In this study, You et al. use a targeted RNAi screen for RNA-binding proteins (RBPs) in mouse ESCs and identify 16 RBPs as important regulators of ESC identity, six of which are part of the small subunit processome (SSUP) that mediates 18S rRNA processing. They then demonstrate that the SSUP enhances translational activity in pluripotent cells and sustains the levels of key regulators such as Nanog, Esrrb, and Tfcp2l1, providing new insights into the control of pluripotency.
Keywords: RNA-binding protein, embryonic stem cell, SSU processome, pluripotency, translational control, ribosome
Abstract
RNA-binding proteins (RBPs) play integral roles in gene regulation, yet only a small fraction of RBPs has been studied in the context of stem cells. Here we applied an RNAi screen for RBPs in mouse embryonic stem cells (ESCs) and identified 16 RBPs involved in pluripotency maintenance. Interestingly, six identified RBPs, including Krr1 and Ddx47, are part of a complex called small subunit processome (SSUP) that mediates 18S rRNA biogenesis. The SSUP components are preferentially expressed in stem cells and enhance the global translational rate, which is critical to sustain the protein levels of labile pluripotency factors such as Nanog and Esrrb. Furthermore, the SSUP proteins are required for efficient reprogramming of induced pluripotent stem cells. Our study uncovers the role of the SSUP and the importance of translational control in stem cell fate decision.
Embryonic stem cells (ESCs) are derived from the inner cell mass of preimplantation embryos and can proliferate indefinitely in an undifferentiated state while maintaining their capacity to differentiate into all cell lineages (Evans and Kaufman 1981; Martin 1981). Extensive efforts have been made to identify the core regulatory network that shapes and maintains the stem cell state. Core transcription factors (TFs), including Pou5f1/Oct4, Sox2, and Nanog, play a central part in establishing and maintaining the pluripotent state, highlighting the importance of transcriptional control in cell fate determination (Young 2011). Subsequent studies unraveled additional layers of regulation, including novel TFs, chromatin modifiers, and cell signaling molecules (Fazzio et al. 2008; Ding et al. 2009; Hu et al. 2009; Chia et al. 2010; Kagey et al. 2010; Yang et al. 2012).
While the transcriptional network has been extensively studied, less attention has been paid to post-transcriptional regulatory pathways. RNA-binding proteins (RBPs) are key players in post-transcriptional regulation through their involvement in RNA processing, localization, translation, and turnover (Keene 2007). However, only a few RBPs such as Fip1, Lin28, Mbnl, and Son have been identified and investigated in the context of ESCs (Ye and Blelloch 2014 and references therein). To gain unbiased insights into the role of RNA-mediated gene regulation in pluripotent stem cells, we applied here an RNAi screen targeting RBPs in mouse ESCs (mESCs) and identified 16 RBPs whose depletion results in a loss of pluripotency. Interestingly, we found the small subunit processome (SSUP or SSU processome), which mediates 18S rRNA biogenesis, to be an important regulator for pluripotent cells. Our results indicate that the SSUP subunits are up-regulated in ESCs, enhancing translation rate, which in turn supports the pluripotency network.
Results and Discussion
RNAi screen for RBPs
A targeted RNAi screen was carried out against RBPs expressed in mESCs. Among 443 genes annotated as RBPs in the mouse Uniprot database, we initially selected 247 genes that rank within the top 50% based on RNA sequencing (RNA-seq) data in mESCs (Supplemental Table S1). As positive controls, we used siRNAs against Pou5f1/Oct4 and Smc1a. The knockdown effects were monitored by measuring multiple markers for pluripotency (Nanog) and differentiation (Fgf5 for embryonic lineage and Cdx2 for extraembryonic differentiation) using quantitative RT–PCR (qRT–PCR) after 4 d (Ivanova et al. 2006). The screen was done twice, and the candidate was considered a positive hit when the Z-score was lower than −1.64 for Nanog or >1.64 for Fgf5 or Cdx2 (Supplemental Table S2; Supplemental Fig. S1A). We obtained 28 genes as the initial hits (Supplemental Fig. S1B; Supplemental Table S3). Of note, we excluded 37 RBPs that cause severe cell death from the subsequent experiments even when the marker expression indicated differentiation (Supplemental Table S4). Among the 28 selected hits, five RBPs (Cnot6, Ddx47, Nifk/Mki67ip, Ptbp1, and Tardbp/Tdp43) were previously identified in different RNAi screens for pluripotency (Fazzio et al. 2008; Ding et al. 2009; Chia et al. 2010; Kagey et al. 2010; Yang et al. 2012).
