Abstract
Background
Iron (Fe) and phosphorus (P) are essential mineral nutrients in plants. Knowledge regarding global changes in the abundance of Fe-responsive genes under Pi deficiency as well as the processes these genes are involved in remains largely unavailable at the genome level. In the current study, we comparatively analyzed RNA sequencing data sets relative to Fe deficiency (NCBI: SRP044814) and Pi starvation (NCBI: SRA050356.1).
Results
Analysis showed a total of 579 overlapping genes that are responsible for both Fe deficiency and Pi starvation in Arabidopsis roots. A subset of 137 genes had greater than twofold changes in transcript abundant as a result of the treatments. Gene ontology (GO) analysis showed that the stress-related processes ‘response to salt stress’, ‘response to oxidative stress’, and ‘response to zinc ion’ were enriched in the 579 genes, while Fe response-related processes, including ‘cellular response to nitric oxide’, ‘cellular response to iron ion’, and ‘cellular iron ion homeostasis’, were also enriched in the subset of 137 genes. Co-expression analysis of the 579 genes using the MACCU toolbox yielded a network consisting of 292 nodes (genes). Further analysis revealed that a subset of 90 genes were up-regulated under Fe shortage, but down-regulated under Pi starvation. GO analysis in this group of genes revealed an increased cellular response to iron ion/nitric oxide/ethylene stimuli. Promoter analysis was performed in 35 of the 90 genes with a 1.5-fold or greater change in abundance, showing that 12 genes contained the PHOSPHATE STARVATION RESPONSE1-binding GNATATNC cis-element within their promoter regions. Quantitative real-time PCR showed that the decreased abundance of Fe acquisition genes under Pi deficiency exclusively relied on Fe concentration in Pi-deficient media.
Conclusions
Comprehensive analysis of the overlapping genes derived from Fe deficiency and Pi starvation provides more information to understand the link between Pi and Fe homeostasis. Gene clustering and root-specific co-expression analysis revealed several potentially important genes which likely function as putative novel players in response to Fe and Pi deficiency or in cross-talk between Fe-deficient responses and Pi-deficient signaling.
Electronic supplementary material
The online version of this article (doi:10.1186/s13104-015-1524-y) contains supplementary material, which is available to authorized users.
Keywords: Iron deficiency, Phosphate deficiency, RNA-seq, Co-expression, Interaction
Background
The evolutionary ability of iron (Fe) to change oxidation states between Fe(III) and Fe(II) renders it irreplaceably important in many essential processes associated with basic redox reactions, such as in photosynthesis, respiration and many vital enzymatic reactions [1–5]. Although Fe is abundant in the earth’s crust, it is one of the least available elements for plants in aerobic soils with neutral to basic pH [1–5]. Approximately 30 % of the land worldwide consists of alkaline soils, leading to a demand in bioavailable Fe for plant fitness [1, 5, 6]. As a consequence, Fe deficiency is a major constraint in crop yield and quality [7]. In contrast, in acidic and anaerobic conditions, accumulation of excess Fe is toxic to plant growth and development due to formation of potentially harmful reactive oxygen species (ROS). Plants therefore must tightly regulate cellular Fe homeostasis to allow for effective acquisition, distribution and utilization of Fe [1, 8, 9].
Under Fe-deficient conditions, Arabidopsis (Arabidopsis thaliana) and other dicotyledonous and non-graminaceous monocotyledonous plants use a reduction strategy, referred to as strategy I [10], to increase Fe bioavailability. In this strategy, acidification of the rhizosphere mediated by the H+-translocating P-type ATPase AHA2 [6, 11] occurs as the first step, which leads to an increase in the concentration of chelated Fe(III). Fe(III) is subsequently reduced to soluble Fe(II) by the root surface-localized ferric chelate reductase FERRIC-REDUCTION OXIDASE2 (FRO2) [12]. Soluble Fe(II) is then transported into epidermal cells by the Fe-REGULATED TRANSPORTER1 (IRT1) [13]. Being the major components of the Fe acquisition system, FRO2 and IRT1 are the major players controlling entry of Fe from the soil into cells. At the transcriptional level, expression of both genes is coordinately regulated by the basic helix-loop-helix (bHLH) transcription factor FER-LIKE Fe DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT), but not the transcription factor POPEYE, which is also involved in Fe homeostasis [14–17]. FIT forms heterodimers with bHLH38 and bHLH39 and positively regulates a subset of Fe-responsive genes, including three key genes required for Fe acquisition [12–14, 18, 19]. Recent studies have shown that the transcription factors bHLH100 and bHLH101, which belong to the Ib sub-group of bHLH proteins, are also involved in Arabidopsis Fe deficiency responses by interacting with FIT [20] or via a FIT-independent manner [21].
Studies have shown that expression of FRO2 and IRT1 is tightly controlled both locally and systemically [22, 23]. However, in some cases disrupted Fe signaling in several mutants, such as frd3 [24, 25], opt3 [26] and the quadruple nicotianamine synthase mutant nas4x-1 [27] in Arabidopsis, dgl and brz mutants [28–30] in pea (Pisum sativum) and the chloronerva mutant chln [31] in tomato (Solanum lycopersicum), constitutively activates expression of Fe acquisition genes even under sufficient Fe conditions. By contrast, FRO2 and IRT1 expression has been documented to be decreased under phosphate (Pi)-deficient conditions [32–36]. Currently, the predominate explanation for decreased expression of Fe acquisition genes under Pi-deficient conditions is that Pi deficiency results in enhanced Fe accessibility to plants in the media, which leads to an over accumulation of Fe in plants, subsequently causing down-regulated expression of Fe-responsive genes. However, if the Pi-deficient media without available Fe or with low concentrations of Fe, does the down-regulated expression of Fe-responsive genes occur? A recent report showed that PHOSPHATE STARVATION RESPONSE1 (PHR1), a major regulator of the Pi deficiency response, could bind the promoter of the Fe storage gene Ferritin1 through the imperfect palindromic sequence motif P1BS (PHR1 binding sequences, GNATATNC), strongly supporting the link between Fe and Pi homeostasis [37]. However, it remains an open question whether this link exists or not in phr1 mutant plants.
Moreover, although down-regulation of Fe deficient-induced Fe acquisition genes under Pi deficient conditions has been documented [33, 38, 39], knowledge regarding genome-wide transcriptional changes of Fe-responsive genes under Pi deficiency remains unavailable, and the processes of the genes involved are largely unknown. To provide systemic information about transcriptional changes in Fe-responsive genes under Pi deficiency and to further extend knowledge of the relationship between Fe and Pi at the transcript level, we mined and re-analyzed previous RNA sequencing (RNA-seq) data sets relative to transcriptome profiling in Fe-deficient [40] and Pi-deficient Arabidopsis roots [36], with an emphasis on 579 overlapping genes that respond to both Fe and Pi deficiency. We revealed that a subset of 137 genes had a twofold or greater change in abundance under either of the treatments. A subset of 90 genes with an increased abundance under Fe deficiency, but a decrease under Pi deficiency, may be critical for Fe responses under Pi-deficient conditions. By gene clustering and root-specific co-expression analysis, we revealed several potentially important genes that likely function as putative novel players in response to Fe and Pi deficiency or in the cross talk between Fe deficient responses and phosphate-deficient signaling, which may be determined in follow-up experiments. Finally, we found that FIT-regulated genes were down-regulated by Pi deficiency, and an extent of Fe in the Pi deficient media is required for this down-regulation, suggesting that, besides FIT, PHR1, Fe itself might be a critical factor involved in the transcriptional regulation under both Pi- and Fe-deficiency.
Results
Genes responsible for Fe and Pi deficiency in Arabidopsis roots
Previously published RNA-seq data sets [36, 40] relative to Fe and Pi deficiency in Arabidopsis roots were re-analyzed, and differentially expressed genes (P < 0.05) upon Fe deficiency were compared with those (P < 0.05) exposed to Pi deficiency. Subsequent analyses focused on the 579 overlapping genes (Additional file l) as shown in Fig. 1. Of the 579 genes, 137 showed an increase or decrease in transcript abundance, with fold changes greater than twofold under either of the treatments (Table 1; Additional file 2). Fe acquisition genes FRO2 and IRT1, copper transporter COPT2, Fe(II)-dependent oxygenase gene AT3G12900, cytochrome P450 CYP82C4 (AT4G31940), mannose-binding lectin protein gene AT1G52120, glutathione transferase lambda 1 GSTL1 and amino acid transporter gene AT5G38820 showed the strongest induction under Fe deficiency and were up-regulated by more than 50-fold (Table 1; Fig. 2a). Excluding AT1G52120, these genes were among the most repressed under Pi deficiency and were down-regulated by two to more than tenfold (Table 1; Fig. 2a). Genes encoding transcriptional factor bHLH039, ZIP9, zinc binding protein (AT1G74770), ATROPGEF10, receptor like protein 24 RLP24, phloem protein 2-B6 and other functionally unknown proteins were among the second group of highly induced genes following Fe deficiency and were up-regulated by more than fivefold (Table 1; Fig. 2a, b). The most induced genes following Pi deficiency were AtOCT1, an unknown protein gene AT5G20790 and a major facilitator protein gene AT1G30560, which were induced by more than 50-fold (Table 1; Fig. 2c). Highly induced genes under Pi deficiency were ATPS3 (phosphate starvation-induced gene 3), SQD2 (sulfoquinovosyl diacylglycerol 2) and U-box domain-containing protein kinase gene AT5G65500, with changes more than fivefold (Table 1; Fig. 2d, e). Interestingly, a subset of genes involved in lignin biosynthesis was induced by both Fe and Pi deficiency (Table 1; Fig. 2f).
