Abstract
The majority of vaccines and several treatments are administered by intramuscular injection. The aim is to engage and activate immune cells, although they are rare in normal skeletal muscle. The phenotype and function of resident as well as infiltrating immune cells in the muscle after injection are largely unknown. While methods for obtaining and characterizing murine muscle cell suspensions have been reported, protocols for nonhuman primates (NHPs) have not been well defined. NHPs comprise important in vivo models for studies of immune cell function due to their high degree of resemblance with humans. In this study, we developed and systematically compared methods to collect vaccine-injected muscle tissue to be processed into single cell suspensions for flow cytometric characterization of immune cells. We found that muscle tissue processed by mechanical disruption alone resulted in significantly lower immune cell yields compared to enzymatic digestion using Liberase. Dendritic cell subsets, monocytes, macrophages, neutrophils, B cells, T cells and NK cells were readily detected in the muscle by the classical human markers. The methods for obtaining skeletal muscle cell suspension established here offer opportunities to increase the understanding of immune responses in the muscle, and provides a basis for defining immediate post-injection vaccine responses in primates.
Keywords: skeletal muscle, nonhuman primate, vaccine administration, flow cytometry
1. Introduction
Normal skeletal muscle contains only a small population of resident immune cells [1-4]. However, during pathophysiological conditions such as contraction or reperfusion-induced insult and injury, endotoxemia or inflammatory myopathies there is a significant infiltration of immune cells [5]. The recruited immune cells play important roles in the regeneration process and resolving the injury or inflammation. Immune cells remove necrotic tissue and secrete soluble factors that contribute to activate muscle satellite cells that differentiate into new muscle cells [6].
Furthermore, several medical treatments are administered by injection into the muscle. The muscle is the most common site for vaccination. Vaccines are intended to target immune cells directly or indirectly but the mechanisms by which immune activation is caused at the site of injection are largely unclear. Inflammatory responses such as the recruitment of immune cells to the site of vaccine delivery are likely central in the initiation of immune responses that subsequently dictate the potency of the vaccine response. There are limitations for performing extensive studies of the presence and function of immune cells in human muscle due to the difficulty of collecting skeletal muscle biopsies. There are few protocols available for obtaining single cell suspensions from human muscle biopsies for the characterization and enumeration of immune cells. Importantly, studies of immune events such as immune cell mobilization to sites injected with vaccines or treatments, definition of target immune cells and degree of inflammation require in vivo studies and cannot be replaced by in vitro model systems. The few in vivo reports that have characterized early immune mechanisms in the muscle after vaccination were performed in mice [7,8]. Rodents and humans differ substantially in their distribution of immune cell populations, phenotype and innate immune responses. In addition, therapeutic doses used in rodents may not be proportionally representative for clinical use. Therefore, nonhuman primates (NHPs) comprise unique in vivo models for immune cell functions. NHP models are therefore commonly used for preclinical and translational studies of vaccines and treatments.
There are numerous publications based on flow cytometric analyses of solid tissues regarding the presence of immune cells and immune activation [9,10]. The accuracy of such analysis is critically dependent on the quality of the cell suspension preparation. It is important to employ methods that allow for isolation and detection of rare and sometimes very delicate cells like infiltrating immune cells to the site inflammation, infection or vaccination. Classical methods for dissociating tissue include enzymatic digestion and manual disaggregation. While tissues such as lymph nodes (LNs) and spleens disaggregate rather easily, firm and tenacious skeletal muscle tissue is more challenging. In this study, we describe strategies to a) define and precisely sample muscle tissue at the injection site of a model vaccine, b) obtain cell suspensions using enzymatic digestion and/or mechanical disruption as well as c) identify and enumerate different immune cells present in the muscle after vaccine injection. The time required for processing, the viability and yields as well as suitability for flow cytometric characterization of isolated immune cell subsets were particularly evaluated.
The protocols defined herein to analyze skeletal muscle tissue from the site of vaccine or treatment delivery will contribute to a greater understanding of the role of immune cells in clinical applications. The methods described are general for obtaining muscle samples and can therefore be applicable for a wide range of investigations.
2. Materials and Methods
2.1. Animals
Approval for this animal study was granted by the Animal Care and Use Committees of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Bethesda, MD, USA. Vietnamese cynomolgus and Indian rhesus macaques recycled from completed studies and scheduled for euthanasia were used in this study. The animals were housed at Bioqual or at NIHAC facility, NIH and handled according to the standards of the American Association for the Accreditation of Laboratory Animal Care.
2. 2. Injections
As a model vaccine in this study we used E1/E3-deleted, replication-incompetent recombinant adenovirus serotype 5 (rAd5) generated in HEK293 cells as previously described [11]. rAd vectors encoding the Zaire Ebola glycoprotein were purified on CsCl gradients and viral titers were determined by measuring the OD at 260nm. A dose of 1 × 1011 vector particles unit suspended in 0.5 ml saline was injected in six different sites (left and right deltoids, quadriceps and calves respectively) using a 1 ml, 25 gauge and 1.6 cm needle (Becton Dickinson, San Jose, CA, USA). The animals were anesthetized with Ketamine and Xylazine (10 mg and 0.5 mg respectively per kg body weight) and injection area were shaved, cleaned with alcohol wipes. The injection site was defined on skin with a permanent marker by encircling an area with 30 mm in diameter. The vaccine was administered at the midpoint of the circle with the needle penetrating 1.5 cm into the muscle in a perpendicular angle without stretching the skin.
2. 3. Sampling of vaccine-injected muscle tissue
Muscle tissue from the injection sites was obtained during necropsy at 72 hrs post-injection. To mark the injection site, a biopsy punch (4 mm, Integra Miltex, Redford, MI, USA) was first pushed through the skin, adipose and connective tissue at the center (needle entry point) of the encircled injection site. After the skin was excised, a puncture wound on the underlying muscle created by the biopsy punch indicated the needle entry point and was used to guide collection of vaccine exposed muscle tissue. An area within 25 mm radius surrounding the puncture wound was considered muscle that was in closest contact with injected vaccine and a cubical piece of muscle tissue of approximately 15 cm3 was dissected with a scalpel, placed in RPMI cell culture media (Gibco, Carlsbad, CA, USA) and stored on ice until processing about 1 hr later.
2.4 Enzymatic digestion and mechanical dissociation
The muscle tissue was weighed and normalized to 3 g by removing adipose tissue, the outermost connective tissue layer (epimysium) and excess muscle tissue farthest away from the punch biopsy wound with scissors. The epimysium and lean muscle were cut separately into 5 mm3 pieces with scissors in petri dish with 10 ml RPMI media. The pieces were drained from excess RPMI media by tilting the dish 45° while gathering the pieces up to the dry part of the tilted dish with forceps. Enzymatic dissociation was done by transferring the drained pieces to 6-well plates (3 g/well) consisting of RPMI media (5 ml/well) supplemented with 0.5 mg/ml DNase (Sigma Aldrich, St. Louis, MO, USA) and 0.25 mg/ml Liberase TL (Roche, Indianapolis, IN, USA), which is a mixture of collagenases, without agitation at +37°C for 2 hrs. In some experiments, Liberase digestion was performed under agitation with orbital shaker set on 12 rpm at +37°C for 1 or 2 hrs as indicated. Epimysium was digested in separate 6-well plates with Liberase (5 ml/well) unless stated otherwise. Liberase activity was inactivated with complete media; RPMI media with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco). The digested pieces of muscle and epimysium were thereafter pooled during filtering through 70 μm cell strainers (BD Bioscience, San Jose, CA, USA) unless otherwise stated. Where indicated, pooled suspension was first passed through a Nitex nylon mesh (500 μm, Wildco, Yulee, FL, USA) before filtering through the cell strainers. Lastly, the cell suspensions were washed at 1500 rpm for 7 min with complete media.
