Abstract
Introduction
Calcium carbonate biominerals participate in diverse physiological functions. Despite intensive studies, little is known about how mineralization is initiated in organisms.
Results
We analyzed the medaka spontaneous mutant, ha, defective in otolith (calcareous ear stone) formation. ha lacks a trigger for otolith mineralization, and the causative gene was found to encode polyketide synthase (pks), a multifunctional enzyme mainly found in bacteria, fungi, and plant. Subsequent experiments demonstrate that the products of medaka PKS, most likely polyketides or their derivatives, act as nucleation facilitators in otolith mineralization. The generality of this novel PKS function is supported by the essential role of echinoderm PKS in calcareous skeleton formation together with the presence of PKSs in a much wider range of animals from coral to vertebrates.
Conclusion
The present study first links PKS to biomineralization and provides a genetic cue for biogeochemistry of carbon and calcium cycles.
Electronic supplementary material
The online version of this article (doi:10.1186/s40851-014-0001-0) contains supplementary material, which is available to authorized users.
Keywords: Biomineralization, Calcium carbonate, Otolith, Polyketide synthase, Medaka, Spicule
Introduction
Biominerals are produced by living organisms through genetically controlled biological processes, and confer stability, rigidity, defences and functionality to organisms; examples include grass opal (rice; silicate), shell (shellfish; calcium carbonate), and bone/teeth (vertebrate; hydroxyl apatite) [1,2]. The addition of organic components determines organism-specific shape, size, hardness and crystallographic axis orientation of biominerals [3]. Among biominerals, calcium carbonate is the most abundant and is known to play an important role in the global biogeochemical cycles of carbon and calcium through reef building and massive precipitation by calcifying algae in the ocean [4-6], and thus has attracted much attention, together with calcium phosphate mineralization of vertebrate skeleton and teeth.
The biomineral crystallization begins with nucleation through the phase of prenucleation clusters [7,8]. Experimental and modeling studies proposed that nucleation from saturated solution is not a simple event, but is preceded by a phase of amorphous precursors, such as amorphous calcium carbonate (ACC) or amorphous calcium phosphate, depending on the type of biominerals. These precursors then coalesce and rearrange to form nuclei, a core of subsequent crystal growth, a process called ‘nucleation’. Although these processes of biomineralization are well described, the underlying mechanism, particularly regarding the initial nucleation step, remains a mystery.
Otoliths (ear stones) in teleost fish are composed of calcium carbonate and organic materials (0.2–10%) [9], and primarily function in gravity and motion sensing, providing excellent systems for biochemical and genetic analyses of the mechanisms of calcium carbonate biomineralization. Factors involved in initial step of mineralization have not been definitively identified thus far, except for a report that glycogens are always found in the core of nascent otoliths [10]. This is in part due to the lack of genetic studies targeting otolith mineralization. Indeed, while most zebrafish otolith mutants exhibit defects in the number, shape, and position, only a few completely lack otoliths, and in the cases where the causal gene was identified, its function in the context of mineralization remained unclear [11,12].
To gain insights into the crystallization of calcium carbonate in organisms, we analyzed the medaka spontaneous and homozygous viable mutant, ha, defective in otolith formation. Positional cloning identified a mutation in a novel gene encoding polyketide synthase (pks), named OlPKS, specifically expressed in the otic vesicle at the onset of mineralization. We demonstrate that the compound synthesized by OlPKS is secreted in the endolymph and it acts as a nucleation facilitator in otolith mineralization. Furthermore, sea urchin pks-2 expressed in skeletogenic cells was found to be indispensable for the formation of spicules, calcareous larval skeletons, indicating the generality of a novel link between calcium carbonate biomineralization and animal polyketide synthase.
Materials and methods
Fish strains and mapping
The medaka (Oryzias latipes) d-rR strain was used as a wild-type. ha was isolated as a spontaneous mutant by Tomita [13]. ki79 was isolated from a ENU-mediated mutant screening (at the Tokyo Institute of Technology, Japan). Mapping of the ha locus was carried out as previously described [14].
Otolith staining
Alizarin Red staining for visualizing mineralized otoliths was essentially done as described [15]. Embryos were fixed with 4% PFA/2× PBS (phosphate buffered saline) containing 1% sodium hydroxide for three hours at room temperature. The samples were then washed with PBS several times, and stained with 2% Alizarin Red S/1% sodium hydroxide solution overnight. After quickly washing with 0.5% KOH, samples were mounted in 80% glycerol.
Transmitted electron microscopy
Embryos were fixed with 2% PFA and 2% glutaraldehyde buffered in 0.1 M cacodylate buffer and postfixed in 2% OsO4 in 0.1 M cacodylate buffer. Uranyl acetate and lead stain solution were used for contrast enhancement. The specimens were embedded in Quetol-812. Ultrathin sections (70 nm) were analyzed and documented with an electron microscope (JEOL, JEM-1400 Plus).
Whole-mount in situ hybridization
All whole-mount in situ hybridization analyses were performed as previously described [16]. cDNAs isolated by RT-PCR were used as the templates for the probes of marker genes (see Additional file 1).
Immunofluorescence
Immunofluorescence for OMP-1 was performed as previously described Murayama et al. [17], whereas staining for acetylated α-tubulin, OlPKS and PKC ζ were done as previously described by Kamura et al. [18] (see Additional file 1). Polyclonal rabbit OlPKS [amino acids 1520–1711] antibodies were raised by immunization of rabbits with bacterially expressed His-tagged truncated proteins.
Morpholino knockdown and mRNA rescue in medaka embryos
The morpholino antisense oligonucleotide (MOs; Gene Tools LLC) for first-Met of OlPKS was as follows: 5'-AACCACGGCTATTCCGTCCTCCATG-3'. in vitro syntheses of mRNAs were conducted as reported previously [19]. Full length of olpks was isolated from the d-rR strain by RT-PCR. The sequences of primers are described in Additional file 1 (Whole-mount in situ hybridization). For injection of olpks mRNA with active site mutation, ki79 fish was used, because it likely carries a null mutation. Amino acid of the active site in each enzymatic domain was substituted as followed (with nucleotide sequences): KS, 173Cys → Asn (TGC → AAC); AT, 608Ser → Ala (TCC → GCA); DH,914His → Ala (CAC → GCC); KR,1859Tyr → Phe(TAC → TTC); ACP, 2010Ser → Ala(TCC → GCC); Loop (interdomain region; control), 435Thr → Ala(ACC → GCC) [20-22]. Microinjection of MO or mRNAs was carried out as previously described [19].
Genomic research
Animal type I PKS sequences were retrieved by TBLASTN against genome sequences of each species or BLASTP search against nr-protein database using the OlPKS sequence. For each candidates, domain search was done by Pfam sequence search, and amino acid sequences having basic PKS domains KS and AT, and additional domains (e.g., DH, KR, ER, ACP etc.) were defined as an animal type I PKS.
Generation the ha chimeric medaka
Transplantation experiments were performed based on the previous works [23]. [Tg (β-actin:DsRed)] were used as wt-cell donors, whereas ha fish were prepared as hosts. Whether otoliths were formed or not and how many fluorescence cell were contained were confirmed by observations around st. 29 using a confocal fluorescence microscope (Zeiss, LSM710). A transplantation experiment was conducted as a negative control in which [ha−/−; Tg (β-actin:DsRed)] animal was used as the donors (detailed in Additional file 1).
OlPKS expression in A. oryzae and medaka rescue assay
The entire OlPKS ORF was amplified and subcloned into a fungal expression vector, pTAex3. The resulting expression plasmid pTA-olpks was used for transformation of A. oryzae M-2-3 according to the protoplast-PEG method described by Gomi et al. [24]. The mycelia of transformants were extracted with acetone, followed purification using partition between ethyl acetate/H2O two times and the ethyl acetate layer was concentrated to dryness, and partitioned between hexane/90% methanol. The methanol layer was concentrated to dryness to give the material for medaka assay. Each extracts re-dissolved in DMSO was added to the medium in which ha medaka embryos were incubated. (detailed in Additional file 1).
Morpholino knockdown in sea urchin embryos
Microinjection of MOs was carried out as described previously with some modifications [25] (see Additional file 1). MOs (Gene Tools LLC) for first-Met blocking and their five-mispair controls were as follows (small letters in control sequences indicate substituted nucleotides).
hppks-1: 5'-CTGGTTTTATTGCTTCCCATGTTGA-3', hppks-2: 5'-CCCTCCAACTATCTTCCATAACTCA-3', hppks-1 control: 5'- CTGcTaTTATTcCTTCCgATcTTGA -3', hppks-2 control: 5'-CCgTgCAAgTATgTTCgATAACTCA-3'.
