Abstract
Aminoglycoside antibiotics, such as gentamicin, may induce premature termination codon (PTC) readthrough and elude the nonsense-mediated mRNA decay mechanism. Because PTCs are frequently involved in lysosomal diseases, readthrough compounds may be useful as potential therapeutic agents. The aim of our study was to identify patients responsive to gentamicin treatment in order to be used as positive controls to further screen for other PTC readthrough compounds. With this aim, fibroblasts from 11 patients affected by 6 different lysosomal diseases carrying PTCs were treated with gentamicin. Treatment response was evaluated by measuring enzymatic activity, abnormal metabolite accumulation, mRNA expression, protein localization, and cell viability. The potential effect of readthrough was also analyzed by in silico predictions. Results showed that fibroblasts from 5/11 patients exhibited an up to 3-fold increase of enzymatic activity after gentamicin treatment. Accordingly, cell lines tested showed enhanced well-localized protein and/or increased mRNA expression levels and/or reduced metabolite accumulation. Interestingly, these cell lines also showed increased enzymatic activity after PTC124 treatment, which is a PTC readthrough-promoting compound. In conclusion, our results provide a proof-of-concept that PTCs can be effectively suppressed by readthrough drugs, with different efficiencies depending on the genetic context. The screening of new compounds with readthrough activity is a strategy that can be used to develop efficient therapies for diseases caused by PTC mutations.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-015-0368-4) contains supplementary material, which is available to authorized users.
Key Words: Gentamicin, aminoglycoside, lysosomal disease, premature stop codon, PTC124, readthrough treatment
Introduction
Nonsense-mediated mRNA decay (NMD) is a quality-control mechanism that selectively degrades mRNAs harboring premature termination codons (PTC). During mRNA processing, exon splice junctions are marked with exon junction complexes (EJC). As protein translation occurs, the ribosome displaces all EJC except in those transcripts harboring a PTC. Thus, when an EJC remains because of the presence of a PTC, NMD machinery is activated to ensure aberrant mRNA degradation, except in those transcripts harboring a PTC greater than around 50 nucleotides upstream of an EJC or in the last exon [1, 2]. This process protects the organism from truncated proteins in recessive inherited disorders and from potential dominant-negative or gain-of-function activities from dominantly inherited disorders that could arise if these proteins were expressed. Therefore, NMD plays a key protective role in a long list of human diseases [3].
Lysosomal storage diseases (LSD) comprise > 50 different genetic disorders involving the accumulation of nondegraded macromolecules into lysosomes and are mainly inherited in an autosomal recessive manner or, in few cases, are X-linked. The global incidence of LSD is approximately 1 in 5000 live births. Most of them are caused by the deficiency of a particular lysosomal enzyme involved in the degradation of a specific substrate, preventing its accumulation [4, 5]. Around two-thirds of patients with LSD will develop brain pathology together with multiorgan involvement. To some extent, enzymatic activity depends on the mutation, and in most cases the severity of the disease correlates with the degree of the enzymatic deficiency. However, residual enzymatic activity of around 10–20 % of the activity detected in healthy individuals may be enough for functional recovery resulting in the wild-type phenotype [6]. Treatment strategies for LSD include bone marrow transplantation, substrate reduction, and enzyme replacement therapies [7]. However, the usefulness of some of these therapies is limited as these compounds are not always able to cross the blood–brain barrier. Therefore, alternative treatment strategies are required to address both the neuropathology and other somatic abnormalities. Although major progress has been made in gene therapy, it is still far from achieving real clinical application. In the last few years, other potential strategies such as pharmacological chaperones [8], or the suppression of pathogenic nonsense mutations through the induction of translational readthrough have emerged [2, 9–18]. To this effect, aminoglycoside antibiotics such as gentamicin [16, 17], and small molecules such as PTC124 [18] and NB84 [19], have been described to induce PTC readthrough, eluding the NMD mechanism and allowing the formation of stable mRNAs encoding for full-length mutant, but probably still quite active, proteins [20, 21]. These products reduce proofreading of codon–anticodon recognition in the ribosome, allowing the suppression of PTCs and, as a general rule, the insertion of glutamine or tryptophan at premature UAG/UAA or UGA codons, respectively, occurs [17]. However, there is increasing evidence that only a small subset of stop codon generating-mutations would benefit from gentamicin or PTC124 treatment depending on their nucleotide context [22, 23]. Floquet et al. [24] described that nucleotides at position –1, –5, +4, and +8 around the premature stop codon are important to determine PTC readthrough efficiency upon gentamicin treatment. However, the strong side effects of gentamicin, the most commonly used aminoglycoside antibiotic, precludes its utilization as a potential therapeutic agent for these diseases because long-term treatment is required [25, 26].
Here, we present the analysis of the effect of treatment with PTC readthrough-inducer drugs on fibroblasts from 11 patients with different LSD, with the aim of identifying responsive mutated cell lines to be used for the development of future therapeutic approaches. Moreover, this work provides additional clues in the knowledge on diseases and mutations able to respond to PTC readthrough treatment.
Patients and Methods
Patients
Primary skin fibroblasts from 11 patients with 6 different LSD (Fabry disease, gangliosidosis type I , Hunter, Hurler, Sanfilippo B and Niemann Pick A/B; see Table 1) were grown in Dulbecco’s Modified Eagle medium with 10 % fetal bovine serum and antibiotics (penicillin and streptomycin) at 37 °C in 5 % CO2 (all PAA Laboratories, Velizy-Villacoublay, France). Patients were selected on the basis of availability of fibroblasts, measurable enzymatic activity, presence of a nonsense or frameshift mutation leading to PTC (preferably in hemi- or homozygosis), and being representative of the most frequent lysosomal diseases. The disease affecting each patient, as well as the genetic characteristics and enzymatic activities, are summarized in Table 1. The use of human samples was approved by the ethical committee of the Hospital Clínic of Barcelona.
Table 1.