Alternatively, the 5 mm3 pieces cut from the muscle biopsies were mechanically dissociated with the Medimachine system (BD Biosciences) without using enzymatic digestion. Epimysium was discarded and approximately, 3-4 muscle pieces at a time were loaded into chambers with rotatable blades called Medicons (50 μm) together with 1 ml complete media and were subsequently mounted to the Medimachine for pulsing with blade rotation speed of 80 rpm for 1 min. Medicons were rinsed with complete media and both disrupted suspension plus rinse was aspirated with 5 ml syringes, filtered through Filcons (70 μm) and washed with complete media.
To combine both enzymatic digestion and mechanical dissociation, we used a commercial murine skeletal muscle dissociation kit together with the GentleMACS system (Miltenyi Biotec, Auburn, CA, USA). Pieces of muscle and epimysium were transferred to C-tubes and digested together with Miltenyi's enzyme cocktail under agitation at +37°C for 30 min, followed by disruption with GentleMACS dissociator running the muscle dissociation program. These steps were repeated once more before filtration through 70 μm cell strainers and washing with complete media. In other experiments combining digestion and disruption, separated muscle and epimysium were digested side-by-side with Liberase instead of Miltenyi's enzymes for 2 hrs without agitation and Liberase activity was inactivated. The Liberase-digested muscle was then disrupted with GentleMACS. Suspensions from Liberase digestion alone or combined with disruption were thereafter pooled during filtration.
The washed cells from all the dissociation methods were resuspended in complete media according to the number of staining panels.
2.5 Flow cytometry
To stain cells for flow cytometric characterization, 1 ml suspensions for each of four staining panels were transferred to 5 ml polypropylene tubes (BD), washed with 3 ml PBS and resuspended in 100 μL staining volume with PBS. Cells were thereafter blocked with 5 μL FcR-blocking reagent (Miltenyi Biotec), incubated with Aqua dye (Molecular Probes, Eugene, OR, USA) for live/dead staining plus a cocktail of fluorescently labeled antibodies (Abs) (Supplementary table 1) for 20 min at RT. Cells were thereafter washed with PBS and fixed with 1% paraformaldehyde. In one of the staining panels, cells were incubated with the biotinylated Abs alone for 20 min, washed and thereafter added Aqua followed by cocktail of antibodies plus streptavidin-Brilliant Violet 650 (Molecular Probes). At least 1×106 events per sample were acquired using BD LSR II flow cytometer and analyzed using FlowJo software (Tree Star).
2.6 Immunohistochemistry
Cryosections of macaque muscle were fixed with 2% paraformaldehyde, blocked with 1% FBS and permeabilized with 0.2% Saponin (Sigma Aldrich). Endogenous peroxidase, avidin and biotin were blocked with 2% hydrogen peroxide and avidin/biotin blocking kit (Vector Laboratories, Burlingame, CA, USA), before incubation with primary mouse anti-HLA-DR (clone: G46-6) Ab (BD). After washing off primary Ab, sections were blocked with 2% rabbit serum (Dako, Glostrup, Denmark) and incubated with biotinylated rabbit-anti mouse Abs (Dako). Staining was visualized by Vectastain Elite ABC standard kit together with 3,3’-diaminobenzidine substrate kit (Vector Laboratories) and counterstaining was done with hematoxylin (Histolab, Gothenburg, Sweden). Images were taken using a Leica DMR-X microscope (Leica Microsystems, Wetzlar, Germany).
2.7 Statistical analyses
Statistical analyses were performed using Student's paired or unpaired t-test with Graph Pad Prism software and considered significant at * for p≤0.05, ** for p≤0.01 and *** for p≤0.001.
3. Results
3.1. Precise sampling of muscle tissue is essential for accurate assessment of immune cells
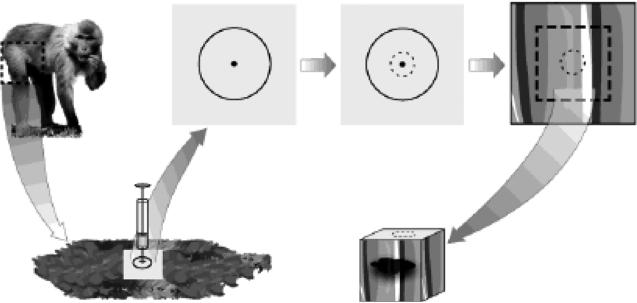
In this study, in order to perform a comprehensive comparison of different dissociation strategies side-by-side and characterization of immune cell subsets large pieces of muscle tissues were needed which prompted for tissue sampling from deltoids, quadriceps and calves during necropsy. However, it is important to note that human, and presumably also macaque, skeletal muscle biopsies with sufficient size for flow cytometric analysis can be obtained with needle biopsy [12] or during surgery [13,14]. In prior mouse studies, the entire quadriceps muscles were collected after antigen injection and euthanasia [7,8,15]. Compared to mice, macaques have considerably larger muscle tissues and it is therefore crucial to only sample the tissue at the site of vaccine/drug delivery to accurately assess the magnitude and phenotype of infiltrating and/or responding immune cells. To be as precise as possible, we carefully marked the injection site on a shaved skin area without stretching the skin before administration of the model vaccine. The injection was done in an upright position to achieve as even distribution of the vaccine formulation as possible within the marked injection site. However, the elasticity of macaque skin makes it difficult for exact sampling of the injected muscle tissue since the marked area easily dislocates from its original position. Therefore, it was critical at the time of muscle sampling that the animal was placed in the same position as when the skin marking and injection were performed. Furthermore, the injection site in the underlying muscle tissue needed to first be identified. To enable this, a biopsy punch was pressed through the skin, adipose and connective tissue at the center of the encircled injection site, where injection of the vaccine occurred (Fig 1). A sharp biopsy punch was essential in order to pass through the tissue layers with minimal moving or stretching of the skin. Using new biopsy punches are therefore recommended for each procedure. After the punch was performed, the skin at the injection area was excised. A readily visible superficial puncture wound on the underlying muscle guided the location of the injection needle entry and thereby the collection of muscle tissue that had the closest exposure to the vaccine. Muscle tissue with an approximate volume of 15 cm3 was dissected.
FIGURE 1. Marking of the injection site for precise sampling.
Schema shows the marking and sample procedure. The injection site was marked on shaved skin with a permanent marker encircling an area of 30 mm in diameter with the center as the needle entry point. The needle was administered in a 90° angle. At the time of muscle sampling, a 4 mm biopsy punch was applied at the center of the marked skin area as depicted by the dotted circle. The puncture wound on the underlying muscle was used as guide to sample the surrounding vaccine exposed muscle tissue by dissecting out a cube-shaped piece of approximately 15 cm3.
