Lee et al. characterized the dynamics of looped enhancer contacts during synchronous transcription elongation. The results support a model in which the gene body changes its position relative to a stable enhancer–promoter complex.
Keywords: chromatin, elongation, enhancer, looping, transcription
Abstract
Enhancers govern transcription through multiple mechanisms, including the regulation of elongation by RNA polymerase II (RNAPII). We characterized the dynamics of looped enhancer contacts during synchronous transcription elongation. We found that many distal enhancers form stable contacts with their target promoters during the entire interval of elongation. Notably, we detected additional dynamic enhancer contacts throughout the gene bodies that track with elongating RNAPII and the leading edge of RNA synthesis. These results support a model in which the gene body changes its position relative to a stable enhancer–promoter complex, which has broad ramifications for enhancer function and architectural models of transcriptional elongation.
Enhancers are regulatory elements that stimulate transcription of genes in response to signaling events or developmental cues. A great deal has been learned about how enhancers function, and it has become clear that their mechanisms of actions are not uniform (Bulger and Groudine 2011; Liu et al. 2013). Enhancers can promote gene expression by altering chromatin structure and histone modifications, influencing the location of the target gene within the nucleus, and aiding in the recruitment of basal/general transcription factors (GTFs). In addition, enhancers have been reported to promote transcription elongation (Sawado et al. 2003; Liu et al. 2013). Since a large fraction of enhancers is occupied by BET family proteins that bind and activate P-TEFb to promote transcription elongation (Yang et al. 2005; Zhang et al. 2012) and since numerous genes display promoter-proximally paused RNA polymerase II (RNAPII) (Adelman and Lis 2012), it is possible that overcoming the pause might be a widespread mechanism of enhancer action (Hargreaves et al. 2009; Zippo et al. 2009; Liu et al. 2013).
Enhancer elements can reside close to the promoters that they control, hundreds of kilobases away, or within the gene bodies. Despite varied positioning, a feature shared among many enhancers is direct physical contact with target promoters via chromatin looping (Sanyal et al. 2012). Looped contacts among regulatory elements can be highly dynamic, as revealed by experiments using conditional activation of transcription factors (Drissen et al. 2004; Vakoc et al. 2005), and can occur during both gene activation and repression (Jing et al. 2008). Another notable feature of a subset of extragenic enhancers is the presence of high levels of GTFs (Szutorisz et al. 2005; Koch et al. 2011) and RNAP (De Santa et al. 2010; Koch et al. 2011; Hah et al. 2013; Schaaf et al. 2013), including its elongation-competent forms (Louie et al. 2003; Wang et al. 2005; Zippo et al. 2009; Zhang et al. 2012). Moreover, there are reports of transcription elongation factors occupying active enhancer elements (Song et al. 2010; Lin et al. 2011, 2013; Kellner et al. 2013). Examples of RNAPII occupancy at active enhancers include the locus control region (LCR) of the β-globin gene (Johnson et al. 2002; Song et al. 2010) and a distal enhancer of the Kit gene (Jing et al. 2008). Besides the presence of RNAPII and GTFs, certain elongation-associated histone modifications found in gene bodies can also be detected at some enhancers, including H3K36me3 (Kim et al. 2007), H3K4me1, and H3K4me2 (Liang et al. 2004; Barski et al. 2007; Heintzman et al. 2007; Kim et al. 2007; Steger et al. 2008). In aggregate, these observations suggest that a subset of enhancers and gene-coding regions resides in similar transcriptional environments.
Here we examined whether enhancers physically contact the gene segments transcribed by RNAPII. Initial results at the β-globin locus are consistent with elongation-dependent LCR–β-globin gene body contacts. To overcome the inherent resolution limitations of chromosome conformation capture (3C) studies associated with a small gene such as β-globin, we carried out our studies at the Kit locus. This locus has a well-characterized enhancer residing 114 kb upstream of the transcription start site (TSS), and its coding region encompasses ∼82 kb, thus lending itself to finely space- and time-resolved 3C analysis (Jing et al. 2008). Using timed elongation block and release experiments, we found that the −114 enhancer forms stable contacts with the promoter-proximal region throughout transcription elongation. Notably, additional dynamic contacts of the enhancer with the coding region were observed at the positions of elongating RNAPII. Similar dynamic enhancer gene body contacts were identified at the CD47 locus. These results are compatible with a dynamic folding pattern during transcription elongation in which the gene body is reeled past a stable enhancer–promoter complex, providing a physical platform upon which enhancers might impact on transcription elongation.
