Abstract
Sprinkled throughout the genome are a million regulatory sequences called transcriptional enhancers that activate gene promoters in the right cells, at the right time. Enhancers endow the brain with its incredible diversity of cell types and also translate neural activity into gene induction. Thanks to rapid advances in genomic technologies, it is now possible to identify thousands of enhancers rapidly, test their transcriptional function en masse, and address their neurobiological functions via genome editing. Enhancers also promise to be a great technological opportunity for neuroscience, offering the potential for cell-type-specific genetic labeling and manipulation without the need for transgenesis. The objective of this review and the accompanying 2015 SfN mini-symposium is to highlight the use of new and emerging genomic technologies to probe enhancer function in the nervous system.
SIGNIFICANCE STATEMENT Transcriptional enhancers turn on genes in the right cells, at the right time. Enhancers are also the genomic sequences that encode the incredible diversity of cell types in the brain and enable the brain to turn genes on in response to new experiences. New technology enables enhancers to be found and manipulated. The study of enhancers promises to inform our understanding of brain development and function. The application of enhancer technology holds promise in accelerating basic neuroscience research and enabling gene therapies to be targeted to specific cell types in the brain.
Introduction
Mammalian genomes contain millions of regulatory sequences called transcriptional enhancers that far outnumber the number of genes and enable the tight control of gene expression in the brain. Enhancers endow the brain with its incredible diversity of cell types, effectively encoding cellular identity in the genome via the control of lineage-specific transcription patterns. They also enable neural activity-regulated gene induction by transducing neuronal activity into genomic responses. Enhancers function by interacting directly with gene promoters to activate gene transcription in the right cells at the right time in response to transcription factor binding (Fig. 1). Whereas promoters are defined biochemically by their ability to encode mRNA transcription start sites (TSSs), enhancers are defined by their ability to activate transcription at promoters (Fig. 2).
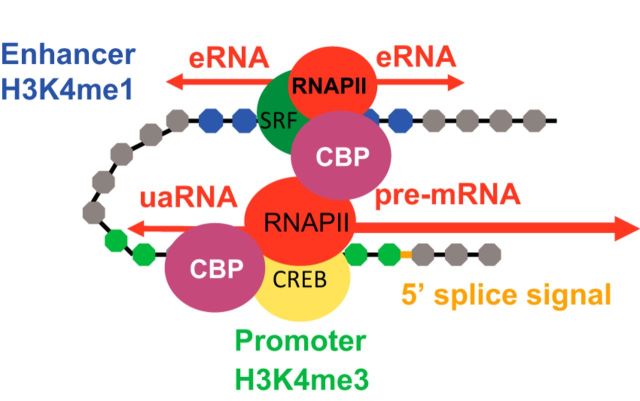
Figure 1.
Enhancers versus promoters. Enhancers juxtapose with their specific target promoters via chromatin looping. Enhancers and promoters both recruit transcription factors and coactivators such as CBP (CREB-binding protein), as well as RNA polymerase II. Whereas enhancers initiate transcription of unspliced, nonpolyadenylated eRNAs, promoters initiate transcription of spliced, polyadenylated mRNAs or long, noncoding RNAs. eRNA transcription typically occurs bidirectionally. In addition to transcribing mRNAs, promoters also drive upstream antisense transcription of transcripts called uaRNAs, which exhibit properties similar to eRNAs. The differences between enhancers and promoters may arise entirely or in part due to the presence of the 5′ splice signal at the end of the first exon of mRNAs. For simplicity, nucleosomes are depicted as gray circles and DNA as a black line, but in reality, DNA is looped around nucleosomes.
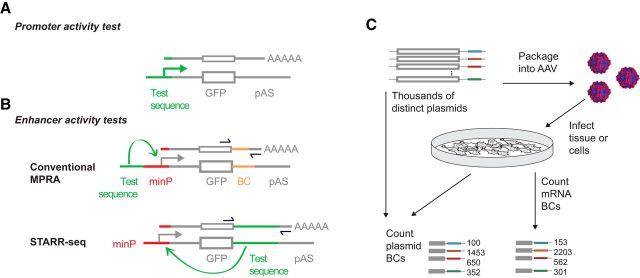
Figure 2.