3.2 Preparation of muscle tissue prior comparison of dissociation strategies and conditions
The dissected muscles were stored in serum free RPMI media prior processing as serum can quench digestion enzyme activity. The muscle samples were weighed and adjusted using sharp scissors to 3 g lean muscle tissue by removal of adipose tissue, the outermost connective tissue layers (epimysium) plus excess muscle tissue farthest away from the puncture wound (Fig 2 a-b). The muscle tissue was further shredded into smaller pieces (approximately 5 mm3) with scissors (Fig 2 c). The pieces were drained from excess media to prevent dilution of the digestion enzyme concentration or the serum content in complete media used with Medimachine.
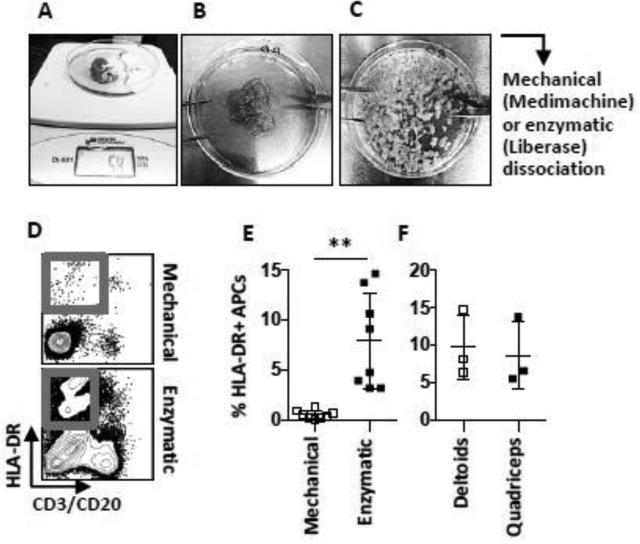
FIGURE 2. Shredding and processing of muscle biopsies into cell suspensions.
Pictures show (A) a collected muscle biopsy on the scale to be normalized for weight and (B) the biopsy after removal of adipose tissue and connective tissue layer (epimysium) plus excess muscle tissue. (C) Muscle pieces of approximately 5 mm3 ready to be further dissociated to obtain single cell suspension. (D) Flow cytometric analysis of cells processed by mechanical (Medimachine) vs. enzymatic (Liberase) dissociation. (E) Graph shows compiled data of the percentages of HLA-DR+ CD3−, CD20− APCs out of all live cells obtained with the different methods and (F) the same population in Liberase-digested muscle obtained from different sites collected from cynomolgus macaques. Mean ± SD is shown. n≤8. **p≤0.01
3.3. Enzymatic dissociation gives higher cell yields compared to mechanical disruption
Enzymatic digestion using different collagenases is a common method to dissociate tissues into single cell suspensions. Muscle is a dense tissue that requires quite rigorous processing to obtain single cell suspensions. There are no commercially available enzymes primarily aimed for primate muscle tissue processing. In this study, we initially tested different collagenases (not shown) and found that Liberase, which is a mix of several collagenases that has been used for various tissues [16,17], was the most efficient for processing of macaque muscle. Therefore, the subsequent experiments were performed using Liberase unless otherwise stated. While enzymatic digestion is known to result in relatively good cell yields, the incubation times required to fully dissociate tissues can be lengthy, and could potentially affect cell viability. To evaluate whether enzymatic digestion could be excluded without compromising on the cell yields, we compared the processing of muscle tissue with Liberase dissociation and with non-enzymatic mechanical disruption. For the mechanical disruption we used the Medimachine system, which has been developed for various solid tissues from both humans [18,19] and mice [20]. For the side-by-side comparison performed here, shredded muscle was divided in two parts where each part equaled about 1.5 g solid tissue. While half of the tissue was being digested with Liberase for 2 hrs, the other half was processed by the Medimachine protocol. For this, the tissue pieces were loaded to Medicon chambers and first pulsed for 10 sec. The time of pulsing was then increased gradually until the dissociation was considered complete after 60 sec when the majority of the tissue was disrupted. We found that neither increasing the pulsing for up to 4 min nor additional pulsing of the semi-disrupted suspensions in new Medicons resulted in more homogenized suspensions. Further, cutting the muscle into even smaller pieces (< 5 mm3) did not result in increased disruption efficiency.
On average, three cell strainers (70 μm) were required to separate the bulk muscle debris from the single cells in suspension obtained by Liberase digestion, while more than five Filcon filters (70 μm) were needed for the suspension generated by the Medimachine. To ensure minimal contamination of muscle debris, the cells were passed through the respective filters by gravity alone without agitating or aspirating the suspension. The filtration step was laborious and time-consuming but was essential to ensure as few contaminating muscle debris as possible in the cell yield. In some separate experiments, the Liberase-digested suspension was first filtered through Nitex nylon mesh prior using the cell strainers, which has been reported for filtration of rat muscle suspension [21]. The pre-filtration with Nitex mesh (500 μm) that has around seven times larger mesh size compared to cell strainers (70 μm), eliminated most debris from the suspension and the subsequent filtration was therefore accelerated. Compared to filtration with cell strainers only, the pre-filtration with Nitex mesh did not significantly alter the frequencies of different cell subsets, expect for higher levels of NKG2A+ NK cell (p <0.01) (Fig S1).
We thereafter evaluated the immune cells in the cell suspensions obtained from the different dissociation strategies. By flow cytometry, we collected equal amounts of events from the cell suspensions and analyzed the proportion of HLA-DR (MHC class II)+ antigen presenting cells (APCs) such as dendritic cells (DCs) and monocytes out of the total live (Aqua-) cells in the suspensions (Fig 2 d). Significantly higher proportions of HLA-DR+ APCs were found in the suspensions digested with Liberase compared to donor-matched suspensions processed with mechanical disruption with Medimachine (p<0.01) (Fig 2 e). This was also observed for T cells and B cells detected by CD3 and CD20 staining. A major population that did not express HLA-DR or CD3/CD20 was observed in all suspensions, which likely represents tissue debris (Fig 2 d). Small numbers of neutrophils were also found within this population after processing of the tissue using Liberase digestion (Fig 4 b). Using the mechanical disruption only, the HLA-DR− and/or CD3/CD20− populations were more frequent than immune cells. Thus, while the Medimachine system is reported to be suitable for efficient dissociation of various solid tissues, we found that this system was not sufficient for processing of macaque skeletal muscle to specifically characterize immune cells. We also analyzed if there were any differences in cell yields depending on if the muscle biopsies were collected from the deltoids or quadriceps muscle. Using Liberase dissociation, we found no significant difference in the proportions of HLA-DR+ APCs between the two sites (Fig 2 f). Finally, we verified by immunohistochemistry that HLA-DR+ cells were present in sections of some muscle biopsies collected from the same animals (Fig S2).