Skin-cultured fibroblasts used in this study
| Patient | Disease | Gene | Genotype | Effect on protein | Passage number | Analysis performed |
|---|---|---|---|---|---|---|
| 1 | Niemman–Pick A/B | SMPD1 | c.[503G > A] + [503G > A] | p.[Trp168*]; [Trp168*] | 4/6 | Enzymatic activity, in silico analysis, immunofluorescent microscopy, mRNA expression analysis |
| 2 | Niemman–Pick A/B | SMPD1 | c.[939A > C] + [939C > A] | p.[Tyr313*]; [Tyr313*] | 5 | Enzymatic activity, in silico analysis |
| 3 | Niemman–Pick A/B | SMPD1 | c.[1159delC] + [1128G > A] | p.[Arg387VfsX7]; [Arg376His] | 4 | Enzymatic activity, in silico analysis |
| 4 | Gangliosidosis type I | GLB1 | c.[1370C > T] + [791 T > C] | p.[Arg457*]; [Leu264Ser] | 6/7 | Enzymatic activity, in silico analysis |
| 5 | Mucopolysaccharidosis type IIIB (Sanfilippo B disease ) | NAGLU | c.[503G > A] + [700C > T] | p.[Trp168*]; [Arg234Cys] | 5/7 | Enzymatic activity, glycosaminoglycan quantification, in silico analysis, mRNA expression analysis |
| 6 | Mucopolysaccharidosis type IIIB (Sanfilippo B disease) | NAGLU | c.[503G > A] + [1696C > T] | p.[Trp168*]; [Q566*] | 5/7 | Enzymatic activity, in silico analysis, immunofluorescent microscopy, glycosaminoglycan quantification and mRNA expression analysis |
| 7 | Fabry | GLA | c.[679C > T] | p.[Arg227*] | 5 | Enzymatic activity, in silico analysis |
| 8 | Mucopolysaccharidosis type I (Hurler disease) | IDUA | c.[1205A > G] + [1205G > A] | p.[Trp402*]; [Trp402*] | 4 | Enzymatic activity, in silico analysis. |
| 9 | Mucopolysaccharidosis type I (Hurler disease) | IDUA | c.[1205A > G] + [1205G > A] | p.[Trp402*]; [Trp402*] | 4 | Enzymatic activity, in silico analysis |
| 10 | Mucopolysaccharidosis type I (Hurler disease) | IDUA | c.[1205A > G] + [1205G > A] | p.[Trp402*]; [Trp402*] | 4 | Enzymatic activity, in silico analysis |
| 11 | Mucopolysaccharidosis type II (Hunter disease) | IDS | c.[1327C > T] | p.[R443*] | 4/7 | Enzymatic activity, in silico analysis, glycosaminoglycan quantification, and mRNA expression analysis |
Readthrough Drug Treatment in Patients’ Fibroblasts
Early passage primary skin fibroblasts were plated in 25-cm2 flasks and grown to 80 % confluence. Cells were treated with gentamicin (Sigma-Aldrich, St. Louis, MO, USA) and PTC124 (Exclusive Chemistry Ltd, Obninsk, Russia) at a concentration previously described to promote a remarkable PTC readthrough (300 μM for gentamicin and 5 μM for PTC124) [27]. However, to ensure the use of the most effective conditions, drug concentration, incubation time, and cell viability were tested in fibroblasts from patients 1, 5, and 6 upon gentamicin treatment, and from patients 5 and 6 upon PTC124 treatment (Figure S1; see Supplementary Material). Nevertheless, cell viability was controlled in all the cell lines (data not shown). Cells were harvested at 72 h. In order to reduce variability, cell passage number in all experiments was also monitored.
Enzymatic Activities
Fibroblasts were harvested and rinsed twice with physiological serum and lysed by 3 freeze–thaw cycles. Protein concentration was measured using the DC Protein Assay (Bio-Rad Laboratories, S.A, Madrid, Spain). Equal amounts of protein lysates were seeded in white 96-well plates, and the corresponding enzymatic activities were determined using a fluorimetric assay with 4-methylumbelliferyl substrates [28]: 6-hexadecanoylamino-4-metylumbeliferyl-P-coline and 4-methylumbelliferyl-α-L-iduronide-2-sulfate (Moscerdam, Oegstgeest, theNetherlands) for Niemann Pick A/B and Hunter disease, respectively; 4-methylumbelliferyl-α-galactopiranoside and 4-methylumbelliferyl-β-galactopiranoside (Sigma-Aldrich) for Fabry disease and gangliosidosis type I, respectively; 4-methylumbelliferyl- α-L-iduronide (Glycosynth, Cheshire, UK) for Hurler disease; and 4-methylumbelliferyl-2-acetamido-2-deoxy-α-D-glucopyranoside (Calbiochem, Whitehouse Station, NJ, USA) for Sanfilippo B. Fluorescence was measured at 365 nm emission and 465 nm excitation with a microplate reader (POLARstar Omega; BMG LABTECH, Offenburg, Germany). Cells were cultured in triplicate, and determination of the enzymatic activity was also performed in triplicate.
Cell Viability
Cell viability was evaluated using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenil tetrazolium bromide assay (Sigma-Aldrich) as described by Sumantran [29]. Briefly, medium from treated and untreated fibroblasts grown in 96-well plates was replaced by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenil tetrazolium bromide solution and incubated for 3 h at 37 °C. Supernatant was removed and crystals were dissolved with dimethyl sulfoxide (Sigma-Aldrich). After 20 min, absorbance was measured at 595 nm with a spectrophotometer (POLARstar Omega; BMG LABTECH).
Immunofluorescence
Immunofluorescence was performed in fibroblasts derived from patient 1 (Niemann Pick A/B) and patient 6 (Sanfilippo B). Fibroblasts were grown on glass coverslips in the presence or absence of 300 μM gentamicin for 72 h. Lysosomes were stained using LysoTracker Red (Life Technologies, Paisley, UK) supplemented in medium for 1 h at 37 °C. Samples were then fixed with acetone and cells were permeabilized with 1 % bovine serum albumin–0.1 % triton solution for 15 min, followed by 1-h incubation with primary anti-α-N-acetylglucosaminidase (NAGLU) antibody (Novus Biological, Littleton, CO, USA) or anti-acid sphingomyelinase (ASM) antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at room temperature. Incubation with fluorescein isothiocyanate-conjugated secondary antibody for 45 min was performed. Coverslips were mounted with UltraCruz Mounting Medium (Santa Cruz Biotechnology) containing 4′,6-diamidino-2-phenylindole staining, and visualized with a fluorescence microscope (Eclipse 50i; Nikon Instruments, Melville, NY, USA).
Glycosaminoglycans Determination
Quantification of glycosaminoglycans (GAGs) was performed using the 1,9-dimethylmethylene blue assay adapted from Barbosa et al. [30]. Cells were cultured in triplicate in 6-well plates and harvested 72 h after treatment with 300 μM gentamicin. 1,9-Dimethylmethylene blue absorbance was measured in duplicate at 656 nm with a microplate reader (POLARstar Omega; BMG LABTECH).
mRNA Expression Analysis
This assay was performed in skin fibroblasts derived from patient 1 (Niemann Pick A/B), patient 5 and patient 6 (Sanfilippo B), and patient 11 (Hunter). mRNA expression was determined by reverse transcriptase polymerase chain reaction (RT-PCR). Patient and control fibroblasts were treated with gentamicin at 300 μM for 72 h or cycloheximide at 500 μg/ml for 5 h [31, 32]. Cycloheximide is a potent inhibitor of protein synthesis that acts in the NMD pathway allowing for the nondegradation of mutated mRNAs carrying PTCs. Total RNA was extracted, followed by DNase I treatment using QIAshredder and RNeasy kits (both from Qiagen, Hilden, Germany). Single-stranded cDNA was obtained using oligo-dT primers and M-MLV Reverse Transcriptase RNase H Minus Point Mutant (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Analysis of cDNA was performed by RT-PCR using SYBR green reagent (Life Technologies) in a Step One plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). IDS, NAGLU, and SMPD1 cDNA was amplified using specific oligonucleotides (available from the authors upon request). Glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control. PCR reactions were carried out in triplicate using 100 ng cDNA from both patient and control samples. Levels of mRNA were relatively quantified by evaluating Ct values using the comparative Ct (ΔCt) method [33].