Detecting enhancer activity using reporter assays and massively parallel reporter assays (MPRAs). A, Promoter activity is detected by fusing a test sequence upstream of a reporter sequence (i.e., GFP) and introducing it into cells or embryos. B, Enhancer activity is detected by fusing a test sequence near a minimal promoter and reporter. Alternatively, enhancer activity may be detected with the test sequence in the 3′ untranslated region (UTR) of a reporter gene (the STARR-seq configuration; Arnold et al., 2013; Cotney et al., 2015). For high-throughput analysis of enhancer activity, identifying sequences (such as barcodes, BCs) are placed in the 3′ UTR to identify individual enhancers so that they may be combined for “massively parallel” or multiplex quantification. C, MPRAs rely on libraries of plasmids in which the enhancer activity of each test sequence is quantified through sequencing of the BCs (or for STARR-seq, the enhancer itself). For MPRAs in primary neurons or in vivo, enhancer libraries can be packaged into AAV and transduced.
The substantial number and biological importance of enhancers is not yet matched by their mindshare in the neuroscience community. A search for “enhancer” from SfN abstracts in 2014 yields 55 hits, of which 23 relate directly to transcriptional enhancers (the others relate to cognitive, performance, and flavor enhancers). However, the complexity and function of the brain may owe as much to transcriptional enhancers as it does to synapses in that the identity of each cell type in the nervous system is likely encoded by the activity of a specific subset of enhancers that control its transcriptional potential. Likewise, enhancer dysfunction, perhaps even more so than mutations in protein-coding sequences, is likely the source of most common genetic risk for mental illnesses such as schizophrenia.
Thanks to rapid technological advances in genomic technologies, it is now possible to identify rapidly and inexpensively thousands of enhancers, perform high-throughput testing of their transcriptional function in a single experiment, and address their neurobiological functions via genome editing. Beyond this scientific opportunity to understand the mechanisms of neural genome regulation, enhancers promise to be a technological opportunity in the form of an ever better toolkit for genetic labeling of cell types and manipulation of functional gene expression in the nervous system.
How can I find transcriptional enhancers?
After the initial discovery of enhancers, it was soon realized that they could confer upon a promoter cell-type-specific activation (Banerji et al., 1983; Gillies et al., 1983; Heintzman et al., 2009); indeed, this is now understood to be a primary function of enhancers in the nervous system and beyond (Heintzman et al., 2009; Nord et al., 2015). Although understanding enhancer biology represents a great scientific opportunity for also understanding how the genome encodes cellular identity, the cell-type specificity of enhancers, along with their ability to act on specific target promoters in orientation and distance-independent manner, have made finding them and characterizing their function a challenging problem. Initially, enhancers were studied using laborious reporter assays (Fig. 2) designed with few clues about which sequences might function as enhancers beyond evolutionary sequence conservation (Nobrega et al., 2003; Pennacchio et al., 2006). However, it is now possible to identify putative enhancers rapidly based on their chromatin signatures in specific cell populations of interest.
Putative enhancers active in a cell type of interest can be identified by first purifying selected populations of neurons from the heterogeneous environment of the brain via fluorescence-activated cell sorting (Jiang et al., 2008; Mellén et al., 2012) or affinity purification of genetically labeled nuclei (Mo et al., 2015). Antibody-based nuclear sorting methods offer a particular advantage for species including humans for which genetic methods for tagging nuclei are not an option (Cheung et al., 2010). As at promoters, chromatin at enhancers is depleted of histones and therefore accessible to DNase (i.e., it is DNase hypersensitive). Therefore, genome-wide quantification of DNase (or transposase) accessibility (Thurman et al., 2012; Buenrostro et al., 2013; Yue et al., 2014), which identifies all accessible chromatin, can be used in a single experiment to find the set of all putative regulatory elements. Chromatin signatures further distinguish enhancers from promoters and other classes of regulatory elements. Nucleosomes surrounding enhancers typically exhibit a high level of histone H3 lysine 4 mono-methylation (H3K4me1) with lower H3K4me3. Active promoters show the opposite pattern of H3K4 methylation (high H3K4me3 and little to no H3K4me1). H3K4me1-enriched enhancers can be in a poised state or fully active. The poised enhancers have high levels of H3K27me3, a mark associated with transcriptionally repressed or inactive loci. Conversely, functionally active enhancers are additionally bound by H3K27ac and also transcribe a class of noncoding RNAs termed enhancer RNAs (eRNAs). Collectively, these marks are strong predictors of enhancer activity (Kim et al., 2010; Wu et al., 2014). Two common methods of putative enhancer identification are to perform chromatin immunoprecipitation sequencing (ChIP-Seq) to quantify genome-wide binding of H3K27ac or to perform ATAC-seq (a method similar to DNase-seq in that it identifies nucleosome-free regions) (Buenrostro et al., 2013; Nord et al., 2013; Markenscoff-Papadimitriou et al., 2014; Mo et al., 2015). Putative enhancers predicted by these signatures are often bona fide enhancers, but the true test of function is still either to manipulate the enhancer genetically or (most commonly) to use enhancer reporter assays in which a reporter such as lacZ or GFP expression depends on enhancer activity (Fig. 2). Techniques in which enzymatically dead Cas9 is fused to transcriptional repressors or activators are highly promising for investigating enhancer function. However, interpreting the results of this kind of manipulation remains a challenge. Targeting a repressor to an enhancer that is in physical proximity with a promoter could bring the repressor to the promoter and therefore may not reveal much about the importance of the enhancer beyond identifying the promoters with which it interacts.
