Abstract
Salmonella enterica is a leading cause of food borne illness. Recent studies have shown that S. enterica is a pathogen capable of causing alterations to the composition of the intestinal microbiome. A recent prospective study of French pork production farms found a statistically significant association between Lawsonia intracellularis and carriage of S. enterica. In the current study the composition of the gut microbiome was determined in pigs challenged with S. enterica serovar Typhimurium and or L. intracellularis and compared to non-challenged control pigs. Principal coordinate analysis demonstrated that there was a disruption in the composition of the gut microbiome in the colon and cecum of pigs challenged with either pathogen. The compositions of the microbiomes of challenged pigs were similar to each other but differed from the non-challenged controls. There also were statistically significant increases in Anaerobacter, Barnesiella, Pediococcus, Sporacetigenium, Turicibacter, Catenibacterium, Prevotella, Pseudobutyrivibrio, and Xylanibacter in the challenged pigs. To determine if these changes were specific to experimentally challenged pigs, we determined the compositions of the fecal microbiomes of naturally infected pigs that were carriers of S. enterica. Pigs that were frequent shedders of S. enterica were shown to have similar fecal microbiomes compared to non-shedders or pigs that shed S. enterica infrequently. In a comparison of the differentially abundant bacteria in the naturally infected pigs compared to experimentally challenged pigs, 9 genera were differentially abundant and each exhibited the same increase or decrease in abundance between the two groups. Thus, there were similar changes in the GI microbiome associated with carriage of S. enterica regardless of whether the pigs were experimentally challenged with S. enterica or acquired it naturally.
Introduction
Salmonella enterica is one of the most common causes of food borne diarrheal disease. The Centers for Disease Control has estimated that S. enterica is responsible for over a 1.5 million cases of food borne illness per year in the United States [1]. S. enterica has been ranked as the leading cause of bacterial food borne disease as measured by the combined cost of illness and Quality Adjusted Life-Year [2]. It has been estimated that the economic losses due to salmonellosis in the US exceed $3.5 billion per year. Pigs are known to be an important source of S. enterica and are frequent asymptomatic carriers of S. enterica. S. enterica is believed to enter the food chain by establishing persistent but asymptomatic infections of pigs. Persistent infections can be experimentally established early in life and once established S. enterica can persist for the life of the animal [3–5]. Persistently infected pigs intermittently shed S. enterica in feces but usually in low numbers (10–100 cells per gram of feces) [5]. Even though pigs show no signs of infection or disease they may sporadically shed the organism in feces. Stresses, including transport and feed withdrawal, promote the resumption of fecal shedding just prior to slaughter, which increases the risk of contaminating meats at the time of processing, and may be responsible for spread to uninfected animals held in lairage [5,6]. Thus, swine can act as a reservoir for the spread of S. enterica throughout the herd, within the packing plant, and during processing to finished product [5,7–9].
There is a growing body of evidence that suggests that interactions between members of the gut microflora (microbiome) of livestock animals contribute to the health and well being of those animals [10,11]. For example, studies employing experimental challenges of mice and chickens with S. enterica have shown that significant alterations of the gastrointestinal (GI) microbiome occur after challenges [12,13]. In one study, Stecher et. al. also showed a link between inflammation caused by S. enterica and alterations of the colonic microbiome, providing S. enterica with a competitive advantage to colonize the microbe rich colon [14].
While a comprehensive description of the pig gut microbiome is just now being accumulated, it has been proposed that microbes in the GI tract form networks of symbiotic relationships that result in exclusion of pathogens. Changes in the structure and function of the microbial ecosystem can lead to microbial dysbiosis and predispose the host to colonization with pathogens. (Dysbiosis is defined as an imbalance in the microbial composition in a specific habitat). It is likely that infection with one pathogen can promote further infections with other pathogens. For example, recent work by Beloeil et al [15] proposed that infection of pigs with the enteric pathogen Lawsonia intracellularis might predispose them to shed S. enterica. In their study of 105 French pork production farms they found a statistically significant association (Odds Ratio 3.2, 90% Confidence Interval 1.4 to 7.2) between infections with L. intracellularis and carriage of S. enterica. This observation is consistent with the hypothesis that L. intracellularis interacts with S. enterica and/or other members of the gut microbiome and that these interactions lead to increased colonization and shedding of S. enterica. Since L. intracellularis, the cause of porcine proliferative enteropathy, colonizes the ileum and S. enterica colonizes the colon and cecum, we assume that L. intracellularis and S. enterica likely have indirect interactions that might be mediated by other members of the GI microbiome.
The work reported here was performed to determine if infection with S. enterica and L. intracellularis individually or together was capable of causing a dysbiosis of the pig gut microbiome. To do this we experimentally challenged pigs with S. enterica and/or L. intracellularis and measured the composition of the gut microbiome at four sites in the GI tract and compared the compositions to non-infected pigs. To determine if the changes in the microbiome were applicable to normal swine rearing practices where natural infections are common but mainly result in low level shedding of S. enterica, we also looked for microbiome changes in the GI tracts of commercial pigs that were naturally infected with S. enterica and compared the observed compositional changes to the changes in the experimentally infected pigs. We found that experimental infections of pigs L. intracellularis, S. Typhimurium, or both lead to changes in the composition of the microbiome in the intestinal tract. Pigs naturally infected with S. enterica have similar changes in their fecal microbiomes.