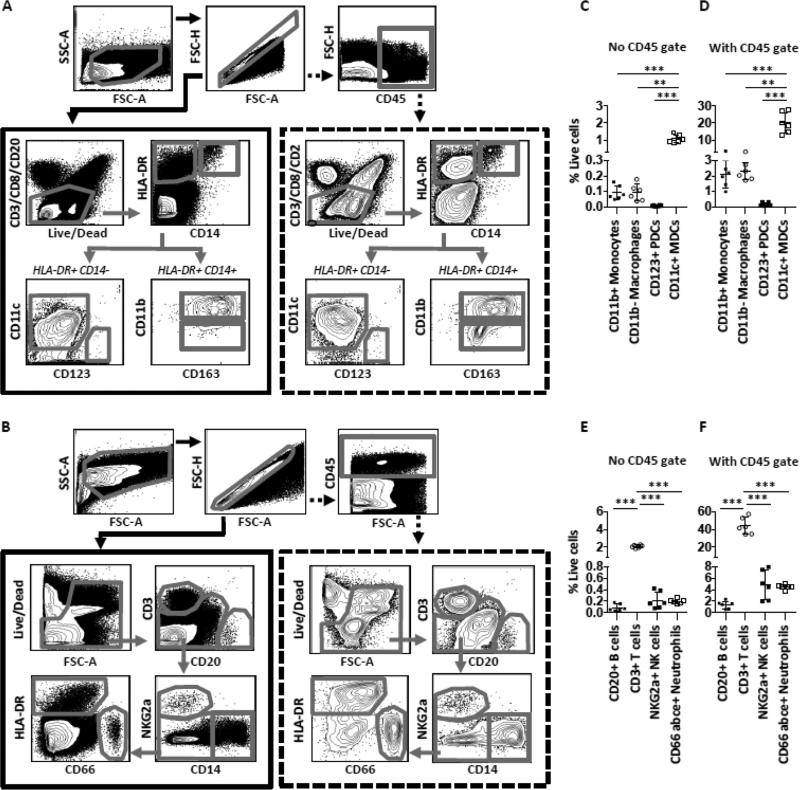
FIGURE 4. Identification of specific immune subsets in the muscle using CD45.
(A-B) Gating strategies for flow cytometric analysis of immune cell populations in the muscle without using (solid lined box), or using (dotted lined box) the CD45 expression as inclusion criteria for immune cells. (C-F) Compiled data of antigen presenting cell populations (C-D) and lymphocyte populations plus neutrophils (E-F) in the muscle without or with CD45 gating strategy. n=6 rhesus macaques. Mean ± SD is shown. *p≤0.05, **p≤0.01, ***p≤0.001.
3.4. Connective tissue contain significant amounts of antigen presenting cells
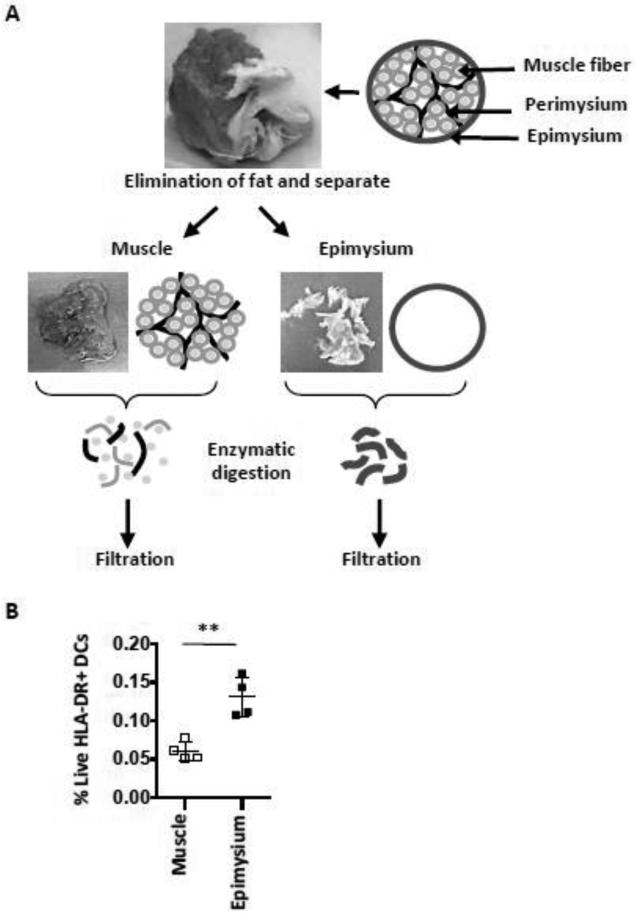
Resident macrophages in interstitial spaces between connective tissues such as epimysium and perimysium surrounding the entire muscle and muscle fascicles, respectively, have been reported to play a critical role during the early stages of acute muscle injury partly by producing chemokines that attract immune cells into the tissue and through supporting differentiation of inflammatory DCs [5,22,23]. We next therefore examined whether the addition of processed epimysium significantly added cell yields, in particular monocytes/macrophages, to the cell suspension. In our experiments, the epimysium was first removed from the muscle tissue at the muscle-epimysium junction. Clean and sharp scissors were essential for both detaching the epimysium from the muscle as well as to efficiently cut both the firm muscle tissue and sinewy epimysium into smaller pieces. Small amount of muscle tissue attached to the separated epimysium was inevitable since these tissues are firmly adhered to each other. However, tearing or peeling off the epimysium should be avoided since this result in more attached muscle tissue. The perimysium between bundles of muscle fibers was not removable with the tools used in the study. Initially, we tried to mechanically disrupt the separated epimysium using the Medimachine. However, the epimysium resulted in being wrapped around the blades, which compromised the blade rotation. The Medimachine dissociation procedure was thereby considered unsuitable for epimysium. In subsequent experiments Liberase digestion only was used unless stated otherwise. Separated epimysium and muscle tissue was digested with Liberase side-by-side (Fig 3 a). We found significantly higher amounts of HLA-DR+ CD14− DCs (p<0.01) in suspensions from the epimysium compared to suspensions from muscle (Fig 3 b). The inclusion of epimysium is therefore an important consideration when studying APCs in the muscle.
FIGURE 3. Inclusion of the epimysium in the muscle processing.
(A) The schema shows the procedure for processing of muscle and epimysium tissue. The cartoon illustrates how the epimysium surrounds the muscle. After the adipose tissue was removed, the epimysium was separated and processed separately alongside the muscle tissue.
(B) Graph shows compiled data of percentages of HLA-DR+ DCs out of all live cells obtained in muscle and epimysium, respectively. n=4 rhesus macaques. Mean ± SD is shown. **p≤0.01.