In silico Analysis of the Predicted Mutations
We conducted an in silico analysis of the predicted missense mutations generated by gentamicin-mediated PTC readthrough using Polyphen-2 (Polymorphism Phenotyping v2; http://genetics.bwh.harvard.edu/pph2/) and SIFT [34]. Polyphen-2 predicts the possible impact of amino acid substitutions on the stability and function of human proteins considering structural and comparative evolutionary data [35]. SIFT prediction is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences [34]. Both predictors allow measuring of the potential pathogenicity of a particular mutation.
Analysis of the Nucleotide Background Surrounding PTCs
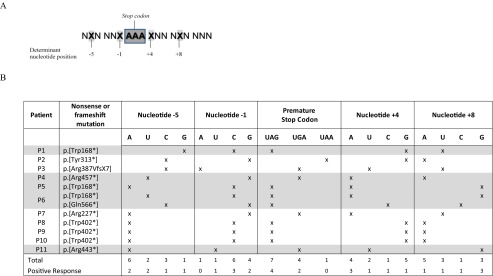
Nucleotides besides the stop codon reported to be determinant for gentamicin readthrough response were compared and annotated for each patient (Table 2) [24].
Table 2.
Nucleotide positions beside the stop codon reported by Floquet et al. [24] to be determinant for gentamicin readthrough response treatment, andnNucleotide context of the different premature termination codon mutations and response to gentamicin treatment
Nucleotide positions described as determinant to gentamicin response are indicated with an X
Patients who responded positively to gentamicin treatment are highlighted in gray
Statistical Analysis
The statistical significance of the data was assessed using the Student’s t test. All data are presented as mean ± SD, with the level of significance set at p < 0.05.
Results
In this study we analyzed the potential effect of gentamicin readthrough in nonsense or frameshift mutations leading to PTCs in 11 patients affected by 6 different lysosomal disorders. In addition, the effect of PTC124 treatment was also evaluated.
Rescue of the Enzymatic Activity After Gentamicin Treatment in Fibroblasts
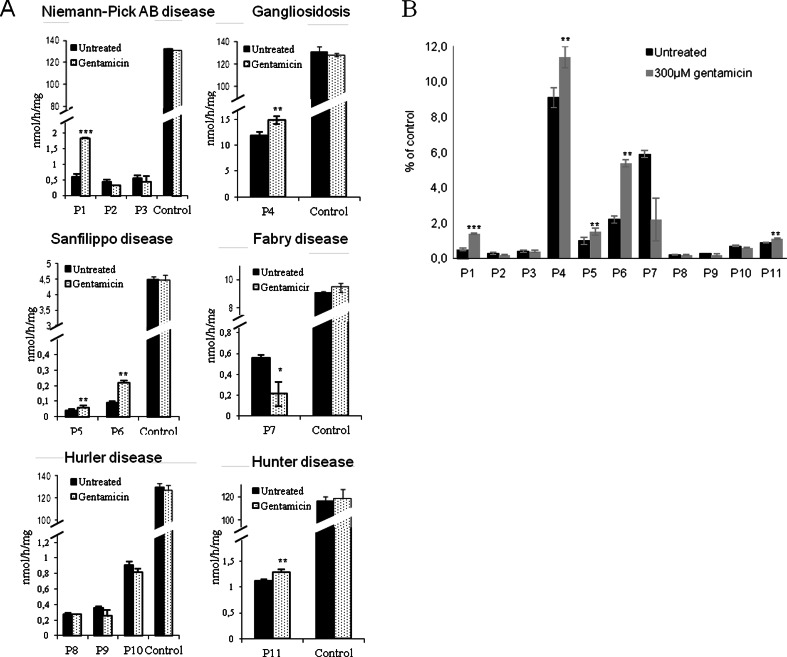
The fibroblasts of 5/11 patients showed an up to 3-fold increase in residual enzymatic activity after gentamicin treatment (Fig. 1A). Patient 1 and patient 6, affected by Niemann–Pick A/B and Sanfilippo B, respectively, showed a slight but statistically significant increase of 2.5–3.0-fold compared with untreated fibroblasts, while patient 4 (gangliosidosis type I), patient 5 (Sanfilippo B), and patient 11 (Hurler) showed a smaller increase of 1.2-fold (Fig. 1A). Cell lines of the remaining 6 patients did not show any increase in enzymatic activity after gentamicin treatment. Cell viability was not altered after treatment in any of the cell lines used in this study (data not shown). Similar results represented as the percentage of residual enzymatic activity relative to controls are shown in Fig. 1B.
Fig. 1.
Residual enzymatic activity in patient fibroblasts upon gentamicin treatment. (A) Equal amounts of protein lysates from cells treated with or without of gentamicin (300 μM) for 72 h were seeded, and the corresponding deficient enzymatic activities for each patient were determined [acid sphingomyelinase for patients 1–3 (P1–3); β-galactosidase for patient 4 (P4); α-N-acetylglucosaminidase for patients 5 and 6 (P5 and P6, respectively); α-galactosidase A for patient 7 (P7); α-L-iduronidase for patients 8–10 (P8–10, respectively), and iduronate-2-sulfatase for patient 11 (P11)]. (B) Residual enzymatic activity in patient fibroblasts upon gentamicin treatment shown as a percentage relative to controls for each disease (*p < 0.05, **p < 0.025, ***p < 0.005)
Rescue of the Enzymatic Activity After PTC124 Treatment in Fibroblasts
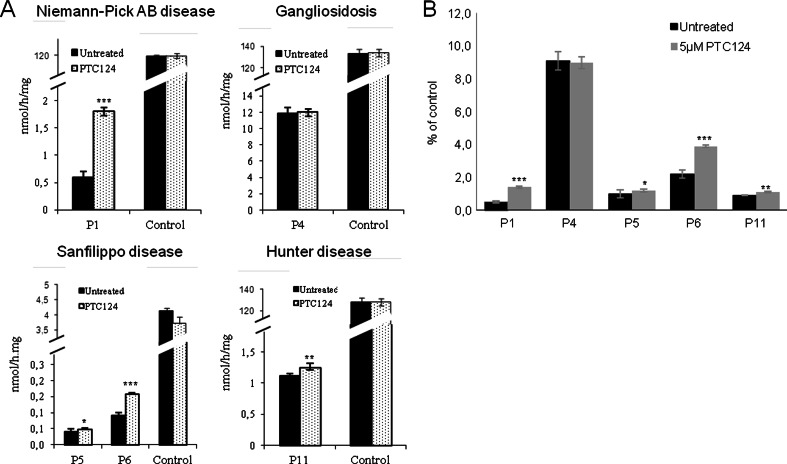
As gentamicin treatment rescued the enzymatic activity in the fibroblasts of some patients, we wondered if PTC124 was also able to restore activity. We treated the gentamicin-responsive fibroblasts with this compound. We first tested which concentration led to the highest readthrough effect without causing any effect on cell viability. This concentration was found to be 5 μM (Fig. S1; see Supplementary Material). Treatment with PTC124 caused a similar pattern of readthrough response in the same cell lines that were already gentamicin responsive, with the exception of those from patient 4 (Fig. 2A). Patients 1, 5, 6, and 11 showed a slight but statistically significant increase of up to 3-fold their residual enzymatic activity (Fig. 2B). Cell viability was not altered after treatment in any of the cell lines used in this study (Fig. S1; see Supplementary Material). Similar results represented as the percentage of residual enzymatic activity relative to controls are shown in Fig. 2B.