Results and Discussion
LCR–β-globin 3′ gene contacts require transcription elongation
Previous evidence hinted at physical contacts between the LCR and sequences downstream from the β-globin TSS. Forcing looped contacts between the LCR and β-globin promoter resulted in efficient RNAPII recruitment to the promoter but submaximal transcription elongation, which was associated with diminished enhancer–gene body contacts (Deng et al. 2012). This suggested the existence of elongation-dependent gene contacts between the LCR and the body of the β-globin gene. To test this directly, we examined LCR–β-globin gene body contacts in the erythroid cell line G1E. G1E is an erythroid precursor cell line that lacks the transcription factor GATA1. Restoration of GATA1 induces erythroid maturation, looping between the LCR and β-globin gene, and induction of β-globin transcription (Weiss et al. 1997; Vakoc et al. 2005). Treatment of GATA1-replete G1E cells with 75 µM RNAPII elongation inhibitor DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) for 3 h inhibited β-globin mRNA production as determined by primary transcript (PT) RT-qPCR (Supplemental Fig. S1B). DRB washout restored transcription. Using anti-pan-RNAPII chromatin immunoprecipitation (ChIP), we examined the amount and location of total RNAPII at the β-globin gene in the presence or absence of DRB and following DRB release. DRB treatment virtually eliminated RNAPII across the β-globin body while retaining a significant fraction of RNAPII at the promoter (Supplemental Fig. S1C). Removal of DRB resulted in partial restoration of RNAPII occupancy (Supplemental Fig. S1C), consistent with mRNA synthesis (Supplemental Fig. S1B).
To examine the impact of elongation blockade on LCR–β-globin gene contacts, we carried out 3C using DNase I-hypersensitive site 2 (HS2) of the LCR as an anchor. In the absence of DRB, HS2 formed contacts with the active β-globin gene (βmaj) but not with the intervening inactive β-type globin genes (εy and βh1) (Supplemental Fig. S1D), consistent with previous reports (Vakoc et al. 2005; Deng et al. 2012). While exposure of cells to DRB did not significantly diminish HS2–promoter interaction, there was a small but reproducible reduction in contacts between HS2 and the 3′ portion of the β-globin gene (Supplemental Fig. S1D). The contacts between HS2 and the 3′ portion of the β-globin gene were restored upon DRB withdrawal (Supplemental Fig. S1D), strongly suggesting that they are formed during transcription elongation and might be functionally linked to the transcription elongation process.