Fig. 1.
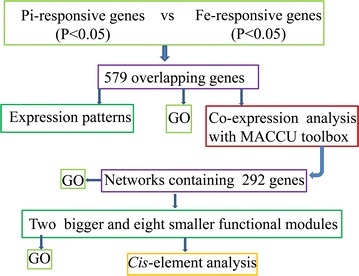
Bioinformatic analysis scheme of the 579 differentially expressed overlapping genes regulated by both iron and phosphate deficiency in Arabidopsis roots
Table 1.
Subset of 137 of the 579 overlapping genes with more than twofold changes in transcript abundance due to either of the treatments
| AGI | Annotation | Mean (−Fe/+Fe) | SD | Mean (−Pi/+Pi) | SD |
|---|---|---|---|---|---|
| At3G12900 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 612.44 | 199.47 | 0.06 | 0.10 |
| At4G31940 | CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 | 184.70 | 3.63 | 0.08 | 0.01 |
| At1G52120 | Mannose-binding lectin superfamily protein | 157.65 | 59.83 | 6.13 | 2.70 |
| At3G46900 | COPT2, copper transporter 2 | 71.15 | 33.39 | 0.40 | 0.36 |
| At1G01580 | ATFRO2, FRD1, FRO2, ferric reduction oxidase 2 | 59.73 | 10.56 | 0.35 | 0.05 |
| At5G02780 | GSTL1, glutathione transferase lambda 1 | 57.98 | 10.87 | 0.69 | 0.06 |
| At4G19690 | ATIRT1, IRT1, iron-regulated transporter 1 | 54.72 | 7.88 | 0.26 | 0.04 |
| At5G38820 | Transmembrane amino acid transporter family protein | 54.35 | 11.69 | 0.43 | 0.09 |
| At3G56980 | BHLH039, ORG3, basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 34.23 | 13.48 | 0.73 | 0.18 |
| At3G13610 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 10.04 | 0.65 | 1.63 | 0.25 |
| At1G73120 | Unknown protein | 8.98 | 2.09 | 0.28 | 0.11 |
| At1G73220 | 1-Oct, AtOCT1, organic cation/carnitine transporter1 | 7.38 | 6.18 | 196.69 | 139.63 |
| At4G33020 | ATZIP9, ZIP9, ZIP metal ion transporter family | 7.09 | 2.01 | 2.21 | 0.97 |
| At5G05250 | Unknown protein | 6.62 | 0.95 | 0.57 | 0.10 |
| At3G61410 | BEST Arabidopsis thaliana protein match is: U-box domain-containing protein kinase family protein (TAIR:AT2G45910.1) | 6.53 | 1.32 | 3.79 | 0.43 |
| At1G74770 | Zinc ion binding | 6.43 | 0.50 | 0.66 | 0.07 |
| At5G19560 | ATROPGEF10, ROPGEF10, ROP uanine nucleotide exchange factor 10 | 5.33 | 0.51 | 2.12 | 0.20 |
| At2G02310 | AtPP2-B6, PP2-B6, phloem protein 2-B6 | 5.30 | 2.01 | 0.49 | 0.12 |
| At2G33020 | AtRLP24, RLP24, receptor like protein 24 | 5.05 | 1.74 | 0.48 | 0.33 |
| At3G59880 | Unknown protein | 4.58 | 2.42 | 2.51 | 0.54 |
| At5G01060 | Protein kinase protein with tetratricopeptide repeat domain | 4.44 | 0.17 | 1.70 | 0.10 |
| At5G04950 | ATNAS1, NAS1, nicotianamine synthase 1 | 4.39 | 0.50 | 0.49 | 0.01 |
| At3G57157 | Other RNA | 4.28 | 0.37 | 9.81 | 1.46 |
| At3G60330 | AHA7, HA7, H(+)-ATPase 7 | 4.27 | 0.69 | 1.79 | 0.21 |
| At3G21500 | DXPS1, 1-deoxy-d-xylulose 5-phosphate synthase 1 | 3.76 | 1.94 | 0.40 | 0.26 |
| At3G21240 | 4CL2, AT4CL2, 4-coumarate:CoA ligase 2 | 3.73 | 0.16 | 1.25 | 0.09 |
| At3G50710 | F-box/RNI-like/FBD-like domains-containing protein | 3.57 | 1.09 | 2.71 | 1.32 |
| At1G18910 | Zinc ion binding | 3.33 | 0.13 | 0.78 | 0.04 |
| At5G48657 | Defense protein-related | 3.33 | 0.71 | 1.36 | 0.23 |
| At1G01380 | ETC1, Homeodomain-like superfamily protein | 3.22 | 1.09 | 3.73 | 1.22 |
| At1G51680 | 4CL.1, 4CL1, AT4CL1, 4-coumarate:CoA ligase 1 | 3.22 | 0.06 | 1.27 | 0.19 |
| At5G54790 | Unknown protein | 3.20 | 0.75 | 2.02 | 0.19 |
| At2G01880 | ATPAP7, PAP7, purple acid phosphatase 7 | 3.14 | 0.21 | 3.31 | 0.81 |
| At5G19970 | Unknown protein | 2.81 | 0.43 | 0.57 | 0.07 |
| At5G65500 | U-box domain-containing protein kinase family protein | 2.73 | 0.60 | 6.26 | 0.42 |
| At4G12735 | Unknown protein | 2.67 | 1.20 | 1.87 | 0.47 |
| At3G57160 | Unknown protein | 2.57 | 0.55 | 1.46 | 0.23 |
| At5G26820 | ATIREG3, IREG3, IREG3, MAR1, RTS3, iron-regulated protein 3 | 2.54 | 0.28 | 0.77 | 0.19 |
| At4G30490 | AFG1-like ATPase family protein | 2.52 | 0.32 | 1.40 | 0.08 |
| At1G78230 | Outer arm dynein light chain 1 protein | 2.51 | 0.57 | 0.59 | 0.22 |
| At3G18290 | BTS, EMB2454, zinc finger protein-related | 2.51 | 0.14 | 0.76 | 0.09 |
| At1G51870 | Protein kinase family protein | 2.42 | 0.42 | 2.25 | 0.83 |
| At5G22555 | Unknown protein | 2.37 | 0.58 | 4.13 | 2.07 |
| At1G53310 | ATPEPC1, ATPPC1, PEPC1, PPC1, phosphoenolpyruvate carboxylase 1 | 2.34 | 0.19 | 2.21 | 0.25 |
| At3G51570 | Disease resistance protein (TIR-NBS-LRR class) family | 2.32 | 0.58 | 2.70 | 1.18 |
| At5G22890 | C2H2 and C2HC zinc fingers superfamily protein | 2.32 | 0.53 | 1.91 | 0.25 |
| At4G22980 | Pyridoxal phosphate (PLP)-dependent transferases superfamily protein (TAIR:AT5G51920.1) | 2.30 | 0.28 | 0.56 | 0.14 |
| At1G14190 | Glucose-methanol-choline (GMC) oxidoreductase family protein | 2.29 | 0.41 | 0.78 | 0.08 |
| At1G48300 | Unknown protein | 2.27 | 0.25 | 0.85 | 0.07 |
| At1G24320 | Six-hairpin glycosidases superfamily protein | 2.26 | 0.27 | 0.78 | 0.09 |
| At1G62422 | Unknown protein | 2.25 | 0.43 | 1.37 | 0.10 |
| At4G38950 | ATP binding microtubule motor family protein | 2.25 | 0.45 | 1.50 | 0.15 |
| At3G47420 | ATPS3, PS3, phosphate starvation-induced gene 3 | 2.24 | 0.27 | 12.82 | 5.53 |
| At5G26320 | TRAF-like family protein | 2.24 | 0.26 | 2.95 | 0.37 |
| At2G43570 | CHI, chitinase, putative | 2.22 | 0.87 | 2.26 | 0.45 |
| At4G26890 | MAPKKK16, mitogen-activated protein kinase kinase kinase 16 | 2.16 | 0.42 | 3.00 | 0.85 |
| At5G13910 | LEP, Integrase-type DNA-binding superfamily protein | 2.14 | 0.15 | 1.25 | 0.16 |
| At5G27920 | F-box family protein | 2.14 | 0.15 | 1.29 | 0.15 |
| At3G15510 | ANAC056, ATNAC2, NAC2, NARS1, NAC domain containing protein 2 | 2.13 | 0.62 | 0.71 | 0.15 |
| At5G53850 | Haloacid dehalogenase-like hydrolase family protein | 2.09 | 0.06 | 0.96 | 0.01 |
| At2G18193 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 2.07 | 0.26 | 1.29 | 0.01 |