We next performed a secondary screen for validation of the initial hits by transfecting cells with individual siRNAs. If more than two out of four siRNAs induced differentiation, we considered the gene to be positive (for examples, see Supplemental Fig. S1C). From the secondary screen, we obtained 16 positive hits (Fig. 1A; Supplemental Table S5). Interestingly, nine out of the 16 genes have been implicated in ribosome biogenesis (Ddx47, Ddx52, Ddx56, Krr1, Nifk, Nol6, Pdcd11, Rbm28, and Rrp7a), while the rest have been documented mainly in splicing (Cwc15, Hnrnpk, Hnrnpu, Ptbp1, Rbm42, Srsf7/9G8, and Tardbp). Some genes were previously implicated in pluripotency. Ddx47 and Hnrnpu were reported as Pou5f1-interacting proteins (Ding et al. 2012). Hnrnpk, Hnrnpu, and Ptbp1 were shown to assist transcriptional control mediated by long noncoding RNAs in ESCs (Xist, lincRNA-p21, and TUNA/megamind, respectively) (Hasegawa et al. 2010; Huarte et al. 2010; Lin et al. 2014). Thus, our screen suggests potentially novel regulatory players involved in the pluripotency network in stem cells.
Figure 1.
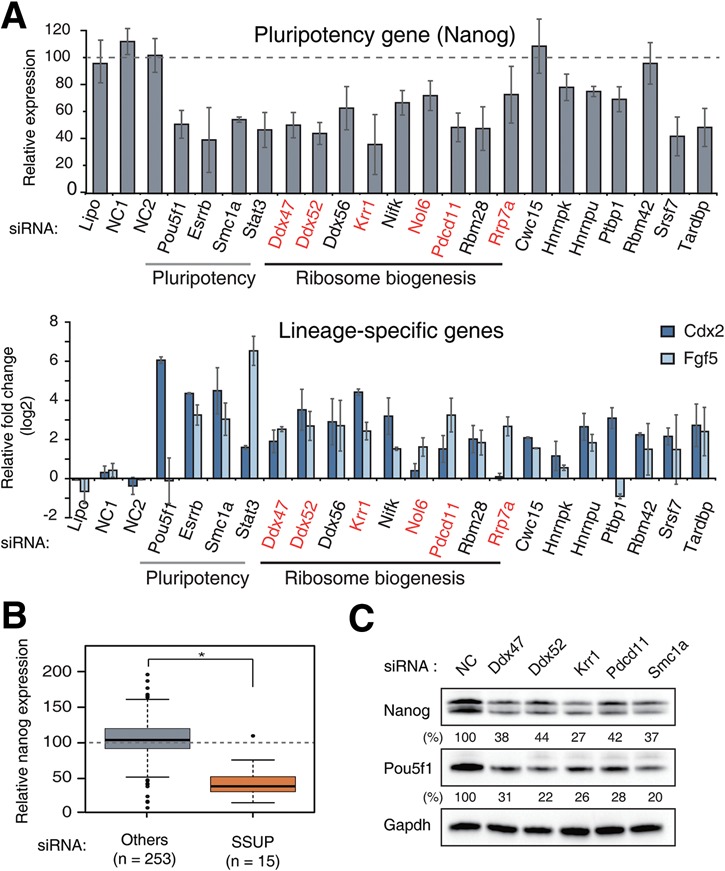
RNAi screen identifying novel RBPs involved in the regulation of pluripotency. (A) Relative expression of Nanog, Cdx2, and Fgf5 mRNA after knockdown of 16 validated RBPs is shown. The SSUP genes are in red letters. All of the error bars represent the mean ± SD of triplicates or two experiments (when indicated). All of the mRNA levels were normalized against Actb mRNA or others (when indicated). (B) A box plot showing reduced expression of Nanog mRNA upon knockdown of SSUP genes. (C) Western blotting shows the relative expression of Nanog and Pou5f1 after depletion of SSUP genes for 3 d. Relative values were normalized to Gapdh levels. (*) P < 1.0 × 10−7 by two-tailed t-test. See also Supplemental Figure S1.
SSUP is necessary for ESC maintenance
It was unexpected that many nucleolar proteins involved in rRNA processing were identified in our screen. Particularly interesting was that, out of the 16 validated genes, six genes (Krr1, Ddx47, Ddx52, Nol6, Pdcd11, and Rrp7a) encode subunits of one particular complex: SSUP (Fig. 1A, red). SSUP is a pre-18S rRNA processing complex composed of U3 snoRNA and ∼54 proteins (Supplemental Table S6; Phipps et al. 2011; Tafforeau et al. 2013). Note that 13 SSUP genes were included in the initial screen, out of which six genes survived the screens.
