERHAAK_AML_WITH_NPM1_MUTATED_UP | 2.006 | .004 |
| GOBERT_OLIGODENDROCYTE_DIFFERENTIATION_UP | 2.004 | .004 |
| MEISSNER_BRAIN_HCP_WITH_H3K27ME3 | 2.001 | .004 |
| KAMIKUBO_MYELOID_CEBPA_NETWORK | 1.998 | .004 |
| VERHAAK_GLIOBLASTOMA_NEURAL | 1.997 | .004 |
| LIANG_SILENCED_BY_METHYLATION_2 | 1.992 | .004 |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_DN | 1.989 | .005 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_3D_UP | 1.980 | .005 |
| MCDOWELL_ACUTE_LUNG_INJURY_UP | 1.974 | .005 |
| Down-Regulated Chemical and Genetic Perturbation Data Sets | ||
|---|---|---|
| Database Web Link: (http://www.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CGP) | ||
| Name | NES | FDR q Value |
| HSIAO_LIVER_SPECIFIC_GENES | −2.829 | .000 |
| LEE_LIVER_CANCER_SURVIVAL_UP | −2.743 | .000 |
| OHGUCHI_LIVER_HNF4A_TARGETS_DN | −2.645 | .000 |
| BILANGES_SERUM_AND_RAPAMYCIN_SENSITIVE_GENES | −2.596 | .000 |
| CAIRO_HEPATOBLASTOMA_DN | −2.601 | .000 |
| CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_DN | −2.526 | .001 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G3_DN | −2.529 | .001 |
| CHNG_MULTIPLE_MYELOMA_HYPERPLOID_UP | −2.483 | .003 |
| SU_LIVER | −2.408 | .011 |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM4 | −2.351 | .025 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G123_DN | −2.327 | .028 |
| HOSHIDA_LIVER_CANCER_SUBCLASS_S3 | −2.331 | .030 |
| SERVITJA_LIVER_HNF1A_TARGETS_DN | −2.310 | .030 |
| WOO_LIVER_CANCER_RECURRENCE_DN | −2.314 | .030 |
| CAIRO_LIVER_DEVELOPMENT_DN | −2.279 | .039 |
| LEE_LIVER_CANCER_ACOX1_DN | −2.241 | .051 |
Note: FDR (false-discovery rate) q value: adjusted P value; NES, normalized enrichment score.
References
- 1.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.S., Chu I.S., Heo J. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 3.Hoshida Y., Nijman S.M., Kobayashi M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshida Y., Villanueva A., Kobayashi M. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchiya M., Parker J.S., Kono H. Gene expression in nontumoral liver tissue and recurrence-free survival in hepatitis C virus-positive hepatocellular carcinoma. Mol Cancer. 2010;9:74. doi: 10.1186/1476-4598-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiodoni C., Colombo M.P., Sangaletti S. Matricellular proteins: from homeostasis to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 2010;29:295–307. doi: 10.1007/s10555-010-9221-8. [DOI] [PubMed] [Google Scholar]
- 7.Yasui N., Mori T., Morito D. Dual-site recognition of different extracellular matrix components by anti-angiogenic/neurotrophic serpin, PEDF. Biochemistry. 2003;42:3160–3167. doi: 10.1021/bi0206558. [DOI] [PubMed] [Google Scholar]
- 8.Uehara H., Miyamoto M., Kato K. Expression of pigment epithelium-derived factor decreases liver metastasis and correlates with favorable prognosis for patients with ductal pancreatic adenocarcinoma. Cancer Res. 2004;64:3533–3537. doi: 10.1158/0008-5472.CAN-03-3725. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald D.P., Subramanian P., Deshpande M. Opposing effects of pigment epithelium-derived factor on breast cancer cell versus neuronal survival: implication for brain metastasis and metastasis-induced brain damage. Cancer Res. 2012;72:144–153. doi: 10.1158/0008-5472.CAN-11-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becerra S.P., Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doll J.A., Stellmach V.M., Bouck N.P. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 12.Chung C., Shugrue C., Nagar A. Ethanol exposure depletes hepatic pigment epithelium-derived factor, a novel lipid regulator. Gastroenterology. 2009;136:331–340.e2. doi: 10.1053/j.gastro.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz J.C., Protiva P., Gattu A.K. Pigment epithelium-derived factor regulates early pancreatic fibrotic responses and suppresses the profibrotic cytokine thrombospondin-1. Am J Pathol. 2011;179:2990–2999. doi: 10.1016/j.ajpath.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho T.C., Chen S.L., Shih S.C. Pigment epithelium-derived factor is an intrinsic antifibrosis factor targeting hepatic stellate cells. Am J Pathol. 2010;177:1798–1811. doi: 10.2353/ajpath.2010.091085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grippo P.J., Fitchev P.S., Bentrem D.J. Concurrent PEDF deficiency and Kras mutation induce invasive pancreatic cancer and adipose-rich stroma in mice. Gut. 2012;61:1454–1464. doi: 10.1136/gutjnl-2011-300821. [DOI] [PubMed] [Google Scholar]
- 16.Mejias M., Coch L., Berzigotti A. Antiangiogenic and antifibrogenic activity of pigment epithelium-derived factor (PEDF) in bile duct-ligated portal hypertensive rats. Gut. 2015;64:657–666. doi: 10.1136/gutjnl-2014-307138. [DOI] [PubMed] [Google Scholar]