Results
The ha embryo fails to mineralize otoliths
Medaka ha is a spontaneous and homo-viable mutant defective in otolith formation [13] (Figure 1A and C). The gross morphology of ha was previously reported as significantly delayed mineralization of otoliths, slightly enlarged otic vesicles (OVs) and malformed semicircular canals [26,27]. We further characterized the ha phenotypes using molecular markers at embryonic stages at which the formation of the OV and otoliths normally takes place (st. 22– 30).
Figure 1.
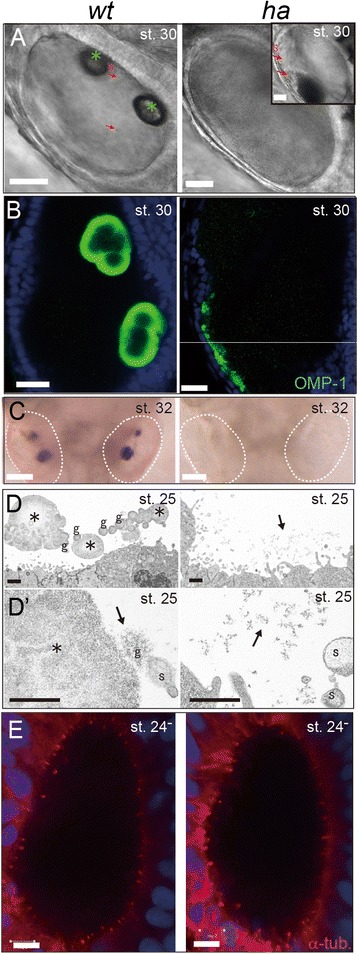
ha embryos fail to mineralize otoliths. (A) DIC images of OVs at st. 30 (dorsal views of the left OV). Grown otoliths are observed in wt OV. Mutant OV contains numerous seeding particles that form a paste-like precipitate (Inset). Red arrows: seeding particles; Asterisks: otoliths. Scale bars: 20 μm. (B) Immunofluorescence of an otolith matrix protein, OMP-1 (dorsal views of the left OV). Anti-Oncorhynchus mykiss (Om) OMP-1 serum is used. In mutant OV, immunoreactive substances cling to the epithelium. Scale bars: 20 μm. (C) Alizarin Red staining for mineralized otolith. Crystal is never observed in mutant OV (dorsal views of the head; white dotted lines show OV). Scale bars: 100 μm. (D) TEM images of the epithelium of the OV at st. 25 when the otolith is forming (the prospective macula region; lateral views). In a wt embryo ‘globules’ coalesce to form the otolith precursor in the posterior end of the OV. In the mutant, by contrast, very fine particles are observed at posterior end of OV (D) and mid-position of the OV (D’). Asterisks: growing otoliths; Black arrows: fine particles; ‘g’:globule; ‘s’:seeding particle. Scale bars: 1 μm. (E) Immunofluorescence of acetylated α-tubulin st. 24− (dorsal views of the left OV). Many short cilia protruded from the epithelium are visible in ha OV as well as wt one. Scale bars: 5 μm.
Otolith formation in medaka proceeds in a way similar to zebrafish [28]; ‘seeding particles’ start to float in the endolymph of the OV (Figure 1A and D) from st. 23 and coalesce into small crystal at st. 24 (Additional file 2K; Upper Left). Otoliths are stereo-microscopically visible as two small crystals at st. 25 and continuously increase in size (Figure 1A and D, and Figure 2D). In zebrafish, essential roles of cilia in otolith formation have been repeatedly shown [28-31]; tethering seeding particles by long kinocilia (5–8 μm) protruded from hair cells in the prospective macula regions and stirring the fluid by shorter motile cilia (1.5–5 μm) lining the entire of the OV epithelium. However, this scenario may not hold true in medaka embryos. Though cilia were found on the OV epithelium of medaka, they are much smaller in size (<1 μm) and their motility was hard to be detected (Figure 1E). Furthermore, probably due to their small size, we failed to identify kinocilia. At least, motile cilia do not contribute to otolith formation in medaka, as the medaka mutant kintoun with paralyzed cilia develops otoliths as normal (Additional file 2K) [32].
Figure 2.
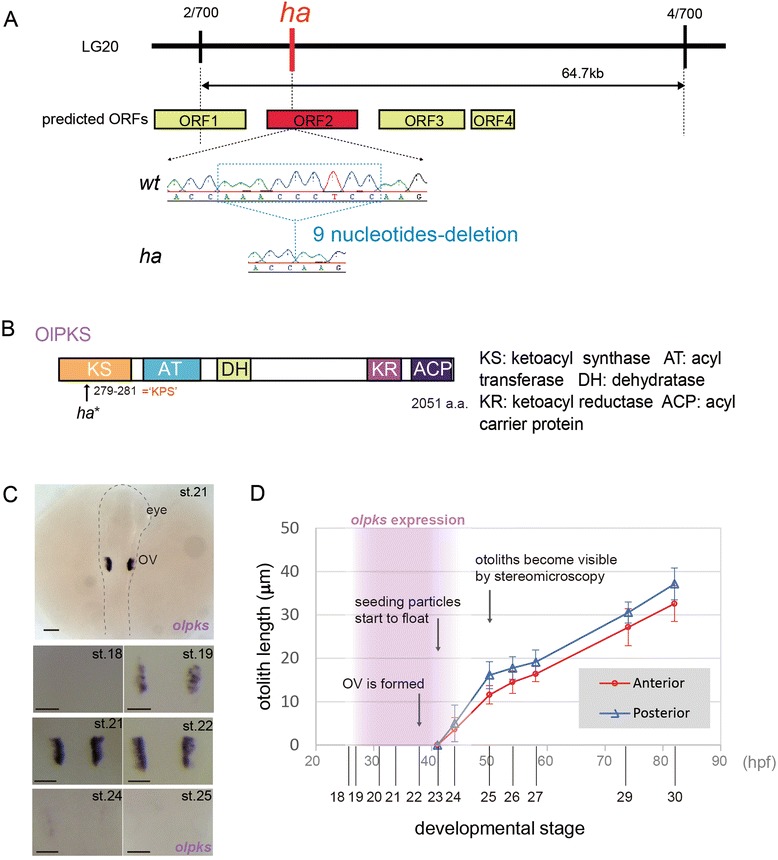
ha gene encodes a polyketide synthase. (A) Positional cloning of the ha mutation in linkage group (LG) 20. The number of recombinants at each marker is shown. Sequencing of ha revealed a 9-nucleotide deletion. ORF: open reading frame. (B) Architecture of OlPKS (2051 amino acid-length) predicted by a Pfam search. Each domain is shown by abbreviation. An arrow indicates mutation site of ha, which is located at 279–281 (K, P and S). (C) Whole-mount in situ hybridization with an antisense RNA probe for olpks at otolith forming developmental stages. A representative picture is shown at st. 21 (Upper; dorsal view; dotted line indicates embryonic body). olpks transcripts detected in various stages are shown at high magnification (Lower; dorsal views of left and right OVs). Scale bars: 50 μm. (D) Period of the expression of olpks in the context of otolith growth. Purple area shows the period of olpks expression. Line graphs show the sizes (longest linear dimensions) of otoliths at some developmental stages. Data are the means and standard deviations of measurements taken of at least 7 specimens each. Some observable changes in the OV during otolith development are described with arrows. Red curcle: anterior otolith; Blue triangle: posterior otolith; hpf: hours post fertilization.
In ha mutant embryos, the OV, macula and cilia develop normally (Additional file 2A to D, I and J), and seeding particles are floating in the OV (Figure 1A and Additional files 3 and 4). Furthermore, organic materials such as OMP-1, a major soluble organic matrix protein, are secreted into the endolymph (Figure 1B) (other known organic components, starmaker-like and sparc are normally expressed in the OV (Additional file 2E and F)). Nevertheless, in ha mutants, mineralized stones never form (Figure 1A and C), but instead OMP-1-positive particles precipitate in the endolymph (Figure 1A Inset and B). This was further supported by TEM observation; fine substances (Figure 1D and D′, arrows), probably organic substances supplied by seeding particles, were accumulated around the epithelium in ha (Figure 1D Right and 1D’ Right), instead of growing otoliths (Figure 1D Left and D′ Left, asterisks). In wild-type (wt) OVs, fine particles were compacted into a round ‘globule’ (Figure 1D Left, ‘g’; [10]), and the globules then form otoliths by coalescing near the macula (Figure 1D asterisks; see also Additional file 2G). Taken together, in ha embryos, otolith mineralization is completely inhibited, even though major organic materials are supplied into the endolymph.