Materials and Methods
Animals and experimental design
The animal protocol used was approved by the University of Minnesota Institutional Care and Uses Committee protocol #0912A75576. For experimental challenges a total of 28 4-week old crossbred, weaned pigs were obtained and randomly allocated into four pens (7 pigs per group). Empty pens were between each of the pens used for the pigs so that the pigs in a group did not have direct contact with pigs in another group. At five weeks of age, four pigs (1 per pen) were removed and necropsied. Ten-centimeter segments of jejunum, ileum (approximately 10 centimeters from the ileocecal junction), cecum and ascending colon were removed. The remaining pigs were treated as follows: pigs in pen numbers three and four were challenged orally with 1 x 1010 CFU L. intracellularis strain PHE/MN1-00. One week later (when the pigs were six weeks of age) pigs in pen numbers 2 and 4 were challenged orally with 2 x 108 CFU of nalidixic acid resistant S. Typhimurium strain 798. Pigs in pen number 1 remained unchallenged and served as controls. When the pigs were 7, 9, and 11 weeks of age two pigs from each pen were euthanatized with an overdose of sodium pentobarbital and necropsied.
For the field-based animal protocol commercial pigs were utilized. Pigs were from a commercial pig farm located in southwestern Minnesota, U.S.A (43°37’26”N95°35’57”W). These pigs were part of a larger study performed to describe microbiome changes in pigs fed the antimicrobial growth promoter tylosin [16]. The production barn contained 20 pens and each pen had 25 pigs. A single pen in the middle of the barn was selected for sampling. Pigs were kept in the same pen for the entire sampling period without introduction of any new pigs. Ten pigs from the total of 25 were randomly selected for sampling and ear tagged for identification. Fresh fecal samples from each of the ear tagged animals were individually collected from the pigs’ rectums. Samples were collected five times over their growth period at 3-week intervals starting when the pigs were 10-weeks old. Pigs did not receive antibiotics in feed or for any therapeutic purposes prior to and during the sampling period.
Growth of bacterial challenge inocula
The nalidixic acid resistant S. enterica serovar Typhimurium strain 798 was prepared by overnight growth in LB broth. L. intracellularis strain PHE/MN1-00 was grown in McCoy cells as previously described [17].
Tissue sampling, DNA extraction from tissues and feces, and DNA sequencing
The 10 cm of cecum and colon samples were opened longitudinally and the mucosal layer scraped with a microscope slide to remove loosely associated contents and most of the mucosal tissue. The fibrous serosal tissue was discarded. The mucosal samples were weighed and 1 gram of each was used to determine the concentrations of S. Typhimurium as described below. DNA was extracted from a second 1 gram sample as previously described [18]. The DNA extraction protocol employed two rounds of bead beating in the presence of NaCl and sodium dodecyl sulfate, followed by sequential ammonium acetate and isopropanol precipitations. The precipitated nucleic acids were then treated with RNase A and proteinase K, and the DNA purified using a QIAgen DNA Mini Stool Kit (Valencia, CA), according to manufacturer's recommendations. For fecal samples, DNA was extracted from the 1 gram of feces using the same extraction protocol.
Detection of S. enterica and L. intracellularis
S. enterica in tissue and fecal samples was detected as previously described [19]. One gram of tissue was suspended in 9 ml tetrathionate broth (TTB) and incubated at 41°C for 24 hours. One hundred μl was then transferred to 900 μl Rappaport-Vassiliadis R10 broth (Remel, Lenexa, KS) and incubated for 24 hours at 41°C. Growth from the Rappaport-Vassiliadis R10 broth was plated on XLT4 agar plates (Becton Dickinson, Baltimore, MD) and incubated at 37°C for 24 hours. Suspect S. enterica colonies were confirmed as S. enterica using API®20E strips (BioMerieux® SA, Marcy-I’Etoiele, France) and by PCR using primers specific for the gene invA (Forward: invA ACAGTGCTCGTTTACGACCTGAAT and Reverse: invA AGACGACTGGTACTGATCGATAAT) [20]. For detection of S. enterica serovar Typhimurium strain 798, colonies from the XLT-4 plates were re-plated on LB agar containing nalidixic acid (100 μg/ml). Template DNA was prepared from S. enterica isolates using a boiling lysis procedure [21]. The PCR mixtures contained 12.5 μl of Taq polymerase (Promega, Madison, WI), 0.2 μM of the primer pairs, and 2 μl of DNA template in a 25μl reaction. Cycling conditions started with an initial denaturation at 95°C for 2 minutes followed by 30 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. A final 7 minute extension was incubated at 72°C.
S. enterica isolates were serotyped by the National Veterinary Services Laboratory (Ames, Iowa).
Detection and quantification of L. intracellularis was performed using a quantitative PCR protocol previously described [17]. The qPCR assay is based on the detection of the aspartate ammonia-lyase (aspA) gene.
Analysis of the intestinal microbiomes of intestinal tissues
PCR primers that flanked the V3 hypervariable region of bacterial 16S rRNAs were designed based on the blueprint for barcodes [18,22]. The oligonucleotide primers included Roche A or B sequencing adapters at the 5' ends and template specific sequences at the 3' end. Barcodes were located in between the sequencing adapter and the template specific sequences of the forward primer. The primer sequences were: 5' (sequence adapter)– 10-base barcode—CCTACGGGAGGCAGCAG 3' (forward) and 5' (sequence adapter)–ATTACCGCGGCTGCTGG 3' (reverse) [22,23]. The amplification mix contained 2.5 units of Taq polymerase, 0.2mM of dNTPs, 0.4μM of each fusion primer, and 50ng of DNA in a reaction volume of 50μl. Cycling conditions started with an initial denaturation at 95°C for 2 minutes followed by 20 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. A final 7 minute extension was incubated at 72°C. For the jejunal and ileal samples, the number of PCR cycles was increased to 25 to compensate for lower bacterial numbers in these samples compared to the cecal and colonic samples. The PCR amplicon products were separated on a 1.5% agarose gels, extracted from the gels and then were cleaned using QIAgen DNA Mini Stool Kits (Valencia, CA). The quality of the product was assessed on a Bioanalyzer 2100 (Agilent, California, USA). Only PCR products without primer dimers and contaminant bands were used for sequencing. The PCR amplicons were sequenced using a Roche 454 GS-FLX sequencer (454 Life Sciences, Branford, CT).