Fig. 2.
Residual enzymatic activity in patient fibroblasts upon treatment with PTC124 treatment. (A) Patients responsive to gentamicin treatment were treated with or without PTC124 (5 μM) for 72 h. Equal amounts of protein lysates, with or without 5 μM PTC124, were seeded and the corresponding enzymatic activities were determined using a fluorimetric assay with 4-methylumbelliferyl substrates [acid sphingomyelinase for patient 1 (P1); β-galactosidase for patient 4 (P4); α-N-acetylglucosaminidase for patients 5 and 6 (P5 and P6), and iduronate-2-sulfatase for patient 11 (P11)]. (B) Residual enzymatic activity in patient fibroblasts upon PTC124 treatment shown as a percentage relative to controls for each disease (*p < 0.05, **p < 0.025, ***p < 0.005)
Immunostaining Analysis After Gentamicin Treatment in Patients’ Fibroblasts
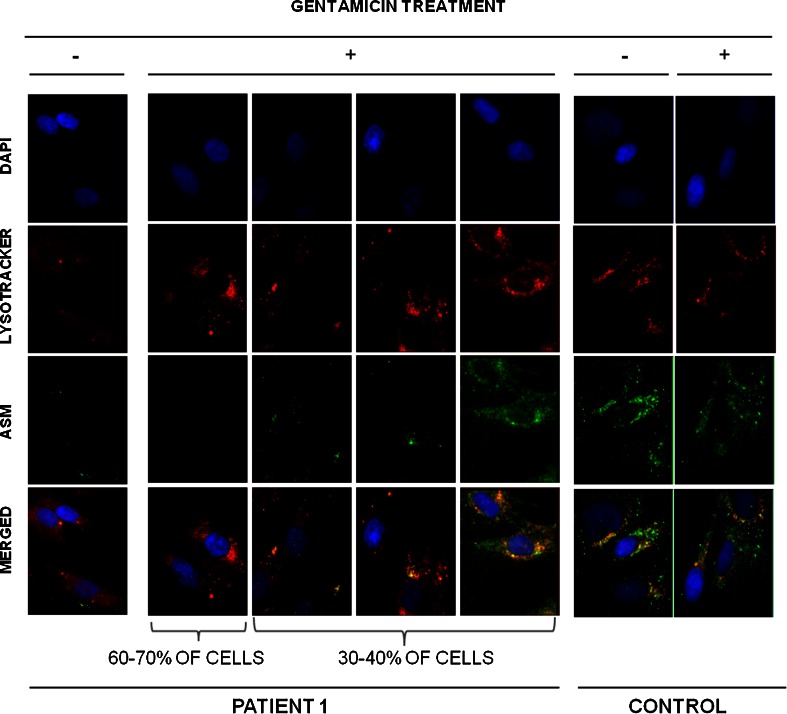
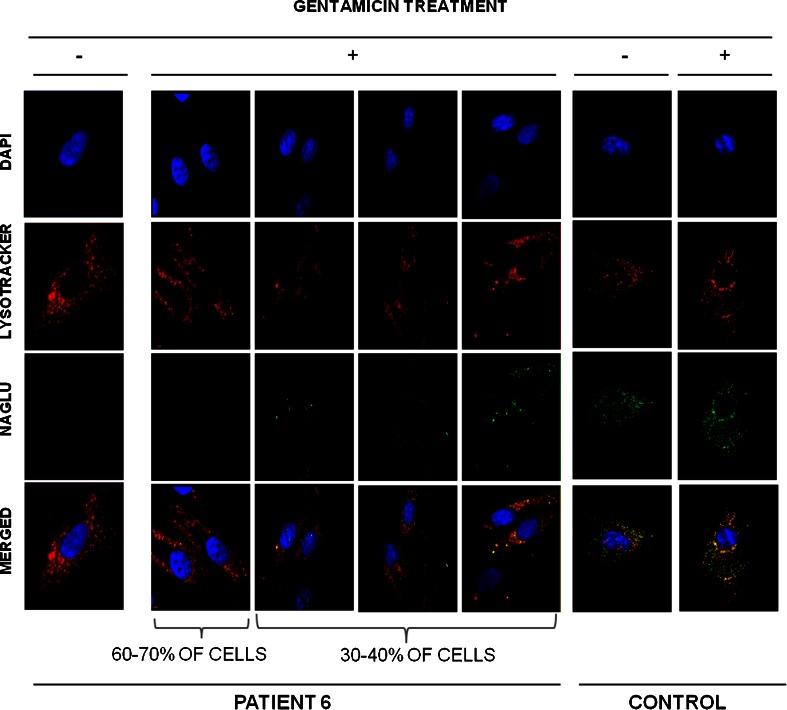
To determine if the rescue of enzymatic activity was due to an increase of protein biosynthesis, we studied the presence of the protein involved in the disease in the fibroblasts that increased their residual enzymatic activity more effectively upon gentamicin treatment (2 patients). We stained acid-sphingomyelinase (the enzyme deficient in Niemann–Pick A/B disease) in patient 1, and α-acetylglucosaminidase (the enzyme deficient in Sanfilippo B disease) in patient 6. Before treatment, almost no protein was observed in patients’ cells (Figs. 3 and 4). However, after treatment about 30–40 % of the cells showed an increase in protein, and properly localized acid-sphingomyelinase and α-acetylglucosaminidase protein into lysosomes (Figs. 3 and 4). It is important to note that the extent of protein expression recovery was not the same in all cells.
Fig. 3.
Immunostaining of fibroblasts from patient 1 and a healthy individual before and after gentamicin treatment. Representative images of patient 1’s cells after gentamicin treatment. Differences among cells are due to the extent of protein expression recovery in each cell. Co-localization of acid-sphingomyelinase with LysoTracker Red (Life Technologies, Paisley, UK) in skin-derived fibroblasts from patient 1 and a healthy control. Photographs of fluorescent microscopy of cells before and after treatment with 300 μM gentamicin are shown (original magnification 100×). Lysosomes were stained with 75 nM LysoTracker Red for 1 h at 37 °C, and a fluorescein isothiocyanate-conjugated secondary antibody was used to stain acid-sphingomyelinase and α-N-acetylglucosaminidase. DAPI = 4′,6-diamidino-2-phenylindole
Fig. 4.