3.5. Gating strategies with or without CD45 result in similar proportions on immune cells
We thereafter expanded the studies by analyzing several immune cell subsets such as DCs, monocytes, macrophages, neutrophils, NK cells, T cells and B cells using classical markers compatible for both macaque and human cells. Muscle tissue comprises of residing nonhematopoietic cells such as myocytes, pericytes, satellite cells, nerve cells, fibroblasts and endothelial cells. Staining for CD45 can help distinguish bone marrow-derived leukocytes from the non-hematopoietic cells. From a technical aspect, designing staining panels for extensive multiparametric flow cytometric analysis on small tissue samples such as muscle biopsies can be challenging due to restrictions in available fluorescence detectors and/or availability of commercial Abs labeled with the preferred fluorophores. It is therefore important to carefully consider the most critical Abs necessary for the analyses. We therefore analyzed the levels of multiple cell subsets in cell suspensions from muscle and epimysium obtained by Liberase treatment and evaluated the need for using CD45 expression in the identification of immune cells. Ab panels were designed to enable simultaneous analyses of the presence of CD11c+ (BDCA-1+ and CD16+) myeloid DCs (MDCs), CD123+ plasmacytoid DCs (PDCs), CD14+ monocytes/macrophages, CD66abce+ neutrophils, NKG2a+ NK cells and CD3+ (CD4 and CD8) T cells (Fig 4 a-b). We found that the percentages of the different immune cell subsets out of total live cells in the cell suspensions were higher when the cells were first selected based on the expression of CD45 (Fig 4 c-f). A considerable proportion of cells in the suspensions therefore consisted of residual nonhematopoietic cells and/or cell debris of such cells as well as immune cells that potentially lost the integrity of their surface markers during tissue processing. Most immune cell populations in the suspensions were enriched by 20-fold when presented within the total CD45+ population as compared to when they were presented without using CD45 gating. However, the proportion of the different immune cells in relation to each other was the same regardless of if CD45 gating was used (Fig 4 c-f). For the most part, the immune cell lineage markers were not expressed on CD45− cells. However, a minor population of CD45− cells showed low expression of HLA-DR and CD11c as also reported earlier in muscle biopsies [24-27]. Autofluorescent cells could contribute to the numbers of HLA-DR+ and CD11c+ cells but this was negligible according to unstained samples (data not shown). However, we found no expression BDCA-1 or CD16 on these cells, which are critical for identification of these MDC subsets (Fig S3 a). There were also low levels of CD3, CD4 and CD8 in the CD45− population (Fig S3 b). Nevertheless, we cannot exclude potential loss of CD45 integrity and/or downregulated CD45 expression by immune cells due to tissue processing and dissociation as well as autofluorescence on cells. We found similar statistically significant differences between populations using either gating strategy, which further solidifies that irrespective of CD45 gating, the hierarchy of the immune cell proportions was the same. These results suggest that inclusion of a CD45 marker in muscle suspensions is not critical to identify specific immune cells but is beneficial to avoid dilution of the immune cell frequencies by non-hematopoetic cells or cell debris. Nonetheless, it is crucial to consistently estimate the immune cells either with or without staining of their CD45 expression throughout a study to enable direct comparison of the cell numbers.
3.6. Optimization of immune cell yields by different conditions for enzymatic dissociation
As shown above, enzymatic digestion by Liberase was found to be superior over mechanical digestion for dissociating macaque muscle. We next evaluated whether the Liberase treatment method could be further refined. We again performed side-by-side comparisons of Liberase digestion under different conditions. We tested whether agitation during incubation with Liberase would increase the yield of immune cells. Pieces of muscle representing approximately 3 g and epimysium were digested separately with Liberase with or without agitation for 1 or 2 hrs before pooling of respective suspensions during filtration with cell strainers. We also investigated if there was any benefit with combining enzymatic and mechanical dissociation. Since the Medimachine system alone was found to be suboptimal for disrupting muscle tissue and epimysium in our earlier experiments, we instead tested a commercially available dissociation kit together with a disruption system (Miltenyi Biotec) that is faster and designed specifically for murine skeletal muscle. Briefly, the dissociation protocol includes repeated incubations of pieces of muscle tissue with a cocktail of enzymes under agitation at +37°C followed by mechanical disruption in containers equipped with rotating blades (C-tubes) run by the GentleMACS dissociator. Separated epimysium and muscle from 1 g tissue were transferred to the same C-tube where they were digested and disrupted together. The reason for the lower amount of muscle tissue processed in Miltenyi's protocol was because each C-tube is recommended for dissociating a maximum of 1 g of muscle tissue. The duration of the enzymatic digestion and pulsing steps were based on the manufacturer's protocol and the suspensions were filtered with cell strainers. In addition, since we had found that Liberase was efficient for macaque muscle, we also examined whether Liberase could replace the enzyme mixture included in the kit (Table 1). Shredded muscle representing 3 g of tissue and epimysium were Liberase-digested side-by-side without agitation for 2 hrs. Only Liberase-digested muscle was disrupted with GentleMACS because of the weight limit for the processing capacity for C-tubes. However, since we found that Liberase had shown such efficient digestion capacity, disruption of up to 3 g Liberase-digested muscle per C-tube was feasible. Therefore, epimysium digested with Liberase was directly pooled with GentleMACS disrupted muscle during filtration. To this end, we found no difference in cell subset frequencies when muscle and epimysium were disrupted in separate C-tubes after side-by-side Liberase digestion compared to when only muscle suspension was disrupted (data not shown).
Table 1.
Enzymatic digestion alone or combined with mechanical disruption of tissues
| Technical Steps | L-1hr-Ag* | L-2hr-Ag† | L-2hr‡ | L-2hr C§ | M¶ |
|---|---|---|---|---|---|
| Muscle weight (g) | 3 | 3 | 3 | 3 | 1 |
| Epimysium | Yes | Yes | Yes | Yes | Yes |
| Enzymatic | Liberase | Liberase | Liberase | Liberase | Miltenyi Cocktail |
| Mechanical | No | No | No | C-tubes | C-tubes |
| Agitation | Yes | Yes | No | No | Yes |
| Incubation (min) | 60 | 120 | 120 | 120 | 2 × 30 |
| Enzyme quench | Yes | Yes | Yes | Yes | No |
| Filtration (70 μm) | Yes | Yes | Yes | Yes | Yes |
| Total time (hrs) | 4:30 | 5:30 | 5:30 | 5:40 | 4:00 |
L-1hr-Ag: Liberase digestion of separated muscle and epimysium side-by-side for 1 hr, with agitation
L-2hr-Ag: Liberase digestion of separated muscle and epimysium side-by-side for 2 hrs, with agitation
L-2hr : Liberase digestion of separated muscle and epimysium side-by-side for 2 hrs, no agitation
L-2hr C : Liberase digestion of separated muscle and epimysium as in L-2hr, followed by disruption of muscle in C-tubes
M: Miltenyi enzyme digestion of separated muscle and epimysium combined in C-tubes followed by disruption in C-tubes