To minimize effects of random sequencing errors, we eliminated (i) sequences that did not appropriately match the PCR primer and the barcode at the beginning of a read, (ii) sequence reads with <50 bases after the proximal PCR primer if they terminated before reaching the distal primer, and (iii) sequences that contained more than one undetermined nucleotide (N). Mothur implemented in Galaxy was used for quality assessments including sequence trimming and assignment of operational taxonomic units using a 97% similarity cutoff [24]. Chimeras were removed using Chimera Slayer implemented in UCHIME [25]. A phylogenetic assessment was conducted using RDP classifier with a bootstrap cutoff of 50%. DNA sequence data is freely available the through the University of Minnesota data conservancy: http://conservancy.umn.edu/handle/11299/172268?show=full.
Statistical analysis
Determination of statistically unique taxa was determined using MetaStats [18]. MetaStats is a tool that was developed to identify differentially abundant species among samples. It uses the nonparametric t-test, Fisher’s exact test, and the false discovery rate to identify prioritized lists of relevant features. Principal coordinate analysis (PCoA) plots were generated by using weighted Fast UniFrac [26].
Results
Microbiome composition in different GI sites of experimentally infected pigs
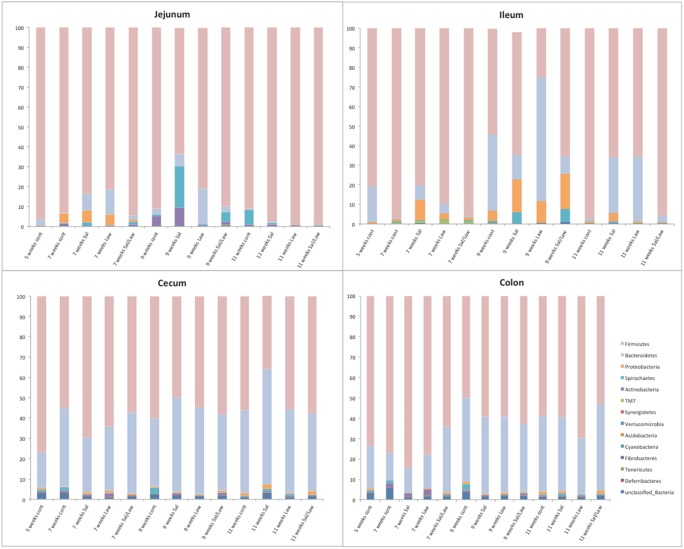
The microbiomes in pigs experimentally challenged with S. enterica serovar Typhimurium (S. Typhimurium), L. intracellularis, or both were determined. S. Typhimurium is known to cause sub-clinical infections with sporadic low level shedding in feces, while L. intracellularis is a pig pathogen that causes proliferative enteropathy in the lower small intestines. To test this hypothesis, groups of pigs were experimentally challenged orally with one or both microbes (S. Typhimurium strain 798 and/or L. intracellularis strain PHE/MN1-00). At multiple times pigs were euthanized and tissue samples collected. Total DNA was extracted from the jejunum, ileum, cecum, and colon of these pigs and the extracted community DNA was PCR amplified and sequenced using primers specific for the V3 region of the 16S rRNA gene. On average, 23,160 DNA sequences were obtained per sample. Taxonomic assignments were made using RDP Classifier and are shown in Fig 1 (phylum) and Fig 2 (class). The taxonomic assignments are shown in S1 Table. The two most common phyla in all samples at all times were Firmicutes followed by Bacteroidetes. These two phyla dominated the cecum and colon samples accounting for greater than 95% of all bacteria. In the cecum and colon tissue samples, Firmicutes were the most common bacteria ranging from 65.4% to 85% in seven-week old pigs, 55.3% to 63.1% in the nine-week old pigs, and 56.4% to 70.7% in the eleven-week old pigs. In the jejunum, Firmicutes and Bacteroidetes were the most predominant phyla observed (greater than 95%). In the ileal samples, Firmicutes was the most predominant phylum (64% to 96% in the seven-week old pigs, 25% to 65.9% in the nine-week old pigs, and 65.6% to 85% in the eleven-week old pigs. They also contained higher levels of Proteobacteria. These results are similar to those obtained by Looft, et. al. who found a similar composition in the ileum of pigs [27].
Fig 1. Distribution of bacterial phyla in pig tissues.
Graph of the distribution of different phyla in four pig tissues (jejunum, ileum, cecum, and colon) at different times post challenge with S. enterica serovar Typhimurium strain 798 (Sal), Lawsonia intracellularis strain PHE/MN1-00 (Law), or both (Sal/Law) as well as non-challenged controls (cont).
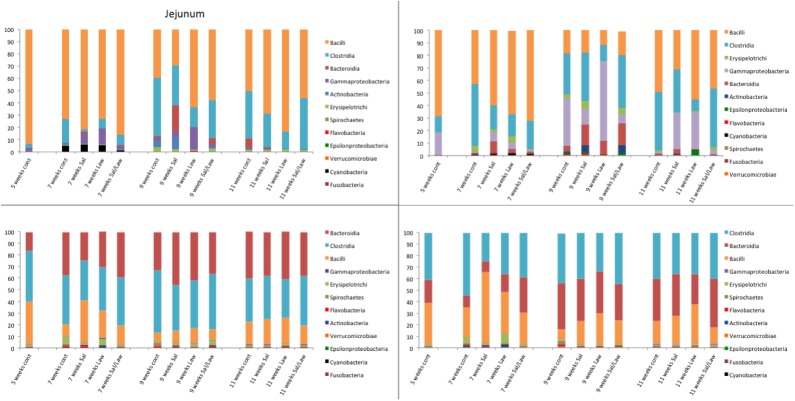
Fig 2. Distribution of bacterial classes in pig tissues.