ha gene encodes a polyketide synthase
Using positional cloning, we narrowed down the ha locus to a 64.7 kb region in linkage group 20, which contains four open reading frames (ORFs 1–4). Sequencing analysis identified a 9-nucleotide deletion (3-amino acid deletion) in ORF 2 that encodes a type I polyketide synthase (PKS) (Figure 2A). We thereafter named this gene Oryzias latipes polyketide synthase (olpks). The olpks transcript is 6153 nt in length, comprises six exons, and encodes a 2051 amino acid protein. PKSs are multifunctional enzymes mainly found in bacteria, fungi and plant, and catalyze the biosynthesis of a diverse group of second metabolite, polyketides, some of which are used for pharmaceuticals with antibiotic and mycotoxic properties [33]. Type I PKS has a set of distinct enzymatic domains that individually catalyze a series of reactions, to produce a final compound. Likewise, OlPKS contains five distinct domains: ketoacyl synthase (KS), acyl transferase (AT), dehydratase (DH), ketoreductase (KR), and acyl carrier protein domain (ACP) (Figure 2B). Essential amino acid sequence motifs for the function of each domain are conserved in OlPKS (Additional file 5B). The vertebrate fatty acid synthase (FAS), essential for all organisms, is thought to be an evolutionary subset of this family [34,35]. Animal FASs and another iterative type I PKSs share a conserved structure that includes KS, AT and ACP domains. FASs contain additional enzymatic domains such as KR, DH, ER and TE, which are present on different PKSs in different combinations. OlPKS possesses the minimal module (KS, AT and ACP) and additional KR and DH domains (Figure 2B and Additional file 5B).
We confirmed that olpks is indeed responsible for the ha phenotype by the following results, (i) phenocopy by injection of an antisense morpholino-oligonucleotide (MO) (Table 1), (ii) identification of a mutation of the olpks locus in another allele, ki79 (Additional file 5A and B) which was isolated from a N-ethyl-N-nitrosourea (ENU)-driven screen (unpublished; screen conducted for medaka mutants with defects in bone or blood development at the Tokyo Institute of Technology, Japan), and (iii) phenotypic rescue by injection of full length olpks mRNA (Table 1). We also confirmed that all domains of OlPKS are indeed required for otolith formation by injecting mRNAs, each of which causes one amino acid substitution at one of the four enzymatic active sites and the essential site of ACP (Additional file 5B and Table 1).
Table 1.
Otolith formation in MO or mRNA of OlPKS
| Experiment | Nucleltide | Fish | 4 otoliths | 1-3 otoliths | No otolith | n |
|---|---|---|---|---|---|---|
| Phenocopy | olpks first-Met MO | wt | 0% | 0% | 100% | 39 |
| Rescue | (uninjected) | ha | 0% | 0% | 100% | 31 |
| olpks mRNA | 73% | 23% | 3% | 30 | ||
| Active site mutation | (uninjected) | ki79 | 0% | 0% | 100% | 14 |
| olpks-mRNA | 65% | 35% | 0% | 43 | ||
| KS*-mRNA | 0% | 0% | 100% | 15 | ||
| AT*-mRNA | 0% | 2% | 98% | 57 | ||
| DH*-mRNA | 0% | 0% | 100% | 18 | ||
| KR*-mRNA | 0% | 0% | 100% | 17 | ||
| ACP*-mRNA | 0% | 0% | 100% | 27 | ||
| Loop*-mRNA | 53% | 40% | 7% | 60 |
Asterisks indicate one amino acid mutation are introduced.
‘Loop’: interdomain region.
Fully rescued: 4 otoliths in one animal.
Partially rescued: 1–3 otoliths in one animal.
Not rescued: no otolith.
The expression of olpks is transient and exclusively restricted to the OV in developing embryos. The expression initiates at the early somite stage (st. 19) and disappears between 16-somite (st. 24) and 19-somite stage (st. 25), a period when otolith mineralization initiates (Figure 2C and D). The expression becomes restricted to the medial and dorsal region of the vesicle at later stages (Figure 2C and Additional file 5C). The medaka genome has three pks related genes including olpks (Table 2) and we confirmed that the other two are not expressed at embryonic stages and adult tissues (Additional file 5D).
Table 2.
Type I PKSs found in animal lineage
| Type 1 PKS? | Species name | General name | Gene name | Accession number | Reference genome | Gene ID | Corresponding RNA (Unigene or RNA)? | Domians |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | - | - | Homo sapiens GRCh37.p13 | - | - | - | |
| Bos taurus | Cattle | - | - | Bos taurus UMD3. | - | - | - | |
| Canis (lupus) familiaris | Dog | - | - | Canis lupus familiaris CanFam3.1 | - | - | - | |
| Tursiops truncatus | Dolphin | - | - | Tursiops truncatus turTur1 | - | - | - | |
| Mus musculus | Mouse | - | - | Mus musculus GRCm38.p1 | - | - | - | |
| ✓ | Monodelphis domestica | Opossum | PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like | XP_001375980 | MonDom5 | LOC100024851 | no | KS-AT-DH-KR-ACP |
| ✓ | Sarcophilus harrisii | Tasmanian devil | mycocerosic acid synthase-like [Sarcophilus harrisii (Tasmanian devil)] | XP_003771909 | Devil_refv7.0 | LOC100922065 | N.A | KS-AT-DH-KR-ACP |
| Ornithorhynchus anatinus | Platypus | LOC100091954 fatty acid synthase-like [Ornithorhynchus anatinus (platypus)] | - | Ornithorhynchus_anatinus-5.0.1 | LOC100091954 (pseudo gene) | N.A | N.A (pseudo gene) | |
| ✓ | Gallus gallus | Chicken | PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like is oformX2 [Gallus gallus]. | XP_418588 | Gallus_gallus-4.0 | LOC420486 | Yes (brain; connective, blood) | KS-AT-DH-KR-ACP |
| ✓ | Taeniopygia guttata | Zebra finch | PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like [Taeniopygia guttata]. | XP_002189754 | Taeniopygia_guttata-3.2.4 | LOC100231542 | No | KS-AT-DH-KR-ACP |
| PREDICTED: Taeniopygia guttata phthioceranic/hydroxyphthioceranic acid synthase-like | XP_002190558 | LOC100222288 | N.A | KS-AT-DH-KR-ACP | ||||
| ✓ | Falco peregrinus | Peregrine falcon | PREDICTED: probable polyketide synthase 1-like [Falco peregrinus]. | XP_005234016 | F_peregrinus_v1.0 | LOC101916009 | N.A | KS-AT-DH-KR-ACP |
| ✓ | Anolis carolinensis | Green anole | PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like [Anolis carolinensis]. | XP_003222100 | AnoCar2.0 | LOC100564455 | No | KS*-AT-DH-KR-ACP |
| PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like [Anolis carolinensis]. | XP_0032222101 | LOC100564655 | No | KS-AT-DH-KR-ACP | ||||
| PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like [Anolis carolinensis]. | XP_0032222102 | LOC100564856 | No | KS-AT-DH-KR-ACP | ||||
| ✓ | Chelonia mydas | Green sea turtle | Phthioceranic/hydroxyphthioceranic acid synthase [Chelonia mydas]. | EMP37033 | CheMyd_v1.0 | locus_tag: UY3_05838 | N.A | KS-AT-DH-KR-ACP |
| Polyketide synthase Pks N [Chelonia mydas] | EMP24664 | locus_tag: UY3_18267 | N.A | KS*-AT-DH-KR-ACP | ||||