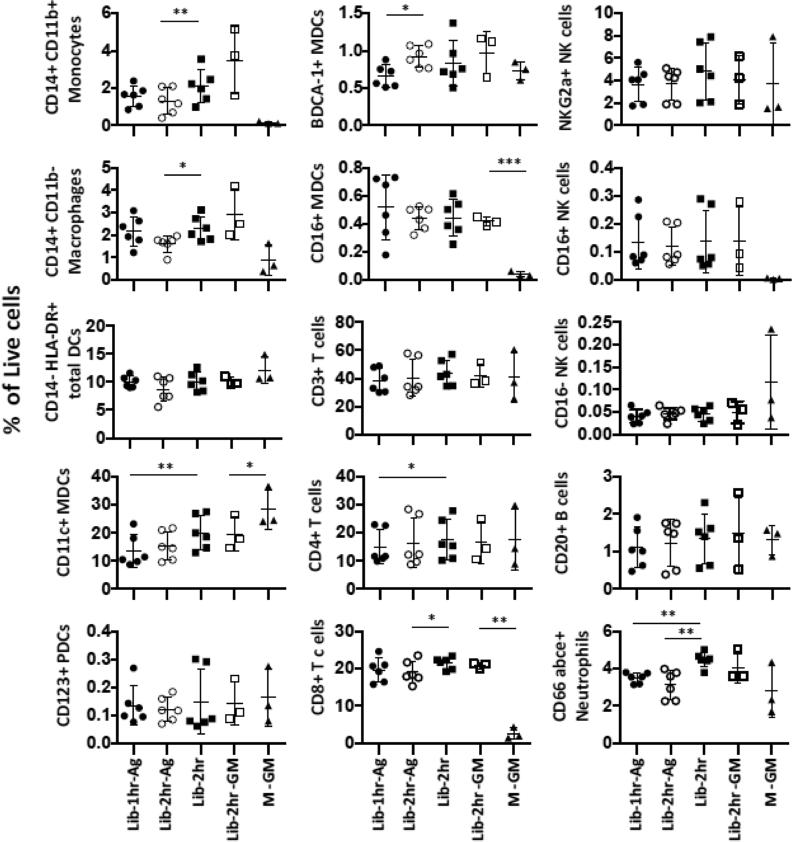
We found that Liberase 2 hrs with agitation has the highest yield of cells acquired per gram of muscle (2.52 × 106 cells) (Table 2). However, Liberase without agitation resulted in the highest number of live cells per gram of muscle (7.32 × 105 cells). When compared to the other protocols, Miltenyi did not performed as well (1.27 × 106 events and 4.98 × 105 live cells). The ratios between the total events acquired and the number of live cells in Liberase with agitation (1 hr or 2 hrs) are the highest (3.76 and 3.87). In addition, the ratios of Liberase 2 hrs without agitation and Miltenyi's protocol are similar (2.9 and 2.6). The suspensions obtained with the different conditions were stained with extended panels to allow further enumeration of specific immune subsets (see Fig S2 for gating strategies). The cell subsets were identified based on the CD45+ population. We found that agitation during Liberase digestion did not result in significant higher yields of the different immune cell subsets compared to the original protocol without agitation (Fig 5). In fact, Liberase digestion without agitation yielded higher frequencies of, CD14+ CD11b+ monocytes, CD14+ CD11b− macrophages, CD11c+ total MDCs, CD4+ and CD8+ T cells as well as CD66 abce+ neutrophils compared to digestion under agitation. The combination of Miltenyi's enzymatic and Miltenyi's mechanical dissociation resulted in cleavage and/or downregulation of some markers defining specific cell subsets. More specifically the frequencies of monocytes, macrophages, CD16+ NK cells and CD8+ T cells were substantially lower compared to the other dissociation conditions. The decreased frequencies were not due to the disruption by GentleMACS itself, since the tissues digested with Liberase prior to the GentleMACS disruption procedure resulted in similar level of immune cells as the suspensions obtained from Liberase digestion alone. The levels of CD11c+ MDCs were significantly higher when both Miltenyi's enzymes plus GentleMACs were used, and it is unlikely that Miltenyi's protocol would preferentially enrich for this subset only. It is also unlikely that this protocol would favor isolation of CD4+ over CD8+ T cells and BDCA-1+ over CD16+ MDCs when the frequencies of total CD3+ T cells and CD11c+ MDCs respectively, were in levels similar or higher than those found with the other conditions. We therefore concluded that Miltenyi's enzyme cocktail interfered with certain cell surface markers rather than enriched for subsets of immune cells.
Table 2.
Yield and viability in suspensions obtained by different dissociation conditions
| Biological Criteria | L-1hr-Ag | L-2hr-Ag | L-2hr | L-2hr C | M |
|---|---|---|---|---|---|
| Samples (n) | 6 | 6 | 6 | 3 | 3 |
| Total acquired events per g muscle | 2.20×106 cells ± 0.30×106 | 2.52×106 cells ± 0.08×106 | 2.14×106 cells ± 0.41×106 | 2.28×106 cells ± 0.22×106 | 1.27×106 cells ± 0.28×106 |
| No. live cells related to acquired events per g muscle | 5.85×105 ± 1.5×105 | 6.51×105 ± 1.6×105 | 7.32×105 ± 1.0×105 | 6.90×105 ± 1.4×105 | 4.98×105 ± 2.5×105 |
| 3.76 | 3.87 | 2.9 | 3.3 | 2.6 |
Total events per g muscle = Sum of total events from each stained cell suspension aliquot / Normalized tissue weight
No. live cells per g muscle = Sum of Aqua- cell counts from each stained cell suspension aliquot / Normalized tissue weight
FIGURE 5. Proportions of immune subsets obtained by different enzymatic dissociation conditions.
The representation of the different immune cells as depicted by the mean percentages ± SD out of all live cells is plotted. The flow cytometry analysis was performed as previously described (Fig 4) and CD45 gating was utilized. n ≥3 side-by-side experiments using rhesus macaques. Lib-1hr-Ag: Liberase treatment for 1 hr with agitation, Lib-2hr-Ag: Liberase treatment for 2 hrs with agitation, Lib-2hr: Liberase treatment for 2 hrs without agitation, Lib-2hr-GM: Liberase treatment for 2 hrs without agitation followed by mechanical disruption with gentleMACS dissociator, M-GM: treatment using Miltenyi's enzyme mix for 2 × 30 min followed by 2 × mechanical disruption.
4. Discussion
Although only a small population of immune cells resides in normal skeletal muscle, it is well known that physiological imbalance including injury, inflammation and infection lead to significant recruitment of immune cells [5]. Infiltrating immune cells create a local inflammation that is part of the muscle repair process and regeneration. In addition, the immune cells are central in initiating immune responses to antigen exposure of the muscle. To this end, vaccine delivery into the muscle is an inefficient process without some degree of inflammation, which presumably is needed to induce recruitment of immune cells into the muscle. While live attenuated vaccine formulations are inherently immunogenic, non-live vaccine formulations often require the addition of an immune-stimulatory adjuvant to induce sufficient innate activation and subsequent vaccine responses. Most vaccines are administered into the muscle but the innate immune activation occurring at the site of injection that likely directs the ensuing vaccine response is largely unclear. Also, the initial target cells for different vaccines are unknown yet represent a fundament in vaccinology. In vitro cell culture systems may be suitable to study aspects of how vaccine components interact with and induce immune cell activation. However, studying functions such as cell mobilization to the site of vaccine administration, defining the local target cells and type of inflammation require in vivo studies. Methods to facilitate studies of the presence and function of immune cells in skeletal muscles are therefore needed for a better understanding of these events. As there is a high degree of resemblance of the immune system between humans and NHPs, we utilized macaque skeletal muscle in this study to develop and evaluate methods for processing and characterizing immune cells by flow cytometry. Earlier studies have mainly reported on immune cell preparation from mouse muscle [7,8,15,28] or isolation of non-hematopoietic cells in human muscle [12-14]. Characterization using in situ imaging on tissue sections has so far been the most common method for evaluating human muscle [29,30]. However, obtaining reproducible data using in situ analyses may require evaluation of multiple sections and simultaneous staining for several markers. In addition, the characteristic morphology of skeletal muscle consisting of the long cylindrical multinucleated myofibers is quite challenging for in situ analyses of rare immune cells located between the fibers (Fig S2). Flow cytometry enables multiparametric analysis of scarce cells.
In this study, to induce mobilization of immune cells into the muscle, the animals received an intramuscular injection of a preclinical vaccine based on a well-characterized adenoviral serotype 5 vector that we and others have used in multiple prior studies [31-33]. The choice of model vaccine was further motivated by that infiltration of immune cells as well as presence of antigen expressed by recombinant adenoviral vectors have previously been reported in human muscle biopsies using immunohistochemistry [34]. We chose to sample muscle tissue at 72 hrs after vaccine administration based on the result of prior mouse studies where significant immune cell infiltration and antigen expression in response to potent vaccine adjuvants was found around this time point [7,8,35]. Also, immune cell infiltration in response to plasmid DNA vaccines has also been shown in mice [36,37] and NHPs [38].