How do enhancers make developmental choices?
Chromatin undergoes dynamic changes during brain development and neuronal differentiation (Stadler et al., 2011; Lister et al., 2013). Two recent studies illustrate the power of genome-wide enhancer identification and the dynamics of enhancer function in the context of brain development. First, ChIP-Seq experiments assessing H3K27ac binding across mouse development identified 50–100 k putative brain developmental enhancers (Nord et al., 2013). A majority of these enhancers are predicted to be transiently activated during development based on detection of H3K27ac at a subset of developmental stages (Fig. 3). The enhancers that activated earlier in development have greater DNA sequence conservation across species, consistent with the greater diversity of phenotypes along development. Reporter data for large numbers of enhancers is available in the VISTA database (http://enhancer.lbl.gov) and enhancers from this database have been used for lineage identification and fate mapping in the brain (Pattabiraman et al., 2014). Presumably, these enhancers serve as docking sites for distinct combinations of both cell-type-specific transcription factors and more general transcription factors that are repurposed for the programming of different cell types through a complex hierarchy of protein–protein and protein–DNA interactions (Mazzoni et al., 2013; Andzelm et al., 2015). The vast number of enhancers engaged during brain development is an important new opportunity to understand how the cellular diversity of the brain is encoded in mammalian genomes. In addition, these enhancers are now a resource for developing toolkits for developmental stage-specific markers.
Figure 3.
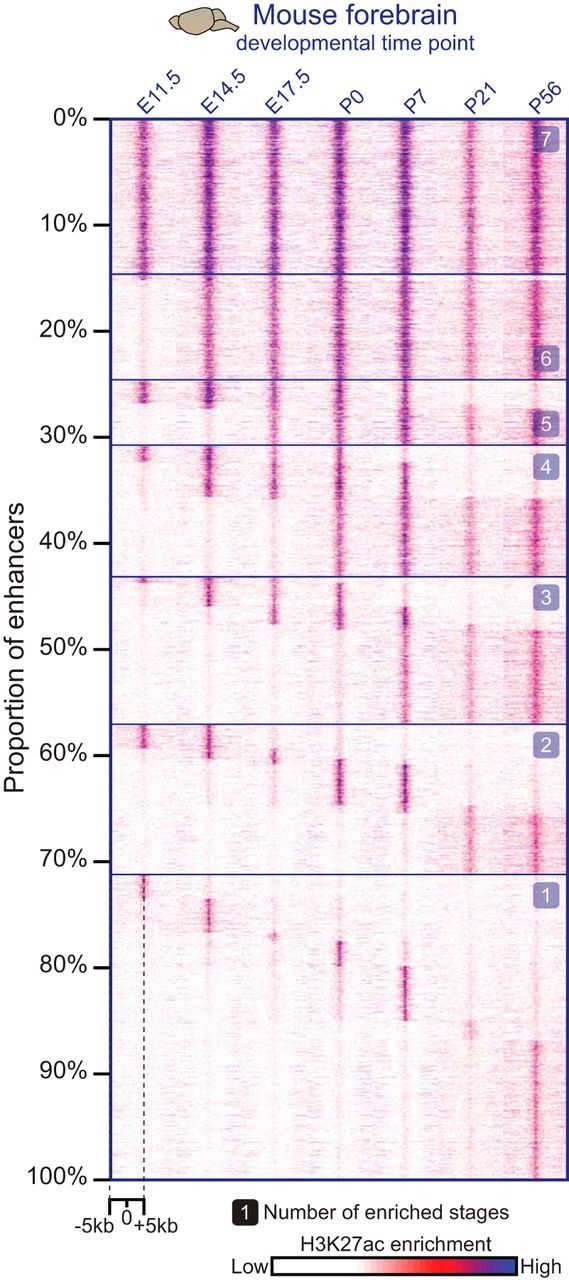
Spatial and temporal specificity of enhancers active in the developing forebrain. Developmental dynamics of the enhancer-associated histone mark H3K27ac at putative enhancers in mouse forebrain analyzed by ChIP-seq across seven stages of brain development. Most sites show temporally restricted H3K27ac marks. Each column of the heat map displays H3K27ac enrichment patterns from a collection of putative enhancers for a given developmental stage, which are arranged from constitutively active enhancers at the top to more developmental-stage-specific enhancers proceeding downward.