| ✓ | Chrysemys picta | Painted turtle | PREDICTED: uncharecterized protien LOC101936604 [Chrysemys picta bellii] | XP_005291085 | Chrysemys_picta_bellii-3.0.1 | LOC101936604 | N.A | KS-AT*-DH*-KR-ACP |
| PREDICTED: uncharecterized protien LOC101937174 [Chrysemys picta bellii] | XP_005291087 | LOC1019371714 | N.A | other-KS-AT*-DH*-KR-ACP | ||||
| ✓ | Pelodiscus sinensis | Chinese soft shell turtle | pep: KNOWN_BY_PROJECTION_protein_coding | Scaffold no. JH209275.1 | PelSin_1.0 | ENSPSIG00000004874 | No | KS*-AT-DH-KR-ACP |
| Xenopus (Silurana) tropicalis | Tropical clawed toad | - | - | Xenopus (Silurana) tropicalis build 1 genome database (v4.2 assembly) | - | - | - | |
| ✓ | Oryzias latipes | Medaka | OIPKS (PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase-like [Oryzias latipes]) | XP_004081385 | Oryzias latipes ASM31367v1 | LOC101169887 | No | KS-AT-DH-KR-ACP |
| OIPKS-2 (PREDICTED: phthioceranic/hydroxyphthioceranic acid synthase type I P ps D-like [Oryzias latipes]) | XP_004081384 | LOC101169644 | No | KS-AT-DH*-KR-ACP | ||||
| OIPKS-3 (PREDICTED: probable polketide synthase 1-like [Oryzias latipes]) | XP_004080917 | LOC101170716 | No | KS-AT-DH*-KR-ACP | ||||
| ✓ | Danio rerio | Zebrafish | Danio rerio wu:fc01d11 (wu: fc01d11),mRNA | XP_682975 | Danio rerio Zv9 | wu:fc01d11 | Yes (muscle) | KS-AT-DH-KR-ACP |
| si: dkey-61p9.11 | NP_001041530 | LOC100000781 | Yes (kidney) | KS-AT-DH-KR-ACP | ||||
| ✓ | Takifugu rubripes | Fugu | phthioceranic/hydroxyphthioceranic acid synthase-like | XP_003968201 | FUGU5 | LOC101079294 | No | KS-AT-DH-KR-ACP |
| PREDICTED: Takifugu rubripes lovastatin nonaketide synthase-like (LOC101079519), mRNA | XP_003968202 | LOC101079519 | No | KS-AT-DH-KR-ACP | ||||
| Petromyzon marinus | Lamprey | - | - | Pmarinus_7.0 | - | - | - | |
| ✓ | Branchiostoma Floridae | Lancelet | hypothetical protien BRAFLDRAFT_205831, partial [Branchiostoma floridae]. | XP002599684 (=EEN55696) | Branchiostoma floridae v1.0 | Gene ID:7231796 | N.A. | KS-AT-DH-KR |
| hypothetical protien BRAFLDRAFT_247081 [Branchiostoma floridae] | XP_002591573 | Gene ID:7219804 | N.A. | KS-AT-DH*-KR | ||||
| hypothetical protien BRAFLDRAFT_90481 [Branchiostoma floridae] | XP_002589799 | Gene ID:7210376 | N.A. | KS-AT-DH*-KR-AMP | ||||
| hypothetical protien BRAFLDRAFT_96868 [Branchiostoma floridae] | XP_002598386 | Gene ID:7254845 | N.A. | KS-AT-DH-KR*-TE-C-AMP | ||||
| hypothetical protien BRAFLDRAFT_96863 [Branchiostoma floridae] | XP_002598380 | Gene ID:7248951 | N.A. | KS-AT-DH-KR*-ACP-TE-C-AMP | ||||
| hypothetical protien BRAFLDRAFT_87472 [Branchiostoma floridae] | XP_002589000 | Gene ID:7246004 | N.A. | KS-AT-DH-KR*-ACP-C-AMP | ||||
| hypothetical protien BRAFLDRAFT_125690 [Branchiostoma floridae] | XP_002610053 | Gene ID:7207083 | N.A. | KS*-AT-DH-KR*-ACP*-TE-C | ||||
| hypothetical protien BRAFLDRAFT_91451 [Branchiostoma floridae] | XP_002608071 | Gene ID:7214024 | N.A. | KS-AT-DH-MT-ADH-KR-ACP-α | ||||
| hypothetical protien BRAFLDRAFT_87410 [Branchiostoma floridae] | XP_002605916 | Gene ID:7243248 | N.A. | KS-AT-DH-MT-ADH-KR-ACP | ||||
| hypothetical protien BRAFLDRAFT_89867 [Branchiostoma floridae] | XP_002610100 | Gene ID:7206066 | N.A. | KS-AT-DH-MT-ADH-KR-ACP | ||||
| hypothetical protien BRAFLDRAFT_87413 [Branchiostoma floridae] | XP_002605913 | Gene ID:7246000 | N.A. | KS-AT-DH-MT-ADH-KR-ACP | ||||
| hypothetical protien BRAFLDRAFT_125650 [Branchiostoma floridae] | XP_002610103 | Gene ID: 7207596 | N.A. | AMP-KS-AT-DH-MT-ADH-KR-ACP | ||||
| hypothetical protien BRAFLDRAFT_71890 [Branchiostoma floridae] | XP_002613500 | Gene ID: 7224978 | N.A. | KS*-AT-DH | ||||
| ✓ | Saccoglossus kowalevskii | Acorn worm | PREDICTED: fatty acids synthase-like | XP_002734101 | Skow_1.1 | LOC100373061 | N.A | KS*-AT-DH-KR-ACP-C-AMP |
| Ciona intestinalis | Ascidian | - | - | Ciona intestinalis KH | - | - | - | |
| ✓ | Strongy locentrotus purpuratus | Purple sea urchin | LOC588806 probable polyketide synthase 1-like [Strongylocentrotus purpuratus (purple sea urchin)] | XP_793564.2 | Spur_3.1 | LOC58806 | Yes | KS-AT*-DH*-MT-ADH-KR*-ACP |
| LOC592147 polyketide synthase 2 | NP_001239013.1 | LOC592147 | Yes | KS-AT-DH*-KR-ACP-TE | ||||
| ✓ | Acropora digitifera | Coral | aug_v2a.12941.t1 aug_v2a.12941scaf5202:2805-26950(−) | aug_v2a.12941 | Adig_1.0 | aug_v2a.12941 | N.A. | other-KS-AT-DH-KR-ACP*-TE |
| aug_v2a.16843.t1 aug_v2a.16843scaf8086:11276-29700(−) | aug_v2a.16843 | aug_v2a.16843 | N.A. | other-KS-AT-DH-KR-TE | ||||
| aug_v2a.16847.t1 aug_v2a.16847scaf8086:95926-115499(−) | aug_v2a.16847 | aug_v2a.16847 | N.A. | other-KS-AT-DH-KR-TE | ||||
| Nematostella vectensis | Sea anemone | - | - | Nematostella vectensis v1.0 | - | - | - | |
| Hydra magnipapillata | Hydra | - | - | Hydra magnipapillataHydra_RP_1.0 | - | - | - | |
| Amphimedon queenslandica | Sponges | - | - | Amphimedon queenslandicav1.0 | - | - | - | |
| ✓ | Caenorhabditis elegans | Nematode | Protein C41A3.1 [Caenorhabditiselegans] | NP_508923 | Caenorhabditis elegansWBcel235 | C41A3.1 | Yes | KS-KS-DH-ACP-KS-ACP-ACP-KR-ACP-AT-DH-KS-KR-KS-AT-ADH-ACP-C-AMP-ACP-TE |
| Drosophila melanogaster | Fruit fly | - | - | Drosophila melanogasterRelease 5 | - | - | - |
Asterisks show domains lacking a residue of the active site that is contained in OlPKS.
Sequence sources are mainly NCBI protein database, except dolphin, Chinese soft shell turtle, lamprey, coral, and sea anemone.
KS: ketoacyl synthase, AT: acyl transferase, DH: dehydratase, KR: ketoreductase, ACP: acyl carrier protein domain, TE: thioesterase, AMP: AMP-binding site, MT: methyltransferase, ADH: alcohol dehydrogenase, C: condensation domain.
Based on above results, we conclude that OlPKS is only required for the early step of otolith mineralization and that the mutation in olpks is responsible for the ha phenotype.
OlPKS produces lipophilic substances secreted into the endolymph
We hypothesized that, similar to other PKSs, OlPKS synthesizes polyketide-related small compounds in the OV epithelium, which are then secreted into the endolymph for the initial step of otolith mineralization.
To test this idea, we first examined subcellular localization of OlPKS. Immunostaining revealed that the medial wall of the OV exclusively expresses OlPKS at st. 23, which is highly localized at the apical of epithelial cells (Figure 3A Left) (a region for antigen is described in Additional file 5B). Double staining with an antibody to PKC ζ (Figure 3A Center), an apical membrane marker, demonstrated that the distribution of OlPKS enzyme partially overlaps with that of PKC ζ but OlPKS signal is detected closer to the lumen (Figure 3A Right). Thus, substances synthesized at the apical surface can be directly secreted to the endolymph, in a way similar to ‘membrane-localized’ PKS observed in bacterial cells [36,37].
Figure 3.