- 17.Filleur S., Volz K., Nelius T. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 18.Crawford S.E., Stellmach V., Ranalli M. Pigment epithelium-derived factor (PEDF) in neuroblastoma: a multifunctional mediator of Schwann cell antitumor activity. J Cell Sci. 2001;114:4421–4428. doi: 10.1242/jcs.114.24.4421. [DOI] [PubMed] [Google Scholar]
- 19.Park K., Lee K., Zhang B. Identification of a novel inhibitor of the canonical Wnt pathway. Mol Cell Biol. 2011;31:3038–3051. doi: 10.1128/MCB.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tombran-Tink J., Mazuruk K., Rodriguez I.R. Organization, evolutionary conservation, expression and unusual Alu density of the human gene for pigment epithelium-derived factor, a unique neurotrophic serpin. Mol Vis. 1996;2:11. [PubMed] [Google Scholar]
- 21.Uhlen M., Fagerberg L., Hallstrom B.M. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Gattu A.K., Swenson E.S., Iwakiri Y. Determination of mesenchymal stem cell fate by pigment epithelium-derived factor (PEDF) results in increased adiposity and reduced bone mineral content. Faseb J. 2013;27:4384–4394. doi: 10.1096/fj.13-232900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gattu A.K., Birkenfeld A.L., Iwakiri Y. Pigment epithelium-derived factor (PEDF) suppresses IL-1beta-mediated c-Jun N-terminal kinase (JNK) activation to improve hepatocyte insulin signaling. Endocrinology. 2014:en20131785. doi: 10.1210/en.2013-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaral J., Becerra S.P. Effects of human recombinant PEDF protein and PEDF-derived peptide 34-mer on choroidal neovascularization. Invest Ophthalmol Vis Sci. 2010;51:1318–1326. doi: 10.1167/iovs.09-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox G., Kable E., Jones A. 3-dimensional imaging of collagen using second harmonic generation. J Struct Biol. 2003;141:53–62. doi: 10.1016/s1047-8477(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 27.Torres R., Vesuna S., Levene M.J. High-resolution, 2- and 3-dimensional imaging of uncut, unembedded tissue biopsy samples. Arch Pathol Lab Med. 2014;138:395–402. doi: 10.5858/arpa.2013-0094-OA. [DOI] [PubMed] [Google Scholar]
- 28.Mikels A.J., Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairo S., Armengol C., De Reynies A. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Chiang D.Y., Villanueva A., Hoshida Y. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavard C., Terris B., Grimber G. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with beta-catenin mutations. Oncogene. 2006;25:599–608. doi: 10.1038/sj.onc.1208860. [DOI] [PubMed] [Google Scholar]
- 32.Kenny P.A., Enver T., Ashworth A. Receptor and secreted targets of Wnt-1/beta-catenin signalling in mouse mammary epithelial cells. BMC Cancer. 2005;5:3. doi: 10.1186/1471-2407-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merle P., de la Monte S., Kim M. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez M., Semela D., Bruix J. Angiogenesis in liver disease. J Hepatol. 2009;50:604–620. doi: 10.1016/j.jhep.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Nejak-Bowen K.N., Monga S.P. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X.F., Tan X., Zeng G. Conditional beta-catenin loss in mice promotes chemical hepatocarcinogenesis: role of oxidative stress and platelet-derived growth factor receptor alpha/phosphoinositide 3-kinase signaling. Hepatology. 2010;52:954–965. doi: 10.1002/hep.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brack A.S., Conboy M.J., Roy S. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J.H., She H., Han Y.P. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 39.DiRocco D.P., Kobayashi A., Taketo M.M. Wnt4/beta-catenin signaling in medullary kidney myofibroblasts. J Am Soc Nephrol. 2013;24:1399–1412. doi: 10.1681/ASN.2012050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker J., Semler O., Gilissen C. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88:362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F., Song N., Tombran-Tink J., Niyibizi C. Pigment epithelium derived factor suppresses expression of Sost/sclerostin by osteocytes: implication for its role in bone matrix mineralization. J Cell Physiol. 2015;230:1243–1249. doi: 10.1002/jcp.24859. [DOI] [PubMed] [Google Scholar]
- 42.Li F., Song N., Tombran-Tink J., Niyibizi C. Pigment epithelium-derived factor enhances differentiation and mineral deposition of human mesenchymal stem cells. Stem Cells. 2013;31:2714–2723. doi: 10.1002/stem.1505. [DOI] [PubMed] [Google Scholar]
- 43.Cruciat C.M., Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brott B.K., Sokol S.Y. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]