A second recent study focused on neural differentiation in the cerebellum, examining chromatin regulation in cerebellar granule neurons (CGNs) in vivo (Frank et al., 2015). Using the identification of DNase-accessible sites as a proxy for all gene regulatory elements, the study identified changes in chromatin regulation that define stages of neuronal maturation. As in the forebrain, chromatin at CGN enhancers was highly dynamic during development, even after exit from the cell cycle. A motif analysis within dynamic DNase-accessible sites identified a binding motif for the Zic family of transcription factors. These data led to experiments showing that Zics play an important role in promoting CGN differentiation, which provides an intriguing complement to their known roles in inhibiting the exit of neuronal precursors from the cell cycle. Finally, this study took advantage of the power of the CRISPR/Cas9 system to investigate the functional importance of the putative enhancer elements identified. Specifically targeting an enzymatically dead Cas9 fused to a transcriptional activator to two putative enhancers near the Grin2c gene was found to be sufficient to significantly and selectively increase Grin2c mRNA expression levels in neurons.
Enhancers are also responsible for making one of the most bizarre and unusual gene expression choices in all of mammalian biology. In each olfactory receptor neuron (ORN), just one allele of one of about a thousand different olfactory receptors is expressed, yet the choice is seemingly random for each ORN. One component of this mechanism involves LSD1-mediated removal of histone methylation (histone H3, lysine 9), which leads to expression of an olfactory receptor (OR) (Lyons et al., 2013). OR expression activates the unfolded protein response, ultimately blocking further LSD1 activity (Dalton et al., 2013). A second component of this mechanism involves a network of interacting enhancers in which numerous enhancers colocalize in an interchromosomal interaction complex with an activated OR (Markenscoff-Papadimitriou et al., 2014). These enhancers bind a transcription factor, BPTF, that is required for OR expression. These results suggest a fascinating and complex structural model in which multiple enhancers are recruited to the activated OR gene, both to activate the OR gene and to sequester the enhancers away from other OR genes (Fig. 4).
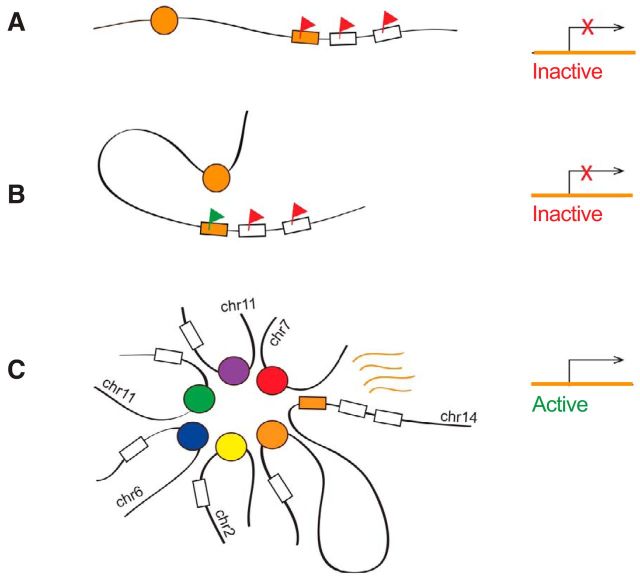
Figure 4.