In our study, emphasis was placed on accurate sampling of the muscle tissue that was in closest proximity to the injection site and therefore most likely had been exposed to the vaccine. Imprecise sampling could result in false low numbers of immune cells, incorrect proportions or phenotypes. Further, processing dense skeletal muscle requires shredding the tissue into small pieces, which is quite laborious. However, quite large muscle biopsies had to be processed for the purpose of this study and conventional needle biopsies would be much more manageable. We also found that enzymatic Liberase digestion for isolation of immune cells from the muscle was superior over mechanical disruption using two different commercially available techniques (Medimachine and GentleMACS) with regard to obtaining suspensions with high proportion and yields of immune cells as well as preserved phenotypes. Thus, while the Medimachine and GentleMACS are highly efficient for processing other types of tissues [18-20,39], they were not found to be optimal for macaque skeletal muscle.
The phenotypes of the immune cells isolated with the Liberase digestion method were comparable to what we have found on macaque PBMC [40,41]. Representative immune cells were detected in the muscle including DCs, monocytes, neutrophils, B cells, T cells and NK cells using the classic cell specific markers used for characterization of leucocytes in blood. With both Liberase and mechanical dissociation of muscle, we found a considerable population in the cell suspensions that did not appear to be immune cells i.e. cells that did not express CD45 or any of the immune cell lineage specific markers. We believe these cells are predominately cell debris presumably from both non-hematopoietic muscle resident cells and hematopoietic cells. Further detail characterization would be needed to elucidate the origin of these events, which was outside the focus in this study. Performing a ficoll density gradient centrifugation may lead to further enrichment of intact immune cells from debris as we and others have observed for other tissues [42-44]. However, since cell suspensions collected from muscle biopsies are rather small we preferred not to risk losing cells by performing additional procedures as well as losing specific immune cells such as neutrophils that are discarded by such centrifugation. We instead focused on characterizing all the immune cell types obtained and whether their identification required initial CD45 gating. We found that inclusion of an anti-CD45 Ab in the staining panel was not critical to identify specific immune cells but it was beneficial to enrich for the immune cell frequencies out of the nonhematopoetic cells or cell debris. On this note, the viability dye Aqua used in this study did not label a large proportion of the cell yield and hence the events representing tissue debris were likely included in our live cell analyses. We suspect that the Aqua negative population were not uniformly consisted of live cells since bright staining of dead cells require presence of cytoplasm. During shredding of the tissue, large muscle cells that can be up to 4 cm in size, likely lost their cytoplasm and thereby not optimally labeled. Therefore, finding a viability dye better at identifying cell debris could result in more accurate results. Nonetheless, it is crucial to note that an analysis has to be consistent throughout a study otherwise cell numbers in suspensions cannot be directly compared.
In conclusion, here we report an extensive comparison of strategies to sample macaque skeletal muscle and obtain cell suspensions using commercially available reagents. Multiparametric flow cytometric protocols including Abs for markers suitable for analyzing multiple immune cell subsets in muscle are also presented. The methods offer opportunities to increase the understanding of immune responses in the muscle as well as the mechanisms of vaccine delivery or other types of applications. Investigations of vaccine or drug exposure of cells in vivo while they are in their tissue environment may generate far more physiologically relevant data than in vitro modeling.
Supplementary Material
Highlights.
High cell yields were obtained from enzymatic dissociated macaque skeletal muscle injected with adenoviral vaccine vector
Multiple immune cell subsets were identified in single cell suspension of macaque skeletal muscle by human CD-markers
CD45 expression was not crucial for phenotypic characterization of immune cell subsets in macaque skeletal muscle
Acknowledgements
We would like to thank Dr. Srinivas Rao and John-Paul Todd at the Vaccine Research Center, NIH and the veterinarians, especially Dr. Michelle Browning and all animal care staff at Bioqual and NIHAC, NIH. This study was supported by the National Institute of Allergy and Infectious Diseases intramural research program (NJS and AP), as well as by the Swedish Research Council (Vetenskapsrådet) and the Swedish Governmental Agency for Innovation Systems (Vinnova) (K.L. funding recipient).
Nonstandard abbreviations
- MDCs
Myeloid dendritic cells
- PDCs
Plasmacytoid DCs
- Ad5
Adenovirus serotype 5
- NK cells
Natural killer Cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Pimorady-Esfahani A, Grounds MD, McMenamin PG. Macrophages and dendritic cells in normal and regenerating murine skeletal muscle. Muscle & nerve. 1997;20:158–166. doi: 10.1002/(sici)1097-4598(199702)20:2<158::aid-mus4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Przybyla B, Gurley C, Harvey JF, Bearden E, Kortebein P, et al. Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp Gerontol. 2006;41:320–327. doi: 10.1016/j.exger.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013;280:4118–4130. doi: 10.1111/febs.12166. [DOI] [PubMed] [Google Scholar]
- 4.Malm C, Sjodin TL, Sjoberg B, Lenkei R, Renstrom P, et al. Leukocytes, cytokines, growth factors and hormones in human skeletal muscle and blood after uphill or downhill running. J Physiol. 2004;556:983–1000. doi: 10.1113/jphysiol.2003.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillon NJ, Bilan PJ, Fink LN, Klip A. Cross-talk between skeletal muscle and immune cells: muscle-derived mediators and metabolic implications. Am J Physiol Endocrinol Metab. 2013;304:E453–465. doi: 10.1152/ajpendo.00553.2012. [DOI] [PubMed] [Google Scholar]
- 6.Gharaibeh B, Chun-Lansinger Y, Hagen T, Ingham SJ, Wright V, et al. Biological approaches to improve skeletal muscle healing after injury and disease. Birth Defects Res C Embryo Today. 2012;96:82–94. doi: 10.1002/bdrc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabro S, Tortoli M, Baudner BC, Pacitto A, Cortese M, et al. Vaccine adjuvants alum and MF59 induce rapid recruitment of neutrophils and monocytes that participate in antigen transport to draining lymph nodes. Vaccine. 2011;29:1812–1823. doi: 10.1016/j.vaccine.2010.12.090. [DOI] [PubMed] [Google Scholar]
- 8.Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, et al. Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayne LJ, Vonderheide RH. Multicolor flow cytometric analysis of immune cell subsets in tumor-bearing mice. Cold Spring Harb Protoc. 2013;2013:955–960. doi: 10.1101/pdb.prot077198. [DOI] [PubMed] [Google Scholar]
- 10.Strauss O, Dunbar PR, Bartlett A, Phillips A. The immunophenotype of antigen presenting cells of the mononuclear phagocyte system in normal human liver--a systematic review. J Hepatol. 2015;62:458–468. doi: 10.1016/j.jhep.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. Development of a preventive vaccine for Ebola virus infection in primates. Nature. 2000;408:605–609. doi: 10.1038/35046108. [DOI] [PubMed] [Google Scholar]
