Abstract
Upon entry into stationary phase, bacteria dimerize 70S ribosomes into translationally inactive 100S particles by a process called ribosome hibernation. Previously, we reported that the hibernation-promoting factor (HPF) of Listeria monocytogenes is required for 100S particle formation and facilitates adaptation to a number of stresses. Here, we demonstrate that HPF is required for the high tolerance of stationary-phase cultures to aminoglycosides but not to beta-lactam or quinolone antibiotics. The sensitivity of a Δhpf mutant to gentamicin was suppressed by the bacteriostatic antibiotics chloramphenicol and rifampin, which inhibit translation and transcription, respectively. Disruption of the proton motive force by the ionophore carbonyl cyanide m-chlorophenylhydrazone or mutation of genes involved in respiration also suppressed the sensitivity of the Δhpf mutant. Accordingly, Δhpf mutants had aberrantly high levels of ATP and reducing equivalents during prolonged stationary phase. Analysis of bacterial uptake of fluorescently labeled gentamicin demonstrated that the Δhpf mutant harbored increased intracellular levels of the drug. Finally, deletion of the main ribosome hibernation factor of Escherichia coli, ribosome modulation factor (rmf), rendered these bacteria susceptible to gentamicin. Taken together, these data suggest that HPF-mediated ribosome hibernation results in repression of the metabolic activity that underlies aminoglycoside tolerance. HPF is conserved in nearly every bacterial pathogen, and the role of ribosome hibernation in antibiotic tolerance may have clinical implications.
INTRODUCTION
Listeria monocytogenes is a Gram-positive, facultative intracellular pathogen that poses serious medical risks for pregnant women, immunocompromised individuals, and infants (1, 2). The significance of L. monocytogenes as a foodborne pathogen was recently highlighted when contaminated cantaloupes caused the deadliest U.S. foodborne disease outbreak since 1924 (3). This bacterium is hardy and able to survive in a variety of environments, including soil, groundwater, and processed meats and cheeses, and within mammalian cells (2, 4–6). L. monocytogenes can also form biofilms that can persist for years in food processing plants, despite repeated exposure to disinfectants (7, 8). Persistent L. monocytogenes contamination of food processing plants remains a significant source of foodborne illness (9–11). Understanding the molecular mechanisms that allow L. monocytogenes to survive and persist in harsh environments is crucial for developing strategies to ensure food safety.
Ribosome dimerization is a highly conserved yet relatively underappreciated arm of the classic ribosome cycle. During stress, bacteria are able to dimerize 70S ribosomes into inactive hibernating 100S ribosome dimers. These dimers lack mRNA and tRNA and have been shown to be translationally silent in vitro (12, 13). Upon introduction of fresh nutrients, 100S ribosomes are immediately dissociated into 70S ribosomes and available for translation (14–16). Thus, the rapid interconversion between 70S and 100S ribosomes is a potentially powerful mechanism for tuning translational capacity to environmental conditions.
Almost all bacteria possess genes that facilitate 100S ribosome formation; however, the distribution of these genes differs between Gram-positive and Gram-negative bacterial species. In Escherichia coli and other gammaproteobacteria, 100S ribosomes are formed through the sequential activities of ribosome modulation factor (RMF) and a short hibernation-promoting factor (HPF) to produce a 90S dimer that is then stabilized into the mature 100S form (17). In Gram-positive bacterial species, ribosome hibernation is mediated solely by a long HPF homolog with an N terminus common to all HPF homologs and a C-terminal extension thought to perform the activity of RMF (18, 19). The temporal profiles of 100S ribosomes formed by short and long HPFs are distinct. In Escherichia coli, the formation of 100S ribosomes is restricted to stationary phase, where they comprise the majority of the ribosome content (19, 20). In contrast, 100S ribosomes formed by long HPF homologs are detected at all phases of growth, with abundance peaking during the transition from logarithmic to stationary-phase growth, where they comprise 50% of the ribosome content (18, 21). The function of translationally silent 100S particles during logarithmic growth remains to be elucidated.