A model for essential and redundant functions of an OR enhancer in cis and in trans, respectively. Schematic representations of different states in the olfactory sensory neuron nucleus (left) and corresponding transcriptional outputs (right) are shown. An OR gene (orange box) located proximal to an enhancer (orange circle) is repressed by H3K9me3 (red flag, A). The cis-proximal enhancer may facilitate derepression of the OR chromatin landscape (green flag, B), but is not sufficient for OR transcription. Multiple trans-interacting enhancers (colored circles) aggregate around the transcribed cis-proximal OR (orange box, C). This model of enhancer–promoter configuration can explain why some enhancers are required in cis but are redundant in trans.
How do enhancers respond to neural activity?
One of the most exciting findings to arise from the profiling of chromatin signatures in neurons is the discovery that thousands of enhancers are engaged by neural activity to regulate transcription (Kim et al., 2010, 2015). Neural-activity-regulated promoter proximal enhancers—those located within 2 kb of a gene's TSS—have been studied since Fos was first found to be regulated by activity (Greenberg et al., 1986). Genome-scale identification of non-promoter-binding sites for the canonical activity-regulated transcription factors CREB and MEF2 first hinted at the potential for activity-dependent transcriptional regulation of enhancers (Impey et al., 2004; Flavell, 2006), and the later observation that regions with the chromatin signature of enhancers were preferential sites of activity-dependent recruitment of CBP, NPAS4, and FOS binding made clear the fundamental importance of this mechanism (Kim et al., 2010; Malik et al., 2014). Enhancers are important for activity-dependent transcription for two reasons. Similar to the function of developmental enhancers in cell-type-specific gene expression, activity-regulated enhancers can define the set of genes that can be activity inducible in any given type of neuron, thus allowing different brain regions to have distinct transcriptional plasticities in response to experience (Spiegel et al., 2014; Whitney et al., 2014). In addition neuronal-activity-regulated enhancers directly mediate gene induction. The enhancer-mediated induction of synaptic plasticity genes suggests a function for activity-regulated enhancer activation in memory formation (Telese et al., 2015).
One of the best understood activity-regulated enhancers is situated 7 kb upstream of the activity-regulated gene Arc and is called the synaptic-activity-regulated element (SARE). This enhancer was the first distal neural-activity-regulated enhancer to be identified and it remains the best characterized (Kawashima et al., 2009, 2014; Schaukowitch et al., 2014). Upon neural depolarization, this enhancer comes into closer physical proximity with the Arc promoter, and SARE eRNAs are synthesized. These eRNAs bind the negative regulator of transcription elongation (NELF), thereby releasing RNA polymerase II to transcribe the Arc gene. When the SARE eRNA is knocked down, chromatin looping between the enhancer and promoter is unaffected. However, NELF release from the Arc promoter is impaired, and Arc gene induction is reduced. Interestingly, not all neuronal-activity-dependent genes are similarly regulated by NELF (Saha and Dudek, 2013), and the specific functional relevance of this mechanism for neural plasticity remains to be explored.
How relevant are enhancers to human diseases of the CNS?
The genomics revolution has led to large-scale identification of genomic loci that confer risk for common neuropsychiatric disease (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015). The causal sequence variants within these risk loci remain almost entirely unidentified, but most of them reside in noncoding regions of the genome. These variants are highly likely to affect enhancers, given that much of the functional noncoding genome is devoted to enhancers (Hemberg et al., 2012). Some of these variants might disrupt transcription factor binding at enhancers, as appears to be the case for a variant in the MEIS1 enhancer locus that is associated with “restless legs syndrome” (Spieler et al., 2014). Alternatively, noncoding mutations outside of enhancers could disrupt long-range promoter–enhancer interactions, as has been suggested for variants near the FKBP5 gene that are associated with posttraumatic stress disorder (Klengel et al., 2013). Risk for diseases of the CNS is influenced not only by variation within enhancers active in the CNS but also by variation in enhancers active in other tissues. Common variants within blood cell enhancers associate with risk for depression, affect cortisol-dependent transcription in blood cells, and predict inappropriate amygdala reactivity, which is a common psychiatric endophenotype for stress-inducible psychiatric disorders (Arloth et al., 2015). An immediate priority is to identify which genetic variants that correlate with disease also affect enhancer function and gene expression—a daunting task given that enhancers often function only in specific cell types.