Immunostaining of fibroblasts from patients 1 and 6, as well as a healthy individual before and after gentamicin treatment. Representative images of patient 6’s cells after gentamicin treatment. Differences among cells are due to the extent of protein expression recovery in each cell. Co-localization of α-N-acetylglucosaminidase with LysoTracker Red (Life Technologies, Paisley, UK) in skin-derived fibroblasts from patient 6 and a healthy control. Photographs of fluorescent microscopy of cells before and after treatment with 300 μM gentamicin are shown (original magnification 100×). Lysosomes were stained with 75 nM LysoTracker Red for 1 h at 37 °C, and a fluorescein isothiocyanate-conjugated secondary antibody was used to stain acid-sphingomyelinase and α-N-acetylglucosaminidase. DAPI = 4′,6-diamidino-2-phenylindole
GAG Accumulation Into Lysosomes in Response to Gentamicin Treatment
We assessed the ability of gentamicin to decrease the accumulation of GAGs in fibroblasts from patients 5, 6, and 11, which were responsive to gentamincin by increasing their residual enzymatic activity. Fibroblasts from patient 6 showed a significant decrease of GAG accumulation—16 % compared with untreated fibroblasts after 48 h of gentamicin incubation. In contrast, fibroblasts from patients 5 and 11 did not show any significant reduction in cellular GAG accumulation (Fig. 5).
Fig. 5.
Glycosaminoglycan (GAG) levels in skin-cultured fibroblasts from patients 5, 6, and 11 (P5, P6, and P11, respectively), and a healthy individual upon gentamicin treatment. Patient fibroblasts affected by a mucopolysaccharidosis and responsive to gentamicin treatment were treated for 72 h with or without 300 μM gentamicin. Total GAG content was assessed and results are shown as μg GAGs per mg of protein (*p < 0.05) and compared with fibroblasts from a healthy individual
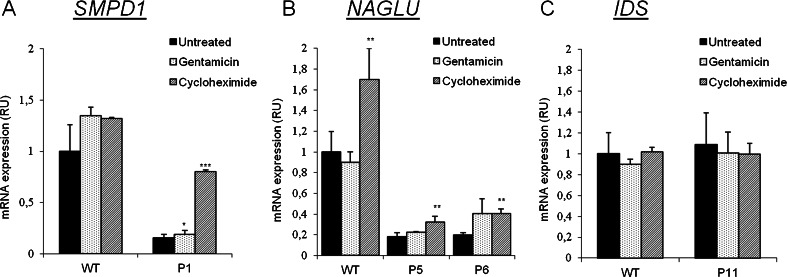
mRNA Expression Analysis in Gentamicin-responsive Fibroblasts
To determine the correlation between the recovery of residual enzymatic activity and mRNA expression in response to gentamicin treatment, we performed RT-PCR analysis of SMPD1 and IDS in fibroblasts from patients 1 and 11, respectively, and of NAGLU in patients 5 and 6. All patients except patient 11 showed levels of mRNA expression of about 20–40 % that of controls. Expression in patient 11, both before and after gentamicin treatment, was similar to that of controls. It is interesting to note that, after treatment, mRNA levels in P6 increased significantly relative to basal levels and almost reached the levels obtained after the addition of cycloheximide addition. The fibroblasts of the remaining patients did not show any increase of mRNA expression after gentamicin treatment (Fig. 6).
Fig. 6.
mRNA expression analyses in patient fibroblasts treated with gentamicin or cycloheximide. Results are shown as relative units compared with the basal values obtained for an untreated control individual (*p < 0.05, **p < 0.025, ***p < 0.005). Cells were treated with gentamicin (300 μM) for 72 h or cycloheximide (500 μg/ml) for 5 h. Cycloheximide allows the observation of the degraded mRNA carrying PTCs. (A) Expression of SMPD1 (Niemann–Pick A/B disease) in patient 1 (P1). (B) Expression of IDUA (Sanfilippo B disease) in patients 5 and 6 (P5 and P6, respectively). (C) Expression of IDS (Hunter disease) in patient 11 (P11). GAPDH expression was used as endogenous control. RU = relative units to control fibroblasts; WT = wild type
Nucleotide Background Around PTC
Floquet et al. [24] reported different gentamicin-induced readthrough responses depending on the nucleotide context (nucleotide positions –5, –1, +4, and +8) around the PTC (Table 2). We also analyzed this in our patients (Tables 2 and 3), suggesting a UAG ≥ UGA > UAA hierarchy of stop codon readthrough and a positive gentamicin readthrough in sequences comprising a uracil or a guanidine at nucleotide position –5, or a uracil at position –1, or an adenosine or a cytosine at position +4, and a guanidine or a cytosine at position +8.
Table 3.
Nucleotide context of positive gentamicin readthrough obtained in comparison with that previously described by Floquet et al. [24]
| Floquet et al. [24] | This report | |
|---|---|---|
| Stop codon | UGA > UAG > UAA | UAG > UGA > UAA |
| Nucleotide –5 | A, C | U, G |
| Nucleotide –1 | U | U > C, G |
| Nucleotide +4 | C | C, A |
| Nucleotide +8 | G | G, C |
In Silico Analysis of the Predicted Substitutions Induced by Gentamicin Treatment
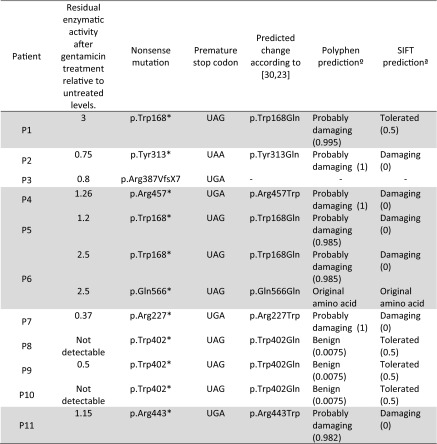
It has been described that glutamine and tryptophan are the 2 most common amino acid insertions when a premature stop codon readthrough occurs. Glutamine is preferably inserted at nonsense UAG or UAA codons, whereas UGA miscode to tryptophan [36–38]. Taking into account these rules, the predicted amino acid changes were analyzed, using an in silico approach, for each patient (Polyphen-2 and SIFT analyses). Then, the possible effect of the “new missense” mutations created by gentamicin readthrough was assessed (Table 4).
Table 4.
In silico analysis of the functional effect of the predicted changes resulting from PTC readthrough. The PTCs: UAG and UAA have been described to miscode to Glutamine (Gln) when readthrough occurs although UGA has been described to miscode to Tryptophan [23]
In silico predictions were performed using Polyphen-2 and SIFT. Patients responsive to gentamicin treatment are indicated in gray
ND = nondetectable
*Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations. Score ranges from 0 (benign) to 1 (damaging)
†SIFT (http://sift.jcvi.org/) prediction is based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences. Score ranges from 0 (damaging) to 1 (tolerated)
Discussion
LSD comprise > 50 different genetic disorders involving the storage of nondegraded macromolecules in lysosomes and are mainly inherited in an autosomal recessive manner or, in a few cases, X-linked. In most cases, the severity of the disease correlates with the levels of the affected enzymatic activity. To some extent, the level of enzyme deficiency depends on the disease-causing mutation and the degree of effect on the protein. Furthermore, a recovery of residual enzymatic activity of only 10–20 % of the activity detected in healthy individuals may be enough for functional relief resulting in the wild-type phenotype [6]. In our study, we focused on nonsense or frameshift mutations leading to PTC resulting in an almost complete absence of particular key proteins involved in lysosomal function and cell homeostasis maintenance.