Synchronizing transcription elongation at the Kit locus
The above experiments point to elongation-dependent enhancer–gene body contacts. However, the small size of the β-globin gene (∼1.4 kb) precludes a more detailed dissection of such contacts under dynamic conditions. Therefore, we chose to study the Kit gene, which is expressed at high levels in parental G1E cells. The Kit locus encompasses a gene body of ∼82 kb, harbors a well-defined distal enhancer at −114 kb from the TSS, and contains a sufficient number of BglII sites for a detailed 3C analysis (Jing et al. 2008). We hypothesized that if transient contacts between the enhancer and segments of the gene body occurred during transcription elongation, they would be more readily detectable if transcription were synchronized across alleles in a cell population. To test this, we treated G1E cells with 75 µM DRB for 3 h, resulting in a nearly complete transcription arrest (≤1.5%) (Supplemental Fig. S2A). Cells were harvested at 3.5-min time intervals following release from DRB, and their RNA was analyzed by PT RT-qPCR (Fig. 1A). Results were normalized to mouse 18S ribosomal RNA, which is transcribed by RNAPIII and is insensitive to DRB (Supplemental Fig. S2A), and were plotted relative to levels of PTs from asynchronous populations. DRB release set off a wave of transcription across the locus (Fig. 1B; Supplemental Fig. S6), with an average elongation rate of 2.17 kb/min ± 0.37 kb/min (Supplemental Fig. S2B,C), which is in the range of reported elongation rates in mammalian cells (Ardehali and Lis 2009). Consistent with previous reports (Danko et al. 2013), late transcription (2.4 kb/min) was faster than early transcription (1.4 kb/min) in the Kit gene, but measurements at the beginning were confounded by the kinetics of the DRB washout (Supplemental Fig. S2C). The first round of transcription was mostly completed at 35 min following release (Fig. 1B). After the 24.5-min time point, PT levels from the 5′ end of the Kit gene declined, presumably as a consequence of transcript processing (Fig. 1B,C). As a means to assess the kinetics of transcription elongation independent of transcript processing, we carried out anti-RNAPII ChIP. RNAPII proceeded through the Kit gene body at a rate matching that of transcript production (represented as heat maps on the X-axes in Fig. 1D; Supplemental Fig. S6). The leading edges of transcript synthesis and RNAPII occupancy were somewhat spread out, probably due to a lack of complete synchrony following DRB washout. Throughout all time intervals, RNAPII occupancy at the enhancer and promoter-proximal region remained largely unchanged (Fig. 1D; Supplemental Fig. S5B).
Figure 1.
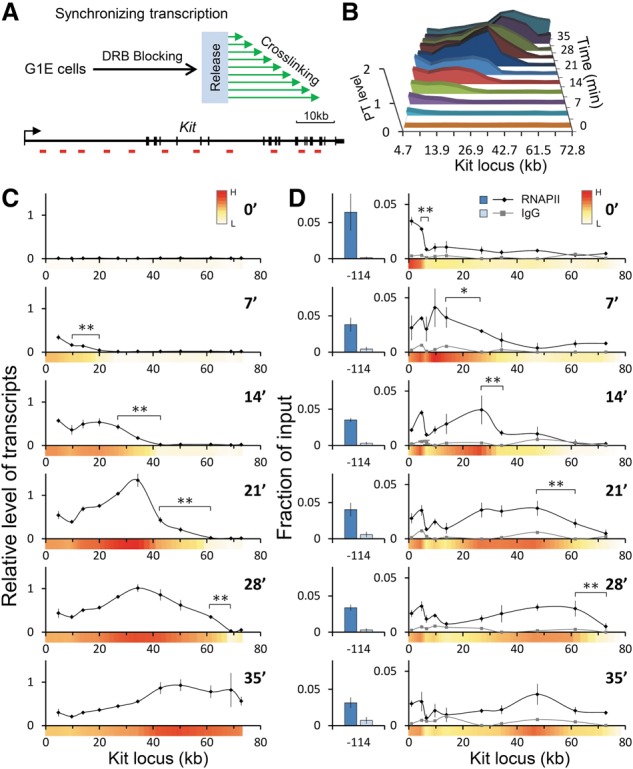
Synchronized PT production and distribution of RNAPII at the Kit locus. (A, top) Experimental scheme for synchronizing transcription. Cells were harvested at the indicated time points following release from the DRB block. (Bottom) Schematic of the Kit gene. Vertical bars demarcate exons. Horizontal red bars denote regions used for RT-qPCR or ChIP. (B) Graph combining the data of Kit PTs. (C) Results were normalized to mouse 18S rRNA and plotted relative to levels obtained from asynchronously transcribing cells. Numbers on the X-axis indicate positions in kilobases with regard to the start site. n = 3. Error bars denote standard deviation. (D) ChIP signal for RNAPII was plotted as a function of input at the indicated time points following DRB release. IgG was used as a control. n = 4. Error bars denote standard deviation. Heat maps were generated by calculating the average normalized signal intensities of two adjacent data points and converting them into a color scale. (Red) High; (white) low. (*) P < 0.05; (**) P < 0.03.