To examine whether SSUP is indeed required for ESC maintenance, we tested 27 additional genes that are known to be involved in various steps of ribosome biogenesis (including six additional SSUP genes). Depletion of five SSUP proteins (Imp4, Mpp10/Mphosph10, Wdr36, Wdr46, and Wdr75) resulted in a reduction of Nanog expression (Supplemental Fig. S1D), while one SSUP component (Cirh1a) caused cell death. Overall, Nanog expression was down-regulated significantly upon depletion of the SSUP genes (∼40% of that of the other genes, on average) (Fig. 1B).
In addition, we tested 27 proteins that we recently identified as novel RBPs in mESCs (Kwon et al. 2013). Three SSUP subunit genes were included in this set of 27 RBP genes, out of which two SSUP genes (Wdr3 and Wdr46) yielded clear differentiation phenotypes upon knockdown (data not shown). Taken together, 20 SSUP genes have been tested in our experiments in total, out of which knockdown of 12 genes induced differentiation and that of four genes caused cell death. Moreover, seven SSUP subunits (Ddx18, Ddx47, Dhx8, Dhx15, Dhx37, Eif4a3, and Utp6) have been reported in previous RNAi screens as potential ESC regulators (Fazzio et al. 2008; Kagey et al. 2010). We also confirmed the relevance of SSUP in maintenance of ESC identity using another mESC line, A3-1 (Supplemental Fig. S2A). Thus, the observed effects of the SSUP depletion are not restricted to the R1 mESC line.
ESCs lost their characteristic dome-like shape and showed reduced expression of Nanog and Pou5f1 2 d after RNAi of SSUP genes (Supplemental Figs. S2B, 1C). Costaining revealed that Nanog expression remained only in cells where Krr1 was still detected (Fig. 2A), indicating that Krr1 may be critical to maintain the Nanog level. Note that we chose Krr1 as a representative component of SSUP because all four tested siRNAs against Krr1 resulted in a clear loss of pluripotency in our experiments (Supplemental Fig. S1C). Furthermore, the Nanog level was rescued by ectopic expression of Krr1 (Fig. 2B), indicating that the observed phenotype is indeed due to Krr1 rather than an off-target effect.
Figure 2.
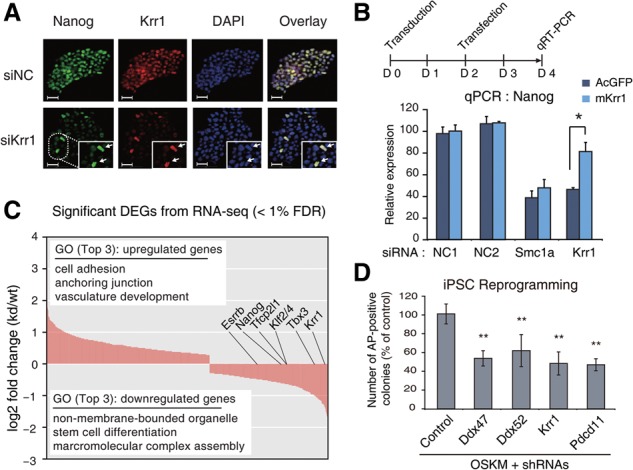
The SSUP is required for the maintenance of ESC identity. (A) Immunostaining shows the expression of Nanog and Krr1 in ESCs transfected with siKrr1 (stained 3 d following knockdown). Bars, 25 μm. (B) Overexpression of krr1 rescues the reduction of Nanog in Krr1-depleted cells. (C) Differentially expressed gene (DEG) analysis of RNA-seq reveals the DEGs after 2 d of Krr1 knockdown (1% false discovery rate [FDR]). (D) Down-regulation of SSUP factors reduces the reprogramming efficiency. Lentiviruses harboring each shRNA were cotransduced with lentivirus containing OSKM (Oct4, Sox2, Klf4, and c-Myc) reprogramming factors. The alkaline phosphatase (AP)-positive colonies were counted 11 d after transduction. (*) P < 0.05; (**) P < 0.005 by two-tailed t-test. See also Supplemental Figures S2–S4.