| At2G32960 | Phosphotyrosine protein phosphatases superfamily protein | 2.07 | 0.23 | 4.79 | 1.23 |
| At1G64590 | NAD(P)-binding Rossmann-fold superfamily protein | 2.06 | 0.13 | 2.39 | 0.70 |
| At5G48930 | HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | 2.04 | 0.17 | 1.16 | 0.07 |
| At2G14210 | AGL44, ANR1, AGAMOUS-like 44 | 2.01 | 0.37 | 0.82 | 0.09 |
| At5G20790 | Unknown protein | 0.32 | 0.13 | 90.27 | 24.26 |
| At1G30560 | Major facilitator superfamily protein | – | – | 55.93 | 32.45 |
| At5G01220 | SQD2, sulfoquinovosyldiacylglycerol 2 | 0.86 | 0.06 | 10.49 | 3.02 |
| At1G72070 | Chaperone DnaJ-domain superfamily protein | 1.72 | 0.62 | 6.60 | 3.34 |
| At3G52720 | ACA1, ATACA1, CAH1, alpha carbonic anhydrase 1 | 0.40 | 0.20 | 5.56 | 3.11 |
| At1G23140 | Calcium-dependent lipid-binding (CaLB domain) family protein | 1.58 | 0.43 | 5.33 | 1.19 |
| At3G52190 | PHF1, phosphate transporter traffic facilitator1 | 1.22 | 0.12 | 5.25 | 0.61 |
| At3G56040 | UGP3, UDP-glucose pyrophosphorylase 3 | 0.74 | 0.18 | 4.98 | 0.71 |
| At3G02870 | VTC4, Inositol monophosphatase family protein | 0.79 | 0.06 | 4.64 | 0.29 |
| At3G16390 | NSP3, nitrile specifier protein 3 | 0.67 | 0.08 | 4.55 | 0.59 |
| At3G07350 | Protein of unknown function (DUF506) | 0.54 | 0.07 | 4.40 | 0.09 |
| At3G19970 | Alpha/beta-hydrolases superfamily protein | 1.58 | 0.08 | 4.37 | 0.76 |
| At1G15040 | Class I glutamine amidotransferase-like superfamily protein | 0.60 | 0.04 | 3.73 | 1.12 |
| At3G12500 | ATHCHIB, B-CHI, CHI-B, HCHIB, PR-3, PR3, basic chitinase | 0.52 | 0.05 | 3.05 | 0.75 |
| At1G18970 | GLP4, germin-like protein 4 | 0.71 | 0.12 | 3.04 | 0.41 |
| At2G29000 | Leucine-rich repeat protein kinase family protein | 1.85 | 0.60 | 3.03 | 0.79 |
| At3G53620 | AtPPa4, PPa4, pyrophosphorylase 4 | 0.82 | 0.03 | 2.96 | 0.19 |
| At1G14220 | Ribonuclease T2 family protein | 0.74 | 0.18 | 2.89 | 0.27 |
| At4G32480 | Protein of unknown function (DUF506) | 1.35 | 0.19 | 2.85 | 1.36 |
| At3G06962 | Other RNA | 1.82 | 0.49 | 2.74 | 0.44 |
| At1G11920 | Pectin lyase-like superfamily protein | – | – | 2.68 | 0.51 |
| At1G08650 | ATPPCK1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 | 1.77 | 0.16 | 2.58 | 0.21 |
| At4G04040 | MEE51, Phosphofructokinase family protein | 0.83 | 0.04 | 2.50 | 0.22 |
| At5G57540 | AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 | 1.53 | 0.11 | 2.33 | 0.36 |
| At5G40860 | Unknown protein | 1.51 | 0.11 | 2.32 | 0.81 |
| At1G68740 | PHO1;H1, EXS (ERD1/XPR1/SYG1) family protein | 0.39 | 0.08 | 2.31 | 0.44 |
| At3G32040 | Terpenoid synthases superfamily protein | 1.32 | 0.21 | 2.30 | 0.11 |
| At2G25240 | Serine protease inhibitor (SERPIN) family protein | 1.53 | 0.15 | 2.27 | 0.08 |
| At2G16430 | ATPAP10, PAP10, purple acid phosphatase 10 | 0.68 | 0.08 | 2.26 | 0.33 |
| At4G30670 | Putative membrane lipoprotein | 1.61 | 0.16 | 2.22 | 0.17 |
| At3G10420 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 1.24 | 0.08 | 2.20 | 0.35 |
| At2G42600 | ATPPC2, PPC2, phosphoenolpyruvate carboxylase 2 | 0.77 | 0.05 | 2.18 | 0.09 |
| At4G11650 | ATOSM34, OSM34, osmotin 34 | 0.67 | 0.10 | 2.16 | 0.38 |
| At2G23960 | Class I glutamine amidotransferase-like superfamily protein | 1.55 | 0.27 | 2.13 | 0.21 |
| At1G05300 | ZIP5, zinc transporter 5 precursor | 0.49 | 0.05 | 2.13 | 0.11 |
| At3G13110 | ATSERAT2;2, SAT-1, SAT-A, SAT-M, SAT3, SERAT2;2, serine acetyltransferase 2;2 | 0.83 | 0.08 | 2.12 | 0.22 |
| At2G22290 | ATRAB-H1D, ATRAB6, ATRABH1D, RAB-H1D, RABH1d, RAB GTPase homolog H1D | 1.91 | 0.66 | 2.10 | 0.59 |
| At4G20160 | RING/U-box superfamily protein (TAIR:AT1G30860.1) | 1.71 | 0.10 | 2.07 | 0.17 |
| At1G20390 | Transposable element gene | 0.56 | 0.11 | 2.07 | 0.04 |
| At5G20280 | ATSPS1F, SPS1F, sucrose phosphate synthase 1F | 0.77 | 0.06 | 2.05 | 0.17 |
| At5G01870 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | 0.71 | 0.10 | 2.05 | 0.51 |
| At3G05858 | Unknown protein | 1.64 | 0.09 | 2.01 | 0.47 |
| At2G17280 | Phosphoglycerate mutase family protein | 1.16 | 0.02 | 2.01 | 0.47 |
| At1G26250 | Proline-rich extensin-like family protein | 0.55 | 0.16 | 2.00 | 0.82 |
| At4G09110 | RING/U-box superfamily protein | – | – | 0.77 | 0.04 |
Change in gene expression is shown as the mean and standard deviation (SD)
No change is indicated as “−” in cases when a gene transcript was not determined (read number = zero) in any of the biological replicates under control conditions
Fig. 2.
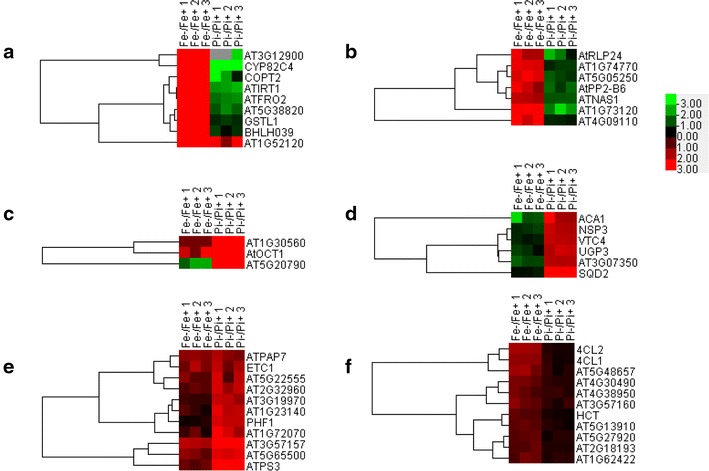
Hierarchical cluster analysis of 137 overlapping genes with greater than twofold changes in transcript abundance in Arabidopsis roots grown under Fe or Pi deficiency. a–f Indicate six representative sub-clusters. Complete clustering results of the 137 overlapping genes can be found in Additional file 2. Results shown are parts of the representative clusters from Additional file 2. Fold change in transcript abundance was defined as transcript level (reads per kilobase per million mapped reads) in Fe-deficient (Fe−) conditions divided by the level in normal conditions (Fe+), with three biological repeats. The same strategy was applied to Pi treatment. The color key indicates log2 transformed intensity; grey indicates that the number is missing
Gene ontology (GO) analysis of the 579 overlapping genes revealed that stress-related processes, including ‘response to salt stress’, ‘response to oxidative stress’ and ‘response to zinc ion’, were enriched (Additional file 3), while analysis of the subset of 137 genes showed that Fe response-related processes, including ‘cellular response to nitric oxide’, ‘cellular response to Fe ion’ and ‘cellular Fe ion homeostasis’, were also enriched (Additional file 4).