To determine the fraction of cells expressing Kit during transcription elongation, we carried out single-molecule RNA FISH using spectrally distinguishable probes that hybridize to the 5′ and 3′ end introns as well as exons. In asynchronous populations, nearly all cells have a signal from the exon probes (marking mRNA) and from the 5′ end and 3′ end intron probes (marking the sites of transcription) (Supplemental Fig.S3). With DRB treatment, most cells have no signal from either the 5′ or 3′ end probe, with a dimmer signal in ∼10% of cells comprised of residual transcription or undegraded intron molecules that may have diffused away from the site of transcription. Upon release, the 5′ end intron signal reaches steady-state maximum by 7 min, whereas the 3′ end intron signal does not emerge until the 35-min time point, which is consistent with transcript quantification by RT-qPCR (Supplemental Fig. S3). Importantly, the uniformity of the signal from the 3′ end intron probe at 35 min (present in 90% of the cells) indicates that, in most cells, polymerase has reached the 3′ end of at least one copy of the gene. These results suggest that the majority of cells respond to the DRB block–release regimen. The consistent kinetics of transcript elongation across the Kit locus in the great majority of cells enable the study of the relationship between higher-order chromatin structure and transcription elongation.
Enhancer–gene body contacts during transcription elongation
To examine physical contacts between the enhancer and gene body, we carried out 3C at 7-min intervals following DRB arrest–release. Seven-minute intervals were chosen because they provide the required minimal lengths of transcribed segments that can be resolved by 3C. The enhancer (−114) region was used as the anchor point. Control experiments showed similar BglII digestion efficiencies among the tested genomic fragments as well as linear amplification and correct sizes of PCR products (Supplemental Fig. S7). During DRB exposure, Kit enhancer–promoter contacts remained stable, consistent with previous results (Jing et al. 2008), indicating that the Kit enhancer–promoter loop is not dependent on transcription elongation (Fig. 2; Supplemental Fig. S5A). Remarkably, during the first round of transcription elongation, the −114 enhancer appeared to make additional contacts within the gene body (Fig. 2), with elevated contact frequencies progressing in a wave similar to those of the PT and RNAPII (Fig. 1; Supplemental Fig. S6). The elongation-associated changes in contact frequencies are especially visible near the 5′ region of Kit. The signals tended to become more spread out and noisier at later time points, perhaps due to a lack of elongation synchrony and an increased elongation rate. Please note that a 10-min cross-linking reaction corresponds to up to ∼20 kb at an elongation rate of 2 kb/min. Two less pronounced contacts between the enhancer and the +24-kb and +63-kb regions in the gene body were observed that appeared stable during DRB treatment (0 min) (Fig. 2). Additionally, a lower-frequency contact occurred between the enhancer and the −26-kb region that seemed to fluctuate during elongation (Fig. 2). However, the significance of these interactions remains uncertain. Some discrepancies in location between elongating RNAPII and 3C contacts are due to the fact that the spatial resolution of 3C is limited by the length of the restriction fragments.
Figure 2.
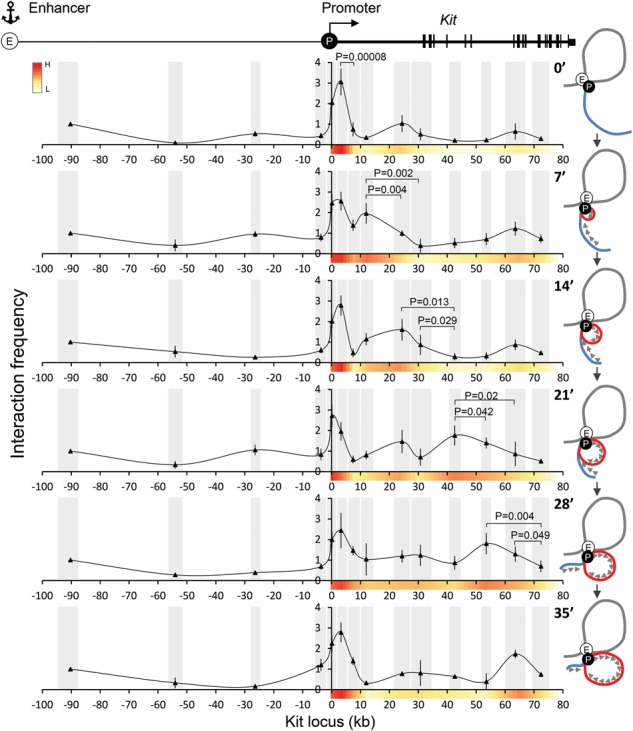
Contacts formed by the Kit −114 enhancer during synchronized transcriptional elongation. (Top) Map of the Kit locus. 3C experiments at the indicated time points using the −114 enhancer as an anchor region. Interaction frequencies were normalized to the intervening region at −93. Gray bars annotate the analyzed BglII fragments, the X-axis indicates genomic distances (in kilobases) from the start site, and the Y-axis indicates interaction frequency. n = 5. Error bars indicate standard deviations. Cartoons at the right illustrate the dynamic architecture compatible with the 3C measurements. (E) Enhancer; (P) promoter; (red line) dynamic loop extrusion during elongation.