To investigate the molecular changes on the transcriptomic scale, we performed RNA-seq 2 d after Krr1 knockdown and identified 1035 genes as differentially expressed genes (DEGs; <1% false discovery rate [FDR]) (Fig. 2C; Supplemental Table S7). In gene ontology analysis, terms related to ribosome (“non-membrane-bounded organelle” and “macromolecular complex assembly”) and stem cell differentiation were enriched for down-regulated genes, which include Esrrb, Klf2/4, Nanog, Tbx3, Tcl1, and Tfcp2l1. As for up-regulated genes, terms such as “cell adhesion” and “cytoskeleton organization,” which are related to embryonic development or differentiation, were significantly enriched (Supplemental Table S8). Additionally, we confirmed the effect of Krr1 knockdown on ESC identity by immunostaining of ESC-specific markers Esrrb, Tfcp2l1, and Ssea1 (Supplemental Fig. S2C).
We also compared the transcriptomic changes of Krr1-depleted cells with those of core TF-depleted cells (Ivanova et al. 2006). DEGs from Krr1 knockdown overlapped significantly with those from core TF depletion (Esrrb, Nanog, Pou5f1, Sox2, Tbx3, and Tcl1) (Supplemental Fig. S3A). Moreover, the majority of the SSUP components, including Krr1, Wdr43, and Mpp10, were reduced upon RNAi of core TFs, suggesting that at least part of the SSUP genes are under the control of core TFs (Supplemental Fig. S3B). These observations indicate that the SSUP may be an integral part of the tightly interconnected regulatory network that governs pluripotency.
We next asked whether the SSUP genes are required for induced pluripotent stem cell (iPSC) formation. We transduced mouse embryonic fibroblasts (MEFs) with a lentivirus expressing Oct4, Sox2, Klf4, and c-Myc (OSKM) along with a lentivirus expressing an shRNA for Krr1, Ddx47, Ddx52, or Pdcd11 (Supplemental Fig. S4A–C). The efficiency of reprogramming, assessed by counting the number of alkaline phosphatase (AP)-positive colonies, was substantially reduced (∼50% of the control) when SSUP components were depleted (Fig. 2D). Furthermore, when we examined temporal changes of marker gene expression during reprogramming, we observed that induction of endogenous Nanog and Esrrb is strongly compromised in Krr1-depleted cells (Supplemental Fig. S4D). Krr1 knockdown does not significantly affect cell cycle regulators such as p53 and p21 (Supplemental Fig. S4E). These results indicate that the SSUP subunits are required for efficient reprogramming into iPSCs. Thus, our data collectively argue that the SSUP may play an important part in the pluripotency regulation circuit.
The SSUP is preferentially expressed in pluripotent stem cells
Although rRNA processing factors are generally considered to be constitutively expressed housekeeping genes, we found that the SSUP components (Krr1, Mpp10, and Wdr46) are down-regulated during embryoid body (EB) differentiation (Fig. 3A). Moreover, according to RNA-seq data (Liu et al. 2011), the majority of SSUP components are down-regulated during EB differentiation (Fig. 3B). We also analyzed the expression profiles of SSUP genes in 89 mouse tissues and cell lines using public data sets (Lattin et al. 2008; Wu et al. 2009). The SSUP genes, including Krr1, are highly expressed in proliferating cells, including stem cells and progenitor cells, especially in ESC lines (Supplemental Fig. S5A,B). Western blotting also confirmed the preferential expression of Ddx47, Krr1, Mpp10, and Wdr46 in mESCs compared with that in NIH3T3 cells and MEFs (Supplemental Fig. S5C). In addition, according to proteome data (Hansson et al. 2012), the SSUP components are mostly up-regulated at the initiation stage of reprogramming, and their increased levels are maintained throughout the reprogramming process (Supplemental Fig. S6). Thus, the SSUP genes are regulated with enhanced expression in stem cells.
Figure 3.
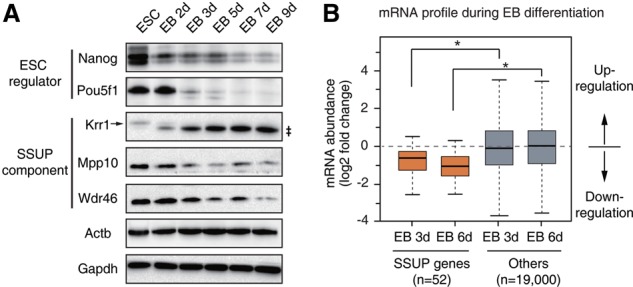
The SSUP is preferentially expressed in pluripotent stem cells. (A) Western blotting determines the levels of SSUP components during EB differentiation. (‡) Nonspecific band. (B) Box plot presenting the change of mRNAs during EB differentiation compared with ESCs (Liu et al. 2011). (*) P < 1.0 × 10−5 by two-tailed t-test. See also Supplemental Figures S5 and S6.