Gene expression patterns of overlapping genes
Expression patterns of the 579 overlapping genes were divided into four types according to changes at the transcript level under two stress conditions (Fig. 3a). Type one was composed of 223 genes (of which 24 genes were down-regulated by more than twofold) with decreased transcription under both Pi and Fe deficiency. GO analysis of this group of genes revealed that the processes of ‘embryo development ending in seed dormancy’, ‘microtubule-based process’ and ‘chloroplast organization’, were most enriched (Fig. 3b). In contrast, transcript abundance of 169 genes in the type two category were shown to be increased following both Pi and Fe deficiency, with processes of ‘glucosinolate and leucine biosynthesis’ and ‘UV response’ being enriched (Fig. 3b). Type three was composed of 97 genes with increased transcript abundance under Pi deficiency, but decreased under Fe deficiency. In contrast, transcript abundance of the 90 genes in type four were decreased under Pi deficiency and increased under Fe deficiency. GO enrichment analysis showed that the zinc-related processes ‘response to zinc ion’, ‘zinc ion transport’ and ‘galactose metabolic process’ were enriched in type three (Fig. 3b). Iron-related processes ‘cellular response to Fe ion’, ‘cellular response to nitric oxide’, ‘cellular response to ethylene stimulus’, ‘cellular Fe ion homeostasis’ and ‘protein import into nucleus’ were enriched in type four (Fig. 3b).
Fig. 3.
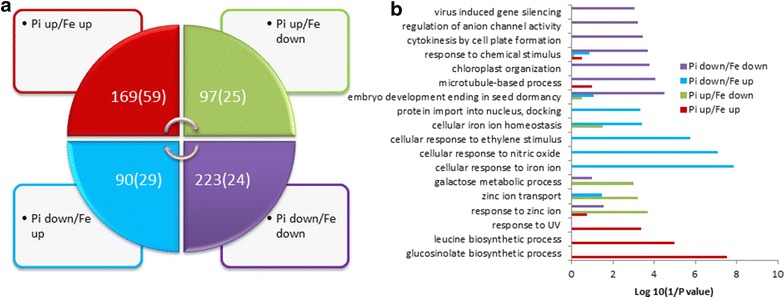
Expression patterns (a) and differential gene ontology (GO) enrichment analysis of the four types of 579 overlapping genes (b). Bracketed numbers in a indicate the number of genes with greater than a twofold change in transcript level under either of the stresses
Identification of overlapping gene modules by co-expression analysis
Stress-specific variability in gene expression may occur at the individual gene level, but can also occur in a coordinated manner. To determine functional modules, co-expression networks (i.e., groups of genes that show similar expression patterns under diverse conditions) of the 579 overlapping genes were generated using MACCU software [41]. Pairwise co-expressed genes were selected with a Pearson correlation coefficient cutoff of 0.7 [36, 41]. The 300 publicly available microarrays that were mined for co-expression analysis discriminated between root-related experiments. As such, the co-expression relationships reported herein are restricted to roots [42–44]. Co-expression relationships between these genes were visualized using Cytoscape (http://www.cytoscape.org). This analysis yielded a network composed of 292 nodes (genes) and 1595 edges (correlations between genes; Additional file 5). The network can be further divided into two large and eight small clusters (modules). The largest module was composed of 210 genes, most of which are associated with stress (Additional file 6). GO enrichment analysis revealed that the biological processes ‘glucosinolate biosynthetic process’, ‘response to cadmium ion’, ‘response to salt stress’ and ‘leucine biosynthetic process’ were most enriched in this module (Additional file 7). Via connection to the zinc binding protein gene AT1G74770, two marker genes strongly induced by Fe deficiency, IRT1 (AT4G19690) and CYP82C4 (AT4G31940), were associated with this module.
Co-expression analysis of the subset of 137 overlapping genes with changes greater than twofold yielded a network consisting of 48 nodes and 56 edges. The Fe deficiency-regulated marker genes IRT1 and CYP82C4 (AT4G31940) remained in the network (Fig. 4a). This network can be divided into one large (26 genes) and six small clusters (Fig. 4a). Detailed expression information of these genes upon Fe or Pi deficiency is shown in Fig. 4b. GO enrichment analysis of the genes involved in the co-expression network revealed that the biological processes ‘cellular response to nitric oxide’, ‘cellular response to Fe ion’ and ‘cellular response to ethylene stimulus’ were enriched (Table 2).
Fig. 4.
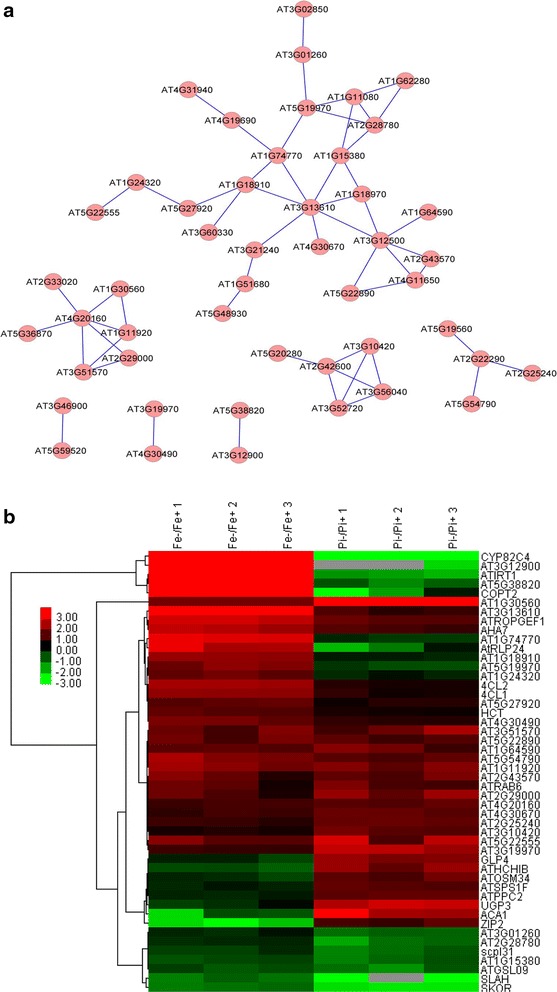
Co-expression relationships of the 137 differentially expressed genes with changes greater than twofold (a), and heat map of genes involved in network construction (b)
Table 2.
Gene ontology enrichment was assessed using GOBUin the 48 genes in the large sub-network shown in Fig. 4a (elim, P < 0.01)
| GOID | P value (elim) | GO name |
|---|---|---|
| GO: 0071732 | 1.51E−06 | Cellular response to nitric oxide |
| GO: 0071281 | 9.74E−06 | Cellular response to iron ion |
| GO: 0009620 | 1.11E−04 | Response to fungus |
| GO: 0071369 | 1.52E−04 | Cellular response to ethylene stimulus |
| GO: 0006829 | 1.54E−04 | Zinc ion transport |
| GO: 0015794 | 0.001428 | Glycerol-3-phosphate transport |
| GO: 0015678 | 0.001428 | High-affinity copper ion transport |
| GO: 0010421 | 0.002855 | Hydrogen peroxide-mediated programmed cell death |
| GO: 0009805 | 0.004279 | Coumarin biosynthetic process |
| GO: 0009871 | 0.004279 | Jasmonic acid and ethylene-dependent systemic resistance, ethylene mediated signaling pathway |
| GO: 0009411 | 0.004536 | Response to UV |
| GO: 0006873 | 0.005599 | Cellular ion homeostasis |
| GO: 0009963 | 0.007122 | Positive regulation of flavonoid biosynthetic process |
| GO: 0009311 | 0.007707 | Oligosaccharide metabolic process |
| GO: 0006828 | 0.009958 | Manganese ion transport |
To search for potentially functional novel modules, co-expression analysis was applied to the subset of 90 genes that were induced by Fe deficiency, but down-regulated by Pi deficiency (Additional file l). A network containing 26 nodes and 17 edges was created using the same criteria (Fig. 5). The network can be divided into 10 small clusters (none with more than ten nodes), with the largest one containing several Fe-responsive marker genes and one transcriptional factor WRKY 17 (Fig. 5). The second largest cluster was composed of four genes, including the Pi homeostasis regulator SIZ1 (Fig. 5). For 97 genes induced by Pi deficiency but repressed by Fe deficiency, co-expression analysis resulted in a network containing 26 nodes and 29 edges that were divided into one large and two small clusters (Additional file 8).