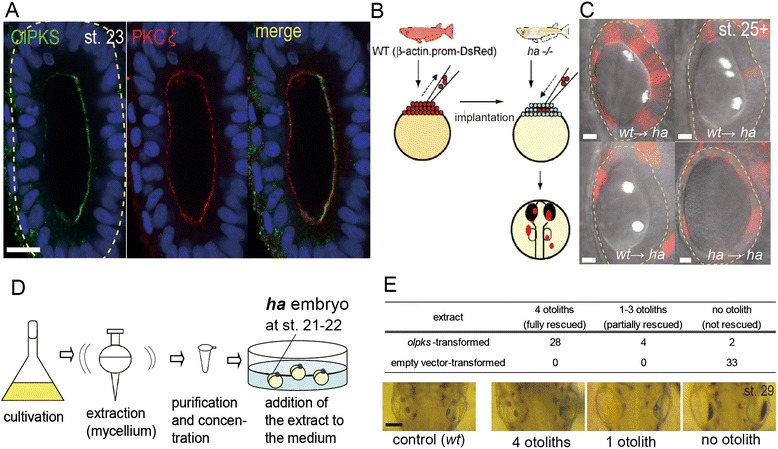
OlPKS produces lipophilic substances secreted into the endolymph. (A) Intracellular localization of OlPKS in the OV epithelium (dorsal views of the left OV). Anti-OlPKS antibody detects the OlPKS protein at the apical region of the epithelial cells. Co-immunostaining with a membrane marker, PKC ζ, shows it localizes near the apical membrane. Yellow dotted line: OV. Scale bar: 10 μm. (B) Schematic procedure of the chimeric experiment. (C) Some images of the chimeric OVs in live embryos. [wt → ha] shows wt cells expressing DsRed are transplanted to an ha embryo. [ha → ha] is a negative control experiment. Yellow dotted lines: OVs. Scale bars: 10 μm. (D) Schematic representation of the heterologous expression system using A. oryzae. (E) Summary of the bioassay in the heterologous expression experiment. Numbers of ha embryos treated by the extract of olpks transfromant or that of empty vector toransformant are shown (Upper table). Grades of recovery of the mineralization: four otoliths (two per OV, fully rescued), 1–3 otoliths (at least one otolith per embryo, partially rescued) and no otolith (not rescued). Representative picture of treated embryo in each category is shown with a picture of wt embryo (Lower). Scale bar: 100 μm.
Next, we performed a chimera experiment in which DsRed-expressing wt cells was transplanted into mutant blastula, and examined for otolith formation when wt donor cells colonized mutant OVs (Figure 3B). Remarkably, irrespective of their number and localization within the OV, wt cells effectively restored otolith formation at the appropriate time and location, the macula region, in ha embryos. Only a few cells, located at any region of the vesicle, were found to be sufficient (Figure 3C and Additional file 6A and B).
Finally, we adopted a heterologous expression system to characterize substances synthesized by OlPKS. Since large-scale expression of PKSs has been established in Aspergillus oryzae [24], we introduced the olpks cDNA into A. oryzae, expecting that exogenous PKS (i.e., OlPKS) could work using endogenous substrates such as acetyl-CoA and malonyl-CoA in fungal cells like their own PKSs (Figure 3D). OlPKS expression in transformed fungi was confirmed by western-blotting (Additional file 6C). Since polyketide-derivatives exhibit moderate hydrophobic nature, we extracted cultivated mycelia with acetone, followed by purification using partition between ethyl acetate/H2O and subsequent purification. We then attempted a simple in vivo assay in which ha mutant embryos were cultured with an aliquot of the extracts (Figure 3D). Remarkably, these extracts restored otolith mineralization in ha embryos, while no such rescue was observed with control extracts (Figure 3E). Taken together, these data demonstrate that it is not OlPKS but ethyl-acetate extractable substances synthesized by OlPKS and secreted into the endolymph that nucleate otolith mineralization.
Broad distribution and conserved roles of polyketide synthases in animals
Animal pks genes were rarely explored, except for two echinoderm pks-1 and pks-2 (isolated from Strongylocentrotus purpuratus) [38,39], and the presence of fish, bird, and nematode pks genes were reported by previous phylogenetic analyses [40-42]. We performed thorough database searches for animal pks genes against current updated genome databases. Identified pks gene candidates were then assessed for a constitution of domains in each predicted amino acid sequence. These searches revealed a remarkably broad distribution of pks genes from Cnidaria to Bilateria, including coral (Acropora digitifera), C. elelgans and reptiles/birds (Figure 4A and Table 2). Usually, 1–3 pks genes are present in each genome, except for the lancelet genome that contains 13 genes. Most vertebrate PKSs have five domains, which are similar to OlPKS (e.g., KS, AT, DH, KR and ACP; Table 2). Zebrafish pks, drpks (wu:fc01d11), expresses in the otic vesicle (Additional file 7B). By contrast, other animal PKSs are not similar to olpks and have versatile domains, especially C. elegans PKS contains 21 domains in the polypeptide (predicted by Pfam search) [42]. The phylogenetic tree constructed using the sequence of KS domain (Additional file 7A) shows that animal PKSs are phylogenetically distinct from animal FASs and rather they are close to microbial PKS genes (Additional file 7A).
Figure 4.

Broad distribution and conserved roles of PKSs in animals. (A) Distribution of the pks genes found by the BLAST searches in the schematic phylogenetic tree of animal kingdom. Red font shows the presence of type I pks gene(s) in the species. Except for fly, frog and mammal, most intensively studied models, pks genes could be overlooked due to incomplete genome information. (B) Whole-mount in situ hybridization of H. pulcherrimus with probes for hppks-1 (Upper Panel) and hppks-2 (Lower Panel). hppks-1 was first detected at the mesenchyme blastula stage in the precursors of the secondary mesenchyme cells (SMCs) at the vegetal pole, and the expression persisted until the prism stage, in the SMCs and then in the ectoderm. The expression was no longer observed in pluteus larvae. hppks-2 expression initiates in PMC precursors at the blastula stage and disappear by late gastrula just after spicule formation starting (mid-gastrulation). (C) Representative results of the MO knockdown experiments in H. pulcherrimus. Images were taken at two stages (24 h and 48 h). Arrows indicate pigment cells. HpPKS-2 first Met MO-injected or its control MO-injected embryos were also observed by a dark-field microscope for visualizing the spicules. Each MO was injected at a concentration of 200 μM. ‘CMO’: Control MO, Scale bars: 50 μm.
We hypothesized that like medaka PKSs, some of the other animal PKSs participate in biomineralization, more specifically calcium carbonate mineralization. To test this idea, we focused on echinoderm pks-2 because its reported expression is in primary mesenchyme cells (PMCs) that give rise to spicules, larval skeletons made of calcium carbonate [38,43]. We first confirmed the PMC-specific expression of pks-2 in our experimental system, Hemicentrotus pulcherrimus (hppks-2). Importantly, the expression of sea urchin pks-2 disappears around late gastrula stage just after PMCs begin to form spicules (Figure 4B Lower Panel). We then examined the function of HpPKS-2 by injecting HpPKS-2 MO. As shown in Figure 4C, HpPKS-2 morphants exhibited severe defects in spicule formation while control MO-injected embryos appeared normal (Figure 4C and Additional file 7C). Another echinoderm pks gene, hppks-1 was found to contribute to pigmentation as previously reported [39] (Figure 4B Upper Panel and C, and Additional file 7C). We thus conclude that echinoderm pks-2 plays a critical role in the formation of calcareous skeletal elements in larva.
Discussion
We here show that olpks encoded by the ha locus is essential for the early step of otolith biomineralization in medaka. In ha mutant embryos, mineralized stones never form, but instead OMP-1-positive particles precipitate in the endolymph. Our TEM observation confirmed that crystal growth is never initiated in ha OV despite the accumulation of otolith materials (Figure 1D), suggesting that the product of OlPKS participates in the initial step, in particular, nucleation of mineralization. The expression pattern of olpks supports its early function; the expression is transient and disappears by the onset of otolith formation (Figure 2D). This pattern of expression contrasts sharply with that of other essential genes identified so far. For instance, otolin-1 and omp-1 expression begins at st. 25 and persists into adulthood as these proteins continue to be deposited on growing otoliths throughout life [44,45]. Furthermore, the expression of medaka starmaker (called starmaker-like), a key regulator of the crystal lattice of calcium carbonates [46], is detected at st. 21 and is again maintained at later stages [47]. Intriguingly, ha fry develop an otolith-like stone (Additional file 2H), and some adult ha fish finally have otolith-like stones, albeit with abnormal shape and size [48]. This does not occur in zebrafish ‘no-otolith’-type mutants such as backstroke and keinstein [49]. These facts further suggest that the endolymph of ha only lacks a trigger. In ha, continuous supply of otolith materials without crystallization could lead to abnormal development of the inner ear and irregular stone formation at later stages. Together with these phenotypic and expression analyses, we conclude that the product of OlPKS functions as a nucleation trigger (facilitator).
Despite our efforts to identify the product of OlPKS, it remains elusive; we failed to detect any specific peak of the synthesized compound in HPLC analysis using extracts of A. oryzae transformed with olpks, probably due to low production of the compound. However, the active substance could have amphiphilic nature like other polyketide (e.g., phenolic lipids [50]) because it was re-solved in medaka culture medium and penetrates into the otic vesicle of the embryo in the rescue experiment (Figure 3D). Given its amphiphilic nature, the product of OlPKS could serve as a bridge between ACC and organic components such as soluble matrix protein (e.g., OMP-1) and/or insoluble scaffold matrices (e.g., Otolin-1)(the possible functions of these organic compounds in biomineralization were proposed by Nagasawa [3]). Supporting this idea, in studies of High-resolution Cryo-TEM imaging, monolayers of fatty acids (arachidic acid or stearic acid) that are also amphiphilic in nature, have been used to artificially induce crystal nucleation [7,51]. Further investigation using cell-free systems [52,53] will be necessary to address at which step of biomineralization PKS products actually work.