Graph of the distribution of different classes in four pig tissues (jejunum, ileum, cecum, and colon) at different times post challenge with S. enterica serovar Typhimurium strain 798 (Sal), Lawsonia intracellularis strain PHE/MN1-00 (Law), or both (Sal/Law) as well as non-challenged controls (cont).
At the class level, Bacilli and Clostridia were dominant in the jejunum and ileum, but depending upon the time of sampling and treatment, Gammaproteobacteria and Bacteroidia were the most dominant classes. In the cecum and colon, Bacteroidia, Clostridia, and Bacilli dominated representing greater than 90% of all bacteria. A summary of the entire taxonomic classification data is available in S2 Table.
Using culture based methods, S. Typhimurium strain 798 was detected in all tissue samples from pigs challenged with S. enterica serovar Typhimurium strain 798 at 7, 9, and 11 weeks of age. The S. Typhimurium challenge strain was not detected prior to challenge (5 week old pigs) or in non-challenged pigs. L. intracellularis was found in samples from pigs challenged with L. intracellularis strain PHE/MN1-00 that were 9 or 11 weeks of age but not in pigs that were 7 weeks of age or that were not challenged. At 9 weeks of age, L. intracellularis was found in the jejunum, ileum, cecum, and colon samples and at 11 weeks of age it was only found in the cecum and colon samples.
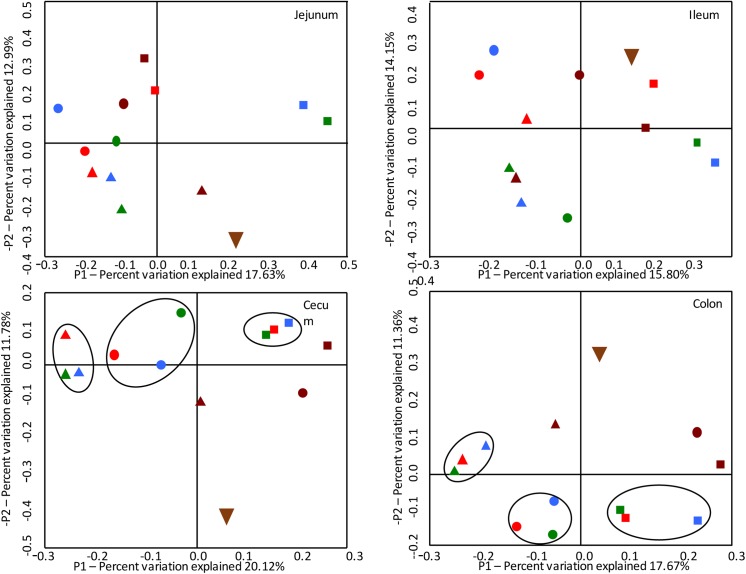
Principal coordinate analysis of all pig tissues
Fast UniFrac [26] was used to perform principal coordinate analysis to compare the overall compositions of the microbiomes in the various treatment groups over time (Fig 3). While the compositions of the microbiomes in the jejunum and ileum did not segregate into distinct sector of the plot based on treatment, segregation was observed in comparing microbiome compositions in the cecum and colon. The group compositions of challenged pigs, regardless of the challenge group, were similar to each other at each time point, but differed from the non-challenged controls. Also, the overall compositions of the microbiome in all challenge groups differed from each other by time rather than treatment.
Fig 3. Principal coordinate analysis graphs comparing the total compositions of the microbiomes in four pig tissues (jejunum, ileum, cecum, and colon) at various times.
The symbol shape associated with each data point represents the time of sample collection (5 (upside down triangle), 7 (triangles), 9 (circles), and 11 (squares) weeks of age). The symbol color reflects the treatment: maroon = non-challenged controls, red = challenged with S. enterica serovar Typhimurium strain 798, green = Lawsonia intracellularis strain PHE/MN1-00, blue = both challenge pathogens.
Microbiome composition differences in the four GI tissues
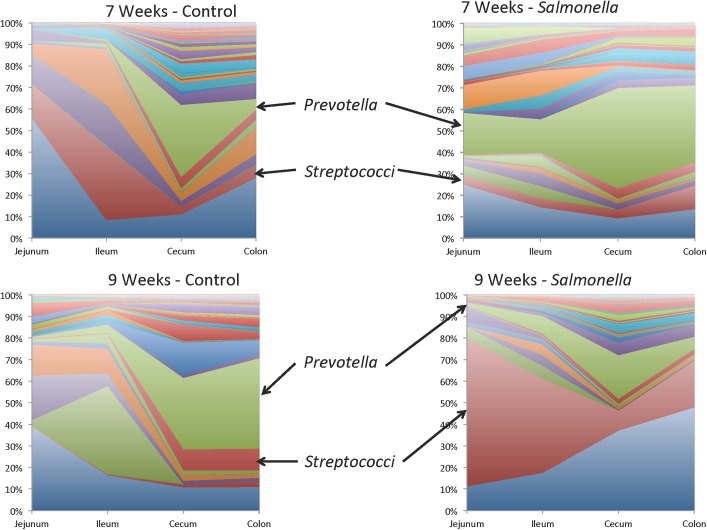
To compare the tissue-specific microbiomes of pigs in the four experimental groups, we plotted the relative concentration of the genus level assignments as the mean of the challenge group (S1, S2, S3 and S4 Figs) at each of the times sampled. Many differences can be seen and one example is shown in Fig 4 where the microbiomes of control and S. Typhimurium challenged pigs at 7 and 9 weeks of age are shown. Compared to control samples, there was an increase in the percentage of Prevotella present in the jejunal and ileal tissue samples at 7 weeks of age (1 week after challenge with S. enterica). By nine weeks of age the levels of Prevotella decreased but there was an increase in the level of Streptococci in the jejunal and ileal tissues. Looking at the non-challenged controls, the compositions of the microbiomes in each tissue within the same pig were shown to vary by tissue (site specific colonization) and time of sampling (microbial succession).