The SSUP contributes to pluripotency by enhancing general translational capacity
To understand the molecular mechanism driving the changes in gene expression following SSUP depletion, we first examined the role of Krr1 in rRNA biogenesis. The yeast homolog of Krr1 is involved in 18S rRNA processing (Sasaki et al. 2000; Zheng et al. 2014). The function of Krr1 is conserved in human cells, as demonstrated by a recent report in which Krr1, Ddx47, Ddx52, and Pdcd11 were identified as ribosome biogenesis genes that mediate the processing of the 5′ external transcribed spacer (ETS) of pre-rRNA (Tafforeau et al. 2013). We observed an accumulation of 30S pre-rRNA intermediates by Northern blotting (Supplemental Fig. S7A) and an alteration of the ratio between 18S and 28S rRNAs in Krr1-depleted mESCs by microfluidics-based electrophoresis (Supplemental Fig. S7B) Thus, Krr1 is indeed involved in the 5′ ETS processing of pre-rRNA in mESCs.
Given that impaired ribosome biogenesis is known to activate p53 through the interaction between surplus ribosomal proteins and Mdm2 (known as “ribosomal stress” or “nucleolar stress”) (for review, see Zhang and Lu 2009; Deisenroth and Zhang 2010), we first tested the possibility of p53-mediated differentiation. A recent study also proposed that fibrillarin, a nucleolar rRNA methyltransferase, modulates pluripotency in a p53-dependent manner (Watanabe-Susaki et al. 2014). Consistent with these findings, depletion of a ribosomal protein, Rpl37, for 2 d increased the expression of a p53 downstream gene, p21/Cdkn1a, and decreased the Nanog level (Supplemental Fig. S8). These changes were blocked when p53 was simultaneously depleted. Under the same conditions, however, knockdown of the SSUP components (Ddx47, Ddx52, and Krr1) did not induce p21 expression. Importantly, the reduction of Nanog upon SSUP knockdown was not influenced by p53 depletion (Supplemental Fig. S8). Therefore, it is unlikely that the p53-dependent ribosomal stress response mediates the early events following SSUP depletion.
Next, we examined the effects of SSUP depletion on translation using polysome profiling, which shows the relative distribution of translating ribosomes. The 40S, 80S, and polysome peaks were reduced relative to the 60S peak (Fig. 4A), which is a characteristic of defective 40S biogenesis (Li et al. 2009). Thus, knockdown of Krr1 decreased small ribosomal subunits and affected translation. The effect on translation was further confirmed by S35-methionine metabolic labeling. Krr1 depletion resulted in reduced protein synthesis by ∼75% of the control (Fig. 4B).
Figure 4.
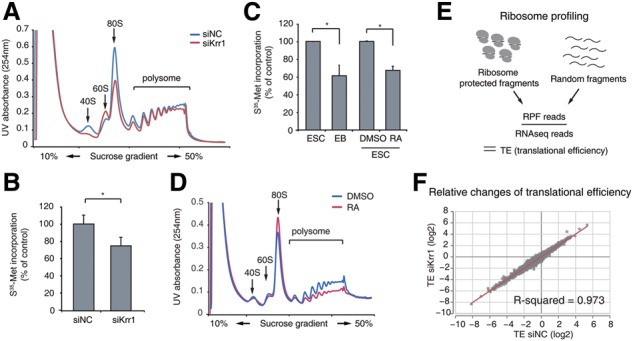
The SSUP contributes to pluripotency by enhancing general translational capacity. (A) Polysome profiles showing the changes in the ribosome pool upon Krr1 knockdown. (B) Metabolic labeling determines the bulk protein synthesis rate in control (siNC) and Krr1-depleted (siKrr1) cells. (C) Metabolic labeling reveals the differences in translational activity of ESCs and differentiating progenies (EBs and retinoic acid [RA]-treated cells). (D) Polysome profiles showing translational repression in RA-treated differentiating cells. (E) Schematic diagram of ribosome profiling, which can measure the translational efficiency (TE) of individual genes. (F) A scatter plot showing the correlation of TE between controls and knockdown samples. The R-value refers to Pearson correlation coefficient. (*) P < 0.05 by two-tailed t-test. See also Supplemental Figures S7 and S9–S11.