In addition to variation in enhancer sequences, disease states may involve misregulation of enhancers in trans. High-throughput sequencing of patient “exomes” (exon-encoding genomes) recently identified mutations in 22 genes underlying autism spectrum disorders (De Rubeis et al., 2014). Surprisingly, a disproportionate number of these genes function to remodel chromatin. Chromatin remodelers (including CHD8, ARID1B, POGZ, and SUV420H1) may alter the histone modifications and positioning of nucleosomes at enhancer sequences (Cotney et al., 2015).
How can we screen thousands of enhancers at a time?
A major challenge for human health will be to determine which of the numerous human genomic sequence variants influence enhancer function. In a big step forward, enhancer reporter assays can now be performed in high-throughput (Fig. 2). High-throughput reporter assays have so far been performed in vitro (Melnikov et al., 2012; Arnold et al., 2013; Shlyueva et al., 2014) and in retinal explants (Kwasnieski et al., 2012). So-called massively parallel reporter assays (MPRAs) are performed much like traditional reporter assays except that a single plasmid with one enhancer is replaced by a library with many enhancers. Instead of measuring GFP fluorescence, high-throughput sequencing is used to quantify cDNA barcodes whose expression reports the activity of a specific enhancer or test sequence. In Drosophila, a related method called STARR-seq has been used to screen the entire genome for enhancer activity (Arnold et al., 2013). These tools promise to make it possible to determine which of the many sequence variants associated with human neuropsychiatric disorders actually influence enhancer activity.
What is the difference between an enhancer and a promoter?
Enhancers and gene promoters have long been considered two separate classes of regulatory sequences. They are now increasingly recognized for their functional and structural similarities and may be differentiated only by the presence of a splicing signal (Almada et al., 2013; Andersson et al., 2014; Core et al., 2014; Andersson et al., 2015). Structurally, both promoters and enhancers can be envisioned as islands of transcription-factor-binding sites in a sea of mostly nonfunctional genomic sequence. The function of sequencing-binding transcription factors is typically to recruit transcriptional machinery to local genomic regions. Therefore, it is not too surprising that from these islands initiate divergent transcription whether those binding sites are near a gene body or distal from that site (Core et al., 2014; Scruggs et al., 2015). Functionally, the very first enhancers to be studied were observed to have promoter and enhancer activity (Moreau et al., 1981; Tuan et al., 1992; Ashe et al., 1997). We now know that, in addition to mRNA transcription, transcription of eRNAs occurs at many or most enhancers (Fig. 1) (De Santa et al., 2010; Kim et al., 2010). Whether an mRNA or eRNA is produced appears to depend on the presence of a 5′ splice site within a few hundred nucleotides of the initiation site (Kaida et al., 2010; Almada et al., 2013; Ntini et al., 2013).
Whether eRNAs are universally required for enhancer function is unknown. When cells differentiate or respond to a stimulus, eRNAs are transcribed before genic mRNAs are transcribed (De Santa et al., 2010; Arner et al., 2015). Enhancer RNA induction at neural-activity-regulated enhancers is greatest near the genes with mRNAs that are also induced the most (Kim et al., 2010). These results suggest the possibility that enhancer transcription or eRNA transcripts are required for enhancer function. Multiple models of eRNA function have been proposed based on experiments outside of the nervous system, including that eRNAs facilitate enhancer–promoter looping or mediate chromatin remodeling near the promoter region (Ørom et al., 2010; Li et al., 2013; Mousavi et al., 2013; Hsieh et al., 2014). Alternatively, transcription of eRNAs rather than eRNA transcripts may be functionally important (Kaikkonen et al., 2013). As mentioned above, in neurons, eRNA-dependent release of a negative regulator (NELF) from the Arc promoter drives neuronal activity-dependent transcription of Arc mRNA (Schaukowitch et al., 2014). From this menagerie of mechanisms, each reported at a different enhancer in a different biological system, we hope that universal mechanisms will eventually emerge.
How should we develop enhancers as tools for neuroscience?