We tested additional conditions of DRB blockage and cross-linking. For example, we increased formaldehyde concentration to 2% and shortened fixation time to 5 min (Supplemental Fig. S8) and also tried 1% for 10 min (Supplemental Fig. S9). The results were similar to those described above, reproducing stable enhancer–promoter contacts throughout the time course and revealing mobile contacts between the enhancer and the gene body (Supplemental Figs. S8, S9). Additionally, to directly compare kinetics of elongation waves of RNAPII and dynamic enhancer–gene body contacts, we performed both RNAPII ChIP and 3C by using the same pool of cross-linked cells. Both waves were well matched, especially at the earlier time points (Supplemental Figs. S6C, S9).
The experimental variability observed in our ChIP and 3C studies is likely related to DRB removal and cross-linking kinetics, leading to incomplete transcriptional synchronization. Moreover, signal to noise ratios are inherently lower when measuring transient and indirect contacts of nuclear proteins with the DNA templates.
Our findings can be interpreted to mean that an enhancer–promoter complex tracks along the chromatin fiber or is stably positioned in the nucleus with the gene being reeled alongside this complex (Papantonis et al. 2010). Our results do not discriminate between these models, but, taking into account the size of such a complex, the latter possibility seems more intuitive. Figure 2 shows cartoons next to each 3C time point that are in line with the experimental observations and accommodate both models.
Promoter–gene body contacts correlate with transcription elongation
The dynamic nature of enhancer–gene body contacts during transcription elongation together with the stability of enhancer–promoter contacts suggest a model that predicts that the promoter itself engages in gene body contacts. Therefore, we performed 3C using the Kit promoter as an anchor and again observed stable promoter–enhancer contacts before and after DRB block–release. Notably, additional contacts with the gene body were detected that correlated with the progression of RNAPII (Fig. 3), similar to promoter–gene body contacts observed at TNFα-inducible genes (Larkin et al. 2012). These results support the idea of intragenic contacts by an enhancer–promoter complex.
Figure 3.