Fig. 5.
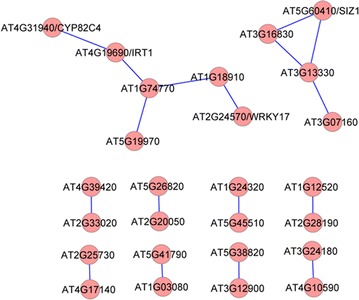
Co-expression relationships of the 90 differentially expressed overlapping genes induced by Fe deficiency but repressed by Pi starvation
Analysis of P1BS motif in 35 genes induced by Fe deficiency but down regulated by Pi deficiency
A subset of 35 genes in the type four category had an increase in transcript abundance under Fe deficiency but a decrease under Pi deficiency, with changes more than 1.5-fold (Additional file 9). A 3000 bp sequence upstream of the translation start (named −3000 bp) of these 35 genes was retrieved from TAIR10 and used to search the PHR1 recognition sequence 5′-GNATATNC-3′ (P1BS motif). Results showed that 12 of the 35 genes contained at least one P1BS motif, and seven of the 12 genes contained the P1BS motif within −1000 bp of their promoter regions (Table 3). In total, 11 P1BS patterns were hit in the 12 genes, with 5′-GTATATGC-3′ and 5′-GTATATTC-3′ being the most frequent (5 and 3 hits out of 18 total hits, respectively).
Table 3.
Distribution of the P1BS motif in promoter regions of 12 genes
| AGI | Matching positions | Hit pattern (5′–3′) | |
|---|---|---|---|
| Start | End | ||
| AT1G01580 | 2660 | 2667 | GTATATTC |
| AT1G01580 | 2701 | 2708 | GTATATTC |
| AT1G18910 | 134 | 141 | GGATATCC |
| AT1G18910 | 359 | 366 | GTATATAC |
| AT1G18910 | 1311 | 1318 | GTATATGC |
| AT1G24320 | 2052 | 2059 | GCATATCC |
| AT1G48300 | 1167 | 1174 | GCATATTC |
| AT2G02310 | 645 | 652 | GCATATAC |
| AT3G12900 | 445 | 452 | GTATATTC |
| AT3G18290 | 129 | 136 | GTATATAC |
| AT3G18290 | 703 | 710 | GCATATGC |
| AT3G56980 | 153 | 160 | GTATATGC |
| AT3G56980 | 2484 | 2491 | GTATATGC |
| AT3G56980 | 2484 | 2491 | GTATATGC |
| AT4G00910 | 2514 | 2521 | GTATATGC |
| AT4G19690 | 838 | 845 | GAATATCC |
| AT4G22980 | 1641 | 1648 | GAATATAC |
| AT5G02780 | 369 | 376 | GAATATGC |
Down-regulation of Fe-acquisition genes upon Pi deficiency is dependent on Fe concentration in the media
To determine how Fe acquisition genes are down-regulated by Pi deficiency and whether this down-regulation is dependent on PHR1, we investigated changes in genes that were most induced by Fe deficiency (including the two Fe acquisition genes FRO2 and IRT1) at the transcript level in wild type and the phr1 mutant under varied growth conditions as follows: Pi deficiency (−Pi, in which the concentration of Fe was 40 µM), Fe deficiency (−Fe), both Pi- and Fe-deficient (−Pi−Fe), Pi deficiency with low Fe concentration (−Pi + 5 µM Fe) and control conditions (+Pi+Fe). Null expression of PHR1 in the phr1 mutant was first verified by quantitative real-time PCR (qPCR) (the ct value of the reference is around 20 cycles while the ct value of the PHR1 is around 34 cycles in the phr1 mutant plants). As shown in Fig. 6a and in agreement with previously reported results [45], transcriptional expression of PHR1 was not significantly regulated by Pi deficiency in wild type plants and could not be detected in phr1 mutant plants under both Pi sufficient and deficient conditions. As a control, the expression of SPX1 [46, 47], a Pi-responsive marker gene, was significantly induced by Pi deficiency. Consistent with our transcriptomic data, transcriptional expression of the Fe acquisition genes IRT1 and FRO2 as well as the Fe deficiency-induced marker gene CYP82C4 was significantly down-regulated under Pi deficiency in both Col-0 and phr1 roots (Fig. 6a). Because all these Fe-responsive genes tested were mainly regulated by transcription factor FIT [14], we thus tested whether the expression of FIT itself was affected or not by Pi deficiency. As shown in Fig. 6a, the expression level of FIT was significantly lower in Pi-deficient roots than in Pi-sufficient roots in wild type plants. In addition to FIT, another transcription factor PYE [34], regulating the expression of another subset of Fe-responsive genes, has been reported to be required for plant Fe homeostasis. However, both PYE and its target AT1G74790 were not affected by Pi deficiency (Fig. 6a). To determine whether this down-regulation is dependent on Fe concentrations in the media, we compared transcript abundance under Pi deficiency with different Fe concentrations in both wild type and mutant plants. In wild type plant roots, all genes evaluated were dramatically induced under Fe deficiency (−Fe) but repressed under Pi deficiency (−Pi) compared to expression under control conditions (+Fe+Pi). Fe deficient-induced up-regulation was not blocked but attenuated by the absence of Pi in Fe-deficient media (−Fe−Pi), while Pi deficient-induced down-regulation was dramatically attenuated by 5 µM Fe (low Fe concentration) in the media (Fig. 6b). Similar to results in wild type plants, these genes were significantly induced under Fe deficiency and Fe and Pi deficiency (−Fe−Pi) in the phr1 mutant roots (Fig. 6b). However, down-regulation of gene expression under Pi deficiency was not all significantly attenuated by 5 µM Fe in Pi-deficient media in the phr1 mutant roots (Fig. 6b).
Fig. 6.
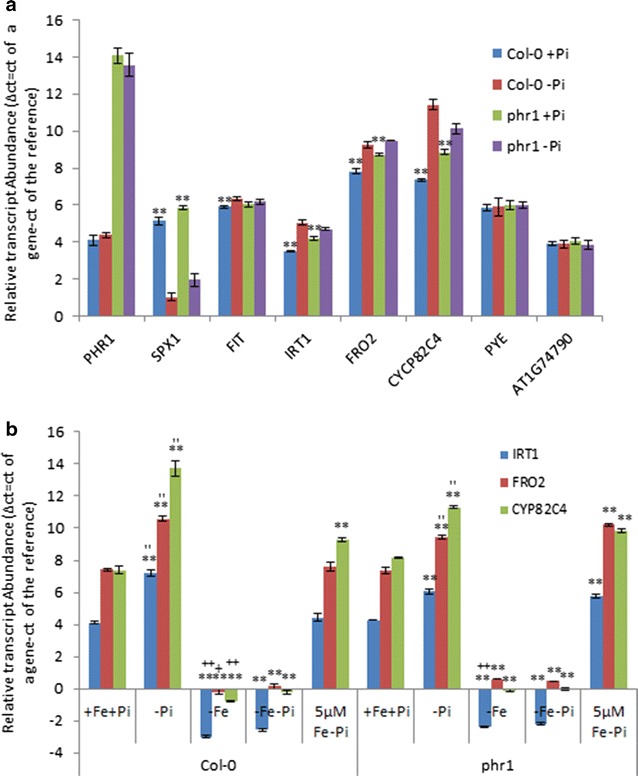
Reverse transcription–quantitative PCR (RT–qPCR) detected expression of Pi- and Fe-responsive marker genes under Pi deficiency (a) or Fe deficiency or Pi deficiency without or with low Fe concentrations (b). Total RNA was isolated from roots in wild type or phr1 mutant plants and qPCR was performed. Expression levels are relative to normal controls. Error bars represent SD of biological replicates from three independent experiments. Data significantly different from the corresponding controls are indicated (‘−Pi’ versus ‘+Pi’, *P < 0.05, **P < 0.01; ‘−Fe’ versus ‘−Fe−Pi’, +P < 0.05, ++P < 0.01; ‘−Pi’ versus ‘5 µMFe−Pi’, ′P < 0.05, ″P < 0.01; Student’s t test)
Discussion
As an essential element for all living organisms, particularly as a major constraint in crop yield and quality, Fe deficiency responses in plants have been extensively studied in the last decade [1, 6, 9]. With the emergence of high throughput research platforms, many genes and proteins have been revealed to be regulated by Fe deficiency [40, 48–55]. Evidence has shown that transcriptional expression of some Fe-responsive genes can be altered due to deficiencies or excesses of mineral elements, including cross-talk between Fe and other mineral elements. For example, the Fe transporter LeIRT1 is reported to be up-regulated by potassium (K) deficiency, as revealed by microarray analysis [56], and expression of the K transporter gene LeKC1 was induced not only by K starvation but also by Fe deficiency [56]. Via comprehensive analysis of Fe-responsive protein kinase (PK) and protein phosphatase (PP) genes, we found that strong over-representation of PK and PP genes that encode proteins is involved in K homeostasis, which supports the link between potassium uptake and Fe deficiency [44]. The ameliorative effect of K supply on Fe-deficient responses was previously reported [57]. Although several lines of evidence have suggested a link between Pi and Fe homeostasis [32–35, 37–39, 58], little genome-wide information on transcriptional expression changes in Fe-responsive genes under Pi deficiency is available, and the biological processes that these genes are involved in remain elusive in Arabidopsis.