Preclinical studies with gentamicin or PTC124 had succeeded by enhancing stop codon readthrough and were able to be used as therapeutic agents for genetic diseases [9–18, 36–42]. PTC124 has been successfully extended into clinical trials [18, 43], although preliminary results are not clear for Duchenne muscular dystrophy [44]. In the present work, in order to develop future therapeutic approaches we studied the potential drug-mediated readthrough to restore the activity of several lysosomal enzymes in 11 patients affected by 6 different LSDs. We found that gentamicin treatment was able to slightly restore (up to 3-fold) the enzymatic activity in fibroblasts of several patients carrying nonsense mutations [Table 1 (patients 1, 4, 5, 6, and 11); Fig. 1]. Interestingly, fibroblasts from patients 1 and 6, which increased their residual enzymatic activity more efficiently, also increased the protein levels as well as their localization into the lysosomes (Figs. 3 and 4). Thus, both enzymatic and immunolocalization criteria suggest that readthrough treatment can partially restore the biochemical alterations of these patients in cultured fibroblasts. However, the fact that protein expression recovery is not equal in all the cells could provide a potential explanation for the slight increase in enzymatic activity (see Figs. 3 and 4). To this effect, although a clear recovery of well-localized protein is detected after gentamicin treatment, differences from cell to cell within patients’ fibroblasts are observed, probably owing to the reduced rates of protein synthesis in potentially low proliferating primary cell lines [45].
It is important to note that, upon gentamicin treatment, only fibroblasts from patient 4 were able to increase residual enzymatic activity up to 10 % of control cells (Fig. 1B), which was described to be sufficient for functional recovery [6]. In LSDs, a correlation among residual enzymatic activity, severity of the clinical presentation, and the biochemical phenotype has been observed [46]. For that reason, we cannot exclude the possibility that the modest increase of residual enzymatic activity detected in fibroblasts from patients 1, 5, 6, and 11 might have a positive impact and improve the clinical and biochemical phenotype of these patients (Fig. 1B). The fact that another readthrough compound, PTC124 [18], also succeeded by slightly enhancing the enzymatic activity of fibroblasts from patients 1, 5, 6, and 11 supports the idea that although not all patients with PTCs are treatable with readthrough therapy, those that are responsive to gentamicin may also be responsive to other readthrough compounds (see Fig. 2). Therefore, it seems worthwhile to perform preliminary approaches with already proven readthrough compounds, such as gentamcin or PTC124, to select responsive mutations before starting a readthrough screening with selected libraries or with other compounds. Surprisingly, and similarly to previous observations with other LSD [14, 15, 23], we found a significant and unexplained reduction of residual enzymatic activity upon gentamicin treatment in fibroblasts from patient 7 (Fabry disease). This reduction cannot be due to cell toxicity as cell viability was normal. Therefore, further experiments are needed to elucidate if gentamicin treatment could directly or indirectly inhibit α-galactosidase A activity, under certain conditions.
By mRNA expression analysis and immunolocalization studies (Figs. 3, 4 and 6), we demonstrated that aminoglycoside therapy may enhance the endogenous synthesis of proteins, as well as their correct localization into the lysosomes. Nevertheless, the percentage of rescue varies among patients. This fact may be due to the different amino acid insertion upon readthough treatment. It has been described that gentamicin induces the insertion of the amino acids glutamine or tryptophan at premature UAG/UAA or UGA termination codons, respectively [47, 48], resulting in a newly synthesized protein with glutamine or tryptophan at the position where the stop mutation occurred (Table 4). This new protein, carrying a missense mutation, if active, may have a reduced half-life due to post-translational surveillance systems, but the recovered enzymatic activity might allow improvement of the biochemical phenotype. To elucidate if a stop codon mutation might be responsive to readthrough treatment we analyzed each mutation with in silico predictors (Polyphen-2 and SIFT) and correlated them with the enzymatic activity data (Table 4). After readthrough treatment, the predicted “new” missense mutations for patients 1 and 6—those that increased more efficiently their residual enzymatic activity—were benign or reversed to the original amino acid, respectively (Table 4). To this effect, patients’ fibroblasts presented an increase of well localized protein after treatment with the subsequent increase in enzymatic activity (Figs. 1, 3 and 4). In addition, patient 6 had a 16 % reduction in glycosaminoglycans in fibroblasts (Fig. 5). However, the other clearly responsive patients (patients 5 and 11) were predicted to produce proteins carrying mutations potentially affecting their function, while patients 8, 9, and 10, who did not show any rescue of enzymatic activity, have a benign mutation prediction (Table 4). Both in silico databases were concordant except for the predicted change in patient 1. The mutation in this patient was predicted to be damaging by Polyphen and tolerable by SIFT analysis, and in this case SIFT prediction was in agreement with the biochemical results (Table 4). Therefore, in silico predictions, although useful, should be interpreted cautiously and cannot be taken as a definitive approach.
Concerning mRNA expression, all responsive patients, except patient 11, experienced an increase in mRNA levels slightly after gentamicin treatment, although these levels did not reach those obtained after cycloheximide treatment, suggesting that gentamicin readthrough was not totally efficient and some mRNA was still being degraded (Fig. 6). Interestingly, patient 11, a male affected by an X-linked disease (Hunter disease), presents mRNA expression levels similar to controls (Fig. 6). He carries a nonsense mutation located in the last exon of IDS (c.1327C > T), so that the resulting mRNA might elude the NMD surveillance mechanism resulting in normal mRNA levels [3]. The same occurs in one of the alleles of patient 6 (c.1696C > T), but in this case mRNA expression in fibroblasts was low (20 % of control values), which might be owing to the fact that this patient carries another nonsense mutation that did not elude the NMD surveillance (Table 1 and Fig. 6). It is also remarkable that in healthy individuals NAGLU mRNA expression increased significantly after cycloheximide treatment (Fig. 6B). This observation could be explained by the fact that 1–10 % of cellular transcripts are upregulated by NMD inactivation as NMD not only controls the expression of aberrant transcripts, but also that of many apparently wild-type mRNAs [49].