Otolith formation shares many features with the formation of vertebrate bones consisting of calcium phosphate (hydroxyapatite) minerals, the best characterized calcification process to date [54]. Intriguingly, in bone formation, phospholipid such as phosphatidylserine (PS) has been implicated in an inducer of hydroxyapatite mineralization and most likely acts as a nucleation facilitator [52,55,56]. Since PS is a small substance with amphiphilic nature, the product of OlPKS could be a functional counterpart in calcium carbonate mineralization, interacting with calcium ions, ACC and matrix proteins.
The phylogenetic analysis based on the KS domain demonstrates the topology in which animal PKSs are separated from the animal FAS clade (Additional file 7A). Unlike the fas gene, the distribution of pks genes in animals is irregular; they are not found in some animal groups, for examples, fly, frog, and mammal, which represent some of the most intensively studied model animals (Figure 4A). These facts complicate the evolutionary origin of animal pks genes, while a single origin of animal fas has been repeatedly supported. Horizontal gene transfer, gene duplications and gene losses could have occurred during evolution, as proposed in bacteria/fungi PKSs [57-59]. Among those animal pks genes, sea urchin pks-2 could be functionally equivalent to olpks because of its early and transient expression in the calcifying cell lineage and its knockdown phenotype, although the domain architecture of sea urchin PKS-2 differs to some degree from that of OlPKS, being KS-AT-DH-ER-KR-ACP-TE and KS-AT-DH-KR-ACP, respectively [38].
Given the conserved role of medaka and sea urchin PKSs, calcium carbonate biomineralization could generally require the products of PKSs for nucleation. PKSs could participate in the production of coral skeletons and calcification of algae in the ocean. For instance, Emiliania huxleyi [58,60] has some pks genes (e.g., fgeneshEH_pg.50__74, protein ID103465, Joint Genome Institute (http://genome.jgi-psf.org/pages/search-for-genes.jsf?organism=Emihu1)). Intriguingly, the presence of PKS in Cnidaria appears to be associated with biomineraliztion; coral (A. digitifera) has a couple of pks genes, whereas Hydra magnipapillata and Nematostella vectensis do not. Furthermore, pks genes of bird and reptile might be involved in production of egg shells. In frogs and mammals, however, pks genes were no longer needed and lost during evolution, although their inner ears still contain calcium carbonate biominerals. This inconsistency may be explained by the fact that their biominerals do not form large stones, but instead small grains called otoconia. However, further intense search in those animals which appear to have lost pks genes will definitely be needed.
Conclusions
The present study addresses the function of the vertebrate pks gene and demonstrates its vital role in calcium carbonate biomineralization. Further functional analyses of newly identified PKSs will uncover a long-overlooked world of polyketides and their derivatives in animals. Our finding also provides genetic and molecular cues for the geochemical study of global carbon and calcium cycles.
Accession numbers
pks genes reported herein have been deposited in GenBank with accession numbers as follow: olpks [AB923905], hppks-1 [AB923906] and hppks-2 [AB923907].
Acknowledgements
We thank Dr. Hiromichi Nagasawa for providing anti- O. mykiss OMP-1 serum; Dr. Joachim Wittbrodt for providing the cDNA of pax2; Dr. Akira Kudo for providing the ki79 fish; Yasuko Ozawa for excellent fish care; Sayaka Tayama for her contribution to the chimeric experiment. Members of the Yasuo Ohnishi laboratory, past and present, are acknowledged, particularly Adeline Muliandi, Takayuki Hayashi and Takuya Makino.
This work was supported in part by Exploratory Research ("Houga Kenkyu") Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology.
Additional files
Materials and Methods.
Phenotypic analyses in ha mutant. (A-F) Whole-mount in situ hybridization of molecular markers expressed in the OVs. (A)(B) Differentiation markers, eya-1 (across the vesicles) and pax-2 (medial region) are normally expressed in ha embryos. All OVs are dorsal views. (C)(D) Normal expression patterns of marker genes. dlx-3b is restricted to the dorsal wall of the OV and bmp-4 marks the neural cristae regions. ac,anterior cristae; lc, lateral cristae; pc, posterior cristae. All OVs are lateral views. Left side shows anterior. (E)(F) The transcripts of otolith matrix proteins, starmaker-like and sparc-1 (are comparably detected in the mutant OV. All OVs are dorsal views. (G) TEM images from a wt specimen and a mutant one (representative images are shown in G to G’’’). Asterisks: growing otolith precursors; Arrows: fine substances; ‘s’: seeding particles; ‘g’: globules. (H) Immunohistochemistry for OMP-1 revealed crystalized ‘otolith’ of ha in hatching stage (st. 40) that looks similar to wt otolith and contains at least some organic materials. (I) double colored-Immunofluorescence of acetylated α-tubulin and γ-tubulin (root of cilia). Short cilia (<1μm) are visible in the OV of ha as well as wt. (J) Scatter plot showing total number of cilia in the OV of wt and ha at various stages. Total number of cilia in the OV is obtained by counting the signal of α-tubulin and γ-butulin. In each stage, there was no significant difference in total number of cilia between wt and ha (>0.05 Student’s t-test). (K) Otolith development in the OV of ktu mutant. In ktu, seeding particles are coalesced at the prospective macula region (Upper; dorsal views of left OV), and otoliths appear normal at later stage just like wt (Middle; dorsal views of left OV and Lower; lateral views of left OV). Asterisks: otoliths, Arrow: paste-like substance.
Additional file 3:
Movement of seeding particles in wt OV at st 25. The movie is recorded at the rate of 1 frame per second and played back 5 times faster than real-time viewing (dorsal view of left OV). A number of seeding particles are floating and otoliths are already mineralized.
Additional file 4:
Movement of seeding particles in ha OV at st 25. The movie is recorded at the rate of 1 frame per second and played back 5 times faster than real-time viewing (dorsal view of left OV). ha OV contains large number of seeding particles due to failure of mineralization.
Mapping analyses and expression pattern of the ha gene. (A) Sequencing of the ha gene in two mutant fish. ki79 fish harbors a nonsense mutation at 3856 nt of the ORF2 locus. (B) Architecture of OlPKS protein predicted from the amino acid sequence (Upper). Critical residues of each domain used for the mRNA injection experiment are shown by arrows. Mutation site of each mutant is shown by arrowhead. Three amino acids corresponding to 9-bp deletion is ‘KPS’. Central proline is found to be a highly conserved residue among PKSs although it is not previously considered as a conserved motif. Based on the structure of mammal fatty acid synthase (FAS), it is possible that the proline contribute to help a substrate to enter the active site of KS. The red bar indicates the region used as an antigen for producing anti-serum of OlPKS. Conserved motifs found in the amino acid sequence of OlPKS (Lower table). Asterisks show residues mutated in mRNA rescue experiments. Underlines show conserved amino acid residues. (C) Whole-mount in situ hybridization with olpks probes at st. 22. The dorsally- and medially- restricted pattern is evidenced by lateral view (Upper) and histology of the OV region (Lower). Yellow dashed lines show OVs. (D) Expression profiles of olpks and paralogous gene candidates in the embryonic stages or adult tissues. A RT-PCR analysis reveals that olpks is expressed but olpks-2 and olpks-3 are not expressed and that olpks is expressed only around embryonic stage 22.
Analyses of property of the OlPKS product. (A)(B) Imaging analyses of the chimeric experiment. (A) All 110 OVs of 55 transplanted animals are observed and sorted to 9 categories depending on their percentage of wt cells in their OV epithelium (Note a chimeric fish has two OVs). The histogram shows that a very small number of wt cells can rescue otolith formation. (B) The same sample set is also analyzed to assess the localization of wt cells within the OV. ‘medial’ means wt cells located around medial wall of the OV region where olpks gene is normally expressed in wt animal; ‘medial and other’ includes medial and any other epithelial region; ‘other’ includes wt cells existing anywhere in the epithelium except the medial wall. (C) Western blot of OlPKS protein produced by A. oryzae. To verify expression of OlPKS in heterologous expression experiment, purified protein from a transformant is analyzed by anti-OlPKS antiserum. The OlPKS protein gave a protein band at a position of ca. 210 kDa on SDS-PAGE. However, some bands (degradade proteins) were also found in this experiment.