Ribosome hibernation is important for adaptation to various conditions, yet the mechanism by which 100S ribosomes mediate these adaptations is not clear. Ribosome hibernation is critical for survival during and recovery from a prolonged stationary phase (18, 20–23). Hibernation-associated factors are important for adaptation to a number of different stresses present during stationary phase, including deprivation of carbon, amino acids, and polyamines, as well as osmotic, heat, and acid stress (23–27). In addition, our lab recently described ribosome hibernation in L. monocytogenes and highlighted for the first time that 100S ribosomes contribute to bacterial pathogenesis (21). Because 100S ribosomes are induced during nutrient starvation, it has been proposed that ribosome hibernation serves to reduce the level of active translation to that commensurate with available nutrients and/or to store and protect ribosomes for use later, when favorable growth conditions return (13, 15, 23, 28). However, these roles have yet to be demonstrated experimentally, and the function that 100S ribosomes play in mediating adaptive processes remains unclear.
L. monocytogenes and many other bacteria naturally reside within complex microbial communities that include antibiotic-producing bacteria (29–33). Bacteria are inherently capable of evading killing by antibiotics (i.e., without acquiring genetic resistance) through various adaptive mechanisms that generally lead to reduced growth and cellular metabolism (34–36). Ribosome hibernation is implicated in bacterial states associated with high levels of tolerance to antibiotics, including stationary-phase, biofilm, and dormant persister cell states (13, 22, 37). We therefore considered the possibility that ribosome hibernation contributes to the natural ability of microbes to survive in the presence of antibiotics.
Here we demonstrate a role for ribosome hibernation in conferring tolerance to antibiotics. An L. monocytogenes Δhpf mutant was sensitive specifically to the ribosome-acting class of aminoglycoside antibiotics but not other classes of bactericidal antibiotics. The susceptibilities of L. monocytogenes and E. coli ribosome hibernation mutants were compared, and the results suggested that the two species have a common mechanism by which 100S ribosomes protect the bacteria from the activity of aminoglycosides. Importantly, 100S ribosomes may be a new target against which therapies against chronic infections caused by other human pathogens that are recalcitrant to traditional antibiotic treatments may be developed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All L. monocytogenes strains used and generated in this study were in the strain 10403S background and are listed in Table 1. L. monocytogenes strains were cultured in filter-sterilized brain heart infusion medium (BHI) buffered to pH 7.3 (Difco, Detroit, MI). E. coli K-12 Keio knockout strains were obtained from the Yale Coli Genetic Stock Center, New Haven, CT. Each strain was transformed with the pCP20 helper plasmid that carries the gene for a flippase recombinase (38) to remove the kanamycin resistance cassette marking the gene deletions. The resulting clean deletion mutant strains are listed in Table 1. E. coli strains were cultured in Luria-Bertani (LB) medium.
TABLE 1.
Strains used in this study
MIC determination.
MICs were determined by a broth microdilution technique in BHI according to the guidelines of Wikler and the Clinical and Laboratory Standards Institute (CLSI) (39).
Long-term antibiotic susceptibility and suppression assay.
Bacterial cells were cultured at 37°C for 16 h with shaking in 5 ml of the appropriate medium. Aliquots from these overnight cultures were removed and subjected to antibiotic treatment. Unless otherwise noted, the antibiotics were used at the following concentrations: 10 μg/ml gentamicin, 200 μg/ml carbenicillin, 30 μg/ml ciprofloxacin, 30 μg/ml norfloxacin, 160 μg/ml amikacin, 150 μg/ml kanamycin, and 20 μg/ml tobramycin. For suppression analyses, overnight aliquots were pretreated for 30 min with the appropriate chemical (20 μg/ml chloramphenicol, 20 μg/ml rifampin, or 2.5 μM carbonyl cyanide m-chlorophenylhydrazone [CCCP]) before gentamicin was added. At specified time points, aliquots of samples were removed, serially diluted, spot plated onto LB agar plates, and incubated overnight at 37°C to determine the numbers of CFU per milliliter.
ATP quantification.
To determine the ATP levels of long-term stationary-phase bacteria, bacterial cells were cultured as described above in buffered BHI at 37°C with shaking for 72 h. The bacterial cells were diluted 100-fold in sterile phosphate-buffered saline (PBS) and combined 1:1 with the BacTiter-Glo reagent (Promega) in white-walled 96-well plates (PerkinElmer). Luminescence was measured on a SpectraMax L luminescence microplate reader (Molecular Devices). Relative ATP levels were calculated by normalizing the luminescence to the numbers of CFU and are reported as the fold change relative to that of the wild type (WT). The experiments were repeated two times, each of which had four biological replicates. The data presented are from a representative experiment. Significance was determined using an unpaired t test.