Another aspect that might explain the differences among patients is the nucleotide context around the PTC. It has been reported that this context might play a crucial role in determining the efficiency of readthrough treatment [24, 34]. Therefore, we analyzed the nucleotide context of the responsive patients and found that fibroblasts that were responsive to treatment carried PTC mutations in a slightly different nucleotide background than previously reported (Table 3). Despite the small sample size of our study, the results presented herein suggest a hierarchy of stop codon readthrough treatment efficacy similar to that reported [20]: UAG ≥ UGA > UAA. Also, in agreement with Floquet et al. [24] we found the same response pattern in nucleotides –1 and +4. Cytosine at the +4 position has been described as determinant for gentamicin-induced readthrough and the relevant parameter for clinical application. In agreement with the study by Floquet et al. [24], patient 6, the only patient presenting a cytosine in this position, was responsive to treatment. In fact, this patient showed a slight increase of residual enzymatic activity and of mRNA expression together with an increase of the correctly localized protein. Nevertheless, in our hands, gentamicin treatment was also effective and sometimes more efficient when uracil/guanidine and adenosine where located at nucleotide positions –5 and +8, respectively, instead of adenosine/cytosine and guanine, as was previously reported (Table 3). In agreement with our findings, several other authors also found that other nucleotide positions were able to induce PTC readthrough [24, 27, 50–54]. However, to confirm this hypothesis analysis of an extended series of patients is required.
To our knowledge, only one knock-in mouse model has been reported for the diseases studied herein [55]. Interestingly, in this particular model (mutation p.Trp392* of IDUA; Hurler disease) no biochemical improvement was observed after gentamicin administration [19]. Accordingly, we observed a similar effect in cultured fibroblasts derived from 3 patients carrying the analogous human mutation (mutation p.Trp402* of IDUA; Hurler disease), as no increase in enzymatic activity was detected.
In conclusion, our results provide a proof of concept that PTCs can be effectively suppressed by readthrough drugs, but with different efficiencies depending on the genetic context. We demonstrated that gentamicin readthrough treatment partially restores some of the biochemical hallmarks of the lysosomal diseases tested here. To this effect, the screening of new compounds with higher readthrough activity is a strategy to develop efficient therapies for diseases caused by PTC mutations. Recently, Taguchi et al. [56] have developed synthetic analogues of natural (+)-negamycin that display potent readthrough activities against nonsense mutations in eukaryotes without exhibiting antimicrobial activities. Thus, these compounds should be analyzed in gentamicin-responsive cell lines.
Finally, all these studies were performed in fibroblasts, which resulted in a useful model to test this therapeutic approach. Therefore, we recommend testing patient fibroblast cell lines prior to attempting other assays.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 1224 kb)
Preliminary studies, testing gentamicin and PTC124 concentration, incubation time and monitoring cell viability in fibroblasts derived from P1 (Niemann-Pick AB), P5 (Sanfilippo B), P6 (Sanfilippo B) and a healthy individual. Monitoring of the residual enzymatic activities after gentamicin treatment during 24h, 48h and 72h (A) and PTC124 during 16h, 24h (B), 48h and 72h (C). Monitoring of the cell viability of P6 and a healthy individual after gentamicin (D) and PTC124 (E,F). NPAB: Niemann-Pick AB disease, SF: Sanfilippo B disease. (PDF 280 kb)
Acknowledgments
This work was supported, in part, by Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), an initiative of the Instituto de Salud Carlos III (Ministerio de Ciencia e Innovación, Spain), grant FIS ADE08/90030 and BCN-PEPTIDES. This work was performed in the context of the Biomedicine PhD programme of the University of Barcelona (UB). We thank all the families involved in this study.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Conflict of Interest
The authors declare no conflict of interest.
Ethics
All the procedures were approved by the ethics committee of the Hospital Clínic, Barcelona. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (Hospital Clínic de Barcelona) and with the Helsinki Declaration of 1975, as revised in 2000.
References
- 1.Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 2.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Ainsworth C. Nonsense mutations: running the red light. Nature. 2005;438:726–728. doi: 10.1038/438726a. [DOI] [PubMed] [Google Scholar]
- 4.Hers HG. The role of lysosomes in the pathogeny of storage diseases. Biochimie. 1972;54:753–757. doi: 10.1016/S0300-9084(72)80179-2. [DOI] [PubMed] [Google Scholar]
- 5.Neufeld EF, Muenzer J. The mucopolysaccharidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. New York: McGraw Hill, 2001, pp. 3421-3452.
- 6.Desnick RJ, Thorpe SR, Fiddler MB. Toward enzyme therapy for lysosomal storage diseases. Physiol Rev. 1976;56:57–99. doi: 10.1152/physrev.1976.56.1.57. [DOI] [PubMed] [Google Scholar]
- 7.Seregin SS, Amalfitano A. Gene therapy for lysosomal storage diseases: progress, challenges and future prospects. Curr Pharm. 2011;17:2558–2574. doi: 10.2174/138161211797247578. [DOI] [PubMed] [Google Scholar]
- 8.Muntau AC, Leandro J, Staudigl M, Mayer F, Gersting SW. Innovative strategies to treat protein misfolding in inborn errors of metabolism: pharmacological chaperones and proteostasis regulators. J Inherit Metab Dis. 2014;37:505–523. doi: 10.1007/s10545-014-9701-z. [DOI] [PubMed] [Google Scholar]
- 9.Pérez B, Rodríguez-Pombo P, Ugarte M, Desviat LR. Readthrough strategies for therapeutic suppression of nonsense mutations in inherited metabolic disease. Mol Syndromol. 2012;3:230–236. doi: 10.1159/000343086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidou L, Hatin I, Perez N, Allamand V, Panthier JJ, Rousset JP. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 2004;11:619–627. doi: 10.1038/sj.gt.3302211. [DOI] [PubMed] [Google Scholar]
- 11.Gunn G, Dai Y, Du M, et al. Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol Genet Metab. 2014;111:374–381. doi: 10.1016/j.ymgme.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Micale L, Augello B, Maffeo C, et al. Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Hum Mutat. 2014;35:841–850. doi: 10.1002/humu.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sleat DE, Sohar I, Gin RM, Lobel P. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur J Paediatr Neurol. 2001;5(Suppl. A):57–62. doi: 10.1053/ejpn.2000.0436. [DOI] [PubMed] [Google Scholar]
- 14.Hein LK, Bawden M, Muller VJ, Sillence D, Hopwood JJ, Brooks DA. alpha-L-iduronidase premature stop codons and potential read-through in mucopolysaccharidosis type I patients. J Mol Biol. 2004;338:453–462. doi: 10.1016/j.jmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Miller JN, Kovács AD, Pearce DA. The novel Cln1(R151X) mouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Hum Mol Genet. 2015;24:185–196. doi: 10.1093/hmg/ddu428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai PC, Davis BD. Triphasic concentration effects of gentamicin on activity and misreading in protein síntesis. Biochemistry. 1979;18:193–198. doi: 10.1021/bi00568a029. [DOI] [PubMed] [Google Scholar]