As the SSUP components are down-regulated during differentiation (Fig. 3), we sought to examine whether general translation is indeed attenuated in cells losing pluripotency. Protein synthesis rates were measured by S35-methionine incorporation in two types of differentiation models: EB formation by hanging drop culture (5 d) or neural differentiation by retinoic acid (RA) treatment (2 d). In both cases, the protein synthesis rate was decreased by 60%–70% compared with that in undifferentiated ESCs (Fig. 4C). Polysome profiles also indicated translational reduction following RA treatment (Fig. 4D). Consistently, NIH3T3 cells and MEFs showed reduced protein synthesis rates compared with mESCs (Supplemental Fig. S9). These data collectively suggest that translational capacity is high in mESCs and reduced during ESC differentiation. Furthermore, time-course experiments during EB differentiation further strengthened our observation that translational repression occurs during differentiation (Supplemental Fig. S10). It is noted that, in contrast to our observations, a previous study proposed that translation is suppressed in mESCs but is enhanced during EB differentiation (Sampath et al. 2008). The reason for this discrepancy is currently unclear. However, multiple lines of evidence support our conclusion. First, in accordance with enhanced protein synthesis, our results and previous transcriptome/proteome data show that SSUP genes are up-regulated in undifferentiated cells compared with their differentiated counterparts. Consistently, the rDNA promoter is unmethylated and kept in an open state in mESCs, producing a high level of pre-rRNA (Savic et al. 2014). Second, as pluripotent stem cells divide rapidly, they have a high demand for protein synthesis to support their unusually accelerated proliferation. Third, Pou5f1 expression alone is sufficient to reprogram triple-knockout MEFs (4E-BP1/2 and p53 knockout) but not the p53 single-knockout MEFs (Tahmasebi et al. 2014). Because 4E-BP1/2 are general translational suppressors, the triple-knockout cells have enhanced translational capacity. These results collectively suggest that enhanced translation may offer a favorable condition for reprogramming.
To further examine whether the SSUP selectively controls a subset of genes in addition to enhancing bulk translation, we conducted ribosome profiling following Krr1 knockdown. This provided quantitative information on translating mRNAs at the genomic scale by sequencing ribosome-protected mRNA fragments (RPFs) (Ingolia et al. 2011). The translational efficiency (TE) of each transcript can be calculated by dividing the RPF read counts by RNA-seq read counts (Fig. 4E). Note that the TE of a gene is “relative” to the other genes in the same library due to global normalization. Therefore, when translational regulation occurs globally (in the same direction and to the same degree), the relative TEs of individual genes do not change. Our data show that relative TEs remained largely unchanged following Krr1 knockdown (Fig. 4F; Supplemental Fig. S11). Thus, we conclude that Krr1 knockdown results in a reduction of bulk translation without substantial specificity.
Enhanced translation is critical for pluripotency maintenance
Our data indicate that protein synthesis is modulated globally and nonspecifically by the SSUP. However, when protein synthesis is globally suppressed, labile proteins are expected to be reduced more rapidly than stable proteins. Many of the key cellular regulators are known to have short half-lives, and their protein levels are dynamically controlled according to the changes in the protein synthesis rate (Schwanhausser et al. 2011). For instance, the Nanog protein is known to have a short half-life (∼2 h) due to ubiquitin-mediated proteolysis (Ramakrishna et al. 2011). To test the possibility that modulation of the global translation rate influences the steady-state level of unstable proteins preferentially, we treated ESCs with a translational inhibitor, 4EGI-1, and measured the level of core ESC TFs. 4EGI-1 interferes with the eIF4E–eIF4G interaction without disrupting ribosome biogenesis (Moerke et al. 2007). The protein levels of Nanog, Esrrb, and Tfcp2l1 decreased rapidly in response to translational repression. The mRNA levels of Nanog, Esrrb, and Tfcp2l1 decreased with a time delay (Fig. 5A), which is expected because these factors are known to be part of an autoregulatory positive feedback loop. Hence, while translational control is nonspecific and global, it can still affect the levels of labile proteins preferentially. As many of the regulatory factors are unstable and part of autoregulatory feedback loops, translational down-regulation is likely to result in rapid changes in the cellular state.
Figure 5.