One of the most valuable uses of the mass of enhancer data gathered in recent years is to improve genetic access to CNS subtypes without the need for transgenesis. Enhancers are small enough that many of them can fit together within an adeno-associated virus (AAV) genome and still leave plenty of room for a promoter, cDNA and other necessary components. Developing enhancer-based genetic labeling tools is a goal that is worth prioritizing because it would enable specific cell types within neural circults to be manipulated (e.g., optogenetically), without the need for time-consuming and costly transgenesis. There are two obvious strategies to achieving this goal. The DESIGNER strategy begins with finding putative enhancers predicted to be active in a particular tissue or cell type (Nord et al., 2013; Vermunt et al., 2014; Mo et al., 2015). Next, it combines those predicted enhancer sequences with promoters (and with each other) and tests them for the desired expression patterns in reporter assays. In contrast, the SCREENING strategy examines the function of random libraries of DNA (or enhancers) in reporter assays and tests for any specific expression pattern in the brain (Pennacchio et al., 2006). Success in the DESIGNER strategy could be an AAV expression system for genetic access to PV+ neurons. The SCREENING strategy instead would produce AAV vectors that label many different, unknown subsets of neurons, analogous to how the GENSAT project produced BAC-transgenic mice (Gerfen et al., 2013). These two strategies can be viewed as two ends of a spectrum in which the emphasis in resource investment is placed on enhancer identification or on reporter assay screening.
The DESIGNER strategy has so far been laborious. There are few AAV-based promoters that drive expression in neural subtypes. The commonly used short CaMKII promoter fragment is an exception (Dittgen et al., 2004) that is nonetheless not 100% specific to excitatory neurons (Nathanson et al., 2009b). In one example of the DESIGNER strategy, regulatory sequences from subtypes of inhibitory neurons drive expression specifically in inhibitory neurons, but not in specific subtypes (Nathanson et al., 2009a). In another example, five tandem copies of the SARE labeled neurons in an activity-dependent manner more effectively than the one copy that suffices in the genomic context (Kawashima et al., 2013, 2014). The biggest challenge in the DESIGNER strategy is that functional testing is always required because regulatory elements, once recombined into plasmids, behave unpredictably. Unfortunately, functional testing has until recently been laborious.
If MPRAs could be extended to work in vivo in a cell-type-specific manner, then they could be used to screen for highly cell-type-specific expression. A hybrid strategy that combines elements of the DESIGNER and SCREENING strategies would be to identify putative cell-type-specific enhancers directly or based on their proximity to cell-type-specific genes, which are now readily identifiable by single-cell drop-seq (Klein et al., 2015; Macosko et al., 2015). Next, the hybrid strategy would use in vivo cell-type-specific MPRA to screen identified sequences for actual cell-type-specific enhancers that function in AAV-transduced tissue. The immediate product, as with the GENSAT project (Gerfen et al., 2013), would be a reagent. A similar screening strategy could prove powerful for screening genetic variants for their potential effect on enhancer function.
Conclusions
These are exciting times for unraveling the complex mechanisms by which gene expression in the brain is controlled. New tools are emerging that promise to revolutionize our ability to dissect gene regulation at level of single cells (Buenrostro et al., 2015), to examine transcriptional dynamics across single cells in the brain at rapidly increasing resolution (Klein et al., 2015; Macosko et al., 2015), to perform targeted genome editing (Li et al., 2014; Meyer et al., 2015), and to generate synthetic transcription factors to probe the activity of enhancers in the endogenous chromatin context (Hilton et al., 2015; Kearns et al., 2015). At the same time, there are large-scale efforts to characterize the role of enhancer function in disorders such as schizophrenia (psychENCODE) and autism (The Simons Autism Genomes Project) and to build our understanding of cell-type-specific enhancer activity during neurodevelopment and after environmental stimulation or challenge. We anticipate that the accompanying 2015 SfN mini-symposium on neural enhancers will mark a new period of increased interest within neuroscience about these critical components of the genome.
Footnotes
J.M.G. acknowledges support from the National Institutes of Health (Grant R01MH101528). T.-K.K. acknowledges support from the National Institutes of Health (NIH-NINDS R01NS085418).