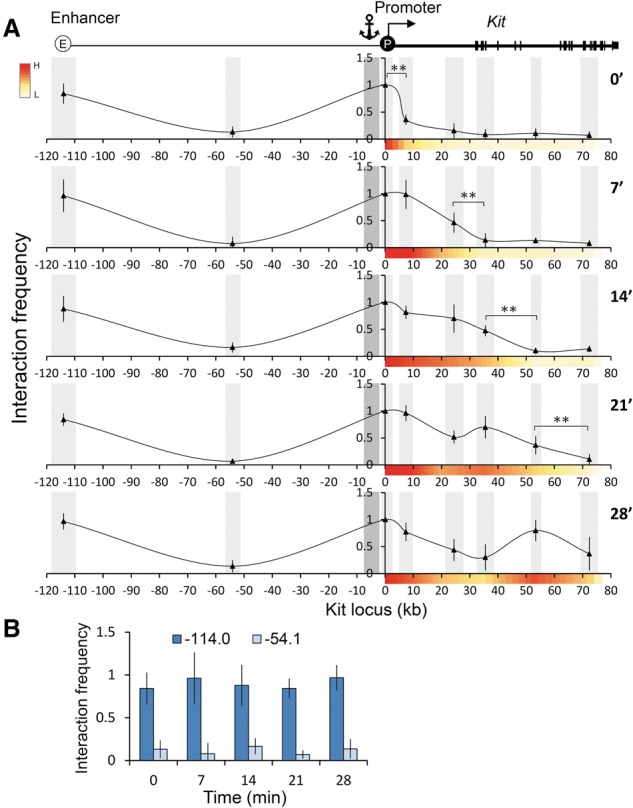
Contacts between the promoter-proximal region and the Kit locus during synchronized transcriptional elongation. (A, top) Map of the Kit locus. Light-gray bars annotate the analyzed BglII fragment, and the dark gray bar indicates the anchor fragment. The X-axis indicates genomic distances (in kilobases) from the start site, and the Y-axis indicates interaction frequency. Interaction frequency was normalized to the +0.1 region. n = 3. Error bars represent standard deviations. (**) P < 0.03 (B) Comparison of interaction frequencies of the promoter-proximal region with the enhancer of Kit (−114) during transcriptional elongation.
Dynamic enhancer–gene body contacts at the CD47 locus
To examine whether our observations are a peculiarity of the Kit locus, we studied the CD47 locus, which harbors a distal enhancer located at −80 kb from the TSS and a 60-kb coding region (Supplemental Fig. S10A). Similar to the Kit locus, we observed stable 3C enhancer–promoter contacts, while enhancer–gene body contacts were dynamic and corresponded to elongating RNAPII (Fig. 4).
Figure 4.
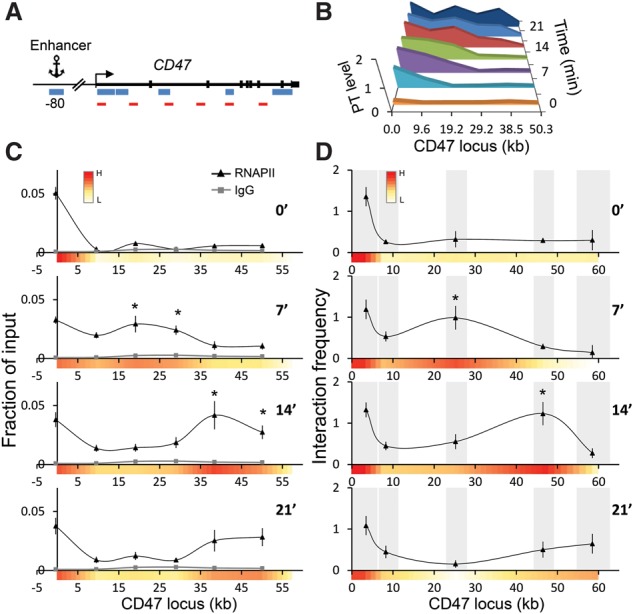
CD47 gene contacts with its distal enhancer at −80 kb during synchronized transcriptional elongation. (A) Map of the CD47 locus. Blue bars indicate fragments of BglII, and red bars indicate the regions used for ChIP and RT-qPCR. (B) PTs of CD47 were measured by RT-qPCR with the denoted primers at the indicated time points after DRB release. (C) RNAPII distribution patterns at the CD47 locus following DRB release. n = 3. Error bars denote standard deviation. IgG served as a negative control (gray line). (*) P < 0.05 (compared with 0 min). (D) Interaction frequencies with −80 kb (anchor) at the CD47 locus were measured by 3C at the indicated time points after DRB release. Gray bars demarcate the analyzed BglII fragments. n = 3. Error bars denote standard deviation. (*) P < 0.05 (compared with 0 min).