By mining previous RNA-seq data sets, we present comprehensive information on transcriptional expression of overlapping genes regulated by Fe and Pi deficiency in Arabidopsis roots. In total, 579 overlapping genes, or less than 20 % of all differentially expressed genes evaluated in each treatment, were responsive to both Fe and Pi deficiency. Only 137 of the 579 genes had greater than twofold changes in transcript abundance (Additional file l; Table 1). Many of the most strongly induced genes under Fe deficiency, such as AT3G12900, IRT1, FRO2, CYP82C4 and AT5G38820 [18, 41, 54], are among the 579 overlapping genes, while most of the strongest induced Pi deficiency-induced marker genes, such as pyridoxal phosphate phosphatase-related protein gene AT1G17710, transposable element gene AT2G04460, ATISP1, SPX3, APT1 and AT4, are not overlapping [33, 35]. GO enrichment analysis of the 137 genes with changes greater than twofold (Additional file 4) showed that Fe response-related processes such as ‘cellular response to Fe ion’ and ‘cellular Fe ion homeostasis’ were enriched, but none of the Pi response-related processes were pronounced, suggesting that plant responses to Fe deficiency might be more specific than responses to Pi deficiency under the conditions presented herein. The most strongly induced Fe-responsive genes were clustered together and down-regulated under Pi deficiency, except for AT1G52120 in which transcript abundance was increased under both stress conditions (Fig. 2a). In this cluster, IRT1, FRO2, and BHLH039 are known to be involved in Fe acquisition and transcriptional regulation, and COPT2 was confirmed to participate in cross talk between Fe deficiency responses and low phosphate signaling in a recent study [59]. Other genes in the group (Fig. 2a), such as AT3G12900, CYP82C4, AT5G38820 and AT1G52120, do not have defined functions currently, but may be involved in responses to Fe deficiency or Pi deficiency or cross-talk between Fe deficiency responses and phosphate-deficient signaling. Another group of interesting genes are AT5G20790, AtOCT1 and AT1G30560 (Fig. 2c) given that their transcriptional expression was among the most highly induced under Pi deficiency. In particular, both AT1G30560 and AtOCT1 were significantly up-regulated upon Fe deficiency, suggesting that these two genes might play important roles in responses to both stresses. In animals, organic cation/carnitine transporters (OCTs) are associated with homeostasis and distribution of various small endogenous amines (e.g. carnitine, choline) and detoxification of xenobiotics like nicotine. AtOCT1 has been reported to be involved in Arabidopsis root development. Knockout of AtOCT1 expression results in a higher degree of root branching compared to the wild type in vitro. This disordered development may be due to an inability to transport carnitine [60]. It has been well established that the number and length of lateral roots are increased under Pi deficiency in Arabidopsis and other plants. Therefore, whether AtOCT1-mediated transport of carnitine or related chemicals is involved in lateral root development under Pi deficiency remains elusive.
GO enrichment analysis of the 579 overlapping genes revealed that these Fe- and Pi-responsive genes were associated with diverse biological processes (Additional file 3), particularly with the GO categories ‘response to salt stress’, ‘response to oxidative stress’ and ‘response to zinc ion’ (Additional file 3). These results imply that acclimation of plants to Fe and Pi deficiency and possibly other nutritional stresses is associated with profound changes in the transcriptome, including stress-specific responses such as alteration of ribosome composition [43] and other general responses. Only four (AT1G27760, AT3G04720, AT4G11650 and AT5G24090) of the 21 genes associated with ‘response to salt stress’ had an increase in transcript abundance greater than 1.5-fold, suggesting that this common response to Fe and Pi deficiency might be less important than Fe response-related processes. GO enrichment analysis of the most responsive genes (i.e., those with greater than 1.5-fold change) revealed that Fe response-related processes, were enriched, but none of the Pi response-related processes were (Additional file 4),suggesting that Pi deficiency has more pronounced effects on Fe homeostasis than Fe deficiency has on Pi homeostasis.
Functional annotation of a given gene is the most important goal in modern molecular biology and is essential for understanding how the cell works. All omics studies are discovery tools and are not capable of defining gene function. The actual functions of differentially expressed genes under certain conditions discovered by high throughput platforms require further experimental evidence. However, current research platforms can discover hundreds to thousands of differentially expressed genes in a single run, and most of them are annotated as function unknown. Functional exploration of every differentially expressed gene without selection would be extremely laborious and impossible. Fortunately, co-expression analysis provides the option to choose genes of interest for further study. The basic idea of co-expression analysis is that genes that show transcriptionally coordinated expression patterns under diverse conditions are often functionally related [61], thus allowing functional predictions regarding genes with unknown functions inferred from their co-expression relationships with genes with known functions [62, 63]. Using co-expression analysis, we discovered ten, six and ten potentially critical regulatory modules with diverse nodes from inputs of the 579 (total overlapping genes), 137 (genes with changes greater than twofold) and 90 Fe deficiency-induced, Pi deficiency-repressed genes (Additional file 5; Figs. 4a, 5). Unexpectedly, only 50, 35 and 29 %, respectively, of the input genes were associated with formation of co-expression networks, suggesting that the majority of overlapping genes are functionally diverse and involved in a variety of biological processes. The network obtained from the group of 90 Fe deficiency-induced, Pi deficiency-repressed genes (Fig. 3a) is of particular interest. In this network, several genes may play important roles in responses to Fe and Pi deficiency. For instance, the gene AT1G74770 annotated with zinc ion binding protein showed a strong relationship with the Fe transporter IRT1, implying that this gene may be required for a Fe response. Another putative zinc ion binding protein encoding gene, AT1G18910, was shown to be connected to AT1G74770 and the transcription factor gene WRKY17, suggesting that these genes may also be involved in plant adaptation to Fe deficiency or zinc toxicity elicited by excess zinc under Fe deficiency.