The authors declare no competing financial interests.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A, Danko CG. A unified architecture of transcriptional regulatory elements. Trends Genet. 2015;31:426–433. doi: 10.1016/j.tig.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arloth J, Bogdan R, Weber P, Frishman G, Menke A, Wagner KV, Balsevich G, Schmidt MV, Karbalai N, Czamara D, Altmann A, Trümbach D, Wurst W, Mehta D, Uhr M, Klengel T, Erhardt A, Carey CE, Conley ED, et al. Genetic differences in the immediate transcriptome response to stress predict risk-related brain function and psychiatric disorders. Neuron. 2015;86:1189–1202. doi: 10.1016/j.neuron.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drabløs F, Lennartsson A, Rönnerblad M, Hrydziuszko O, Vitezic M, Freeman TC, Alhendi AM, Arner P, Axton R, Baillie JK, Beckhouse A, Bodega B, Briggs J, Brombacher F, Davis M, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, Akbarian S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, Reilly SK, Tebbenkamp AT, Bichsel C, Pletikos M, Sestan N, Roeder K, State MW, Devlin B, Noonan JP. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of mef2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, Vockley CM, Safi A, Gersbach CA, Crawford GE, West AE. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18:647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberg M, Gray JM, Cloonan N, Kuersten S, Grimmond S, Greenberg ME, Kreiman G. Integrated genome analysis suggests that most conserved non-coding sequences are regulatory factor binding sites. Nucleic Acids Res. 2012;40:7858–7869. doi: 10.1093/nar/gks477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GSM, Chen S, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci U S A. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, Bito H. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Okuno H, Bito H. A new era for functional labeling of neurons: activity-dependent promoters have come of age. Front Neural Circuits. 2014;8:37. doi: 10.3389/fncir.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol. 2015;7:a018622. doi: 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci U S A. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rivera CM, Ishii H, Jin F, Selvaraj S, Lee AY, Dixon JR, Ren B. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, Mosley CP, Ahituv N, Lomvardas S. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci. 2013;16:1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr, Kinney JB, Kellis M, Lander ES, Mikkelsen TS. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. Selective distal enhancer control of the Mmp13 gene identified through clustered regularly interspaced short palindromic repeat (CRISPR) genomic deletions. J Biol Chem. 2015;290:11093–11107. doi: 10.1074/jbc.M115.648394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, Nathans J. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Jappelli R, Scheeff ED, Manning G, Obata K, Brenner S, Callaway EM. Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front Neural Circuits. 2009a;3:19. doi: 10.3389/neuro.04.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009b;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Rubenstein JL, Rubin EM, Pennacchio LA, Visel A. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JL. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, Andersen PK, Preker P, Valen E, Zhao X, Pelechano V, Steinmetz LM, Sandelin A, Jensen TH. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman K, Golonzhka O, Lindtner S, Nord AS, Taher L, Hoch R, Silberberg SN, Zhang D, Chen B, Zeng H, Pennacchio LA, Puelles L, Visel A, Rubenstein JL. Transcriptional regulation of enhancers active in protodomains of the developing cerebral cortex. Neuron. 2014;82:989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Saha RN, Dudek SM. Splitting hares and tortoises: a classification of neuronal immediate early gene transcription based on poised RNA polymerase II. Neuroscience. 2013;247:175–181. doi: 10.1016/j.neuroscience.2013.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs BS, Gilchrist DA, Nechaev S, Muse GW, Burkholder A, Fargo DC, Adelman K. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieler D, Kaffe M, Knauf F, Bessa J, Tena JJ, Giesert F, Schormair B, Tilch E, Lee H, Horsch M, Czamara D, Karbalai N, von Toerne C, Waldenberger M, Gieger C, Lichtner P, Claussnitzer M, Naumann R, Müller-Myhsok B, Torres M, et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Telese F, Ma Q, Perez PM, Notani D, Oh S, Li W, Comoletti D, Ohgi KA, Taylor H, Rosenfeld MG. LRP8-Reelin-regulated neuronal enhancer signature underlying learning and memory formation. Neuron. 2015;86:696–710. doi: 10.1016/j.neuron.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci U S A. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermunt MW, Reinink P, Korving J, de Bruijn E, Creyghton PM, Basak O, Geeven G, Toonen PW, Lansu N, Meunier C, van Heesch S, van Heesch S, Clevers H, de Laat W, Cuppen E, Creyghton MP. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 2014;9:767–779. doi: 10.1016/j.celrep.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, Wang R, Audet JN, Kellis M, Mukherjee S, Sinha S, Hartemink AJ, West AE, Jarvis ED. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346:1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA, Visel A. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genetics. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, et al. Mouse ENCODE Consortium. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]