- 17.Manuvakhova M, Keeling K, Bedwell D. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/S1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Belakhov V, Kandasamy J, et al. The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol Genet Metab. 2012;105:116–125. doi: 10.1016/j.ymgme.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedwell DM, Kaenjak A, Benos DJ, et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 21.Keeling KM, Brooks DA, Hopwood JJ, Li P, Thompson JN, Bedwell DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10:291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 22.Buck NE, Wood LR, Hamilton NJ, Bennett MJ, Peters HL. Treatment of a methylmalonyl-CoA mutase stop codon mutation. Biochem Biophys Res Commun. 2012;427:753–757. doi: 10.1016/j.bbrc.2012.09.133. [DOI] [PubMed] [Google Scholar]
- 23.Bartolomeo R, Polishchuk EV, Volpi N, Polishchuk RS, Auricchio A. Pharmacological read-through of nonsense ARSB mutations as a potential therapeutic approach for mucopolysaccharidosis VI. J Inherit Metab Dis. 2013;36:363–371. doi: 10.1007/s10545-012-9521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floquet C, Hatin I, Rousset JP, Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob Agents Chemother. 1994;38:2517–2520. doi: 10.1128/AAC.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mingeot-Leclercq M-P, Tulkens PM. Aminoglycosides: nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003–1012. doi: 10.1128/aac.43.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan L, Narayan SB, Chen J, Meyers GD, Bennett MJ. PTC124 improves readthrough and increases enzymatic activity of the CPT1A R160X nonsense mutation. J Inherit Metab Dis. 2011;34:443–447. doi: 10.1007/s10545-010-9265-5. [DOI] [PubMed] [Google Scholar]
- 28.Annunziata P, Dimatteo G. Study of influence of sex and age on human serum lysosomal enzymes by using 4-methylumbelliferyl substrates. Clin Chim Acta. 1978;90:101–106. doi: 10.1016/0009-8981(78)90510-7. [DOI] [PubMed] [Google Scholar]
- 29.Sumantran VN. Cellular chemosensitivity assays: an overview. Methods Mol Biol. 2011;731:219–236. doi: 10.1007/978-1-61779-080-5_19. [DOI] [PubMed] [Google Scholar]
- 30.Barbosa I, Garcia S, Barbier-Chassefière V, Caruelle JP, Martelly I, Papy-García D. Improved and simple micro assay for sulfated glycosaminoglycans quantification in biological extracts and its use in skin and muscle tissue studies. Glycobiology. 2003;13:647–653. doi: 10.1093/glycob/cwg082. [DOI] [PubMed] [Google Scholar]
- 31.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 32.Noensie EN, Dietz HC. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol. 2001;19:434–439. doi: 10.1038/88099. [DOI] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 36.Wilschanski M, Famini C, Blau H. A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations. Am J Respir Crit Care Med. 2000;161:860–865. doi: 10.1164/ajrccm.161.3.9904116. [DOI] [PubMed] [Google Scholar]
- 37.Keeling KM, Wang D, Dai Y, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks DA, Muller VJ, Hopwood JJ. Stop-codon read-through for patients affected by a lysosomal storage disorder. Trends Mol Med. 2006;12:367–373. doi: 10.1016/j.molmed.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Barton-Davis ER, Cordier L, Shoturma DI, Le- land SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai CH, Chun HH, Nahas SA, et al. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci U S A. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grayson C, Chapple JP, Willison KR, Webster AR, Hardcastle AJ, Cheetham ME. In vitro analysis of aminoglycoside therapy for the Arg120stop nonsense mutation in RP2 patients. J Med Genet. 2002;39:62–67. doi: 10.1136/jmg.39.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du M, Liu X, Welch EM, Hirawat S, Peltz SW, Bedwell DM. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci U S A. 2008;105:2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerem E, Hirawat S, Armoni S, et al. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- 44.Sheridan C. Doubts raised over ‘read-through’ Duchenne drug mechanism. Nat Biotechnol. 2013;31:771–773. doi: 10.1038/nbt0913-771. [DOI] [PubMed] [Google Scholar]
- 45.Cooper GM. The cell: a molecular approach. 2nd ed. Available at: http://www.ncbi.nlm.nih.gov/books/NBK9876/ (accessed 26 June 2015).
- 46.Boustany RM. Lysosomal storage diseases--the horizon expands. Nat Rev Neurol. 2013;9:583–588. doi: 10.1038/nrneurol.2013.163. [DOI] [PubMed] [Google Scholar]
- 47.Harrell L, Melcher U, Atkins JF. Predominance of six different hexanucleotide recoding signals 3' of read-through stop codons. Nucleic Acids Res. 2002;30:2011–2017. doi: 10.1093/nar/30.9.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF. Sequence specificity of aminoglycoside-induced stop condon readthrough: potencial implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48:164–169. doi: 10.1002/1531-8249(200008)48:2<164::AID-ANA5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 49.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson M, Rydén-Aulin M. Glutamine is incorporated at the nonsense codons UAG and UAA in a suppressor-free Escherichia coli strain. Biochim Biophys Acta. 2003;1627:1–6. doi: 10.1016/S0167-4781(03)00050-2. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar C, Zhang Z, Mukherjee AB. Stop codon read-through with PTC124 induces palmitoyl-protein thioesterase-1 activity, reduces thioester load and suppresses apoptosis in cultured cells from INCL patients. Mol Genet Metab. 2011;104:338–345. doi: 10.1016/j.ymgme.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sánchez-Alcudia R, Pérez B, Ugarte M, Desviat LR. Feasibility of nonsense mutation readthrough as a novel therapeutical approach in propionic acidemia. Hum Mutat. 2012;33:973–980. doi: 10.1002/humu.22047. [DOI] [PubMed] [Google Scholar]
- 53.Ho G, Reichardt J, Christodoulou J. In vitro read-through of phenylalanine hydroxylase (PAH) nonsense mutations using aminoglycosides: a potential therapy for phenylketonuria. J Inherit Metab Dis. 2013;36:955–959. doi: 10.1007/s10545-013-9602-6. [DOI] [PubMed] [Google Scholar]
- 54.Loudon JA. Ataluren: a ‘no-nonsense’ approach for pulmonary diseases. Pulm Pharmacol Ther. 2013;26:398–399. doi: 10.1016/j.pupt.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Shukla C, Liu X, et al. Characterization of an MPS I-H knock-in mouse that carries a nonsense mutation analogous to the human IDUA-W402X mutation. Mol Genet Metab. 2010;99:62–71. doi: 10.1016/j.ymgme.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taguchi A, Hamada K, Kotake M, et al. Discovery of natural products possessing selective eukaryotic readthrough activity: 3-epi-deoxynegamycin and its leucine adduct. ChemMedChem. 2014;9:2233–2237. doi: 10.1002/cmdc.201402208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)
Preliminary studies, testing gentamicin and PTC124 concentration, incubation time and monitoring cell viability in fibroblasts derived from P1 (Niemann-Pick AB), P5 (Sanfilippo B), P6 (Sanfilippo B) and a healthy individual. Monitoring of the residual enzymatic activities after gentamicin treatment during 24h, 48h and 72h (A) and PTC124 during 16h, 24h (B), 48h and 72h (C). Monitoring of the cell viability of P6 and a healthy individual after gentamicin (D) and PTC124 (E,F). NPAB: Niemann-Pick AB disease, SF: Sanfilippo B disease. (PDF 280 kb)