Fig 4. Comparison (mean of the group) of the tissue (jejunum, ileum, cecum, and colon) microbiomes of pigs at 7 or 9 weeks of age that were not challenged (control) or were challenged with S. enterica serovar Typhimurium strain 798 (Salmonella).
The colors represent the same genus in all four panels. A figure legend can be found in S1 Table.
Differentially abundant bacteria in the colon of experimentally infected pigs
To identify phyla that were differentially abundant, MetaStats [18] was used with a probability cut off of P< 0.05. Because cecum and colon tissues yielded similar PCoA plots and because no association between treatment group and jejunum and ileum were observed, only the colon samples were included in this analysis. In the seven-week old pigs (2 weeks post challenge with L. intracellularis and 1 week post challenge with S. Typhimurium) the colon of the dual infected group contained fewer members of the phylum Bacteroidetes (Table 1). This difference was compared to the control and to the single pathogen infection groups. Pigs challenged with L. intracellularis had increased levels of Actinobacteria compared to the other groups. In pigs that were 9 weeks of age, there was a significant decrease in the number of Spirochaetes in all challenge groups compared to the control group. In the 11 week old pigs, members of the phylum Verrucomicrobia were decreased in each of the challenge groups although the decrease in the group challenged with L. intracellularis alone was not statistically significant. Members of the phylum Proteobacteria were reduced in the group challenged with L. intracellularis only.
Table 1. Distribution of phyla in the colon of pigs that are statistically unique (≤0.05)*.
| Phylum | Control | Salmonella | Lawsonia | Salmonella/Lawsonia | |
|---|---|---|---|---|---|
| 7 weeks | Bacteroidetes | 79.79 | 85.01 | 78.78 | 65.38 a |
| Actinobacteria | 1.89 | 1.49 | 3.13 c | 0.75 | |
| 9 weeks | Spirochaetes | 2.67 | 0 b | 0.33 b | 0.16 b |
| Fibrobacteres | 0.19 | 0 b | 0.02 | 0 b | |
| 11 weeks | Verrucomicrobia | 0.18 | 0 b | 0.05 | 0 b |
| Proteobacteria | 1.23 | 0.12 | 0.29 c | 0.91 |
* Numbers represent the percent of each phylum in the group
a Compared to control, Salmonella, and Lawsonia
b Compared to control
c Compared to control, Salmonella, and Salmonella/Lawsonia
Differentially abundant bacteria at the genus level also were identified using MetaStats [28]. The results shown in S2 Table demonstrate numerous differences between the various groups. The differences are expressed as an overall increase or decrease in the relative abundance of the statistically unique genera compared to controls and the three treatment groups (S. Typhimurium, L. intracellularis, or S. Typhimurium and L. intracellularis) and then between each challenge group as well. Of particular interest was the consistent increase in Anaerobacter in seven-week old pigs receiving infectious challenges compared to the controls, increases in Lactobacillus in the pigs challenged with L. intracellularis and decreases in Prevotella in dual challenged pigs. In the nine-week old pigs, there were consistent increases in Oribacterium compared to control pigs. In the eleven-week old pigs, there was a consistent increase in Paraprevotella in pigs challenged with L. intracellularis and S. Typhimurium. These results demonstrate numerous perturbations of the colonic microbiome based on the challenge and time post challenge.
Analysis of the fecal microbiome composition of commercial pigs that shed or did not shed S. enterica
Three levels of S. enterica shedding were defined for the commercial, naturally infected pigs based on S. enterica isolation from feces and PCR: non shedder (pigs 1, 3, 4, 5, and 7 were negative by both isolation and invA PCR at all sampling time points), moderate shedder (pigs 2 and 6 were positive by both tests at no more than two sampling times), and active shedder (pigs 8, 9, and 10 were positive by both tests at more than 3 sampling times) (Table 2). Pig 10 was shedding S. enterica at all time points. All but one of the S. enterica isolates were identified as serovar Manhattan. Pig 9 also shed serovar Typhimurium at the 16-weeks of age sampling time. The overall phylogenetic community composition was compared between active, moderate and non-shedding pigs using Fast UniFrac distance metric. Overall, the active shedders did appear to have a unique fecal microbiome composition, which was different from the moderate shedders and non-shedder. However, the fecal microbiome compositions of pigs sampled at 10 weeks of age did not segregate by shedding group. At sampling times 13, 16, and 19 weeks of age, pigs in the active S. enterica shedding group shared a similar community membership compared to pigs in the moderate and no-shedders group (Fig 5). Pig 10 was shedding S. enterica during the entire sampling period and it had a different microbiome composition compared to the other pigs when they were 22 weeks of age. By 22 weeks of age all of the other pigs had stopped shedding S. enterica (Table 2).