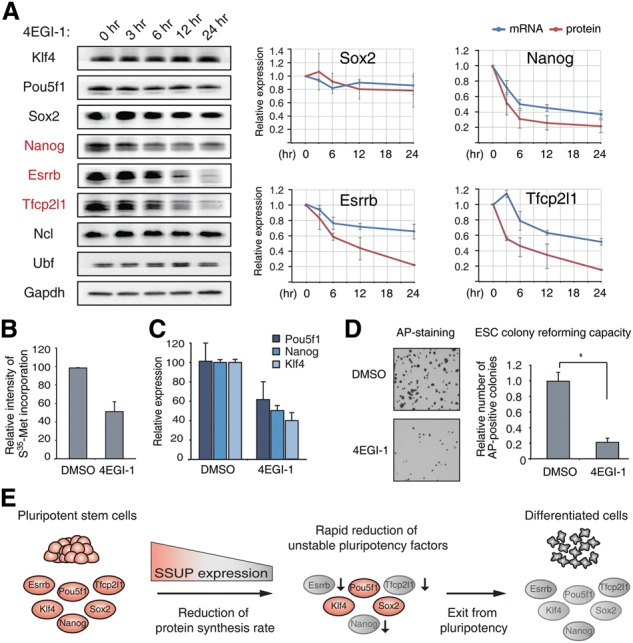
Regulation of global protein synthesis influences ESC identity. (A) Translational repression affects the level of several core ESC TFs. Ncl and Ubf were used as control nuclear proteins. Quantitation of protein levels by Western blotting and of mRNA levels by qRT–PCR is shown at the right. (B) Metabolic labeling indicates the changes in protein synthesis rate after treatment with 50 μM 4EGI-1 for 3 d. (C) Treatment of 4EGI-1 as in B affects the expression of pluripotent genes. (D) The same number of 4EGI-1-treated cells was reseeded, and, after 3 d of incubation in ESC culture conditions, the AP-positive colonies were counted. (E) Model depicting the mechanism of SSUP regulation and stem cell control. (*) P < 1.0 × 10−7 by two-tailed t-test.
Consistent with this notion, when we treated mESCs with 4EGI-1 at a low concentration for 3 d, mESCs lost their ESC identity. 4EGI-1 treatment reduced the protein synthesis rate by ∼50% (Fig. 5B) and the expression of Nanog, Pou5f1, and Klf4 by 40%–50% (Fig. 5C). Furthermore, when we reseeded the equal number of cells in the fresh ESC medium, 4EGI-1-treated cells showed a reduced capacity to re-establish AP-positive colonies (∼20% of control) (Fig. 5D). These results collectively suggest that enhanced translational activity is critical for ESC maintenance.
In summary, our unbiased screen uncovered a yet unappreciated contribution of the SSUP to the pluripotency program (Fig. 5E). As there are ∼54 subunits in the SSUP, we do not exclude a possibility that a subset of SSUP components may play a more critical (and specific) role than other components. Differential functions of individual SSUP components will need to be investigated in future studies.
While ribosome biogenesis factors are generally thought to be constitutively expressed and play “housekeeping” roles, our analyses indicate that at least part of the SSUP subunits is under tight regulation of core TFs and that the SSUP preferentially modulates the levels of labile regulatory factors. Core pluripotency TFs such as Nanog, Pou5f1, and Sox2 are known to form a highly interconnected network that generates a bistable state for ESCs (Young 2011); that is, ESCs stay in a pluripotency program when the master TFs are sustained at appropriate levels, while the cells enter into a differentiation program when any of the TFs are no longer available. Thus, a rapid loss of unstable TFs (such as Nanog) caused by translational suppression will lead to a silencing of the master TF circuit and the subsequent induction/repression of a wide spectrum of downstream genes, reconfiguring the regulation program (Fig. 5E).
Developmental cues may take advantage of this regulatory paradigm, which facilitates transition into differentiated states. Indeed, the importance of global translational control has been implicated in various animal models. Mouse haematopoietic stem cells (HSCs) require highly regulated protein synthesis to maintain their cellular identity; the Pten deletion that enhances protein synthesis blocks HSC differentiation, while the Rpl24 mutation that reduces translation impairs self-renewal (Signer et al. 2014). Additionally, a recent study in Drosophila found that down-regulation of RNA polymerase I activity promotes germline stem cell (GSC) differentiation by reducing the abundance of specific proteins of BMP signaling (Zhang et al. 2014). This result suggested that modulation of ribosome biogenesis affects the expression of specific proteins that regulate cell fate decision. This is in line with an earlier finding that Wicked (an SSUP component Utp18 homolog) is asymmetrically distributed during mitosis of GSCs and neural stem cells (Fichelson et al. 2009). The daughter cell that inherits more Wicked remains as a stem cell, while the other cell differentiates into cysts. Genetic depletion of Wicked induces differentiation of GSCs and neural stem cells. Thus, precise regulation of global translation rates may critically influence cell fate decisions in animal development.