Broad distribution of animal PKSs. (A) Maximum parsimony phylogenetic estimate of relationships among animal type I PKSs and FASs, based on an alignment of amino acid sequences of the KS domain. Animal PKSs containing OlPKS are not divided from other bacterial or fungal PKSs, as indicated by low values of the bootstrap. By contrast, the animal FAS clade is clearly separated from animal PKSs. Protein accession number of Genbank or other database is shown with species name. Numbers described around branches indicate percentage of the bootstrap value supporting each clade. Branch length indicates number of inferred amino acid changes. (B) Representative image of whole-mount in situ hybridization of zebrafish embryo using probes for pks homologue. The transcript of a pks homologue, drpks(wu:fc01d11), is exclusively expressed in zebrafish OV at the 20 somite-stage (19hpf). Arrows: OVs. (C) Summary of knockdown experiment using MOs. Injected embryos are categorized by the phenotype of spicule. HpPKS-1 knockdown animal lacked pigment while HpPKS-1 control MO (CMO)-injected animals showed normal pigmentation. These two morphants didn’t show any spicule abnormality. Injection of HpPKS-1 MO and HpPKS-1 CMO at high concentration (200 μM) caused developmental arrest at the gastrulation. Various concentrations of HpPKS-2 MO injection (three levels; 50, 100 and 200 μM) shows dose dependency. All HpPKS-2 CMO-injected embryos showed no spicule defect. Standard control MO did not give any effect.
Footnotes
Competing interests
We declare that no actual or potential competing interests in relation to this article exist.
Authors’ contributions
MH carried out the embryological studies, biochemical analysis in the heterologous expression experiment and phylogenetic analysis, and drafted the manuscript. AO carried out the mapping analysis and participated in the expression analysis in medaka embryos. GH carried out the knockdown experiment in sea urchin. KS carried out the analytical chemistry in the heterologous expression experiment. AS carried out the transplant experiment for producing chimaera medaka fish. MK carried out the expression analysis of sea urchin genes. TN participated in the mapping analysis. MK participated in the design of the study using sea urchin embryos. YK and YO participated in the design of the heterologous expression experiment. NI participated in the design of the phylogenetic study. HT conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Motoki Hojo, Email: Motoki_Hojo@member.metro.tokyo.jp.
Ai Omi, Email: ai.omi@riken.jp.
Gen Hamanaka, Email: hamanaka.gen@ocha.ac.jp.
Kazutoshi Shindo, Email: kshindo@fc.jwu.ac.jp.
Atsuko Shimada, Email: kirita@bs.s.u-tokyo.ac.jp.
Mariko Kondo, Email: konmari@mmbs.s.u-tokyo.ac.jp.
Takanori Narita, Email: narita.takanori@nihon-u.ac.jp.
Masato Kiyomoto, Email: kiyomoto.masato@ocha.ac.jp.
Yohei Katsuyama, Email: ayasuo@mail.ecc.u-tokyo.ac.jp.
Yasuo Ohnishi, Email: aykatsu@mail.ecc.u-tokyo.ac.jp.
Naoki Irie, Email: irie@bs.s.u-tokyo.ac.jp.
Hiroyuki Takeda, Email: htakeda@bs.s.u-tokyo.ac.jp.
References
- 1.Addadi L, Weiner S. Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc Natl Acad Sci U S A. 1985;82:4110–4114. doi: 10.1073/pnas.82.12.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiner S, Hood L. Soluble protein of the organic matrix of mollusk shells: a potential template for shell formation. Science. 1975;190:987–989. doi: 10.1126/science.1188379. [DOI] [PubMed] [Google Scholar]
- 3.Nagasawa H. The molecular mechanism of calcification in aquatic organisms. Biosci Biotechnol Biochem. 2013;77:1991–1996. doi: 10.1271/bbb.130464. [DOI] [PubMed] [Google Scholar]
- 4.Lowenstam HA, Weiner S. On Biomineralization. New York: Oxford University Press; 1989. [Google Scholar]
- 5.Westbroek P, Brown CW, Van Bleijswijk J, Brownlee C, Brummer GJ, Conte M, Egge J, Fernandez E, Jordan R, Knappertsbusch M, Stefels J, Veldhuis M, Van der Wal P, Young J. A model system approach to biological climate forcing. The example of Emiliania huxleyi. Glob Planet Change. 1993;8:27–46. doi: 10.1016/0921-8181(93)90061-R. [DOI] [Google Scholar]
- 6.Van CP. Biomineralization and Global Biogeochemical Cycles. Rev Mineral Geochem. 2003;54:357–381. doi: 10.2113/0540357. [DOI] [Google Scholar]
- 7.Dey A, Bomans PH, Muller FA, Will J, Frederik PM, de With G, Sommerdijk NA. The role of prenucleation clusters in surface-induced calcium phosphate crystallization. Nat Mater. 2010;9:1010–1014. doi: 10.1038/nmat2900. [DOI] [PubMed] [Google Scholar]
- 8.Gebauer D, Volkel A, Colfen H. Stable prenucleation calcium carbonate clusters. Science. 2008;322:1819–1822. doi: 10.1126/science.1164271. [DOI] [PubMed] [Google Scholar]
- 9.Campana SE. Chemistry and composition of fish otoliths: pathway, mechanisms and applications. Mar Ecol Prog Ser. 1999;188:263–297. doi: 10.3354/meps188263. [DOI] [Google Scholar]
- 10.Pisam M, Jammet C, Laurent D. First steps of otolith formation of the zebrafish: role of glycogen? Cell Tissue Res. 2002;310:163–168. doi: 10.1007/s00441-002-0622-z. [DOI] [PubMed] [Google Scholar]
- 11.Whitfield TT, Granato M, van Eeden FJ, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Mutations affecting development of the zebrafish inner ear and lateral line. Dev. 1996;123:241–254. doi: 10.1242/dev.123.1.241. [DOI] [PubMed] [Google Scholar]
- 12.Schibler A, Malicki J. A screen for genetic defects of the zebrafish ear. Mech Dev. 2007;124:592–604. doi: 10.1016/j.mod.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Tomita H. Biology of the Medaka (Egami, N ed University of Tokyo Press) 1990. Strains And Mutants; pp. 111–127. [Google Scholar]
- 14.Kimura T, Jindo T, Narita T, Naruse K, Kobayashi D, Shin IT, Kitagawa T, Sakaguchi T, Mitani H, Shima A, Kohara Y, Takeda H. Large-scale isolation of ESTs from medaka embryos and its application to medaka developmental genetics. Mech Dev. 2004;121:915–932. doi: 10.1016/j.mod.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsuka M, Kikuchi N, Yokoi H, Kinoshita M, Wakamatsu Y, Ozato K, Takeda H, Inoko H, Kimura M. Possible roles of zic1 and zic4, identified within the medaka Double anal fin (Da) locus, in dorsoventral patterning of the trunk-tail region (related to phenotypes of the Da mutant) Mech Dev. 2004;121:873–882. doi: 10.1016/j.mod.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Hojo M, Takashima S, Kobayashi D, Sumeragi A, Shimada A, Tsukahara T, Yokoi H, Narita T, Jindo T, Kage T, Kitagawa T, Kimura T, Sekimizu K, Miyake A, Setiamarga D, Murakami R, Tsuda S, Ooki S, Kakihara K, Naruse K, Takeda H. Right-elevated expression of charon is regulated by fluid flow in medaka Kupffer's vesicle. Develop Growth Differ. 2007;49:395–405. doi: 10.1111/j.1440-169X.2007.00937.x. [DOI] [PubMed] [Google Scholar]
- 17.Murayama E, Herbomel P, Kawakami A, Takeda H, Nagasawa H. Otolith matrix proteins OMP-1 and Otolin-1 are necessary for normal otolith growth and their correct anchoring onto the sensory maculae. Mech Dev. 2005;122:791–803. doi: 10.1016/j.mod.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Kamura K, Kobayashi D, Uehara Y, Koshida S, Iijima N, Kudo A, Yokoyama T, Takeda H. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Dev. 2011;138:1121–1129. doi: 10.1242/dev.058271. [DOI] [PubMed] [Google Scholar]
- 19.Yokoi H, Shimada A, Carl M, Takashima S, Kobayashi D, Narita T, Jindo T, Kimura T, Kitagawa T, Kage T, Sawada A, Naruse K, Asakawa S, Shimizu N, Mitani H, Shima A, Tsutsumi M, Hori H, Wittbrodt J, Saga Y, Ishikawa Y, Araki K, Takeda H. Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships. Dev Biol. 2007;304:326–337. doi: 10.1016/j.ydbio.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 20.Witkowski A, Joshi A, Smith S. Fatty acid synthase: in vitro complementation of inactive mutants. Biochemistry. 1996;35:10569–10575. doi: 10.1021/bi960910m. [DOI] [PubMed] [Google Scholar]
- 21.Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res. 2003;42:289–317. doi: 10.1016/S0163-7827(02)00067-X. [DOI] [PubMed] [Google Scholar]