Table 2. Salmonella shedding status evaluated by PCR and isolation.
| Weeks of age | |||||||
|---|---|---|---|---|---|---|---|
| Pig ID | Shedding status | Serotype | 10 | 13 | 16 | 19 | 22 |
| 1 | No | - | - | - | - | - | |
| 2 | Moderate | Manhattan | + | + | - | - | - |
| 3 | No | - | - | - | - | - | |
| 4 | No | - | - | - | - | - | |
| 5 | No | - | - | - | - | - | |
| 6 | Moderate | Manhattan | + | - | - | - | - |
| 7 | No | - | - | - | - | - | |
| 8 | Active | Manhattan | + | + | + | + | - |
| 9 | Active | Manhattan | + | + | * + | - | - |
| 10 | Active | Manhattan | + | + | + | + | + |
* Salmonella typhimurium
Fig 5. Principal coordinate analysis graphs comparing the total compositions of the fecal microbiome in 10 pigs at five times (10, 13, 16, 19, and 22 weeks of age) in pigs that were shedding or not shedding S. enterica.
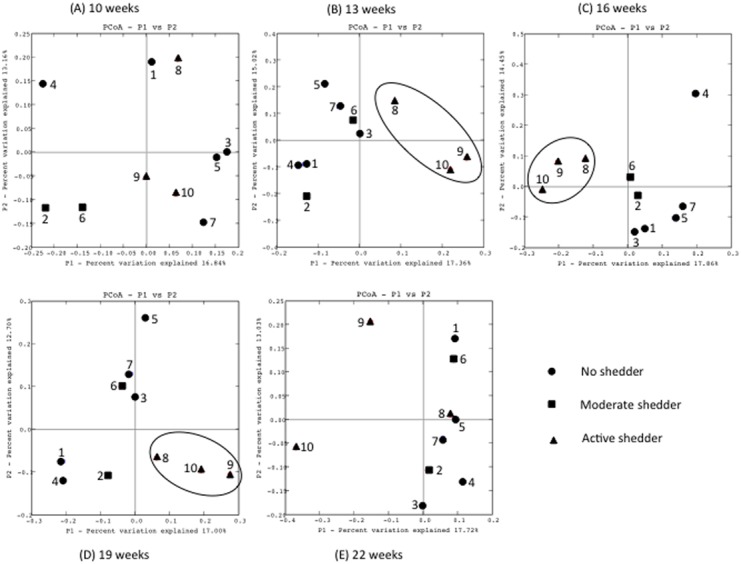
● = Non-shedding pigs, ■ = moderate shedding pigs, ▲ = active shedding pigs.
Using MetaStats, differentially abundant bacteria present in pigs infected with S. enterica were identified by comparison to pigs that never shed S. enterica. The results are shown in S3 Table. The top 10 most abundant bacteria at the 10 week time point were used as a point of reference to compare to the other sampling times. Of the top 10, Fastidiosipela was present in reduced numbers in all five samples, while Pseudobutyriovibrio was present at higher levels. Prevotella, Lactobacillus, Anaerobacter, Treponema, and Roseburia were present at reduced levels at four of the five sampling times while Barnsiella was at higher levels. Campylobacter an important food borne pathogen was present at lower levels at three of the five sampling times in S. enterica shedding pigs.
Discussion
The work presented here was initiated to determine if specific pathogens caused a dysbiosis in the composition of the gut microbiome of pigs. Two approaches were used in the work. The first was to experimentally challenge pigs with S. Typhimurium, an important food borne pathogen, and/or L. intracellularis, the cause of proliferative enteropathy in pigs and then measure microbiome compositions over time and within different sites in the pig intestinal tract. The second approach was to analyze fecal samples collected from commercial pigs that were naturally infected and shedding low levels S. enterica in their feces and to compare the fecal microbiomes to pigs that were not shedding S. enterica. Using both approaches, the results demonstrated alterations in the composition of the gut microbiomes of pigs infected with S. enterica either experimentally or naturally. In addition, we showed that the pig pathogen, L. intracellularis, was able to cause perturbations to the pig gut microbiome.
Using two separate tools for data analysis, PCoA and MetaStats, the composition of the gut microbiome in the cecum and colon was shown to be perturbed by challenge with either S. Typhimurium or L. intracellularis or both. PCoA was used to compare the overall microbiome compositions and structures while MetaStats was used to identify statistically significant differentially abundant bacteria. Based on PCoA, the compositions and structures of the colonic or cecal microbiomes of experimentally infected pigs were more similar to each other compared to the non-challenged control pigs, regardless of the challenge organisms. PCoA demonstrated a clustering of the microbiome compositions in infected animals when they were 7, 9, and 11 weeks of age, while the non-challenged controls segregated in different sectors of the PCoA plots. However, similar changes were not observed in the jejunal or ileal tissues. Since S. Typhimurium preferentially colonizes the cecum and colon, it isn’t surprising that dysbiosis of the jejunum and ileum did not occur in the S. Typhimurium infected pigs.
Using MetaStats, perturbations of the microbiome in the cecum and colon of the experimentally infected pigs were identified at the phylum and genus levels. For example (see Table 1), at 7 weeks of age (1 week post challenge with S. Typhimurium and 2 weeks post challenge with L. intracellularis) overall levels in the phyla Bacteroidetes and Actinobacteria were reduced in the dual challenged or L. intracellularis challenged pigs, respectively. Using a mouse model, Deatherage et al. showed similar reductions in bacteria in the phylum Bacteroidetes in fecal samples after challenge with S. enterica (12). However, while they also saw reductions in Firmicutes, our study in pigs did not show a similar decrease. At 9 weeks of age there were reductions in Spirochaetes for all challenged pigs and reductions in Fibrobacteres for pigs challenged with S. Typhimurium. At 11 weeks of age, there were reductions in Verrucomicrobia in pigs challenged with S. Typhimurium while pigs challenged with L. intracellularis exhibited reductions in Proteobacteria. In addition, numerous genera were identified that became more abundant after the challenges or less abundant when compared to the non-challenged control pigs (Table 2). These data taken together demonstrate that both pathogens are able to perturb the cecal and colonic microbiomes of pigs. L. intracellularis is known to mainly colonize the ileum of pigs and causes localized tissue damage [29] although L. intracellularis also can colonize anterior portions of the colon [30]. Based on the data collected in this experimental protocol it was not possible to determine if the L. intracellularis induced microbiome changes in the cecum and colon were a direct result of this pathogen or due to effects that occurred in the ileum that resulted in indirect effects in the cecum and colon. However, since L. intracellularis was found in the cecum and colon samples of challenged pigs when the pigs were 9 and 11 weeks of age, direct interactions between these two microbes is possible.