It is generally accepted that a group of genes with similar expression patterns might be positively and/or negatively regulated by the same regulator(s). In Arabidopsis, the PHR1 transcription factor (TF) and its homolog PHL1 consist of the central regulatory system controlling transcriptional expression of a subset of Pi deficiency response genes by binding to the P1BS motif in promoter regions; while FIT and PYE are two major TFs regulating transcriptional expression of two subsets of Fe deficiency response genes. 35 out of the 579 genes, including Fe acquisition genes IRT1 and FRO2 and Fe responsive marker gene CYP82C4, were induced under Fe deficiency but down-regulated under Pi deficiency with changes greater than 1.5-fold. qPCR examination (Fig. 6a, b) confirmed that both IRT1 and FRO2 as well as CYP82C4, mainly regulated by FIT in response to Fe deficiency, were down-regulated by Pi deficiency in wild type plants, probably due to the decreased abundance of FIT (Fig. 6a). By contrast, the transcriptional expression of both PYE and its target AT1G74790 was not altered in response to Pi deficiency in both wild type and phr1 mutant plants. Taken together, these results suggest that FIT-regulated but not PYE-regulated Fe-response genes are affected by Pi deficiency and the down-regulation by Pi deficiency might be partially due to the down-regulation of FIT (since although the transcript abundance of FIT was not significantly different between Pi-sufficient and -deficient conditions in phr1 mutant plants, the transcriptional expression of IRT1 and FRO2 as well as CYP82C4 was still significantly down-regulated by Pi deficiency). In addition, under Pi sufficiency, although the transcript level of FIT was not different between wild type and phr1 mutant plants, the transcript abundance of IRT1 and FRO2 as well as CYP82C4 was still significantly down-regulated in the mutant plants (Fig. 6a, b). These results indicate that, besides FIT and P1BS motif (CYP82C4 doesn’t contain a P1BS motif in the −3000 bp sequence of its promoter region), some other factors may be involved in the down-regulation of gene expression under Pi deficiency. Indeed, only 34 % of the genes (12 of 35) contained at least one P1BS motif in promoter regions (−3000 bp sequence upstream of the translation start) and only 20 % (7 of 35) had a P1BS motif within −1000 bp of their promoter regions, further suggesting that other positive or negative regulators might be involved in down-regulation of these Pi-responsive genes. One of these regulators may be Fe itself. It has been reported that Pi deficiency results in enhanced Fe accessibility to plants in the media, which leads to an over accumulation of Fe in plants, subsequently causing down-regulated expression of Fe-responsive genes. This point of view was confirmed by supply of different Fe concentrations in the Pi deficient media (Fig. 6b). If no additional Fe was supplied to the Pi deficient media (−Fe−Pi), the transcriptional expression of all tested genes was induced both in wild type and phr1 mutant plants, an expression pattern similar to the one of Fe deficiency (Fig. 6b). This result suggests that an extent of Fe in the Pi deficient media is required for the down-regulation of Fe-responsive genes under Pi deficiency. Indeed, Pi-deficiency caused down-regulation was much enhanced by supply of 5 µM Fe in the Pi deficient media both in wild type and phr1 mutant plants (Fig. 6b). In the future, the dose effects of Fe in the Pi-deficient media on the transcriptional expression of Fe-responsive genes need further validation.
Conclusions
In summary, we provide genome-wide information on the transcriptional expression of 579 overlapping genes that responded to both Fe and Pi deficiency in Arabidopsis roots and the biological processes that the genes are involved in. Gene clustering and root-specific co-expression analysis revealed several potentially important genes, including CYP82C4 and AT5G38820, which likely function as putative novel players in response to Fe and Pi deficiency or in cross-talk between Fe-deficient responses and Pi-deficient signaling. These results imply that Pi deficiency has more pronounced effects on Fe homeostasis than Fe deficiency has on Pi homeostasis.
Materials and methods
Plant growth and treatments
Arabidopsis (Arabidopsis thaliana) seeds from the Columbia ecotype obtained from the Arabidopsis Biological Resource Center (ABRC) were used in this study. Phr1 mutant seeds (SALK_067629C) were a gift from Professor Tzyy-Jen Chiou as previously described [64]. Seeds were surface sterilized by immersion in 5 % (v/v) NaOCl for 5 min and 70 % ethanol for 7 min, followed by four rinses in sterile water. Seeds were placed into Petri dishes and stored for 1 day at 4 °C in the dark. Plates were then transferred to a growth chamber and grown at 21 °C under continuous illumination (50 µmol m−2 s−1; Philips TL lamps). The agar-based medium [65] was composed of (mM): KNO3 (5), MgSO4 (2), Ca(NO3)2 (2), KH2PO4 (2.5); (µM): H3BO3 (70), MnCl2 (14), ZnSO4 (1), CuSO4 (0.5), NaCl (10), Na2MoO4 (0.2); and 40 µM Fe-EDTA solidified with 0.8 % agar (Sigma-Aldrich). Sucrose (43 mM) and 4.7 mM MES were included, and the pH was adjusted to 5.8. After 10 d of precultivation, plants were transferred either to fresh agar medium with 100 µM 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate without Fe, medium without Pi, medium without both Fe and Pi, medium without Pi containing 5 µm Fe or fresh control medium and grown for another 3 d. Lower potassium concentrations due to the absence of KH2PO4 in the Pi-free medium was corrected by addition of KCl.
Quantitative reverse transcription-PCR
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase using the TURBO DNA-free Kit (Ambion) as suggested by the manufacturer. cDNA was synthesized and qPCR was performed according to a previous report [40] using the SYBR Green PCR Master Mix (Applied Biosystems) with programs recommended by the manufacturer in the ABI Prism 7500 Sequence Detection System (Applied Biosystems). The melting temperature of the primers ranged from 58 to 62 °C. Primer pairs were selected using Primer3 (http://primer3.sourceforge.net/). Elongation factor1-β2 (At5g19510) and Tubulin3 (At5g19770) were used as internal controls (transcript abundance of both genes did not change under Fe and Pi deficiency) for transcript normalization. The primers used in this study are listed in Additional file 10. Three independent replicates were performed for each sample. The delta threshold cycle (∆ct = the ct of a gene−the ct of the reference) was used to determine the relative amount of gene expression. Student’s t test (P < 0.05) was used to compare differences between samples grown under treatment and control conditions.
Data collection and processing
Transcriptomic data sets of roots from 13-day-old Arabidopsis seedlings grown in the presence or absence of Fe or Pi by RNA-seq were downloaded from a public database (NCBI: SRP044814, SRA050356.1). The 3106 and 3296 differentially expressed genes (P < 0.05) upon Pi and Fe deficiency were compared, and the resulting 579 overlapping genes were subsequently analyzed as shown in Fig. 1. Microarray data of 2671 ATH1 arrays from the NASCarray database (http://affymetrix.arabidopsis.info/) were downloaded and normalized using the RMA function in the Bioconductor Affy package software. Three hundred root-related arrays were manually identified as described [41] and were used as a database for co-expression analysis.
Gene ontology analysis
The gene ontology browsing utility (GOBU) [66] was adopted for gene ontology (GO) enrichment analysis using the TopGo ‘elim’ method [67]. The elim algorithm iteratively removes the genes mapped to significant terms from higher level GO terms, thus avoiding enrichment of unimportant functional categories.
Generation of co-expression networks using the MACCU toolbox
Gene functional networks were constructed based on 300 publicly available root-related microarrays using the MACCU toolbox [41], with a Pearson correlation threshold of 0.7. The generated co-expression networks were visualized by Cytoscape (http://www.cytoscape.org). If one cluster of genes did not have any connection (edges) to any other cluster in the co-expression network, it was referred to as a module.
Authors’ contributions
WL and PL performed the data analysis and drafted the manuscript. PL conceived the study. Both authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB15030103), the National Key Basic Research Program of China (No. 2015CB150501), the Natural Science Foundation of China (31470346, 31370280) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
10.1186/s13104-015-1524-y Subset of 579 overlapping genes between phosphate-deficiency regulated and iron-deficiency regulated in the Arabidopsis roots (P < 0.05). The fold change of the gene expression was indicated as mean with standard deviation (SD).
10.1186/s13104-015-1524-y Hierarchical cluster analysis of 579 overlapping genes with greater than twofold changes in transcript abundance in Arabidopsis roots grown under Fe- or Pi-deficient conditions.
10.1186/s13104-015-1524-y Gene Ontology enrichment was assessed using GOBU (Lin et al. [66]) in the 579 overlapping genes (elim, P < 0.01). In the term type column, P, F and C indicate biological process, functional process and subcellular localization, respectively.
10.1186/s13104-015-1524-y Representative Gene Ontology categories (in biological process) enriched in the 137 overlapping genes with twofold change in expression.
10.1186/s13104-015-1524-y Co-expression relationships of the 579 differentially expressed overlapping genes.
10.1186/s13104-015-1524-y Subset of 210 overlapping genes consists of the largest co-expression module. The fold change of the gene expression was indicated as mean with standard deviation (SD).
10.1186/s13104-015-1524-y Gene ontology enrichment was assessed using GOBU (Lin et al. [66]) in the 210 overlapping genes comprising the largest model in Figure S2 (elim, P < 0.01). In the term type column, P, F and C indicate biological process, functional process and subcellular localization, respectively.
10.1186/s13104-015-1524-y Co-expression relationships of the 97 differentially expressed overlapping genes induced by Pi starvation but repressed by Fe deficiency.
10.1186/s13104-015-1524-y Subset of 35 overlapping genes induced by iron deficiency with fold change more than 1.5-fold but down-regulated by Pi starvation the Arabidopsis roots (P < 0.05). The fold change of the gene expression was indicated as mean with standard deviation (SD).
10.1186/s13104-015-1524-y Primers used in this study.
Contributor Information
Wenfeng Li, Email: wfli@njfu.edu.cn.
Ping Lan, Email: plan@issas.ac.cn.