The ability of small molecules such as formaldehyde to cross-link regions that are far apart on the linear genome reflects close physical proximity due to chromatin folding. However, we cannot rule out cross-linking biases that might result from variation in protein composition along the chromatin fiber; for example, at promoter regions or DNase I-accessible versus DNase I-inaccessible chromatin. We addressed this point by measuring the relative interaction frequency between the CD47 enhancer and the IFT57 promoter, which is located ∼77 kb in the opposite direction from the CD47 enhancer (−80 kb) and is marked by a strong DNase I-hypersensitive site, high levels of H3K4me3, and RNAPII (Supplemental Fig. S10A). When compared with the interaction of the −80 kb CD47 enhancer and the CD47 promoter, the contact intensity with the IFT57 promoter was minimal, and even lower signals were observed in the IFT57 gene body (Supplemental Fig. S10B). The interactions between the CD47 enhancer and the IFT57 gene were even lower than those between the CD47 enhancer and the +46.5 kb region of the CD47 gene, representing a total genomic distance of ∼126.5 kb (Supplemental Fig. S10B). While it is impossible to rule out additional confounding method-related biases, these results support the idea that enhancers and promoters can form contacts with the transcribed portions of genes during transcription elongation.
Distribution of transcription elongation factors along the Kit locus
If the Kit enhancer contacts the gene body during transcription elongation, elongation factors that traverse the gene should remain in constant proximity with both the enhancer and the promoter-proximal regions. We tested this by carrying out ChIP experiments for Spt5, the component of DSIF that travels along the coding region (Peterlin and Price 2006). Spt5 levels were low throughout the coding region upon DRB treatment as expected but remained high at the enhancer and promoter-proximal region (Supplemental Figs. S4, S5B). Release from DRB block triggered progressive Spt5 occupancy across the gene body, with sustained enrichment at both the enhancer and promoter-proximal region (Supplemental Figs. S4, S5B). Within the Kit gene body, Spt5 dynamics were similar to those of transcript synthesis, RNAPII progression, and enhancer contact frequencies (Supplemental Fig. S6A). Hence, Spt5 behaves in a manner that would be predicted by our model in which an elongation-competent complex consisting of promoter and enhancer elements and their associated factors promotes transcriptional elongation by physically contacting the transcribed portion of the gene (Supplemental Fig. S11, right).
We suspect that enhancer–gene body contacts could be a widespread phenomenon. Detection of such transient interactions by 3C-based methods might require synchronous transcription elongation, as in the case of our experiments on the Kit and CD47 genes, or very high expression levels, as in the case of the β-globin gene. Our results further suggest that the presence of elongation-competent forms of RNAPII at numerous enhancers might not simply be a result of enhancer transcription but instead reflect proximity to the gene body (Song et al. 2010; Zhang et al. 2012). In this context, it is worth noting that cohesins, which contribute to looped chromatin interactions, are not only found at the majority of enhancers but can also occupy the transcribed portion of some genes (Kagey et al. 2010; Schaaf et al. 2013), and their depletion can lead to failure to overcome promoter-proximal elongation pauses (Schaaf et al. 2013). Additional characteristics, such as the presence of DNase I-hypersensitive exons, might contribute to these types of contacts at actively transcribed genes (Mercer et al. 2013).
In summary, our results favor a model in which the gene body changes its relative position with regard to a stable enhancer–promoter complex (Supplemental Fig. S11, right) through a dynamic folding mechanism rather than the RNAPII machinery separating from the complex and tracking along the gene body (Supplemental Fig. S11, left). It has been proposed that contacts between promoters and gene bodies reflect the proximity to structures termed transcription factories, immobile sites of active transcription involving two or more genes (Papantonis et al. 2010; Larkin et al. 2012). Our results are not informative with regard to the transcription factory model but do support a scenario of elongation-associated positional changes of the gene body relative to both the enhancer and the promoter, with the latter two maintaining stable contact with each other. Given the multitude of sizeable protein complexes that control transcription initiation, elongation, chromatin structure, and RNA processing, an attractive possibility is that these complexes are stationary, while the chromatin fiber itself is mobile and flexible. However, such a model needs to accommodate varying aggregation dynamics of RNAPII and elongation factors (Cisse et al. 2013; Ghamari et al. 2013). Whether the entire assembly of enhancer- and promoter-associated components moves along the gene or whether the gene is reeled along this complex remains an open question that might ultimately be addressable by live-cell imaging.