In addition to changes in the cecal and colonic microbiomes, the experimental approach also allowed us to visualize differences in the microbial compositions of the microbiomes of the jejunum and ileum compared to the cecum and colon. While there were similarities in which bacteria were present in each GI location, their relative abundances were quite different. For example, Proteobacteria were relatively minor components of the cecum and colon but were more abundant in the jejunum and ileum (particularly the ileum). In addition, the data presented demonstrated the phenomenon of bacterial succession. Over time, the relative composition of all tissue sites changed as the microbiome progressed to a climax community composition [31].
This mode of visualization did show some large variations of microbiome compositions after challenge with S. enterica serovar Typhimurium. In the jejunum and ileum of challenged pigs at 7 weeks of age (1 week post challenge with S. enterica) there was an increase in Prevotella and a decrease in Streptococci (Fig 4). By 9 weeks of age, the levels of Prevotella in the jejunum and ileum were reduced to similar to that seen in uninfected controls, but the levels of Streptococci remained high relative to other members of the microbiome.
Taken together, the interpretation of the data from the experimental challenge experiments was that pathogens do perturb the gut microbiomes of pigs. However, to determine if this was a phenomenon restricted to experimentally challenged pigs we also examined the microbiomes of pigs naturally infected with S. enterica by using commercially raised pigs. The herd used contained pigs that shed S. enterica (serovars Manhattan and Typhimurium) at low levels. By measuring shedding over time, we were able to segregate pigs into groups that never shed S. enterica at any of the sampling times, those that shed S. enterica one or two times, and pigs that shed S. enterica three or more times. Pigs in the highest shedding group were shown to have similar microbiome compositions and structures using PCoA but were different than pigs that were not shedding S. enterica or those that shed S. enterica only one or two times. It should be noted, that the classification of these pigs into shedding categories was based on the five discrete sampling times and it was possible that if other times had been selected for sampling that other pigs would been have found to be shedders. Nonetheless, there was a correlation between high shedders and their microbiome compositions.
To investigate the compositions of the naturally infected pigs further, we used MetaStats to identify differentially abundant genera (see S3 Table). The microbiomes of the pigs that were shedding at the time of sampling were compared to the non-shedding pigs. Several observations were of note. Firstly, there was an overall increase in the number of differentially abundant genera over time. Secondly, pigs actively shedding S. enterica at 22 weeks of age had elevated levels of Akkermansia although this was not the case at earlier times of sampling. This might be significant because Akkermansia is known to enhance gut inflammation and in doing so may enhance infections caused by S. enterica [32]. Given that pigs that are 22 weeks of age are approaching market weight, any factors that influence S. enterica infections might also increase the risk of cross contamination at the time of transport to slaughter plants. However, it should be noted that at 22 weeks of age, only one pig was shedding S. enterica and thus, this might not apply to other animals shedding Akkermansia at the same age. It also was observed that the relative level of Campylobacter was reduced at three of the five sampling times. This result could suggest an incompatibility between carriage of S. enterica and Campylobacter.
Thirdly, there were commonalities between the differentially abundant bacteria found in the pigs experimentally challenged with S. enterica and those that were naturally infected. The list of microbes differentially present in the colon of pigs at any of the three experimental times to those differentially present in naturally infected pig feces at 10 weeks of age were compared. We picked those time points because they were the most similar times of sampling in both protocols. Note, that the comparison is between colonic tissues versus feces. A total of nine genera were in common between these two groups of pigs: Barnesiella, Pseudobutyrivibrio, Prevotella, Lactobacillus, Anaerobacter, Roseburia, Fastidiosipila, Campylobacter, and Succinivibrio. Barnesiella and Pseudobutyrivibrio were present in increased levels and the other genera were present at reduced levels. Since the direction of abundance was the same between experimentally infected pigs and naturally infected pigs, it is likely that these represent true effects of being infected with S. enterica and further suggested that this pathogen is able to direct changes in the overall composition of the gut microbiome.
Conclusions
In summary, the data presented here demonstrated that the pathogen S. enterica can cause disruptions in the gut microbiomes of pigs whether experimentally introduced into pigs or through natural infections. A similar conclusion was drawn from other studies that used chickens [13] and mice [12]. The major changes in the gut microbiome occurred in the lower intestinal tract: the cecum, colon, and feces. However, changes could be detected in the jejunum and ileum of these pigs. There was a correlation between the changes found between experimentally challenged pigs and naturally infected commercial pigs. While not unexpected, the data from the experimentally infected pigs also showed that there was a shift in the composition of the gut microbiome over time (in non-infected pigs) and that there were differences in the microbiome compositions based on anatomic location in the intestinal tract.
Supporting Information
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
(XLSX)
(DOCX)
(DOCX)
Data Availability
All DNA sequence data is now available through the University of Minnesota data conservancy: http://conservancy.umn.edu/handle/11299/172268?show=full.
Funding Statement
This work was supported by a grant from the Emerging Infectious Diseases signature research program, College of Veterinary Medicine, University of Minnesota and by a grant from the USDA/AFRI #2007-35212-18046. REI was the principal investigator on the two grants. The funders did not play a role in the experimental design or protocols.