References
- 1.Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Millan AF, Grusak MA, Abadia A, Abadia J. Iron deficiency in plants: an insight from proteomic approaches. Front Plant Sci. 2013;4:254. doi: 10.3389/fpls.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samira R, Stallmann A, Massenburg LN, Long TA. Ironing out the issues: integrated approaches to understanding iron homeostasis in plants. Plant Sci Int J Exp Plant Biol. 2013;210:250–259. doi: 10.1016/j.plantsci.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2014;20:124–133. doi: 10.1016/j.tplants.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Briat J, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015;20(1):33–40. doi: 10.1016/j.tplants.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov R, Brumbarova T, Bauer P. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant. 2012;5(1):27–42. doi: 10.1093/mp/ssr065. [DOI] [PubMed] [Google Scholar]
- 7.Rellan-Alvarez R, El-Jendoubi H, Wohlgemuth G, Abadia A, Fiehn O, Abadia J, Alvarez-Fernandez A. Metabolite profile changes in xylem sap and leaf extracts of strategy I plants in response to iron deficiency and resupply. Front Plant Sci. 2011;2:66. doi: 10.3389/fpls.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong J, Guerinot ML. Homing in on iron homeostasis in plants. Trends Plant Sci. 2009;14(5):280–285. doi: 10.1016/j.tplants.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Hindt MN, Guerinot ML. Getting a sense for signals: regulation of the plant iron deficiency response. Biochim Biophys Acta. 2012;1823(9):1521–1530. doi: 10.1016/j.bbamcr.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986;80(1):175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183(4):1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- 12.Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397(6721):694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 13.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93(11):5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16(12):3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer P, Ling HQ, Guerinot ML. FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem PPB/Soc Fr de Physiol Veg. 2007;45(5):260–261. doi: 10.1016/j.plaphy.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22(7):2219–2236. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt W, Buckhout TJ. A hitchhiker’s guide to the Arabidopsis ferrome. Plant Physiol Biochem PPB/Soc Fr de Physiol Veg. 2011;49(5):462–470. doi: 10.1016/j.plaphy.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14(6):1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18(3):385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 20.Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant. 2013;6(2):503–513. doi: 10.1093/mp/sss089. [DOI] [PubMed] [Google Scholar]
- 21.Sivitz AB, Hermand V, Curie C, Vert G. Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS One. 2012;7(9):e44843. doi: 10.1371/journal.pone.0044843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vert GA, Briat JF, Curie C. Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 2003;132(2):796–804. doi: 10.1104/pp.102.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giehl RF, Lima JE, von Wiren N. Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell. 2012;24(1):33–49. doi: 10.1105/tpc.111.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers EE, Guerinot ML. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell. 2002;14(8):1787–1799. doi: 10.1105/tpc.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144(1):197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G. The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 2008;146(2):589–601. doi: 10.1104/pp.107.108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuler M, Rellan-Alvarez R, Fink-Straube C, Abadia J, Bauer P. Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell. 2012;24:2380–2400. doi: 10.1105/tpc.112.099077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grusak MA, Welch RM, Kochian LV. Physiological characterization of a single-gene mutant of pisum sativum exhibiting excess iron accumulation: I. Root iron reduction and iron uptake. Plant Physiol. 1990;93(3):976–981. doi: 10.1104/pp.93.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grusak MA, Pezeshgi S. Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 1996;110(1):329–334. doi: 10.1104/pp.110.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kneen BE, Larue TA, Welch RM, Weeden NF. Pleiotropic effects of brz: a mutation in Pisum sativum (L.) cv; sparkle’ conditioning decreased nodulation and increased iron uptake and leaf necrosis. Plant Physiol. 1990;93(2):717–722. doi: 10.1104/pp.93.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker R, Grun M, Scholz G. Nicotianamine and the distribution of iron into the apoplasm and symplasm of tomato (Lycopersicon esculentum Mill.): I. Determination of the apoplasmic and symplasmic iron pools in roots and leaves of the cultivar Bonner Beste and its nicotianamine-less mutant chloronerva. Planta. 1992;187(1):48–52. doi: 10.1007/BF00201622. [DOI] [PubMed] [Google Scholar]
- 32.Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 2011;156(3):1116–1130. doi: 10.1104/pp.110.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102(33):11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell. 2013;25(11):4285–4304. doi: 10.1105/tpc.113.117325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J Cell Mol Biol. 2010;64(5):775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 36.Lan P, Li W, Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteom MCP. 2012;11(11):1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bournier M, Tissot N, Mari S, Boucherez J, Lacombe E, Briat JF, Gaymard F. Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J Biol Chem. 2013;288(31):22670–22680. doi: 10.1074/jbc.M113.482281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie. 2006;88(11):1767–1771. doi: 10.1016/j.biochi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147(3):1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Lin WD, Ray P, Lan P, Schmidt W. Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiol. 2013;162(3):1750–1763. doi: 10.1104/pp.113.217778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin WD, Liao YY, Yang TJ, Pan CY, Buckhout TJ, Schmidt W. Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 2011;155(3):1383–1402. doi: 10.1104/pp.110.166520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan P, Li W, Schmidt W. Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genom. 2013;14:210. doi: 10.1186/1471-2164-14-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Lan P, Gao H, Zheng L, Li W, Schmidt W. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genom. 2013;14:783. doi: 10.1186/1471-2164-14-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan P, Li W, Schmidt W. A digital compendium of genes mediating the reversible phosphorylation of proteins in fe-deficient Arabidopsis roots. Front Plant Sci. 2013;4:173. doi: 10.3389/fpls.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15(16):2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu TY, Aung K, Tseng CY, Chang TY, Chen YS, Chiou TJ. Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol. 2011;156(3):1176–1189. doi: 10.1104/pp.111.175257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodriguez J, et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(41):14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamboni A, Zanin L, Tomasi N, Pezzotti M, Pinton R, Varanini Z, Cesco S. Genome-wide microarray analysis of tomato roots showed defined responses to iron deficiency. BMC Genom. 2012;13:101. doi: 10.1186/1471-2164-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urzica EI, Casero D, Yamasaki H, Hsieh SI, Adler LN, Karpowicz SJ, Blaby-Haas CE, Clarke SG, Loo JA, Pellegrini M, et al. Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell. 2012;24(10):3921–3948. doi: 10.1105/tpc.112.102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan P, Li W, Wen TN, Schmidt W. Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiol. 2012;159(1):403–417. doi: 10.1104/pp.112.193987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W. iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol. 2011;155(2):821–834. doi: 10.1104/pp.110.169508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bauer P, Blondet E. Transcriptome analysis of ein3 eil1 mutants in response to iron deficiency. Plant Signal Behav. 2011;6(11):1669–1671. doi: 10.4161/psb.6.11.17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang TJ, Lin WD, Schmidt W. Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 2010;152(4):2130–2141. doi: 10.1104/pp.109.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buckhout TJ, Yang TJ, Schmidt W. Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genom. 2009;10:147. doi: 10.1186/1471-2164-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science (New York, NY) 2008;320(5878):942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 56.Wang YH, Garvin DF, Kochian LV. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 2002;130(3):1361–1370. doi: 10.1104/pp.008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barak P, Chen Y. The effect of potassium fertilization on iron deficiency. Commun Soil Sci Plant Anal. 1983;14(10):945–950. doi: 10.1080/00103628309367422. [DOI] [Google Scholar]
- 58.Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 2009;151(1):262–274. doi: 10.1104/pp.109.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perea-Garcia A, Garcia-Molina A, Andres-Colas N, Vera-Sirera F, Perez-Amador MA, Puig S, Penarrubia L. Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol. 2013;162(1):180–194. doi: 10.1104/pp.112.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lelandais-Briere C, Jovanovic M, Torres GA, Perrin Y, Lemoine R, Corre-Menguy F, Hartmann C. Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. Plant J Cell Mol Biol. 2007;51(2):154–164. doi: 10.1111/j.1365-313X.2007.03131.x. [DOI] [PubMed] [Google Scholar]
- 61.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32(12):1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 63.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48(3):381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- 64.Kuo HF, Chang TY, Chiang SF, Wang WD, Charng YY, Chiou TJ. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J Cell Mol Biol. 2014;80(3):503–515. doi: 10.1111/tpj.12650. [DOI] [PubMed] [Google Scholar]
- 65.Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet. 1987;206(2):200–206. doi: 10.1007/BF00333575. [DOI] [Google Scholar]
- 66.Lin W-D, Chen Y-C, Ho J-M, Hsiao C-D. GOBU: toward an integration interface for biological objects. J Inf Sci Eng. 2006;22(1):19. [Google Scholar]
- 67.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics (Oxford, England). 2006;22(13):1600–07. [DOI] [PubMed]