- 22.Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 23.Shimada A, Yabusaki M, Niwa H, Yokoi H, Hatta K, Kobayashi D, Takeda H. Maternal-zygotic medaka mutants for fgfr1 reveal its essential role in the migration of the axial mesoderm but not the lateral mesoderm. Dev. 2008;135:281–290. doi: 10.1242/dev.011494. [DOI] [PubMed] [Google Scholar]
- 24.Gomi K, Iimura Y, Hara S. Integrative Transformation of Aspergillus oryzae with a Plasmid Containing the Aspergillus nidulans argB Gene. Agric Biol Chem. 1987;51:2549–2555. doi: 10.1271/bbb1961.51.2549. [DOI] [Google Scholar]
- 25.Katow H, Katow T, Abe K, Ooka S, Kiyomoto M, Hamanaka G. Mesomere-derived glutamate decarboxylase-expressing blastocoelar mesenchyme cells of sea urchin larvae. Biology open. 2014;3:94–102. doi: 10.1242/bio.20136882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ijiri K. Vestibular and visual contribution to fish behavior under microgravity. Adv Space Res. 2000;25:1997–2006. doi: 10.1016/S0273-1177(99)01005-4. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno R, Ijiri K. Otolith formation in a mutant medaka with a deficiency in gravity-sensing. Adv Space Res. 2003;32:1513–1520. doi: 10.1016/S0273-1177(03)90389-9. [DOI] [PubMed] [Google Scholar]
- 28.Riley BB, Zhu C, Janetopoulos C, Aufderheide KJ. A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Dev Biol. 1997;191:191–201. doi: 10.1006/dbio.1997.8736. [DOI] [PubMed] [Google Scholar]
- 29.Colantonio JR, Vermot J, Wu D, Langenbacher AD, Fraser S, Chen JN, Hill KL. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature. 2009;457:205–209. doi: 10.1038/nature07520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu D, Freund JB, Fraser SE, Vermot J. Mechanistic basis of otolith formation during teleost inner ear development. Dev Cell. 2011;20:271–278. doi: 10.1016/j.devcel.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stooke-Vaughan GA, Huang P, Hammond KL, Schier AF, Whitfield TT. The role of hair cells, cilia and ciliary motility in otolith formation in the zebrafish otic vesicle. Dev. 2012;139:1777–1787. doi: 10.1242/dev.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O'Toole E, Hara C, Mizuno H, Kawano H, Fliegauf M, Yagi T, Koshida S, Miyawaki A, Zentgraf H, Seithe H, Reinhardt R, Watanabe Y, Kamiya R, Mitchell DR, Takeda H. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolouli K, Mossialos D. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol Lett. 2012;34:1393–1403. doi: 10.1007/s10529-012-0919-2. [DOI] [PubMed] [Google Scholar]
- 34.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 35.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 2005;1:12–19. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beeble A, Calestani C. Expression pattern of polyketide synthase-2 during sea urchin development. Gene Expr Patterns. 2012;12:7–10. doi: 10.1016/j.gep.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Dev. 2003;130:4587–4596. doi: 10.1242/dev.00647. [DOI] [PubMed] [Google Scholar]
- 40.Castoe TA, Stephens T, Noonan BP, Calestani C. A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs. Gene. 2007;392:47–58. doi: 10.1016/j.gene.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Leibundgut M, Maier T, Jenni S, Ban N. The multienzyme architecture of eukaryotic fatty acid synthases. Curr Opin Struct Biol. 2008;18:714–725. doi: 10.1016/j.sbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien RV, Davis RW, Khosla C, Hillenmeyer ME. Computational identification and analysis of orphan assembly-line polyketide synthases. J Antibiot (Tokyo) 2014;67:89–97. doi: 10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rafiq K, Cheers MS, Ettensohn CA. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Dev. 2012;139:579–590. doi: 10.1242/dev.073049. [DOI] [PubMed] [Google Scholar]
- 44.Nemoto Y, Chatani M, Inohaya K, Hiraki Y, Kudo A. Expression of marker genes during otolith development in medaka. Gene Expr Patterns. 2008;8:92–95. doi: 10.1016/j.modgep.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Murayama E, Takagi Y, Nagasawa H. Immunohistochemical localization of two otolith matrix proteins in the otolith and inner ear of the rainbow trout, Oncorhynchus mykiss: comparative aspects between the adult inner ear and embryonic otocysts. Histochem Cell Biol. 2004;121:155–166. doi: 10.1007/s00418-003-0605-5. [DOI] [PubMed] [Google Scholar]
- 46.Sollner C, Burghammer M, Busch-Nentwich E, Berger J, Schwarz H, Riekel C, Nicolson T. Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science. 2003;302:282–286. doi: 10.1126/science.1088443. [DOI] [PubMed] [Google Scholar]
- 47.Bajoghli B, Ramialison M, Aghaallaei N, Czerny T, Wittbrodt J. Identification of starmaker-like in medaka as a putative target gene of Pax2 in the otic vesicle. Dev Dyn. 2009;238:2860–2866. doi: 10.1002/dvdy.22093. [DOI] [PubMed] [Google Scholar]
- 48.Noro S, Yamamoto N, Ishikawa Y, Ito H, Ijiri K. Studies on the morphology of the inner ear and semicircular canal endorgan projections of ha, a medaka behavior mutant. Fish Biol J Medaka. 2007;11:31–41. [Google Scholar]
- 49.Sollner C, Schwarz H, Geisler R, Nicolson T. Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol. 2004;214:582–590. doi: 10.1007/s00427-004-0440-2. [DOI] [PubMed] [Google Scholar]
- 50.Kozubek A, Tyman JH. Resorcinolic Lipids, the Natural Non-isoprenoid Phenolic Amphiphiles and Their Biological Activity. Chem Rev. 1999;99:1–26. doi: 10.1021/cr970464o. [DOI] [PubMed] [Google Scholar]
- 51.Pouget EM, Bomans PH, Goos JA, Frederik PM, de With G, Sommerdijk NA. The initial stages of template-controlled CaCO3 formation revealed by cryo-TEM. Science. 2009;323:1455–1458. doi: 10.1126/science.1169434. [DOI] [PubMed] [Google Scholar]
- 52.Boskey AL. Hydroxyapatite formation in a dynamic collagen gel system: effects of type i collagen, lipids, and proteoglycans. J Phys Chem. 1989;93:1628–1633. doi: 10.1021/j100341a086. [DOI] [Google Scholar]
- 53.Tohse H, Saruwatari K, Kogure T, Nagasawa H, Takagi Y. Control of polymorphism and morphology of calcium carbonate crystals by a matrix protein aggregate in fish otoliths. Cryst Growth Des. 2009;9:4897–4901. doi: 10.1021/cg9006857. [DOI] [Google Scholar]
- 54.Hughes I, Thalmann I, Thalmann R, Ornitz DM. Mixing model systems: using zebrafish and mouse inner ear mutants and other organ systems to unravel the mystery of otoconial development. Brain Res. 2006;1091:58–74. doi: 10.1016/j.brainres.2006.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu LN, Genge BR, Dunkelberger DG, LeGeros RZ, Concannon B, Wuthier RE. Physicochemical characterization of the nucleational core of matrix vesicles. J Biol Chem. 1997;272:4404–4411. doi: 10.1074/jbc.272.7.4404. [DOI] [PubMed] [Google Scholar]
- 56.Satsangi A, Satsangi N, Glover R, Satsangi RK, Ong JL. Osteoblast response to phospholipid modified titanium surface. Biomaterials. 2003;24:4585–4589. doi: 10.1016/S0142-9612(03)00330-2. [DOI] [PubMed] [Google Scholar]
- 57.Jenke-Kodama H, Dittmann E. Evolution of metabolic diversity: insights from microbial polyketide synthases. Phytochemistry. 2009;70:1858–1866. doi: 10.1016/j.phytochem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 58.John U, Beszteri B, Derelle E, Van de Peer Y, Read B, Moreau H, Cembella A. Novel Insights into Evolution of Protistan Polyketide Synthases through Phylogenomic Analysis. Protist. 2008;159:21–30. doi: 10.1016/j.protis.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Foerstner KU, Doerks T, Creevey CJ, Doerks A, Bork P. A computational screen for type I polyketide synthases in metagenomics shotgun data. PLoS One. 2008;3:e3515. doi: 10.1371/journal.pone.0003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones BM, Edwards RJ, Skipp PJ, O'Connor CD, Iglesias-Rodriguez MD. Shotgun proteomic analysis of Emiliania huxleyi, a marine phytoplankton species of major biogeochemical importance. Mar Biotechnol. 2011;13:496–504. doi: 10.1007/s10126-010-9320-0. [DOI] [PubMed] [Google Scholar]