References
- 1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. (2011) Foodborne illness acquired in the United States—major pathogens. Emerging Infectious Diseases 17: 7–15. 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batz MB, Hoffmann S, Morris JG Jr. (2011) Ranking the risks: The 10 pathogen-food combinations with the greatest burden on public health Gainesville, FL: University of Florida; 1–68 p. [Google Scholar]
- 3. Wood RL, Pospischil A, Rose R (1989) Distribution of persistent Salmonella typhimurium infection in internal organs of swine. American Journal of Veterinary Research 50: 1015–1021. [PubMed] [Google Scholar]
- 4. Kampelmacher EH, Edel W, Guinee PAM (1969) Experimental Salmonella infections in pigs. Zentralbl Veterinarmed (B) 16: 717–724. [DOI] [PubMed] [Google Scholar]
- 5. Isaacson RE, Firkins LD, Weigel RM, Zuckermann FA, DiPietro JA (1999) Effect of transportation and feed withdrawal on shedding of Salmonella typhimurium among experimentally infected pigs. American Journal of Veterinary Research 60: 1155–1158. [PubMed] [Google Scholar]
- 6. Hurd HS, McKean JD, Wesley IV, Karriker LA (2001) The effect of lairage on Salmonella isolation from market swine. Journal of Food Protection 64: 939–944. [DOI] [PubMed] [Google Scholar]
- 7. Fedorka-Cray PJ, Whipp SC, Isaacson RE, Nord N, Lager K (1994) Transmission of Salmonella typhimurium to swine. Veterinary Microbiology 41: 333–344. [DOI] [PubMed] [Google Scholar]
- 8. Letellier A, Messier S, Pare J, Menard J, Quessy S (1999) Distribution of Salmonella in swine herds in Quebec. Veterinary Microbiology 67: 299–306. [DOI] [PubMed] [Google Scholar]
- 9. Gray JT, Stabel TJ, Fedorka-Cray PJ (1996) Effect of dose on the immune response and persistence of Salmonella choleraesuis infection in swine. American Journal of Veterinary Research 57: 313–319. [PubMed] [Google Scholar]
- 10. Isaacson R, Kim HB (2012) The intestinal microbiome of the pig. Animal Health Research Reviews 13: 100–109. 10.1017/S1466252312000084 [DOI] [PubMed] [Google Scholar]
- 11. Brogden KA, Guthmiller JM, Taylor CE (2005) Human polymicrobial infections. Lancet 365: 253–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deatherage Kaiser BL, Li J, Sanford JA, Kim YM, Kronewitter SR, et al. (2013) A multi-omic view of host-pathogen-commensal interplay in -Mediated Intestinal Infection. PLoS One 8: e67155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Videnska P, Sisak F, Havlickova H, Faldynova M, Rychlik I (2013) Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Veterinary Research 9: 140–147. 10.1186/1746-6148-9-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stecher Br, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. (2007) Salmonella enterica Serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5: e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beloeil PA, Fravalo P, Fablet C, Jolly JP, Eveno E, et al. (2004) Risk factors for Salmonella enterica subsp. Enterica shedding by market-age pigs in French farrow-to-finish herds. Preventive Veterinary Medicine 63: 103–120. [DOI] [PubMed] [Google Scholar]
- 16. Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, et al. (2012) Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proceedings of the National Academy of Sciences 109: 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wattanaphansak S, Gebhart CJ, Anderson JM, Singer RS (2010) Development of a Polymerase Chain Reaction Assay for Quantification of Lawsonia Intracellularis . Journal of Veterinary Diagnostic Investigation 22: 598–602. [DOI] [PubMed] [Google Scholar]
- 18. Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, et al. (2011) Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Veterinary Microbiology 153: 124–133. 10.1016/j.vetmic.2011.05.021 [DOI] [PubMed] [Google Scholar]
- 19. Kim HB, Singer RS, Borewicz K, White BA, Sreevatsan S, Johnson TJ, et al. (2014) Effects of tylosin administration on C-reactive protein concentration and carriage of Salmonella enterica in pigs. American Journal of Veterinary Research 75: 460–467. 10.2460/ajvr.75.5.460 [DOI] [PubMed] [Google Scholar]
- 20. Singer RS, Cooke CL, Maddox CW, Isaacson RE, Wallace RL (2006) Use of pooled samples for the detection of Salmonella in feces by polymerase chain reaction. Journal of Veterinary Diagnostic Investigations 18: 319–325. [DOI] [PubMed] [Google Scholar]
- 21. Johnson JR, Clabots C (2000) Improved repetitive-element PCR fingerprinting of Salmonella enterica with the use of extremely elevated annealing temperatures. Clinical Diagnostic and Laboratory Immunology 7: 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, et al. (2007) A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35: e130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology 75: 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. International Society for Microbial Ecology Journal 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedfeld RD, et al. (2012) In-feed antibiotic effects on the swine intestinal microbiome. Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS computational biology 5: e1000352 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawson GH, Gebhart CJ (2000) Proliferative enteropathy. Journal of Comparative Patholology 122: 77–100. [DOI] [PubMed] [Google Scholar]
- 30. Jacobson M, Fellstrom C, Jensen-Waern M (2010) Porcine proliferative enteropathy: an important disease with questions remaining to be solved. Veterinary Journal 184: 264–268. [DOI] [PubMed] [Google Scholar]
- 31. Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annual Reviews in Microbiology 31: 107–133. [DOI] [PubMed] [Google Scholar]
- 32. Ganesh BP, Klopfleisch R, Loh G, Blaut M (2013) Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella typhimurium-infected gnotobiotic mice. PLoS One 8: e74963 10.1371/journal.pone.0074963 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
Each sample is expressed as the mean for the group.
(PDF)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All DNA sequence data is now available through the University of Minnesota data conservancy: http://conservancy.umn.edu/handle/11299/172268?show=full.