Materials and methods
Cell culture
G1E cells were cultured as described (Weiss et al. 1997). To synchronize transcription, cells were incubated with 75 µM DRB (Sigma, D1916) for 3 h, rinsed in 1× cold PBS, and resuspended in prewarmed medium for release from DRB.
PT RT-qPCR
Total RNA was extracted with Trizol (Invitrogen) from 0.5 × 106 cells. Reverse transcription reactions were performed with iScript (Bio-Rad, 170–8840). Real-time PCR with SYBR Green dye was performed on an ABI Prism 7000 system. Primers are listed in the Supplemental Material. Data were normalized to mouse 18S rRNA and steady-state PT levels.
ChIP
ChIP was performed as described (Deng et al. 2012). The antibodies used were Pan-RNAPII (N20; sc-399) and SPT5 (H300; sc-28678) from Santa Cruz Biotechnology. DNA was quantified by real-time PCR with SYBR Green dye on an ABI Prism 7000 system. Primers are listed in the Supplemental Material.
3C
The 3C assay was performed as described (Deng et al. 2012). G1E cells (1 × 107) were cross-linked in 1%, 1.5%, or 2% formaldehyde in 1× PBS for 5 or 10 min at room temperature and quenched with 0.25 M glycine for 5 min. We used highly concentrated BglII (New England Biolabs, R0144M) and T4 ligase (New England Biolabs, M0202L). Proximity ligation products were quantified in triplicate samples by quantitative TaqMan real-time PCR. TaqMan probes and 3C primers were designed using Primer Express 2.0 software (Applied Biosystems) and tested for linear amplification and specificity using serial dilutions of digested and randomly ligated BAC DNA containing the Kit (#RP23-156C14, Children's Hospital Oakland Research Institute) or CD47 (#RP23-230M1) locus and gel electrophoresis (Supplemental Fig.S7A,B). Digestion efficiencies were measured (Supplemental Fig.S7D) using qPCR primer pairs listed in the Supplemental Material. For the β-globin locus, 3C interactions were analyzed with published TaqMan probes and primers (Deng et al. 2012). The ERCC3 locus served as a control.
Supplementary Material
Acknowledgments
We thank members of the laboratory for critical comments on the manuscript. We are grateful to Chris Vakoc and Danny Reinberg for helpful discussions. This work was supported by National Institutes of Health grant 5R37DK058044.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.255265.114.
References
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT. 2009. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol 16: 1123–1124. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667. [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. 2004. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamari A, van de Corput MP, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, Soler E, Eick D, Lenhard B, Grosveld FG. 2013. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev 27: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. 2013. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 23: 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci 99: 11760–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner WA, Van Bortle K, Li L, Ramos E, Takenaka N, Corces VG. 2013. Distinct isoforms of the Drosophila Brd4 homologue are present at enhancers, promoters and insulator sites. Nucleic Acids Res 41: 9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Kiefer CM, Dean A. 2007. Distinctive signatures of histone methylation in transcribed coding and noncoding human β-globin sequences. Mol Cell Biol 27: 1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963. [DOI] [PubMed] [Google Scholar]
- Larkin JD, Cook PR, Papantonis A. 2012. Dynamic reconfiguration of long human genes during one transcription cycle. Mol Cell Biol 32: 2738–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci 101: 7357–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. 2013. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152: 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. 2013. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie MC, Yang HQ, Ma AH, Xu W, Zou JX, Kung HJ, Chen HW. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc Natl Acad Sci 100: 2226–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Edwards SL, Clark MB, Neph SJ, Wang H, Stergachis AB, John S, Sandstrom R, Li G, Sandhu KS, et al. 2013. DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nat Genet 45: 852–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. 2010. Active RNA polymerases: mobile or immobile molecular machines. PLoS Biol 8: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M. 2003. The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. 2013. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet 9: e1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. 2010. Multiple functions of Ldb1 required for β-globin activation during erythroid differentiation. Blood 116: 2356–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N, Tora L. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci 30: 593–599. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17: 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545. [DOI] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, et al. 2012. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem 287: 43137–43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138: 1122–1136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