- 12.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 13.Bareja A, Holt JA, Luo G, Chang C, Lin J, et al. Human and mouse skeletal muscle stem cells: convergent and divergent mechanisms of myogenesis. PLoS One. 2014;9:e90398. doi: 10.1371/journal.pone.0090398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng B, Cao B, Crisan M, Sun B, Li G, et al. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nat Biotechnol. 2007;25:1025–1034. doi: 10.1038/nbt1334. [DOI] [PubMed] [Google Scholar]
- 15.Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lierman S, Tilleman K, Cornelissen M, De Vos WH, Weyers S, et al. Follicles of various maturation stages react differently to enzymatic isolation: a comparison of different isolation protocols. Reprod Biomed. 2014 doi: 10.1016/j.rbmo.2014.10.009. Online. [DOI] [PubMed] [Google Scholar]
- 17.Lund T, Korsgren O, Aursnes IA, Scholz H, Foss A. Sustained reversal of diabetes following islet transplantation to striated musculature in the rat. J Surg Res. 2010;160:145–154. doi: 10.1016/j.jss.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Brockhoff G, Fleischmann S, Meier A, Wachs FP, Hofstaedter F, et al. Use of a mechanical dissociation device to improve standardization of flow cytometric cytokeratin DNA measurements of colon carcinomas. Cytometry. 1999;38:184–191. doi: 10.1002/(sici)1097-0320(19990815)38:4<184::aid-cyto5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Novelli M, Savoia P, Cambieri I, Ponti R, Comessatti A, et al. Collagenase digestion and mechanical disaggregation as a method to extract and immunophenotype tumour lymphocytes in cutaneous T-cell lymphomas. Clin Exp Dermatol. 2000;25:423–431. doi: 10.1046/j.1365-2230.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 20.Hedrick MN, Lonsdorf AS, Shirakawa AK, Richard Lee CC, Liao F, et al. CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J Clin Invest. 2009;119:2317–2329. doi: 10.1172/JCI37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragusin M, Wehner S, Kelly S, Wang E, Merrill AH, Jr., et al. Effects of sphingosine-1-phosphate and ceramide-1-phosphate on rat intestinal smooth muscle cells: implications for postoperative ileus. FASEB J. 2006;20:1930–1932. doi: 10.1096/fj.05-5518fje. [DOI] [PubMed] [Google Scholar]
- 22.Brigitte M, Schilte C, Plonquet A, Baba-Amer Y, Henri A, et al. Muscle resident macrophages control the immune cell reaction in a mouse model of notexin-induced myoinjury. Arthritis Rheum. 2010;62:268–279. doi: 10.1002/art.27183. [DOI] [PubMed] [Google Scholar]
- 23.Chazaud B, Brigitte M, Yacoub-Youssef H, Arnold L, Gherardi R, et al. Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc Sport Sci Rev. 2009;37:18–22. doi: 10.1097/JES.0b013e318190ebdb. [DOI] [PubMed] [Google Scholar]
- 24.Jain A, Sharma MC, Sarkar C, Bhatia R, Singh S, et al. Major histocompatibility complex class I and II detection as a diagnostic tool in idiopathic inflammatory myopathies. Arch Pathol Lab Med. 2007;131:1070–1076. doi: 10.5858/2007-131-1070-MHCCIA. [DOI] [PubMed] [Google Scholar]
- 25.Englund P, Lindroos E, Nennesmo I, Klareskog L, Lundberg IE. Skeletal muscle fibers express major histocompatibility complex class II antigens independently of inflammatory infiltrates in inflammatory myopathies. Am J Pathol. 2001;159:1263–1273. doi: 10.1016/S0002-9440(10)62513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinjo SK, Sallum AM, Silva CA, Marie SK. Skeletal muscle major histocompatibility complex class I and II expression differences in adult and juvenile dermatomyositis. Clinics (Sao Paulo) 2012;67:885–890. doi: 10.6061/clinics/2012(08)05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young HE, Steele TA, Bray RA, Hudson J, Floyd JA, et al. Human reserve pluripotent mesenchymal stem cells are present in the connective tissues of skeletal muscle and dermis derived from fetal, adult, and geriatric donors. Anat Rec. 2001;264:51–62. doi: 10.1002/ar.1128. [DOI] [PubMed] [Google Scholar]
- 28.Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 29.Malm C, Nyberg P, Engstrom M, Sjodin B, Lenkei R, et al. Immunological changes in human skeletal muscle and blood after eccentric exercise and multiple biopsies. J Physiol 529 Pt. 2000;1:243–262. doi: 10.1111/j.1469-7793.2000.00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozden S, Huerre M, Riviere JP, Coffey LL, Afonso PV, et al. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2:e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barouch DH, Nabel GJ. Adenovirus vector-based vaccines for human immunodeficiency virus type 1. Human gene therapy. 2005;16:149–156. doi: 10.1089/hum.2005.16.149. [DOI] [PubMed] [Google Scholar]
- 32.Geisbert TW, Bailey M, Hensley L, Asiedu C, Geisbert J, et al. Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. J Virol. 2011;85:4222–4233. doi: 10.1128/JVI.02407-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledgerwood JE, Costner P, Desai N, Holman L, Enama ME, et al. A replication defective recombinant Ad5 vaccine expressing Ebola virus GP is safe and immunogenic in healthy adults. Vaccine. 2010;29:304–313. doi: 10.1016/j.vaccine.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 34.Mueller C, Chulay JD, Trapnell BC, Humphries M, Carey B, et al. Human Treg responses allow sustained recombinant adeno-associated virus-mediated transgene expression. J Clin Invest. 2013;123:5310–5318. doi: 10.1172/JCI70314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis M, Denis-Mize K, Woo C, Goldbeck C, Selby MJ, et al. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 37.Mann CJ, Anguela XM, Montane J, Obach M, Roca C, et al. Molecular signature of the immune and tissue response to non-coding plasmid DNA in skeletal muscle after electrotransfer. Gene Ther. 2012;19:1177–1186. doi: 10.1038/gt.2011.198. [DOI] [PubMed] [Google Scholar]
- 38.Martinon F, Kaldma K, Sikut R, Culina S, Romain G, et al. Persistent immune responses induced by a human immunodeficiency virus DNA vaccine delivered in association with electroporation in the skin of nonhuman primates. Human gene therapy. 2009;20:1291–1307. doi: 10.1089/hum.2009.044. [DOI] [PubMed] [Google Scholar]
- 39.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res. 2012;38:363–373. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 40.Gujer C, Sundling C, Seder RA, Karlsson Hedestam GB, Lore K. Human and rhesus plasmacytoid dendritic cell and B-cell responses to Toll-like receptor stimulation. Immunology. 2011;134:257–269. doi: 10.1111/j.1365-2567.2011.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lore K. Isolation and immunophenotyping of human and rhesus macaque dendritic cells. Methods Cell Biol. 2004;75:623–642. doi: 10.1016/s0091-679x(04)75026-8. [DOI] [PubMed] [Google Scholar]
- 42.Lore K, Sonnerborg A, Spetz AL, Andersson U, Andersson J. Immunocytochemical detection of cytokines and chemokines in Langerhans cells and in vitro derived dendritic cells. J Immunol Methods. 1998;214:97–111. doi: 10.1016/s0022-1759(98)00040-4. [DOI] [PubMed] [Google Scholar]
- 43.Bond E, Adams WC, Smed-Sorensen A, Sandgren KJ, Perbeck L, et al. Techniques for time-efficient isolation of human skin dendritic cell subsets and assessment of their antigen uptake capacity. J Immunol Methods. 2009;348:42–56. doi: 10.1016/j.jim.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Coquery CM, Loo W, Buszko M, Lannigan J, Erickson LD. Optimized protocol for the isolation of spleen-resident murine neutrophils. Cytometry A. 2012;81:806–814. doi: 10.1002/cyto.a.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.