Materials and methods
Details of the materials and methods used are described in the Supplemental Material, including cell culture and differentiation, RNAi screens, qRT–PCR, construction of RNA-seq and ribosome-profiling libraries, classification of SSUP components, bioinformatics analyses, S35-methionine metabolic labeling, reprogramming assays, AP staining, Western blotting, polysome profiling, Northern blotting, and immunostaining. Below is a simple description of the major experimental procedures.
Cell culture
R1 mouse embryonic stem cells were maintained on 0.1% gelatin-coated dishes with DMEM containing 15% FBS (Gibco), 1× nonessential amino acids (Gibco), 100 µM 2-mercaptoethanol (Sigma), and 1000 U/mL LIF (Millipore).
RNAi library construction and RNAi screen
For each RBP, four different siRNAs were designed using Block-It RNAi designer (Invitrogen). R1 cells were reverse-transfected using Lipofectamine 2000 mixed with siRNAs to a final concentration of 20 nM. Cells were incubated in ESC medium for 4 d and harvested for analysis of gene expression.
Construction of RNA-seq and ribosome-profiling libraries
RNA-seq and ribosome profiling libraries were prepared using the protocol of the ARTseq ribosome-profiling kit (Epicentre) with minor modifications. Cell lysates were digested by RNase I (Ambion) and gel-purified in the range of 30 nucleotides (nt) for ribosome profiling or 40–60 nt for RNA-seq libraries. The libraries were sequenced on an Illumina HiSeq 2000 (multiplexing 50-base single-end). All adapters and primers were synthesized by IDT.
Accession numbers
Sequencing data are available at NCBI Gene Expression Omnibus (GEO) under accession number GSE73369.
Supplementary Material
Acknowledgments
We thank Sung Chul Kwon, Mihye Lee, Yoosik Kim, Boseon Kim, and members of our laboratory for critical reading of this manuscript and helpful discussions; Jun Cho for assistance with ribosome profiling; and Ahyoung Cho, Eunji Kim, and Jihye Yang for technical assistance. This work was supported by IBS-R008-D1 of the Institute for Basic Science from the Ministry of Science, ICT, and Future Planning of Korea, and the BK21 Research Fellowships from the Ministry of Education of Korea. K.T.Y. designed and performed all of the experiments. J.P. analyzed the RNA-seq and RPF data. V.N.K. designed and supervised this project. K.T.Y., J.P., and V.N.K. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.267112.115.
Freely available online through the Genes & Development Open Access option.
References
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320. [DOI] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. 2010. Ribosome biogenesis surveillance: probing the ribosomal protein–Mdm2–p53 pathway. Oncogene 29: 4253–4260. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. 2009. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4: 403–415. [DOI] [PubMed] [Google Scholar]
- Ding J, Xu H, Faiola F, Ma'ayan A, Wang J. 2012. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res 22: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. 2009. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol 11: 685–693. [DOI] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. 2012. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep 2: 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genetics 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. 2008. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 7: e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al. 2014. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53: 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R. 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267. [DOI] [PubMed] [Google Scholar]
- Phipps KR, Charette J, Baserga SJ. 2011. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA 2: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Suresh B, Lim KH, Cha BH, Lee SH, Kim KS, Baek KH. 2011. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev 20: 1511–1519. [DOI] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2: 448–460. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Toh EA, Kikuchi Y. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol Cell Biol 20: 7971–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, et al. 2014. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15: 720–734. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51: 539–551. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Alain T, Rajasekhar VK, Zhang JP, Prager-Khoutorsky M, Khoutorsky A, Dogan Y, Gkogkas CG, Petroulakis E, Sylvestre A, et al. 2014. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14: 606–616. [DOI] [PubMed] [Google Scholar]
- Watanabe-Susaki K, Takada H, Enomoto K, Miwata K, Ishimine H, Intoh A, Ohtaka M, Nakanishi M, Sugino H, Asashima M, et al. 2014. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 32: 3099–3111. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW III, et al. 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Kalkan T, Morrisroe C, Smith A, Sharrocks AD. 2012. A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet 8: e1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Blelloch R. 2014. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shalaby NA, Buszczak M. 2014. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Lan P, Liu X, Ye K. 2014. Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast. J Biol Chem 289: 22692–22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