Tetrazolium salt (XTT) reduction assay.
To determine the metabolic reducing power of long-term stationary-phase bacteria, the samples collected for ATP quantification (as described above) were subjected in parallel to an 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay. The XTT assay methodology was essentially the same as that described previously (40). Briefly, the bacterial cells were diluted 100-fold in sterile PBS and aliquoted into a 96-well microtiter plate (Sarstedt). A 1/10 volume of reagent (9:1 solution of XTT reagent [11.1 mM] and phenazine methosulfate [8 mM]) was added to the samples, and the resulting mixtures were incubated at 37°C in the dark for 1 h. The reduction of XTT to formazan was determined by measurement of the absorbance at 492 nm on an Infinite M1000 Pro microplate reader (Tecan). Values were normalized to the number of CFU and are reported as a multiple of the wild-type level. The experiments were repeated two times, each of which had four biological replicates. The data presented are from a representative experiment. Significance was determined using an unpaired t test.
Texas Red-gentamicin uptake assay.
A Texas Red-gentamicin mixture was made as described previously (41), and Texas Red-gentamicin uptake was measured by flow cytometry as described previously (42). Briefly, the Texas Red fluorescent dye was conjugated to gentamicin sulfate and purified. The resulting Texas Red-gentamicin reagent was incubated with stationary-phase samples for 24 h in the same manner described below for the long-term gentamicin assay. Samples were then washed, resuspended in PBS, analyzed on a BD LSR Fortessa flow cytometer, and plated for determination of the numbers of CFU. Gentamicin uptake was calculated as the mean fluorescence intensity normalized to that for the untreated samples. Results are from two independent experiments, each of which had five biological replicates. Significance was determined using an unpaired t test.
Long-term gentamicin susceptibility of E. coli hibernation mutants.
Bacterial cells were cultured at 37°C for 16 h with shaking in LB medium. Aliquots from these overnight cultures were removed and subjected to gentamicin (5 μg/ml) treatment. At specified time points, aliquots of samples were removed, serially diluted, spot plated onto LB agar plates, and incubated overnight at 37°C to determine the numbers of CFU per milliliter.
RESULTS
Stationary-phase Δhpf mutants are susceptible to prolonged gentamicin exposure.
Antibiotic-tolerant cells are characterized as phenotypic variants able to withstand antibiotic treatment without acquiring genetic resistance (37). Mutants with mutations that affect the frequency of antibiotic-tolerant cells therefore do not display MICs different from those of their parental strains (43). To determine whether deletion of hpf affected the MIC of L. monocytogenes, several antibiotics from each of the bactericidal classes were tested. Deletion of hpf had no significant (greater than 2-fold) effect on the MIC of any of the antibiotics tested (Table 2).
TABLE 2.
MICs of different classes of antibiotics for L. monocytogenes 10403S WT and Δhpf mutant strains
| Antibiotica | MIC (μg/ml) |
||
|---|---|---|---|
| WT strain 10403S | Δhpf mutant | Δhpf mutant carrying pPL2-HPF | |
| Ampicillin | 0.06–0.12 | 0.06–0.12 | 0.06–0.12 |
| Carbenicillin | 1–2 | 1–2 | 1–2 |
| Vancomycin | 1 | 1 | 1 |
| Ciprofloxacin | 1 | 1 | 1 |
| Norfloxacin | 8–16 | 8–16 | 8–16 |
| Gentamicin | 0.5–1 | 0.5–1 | 0.5–1 |
| Kanamycin | 4–8 | 4 | 4–8 |
| Tobramycin | 0.5–1 | 1 | 1 |
| Amikacin | 2–8 | 2–8 | 2–8 |
| Chloramphenicol | 4 | 4 | 4 |
| Rifampin | 0.03–0.06 | 0.03–0.06 | 0.03–0.06 |
Boldface indicates a bactericidal antibiotic.
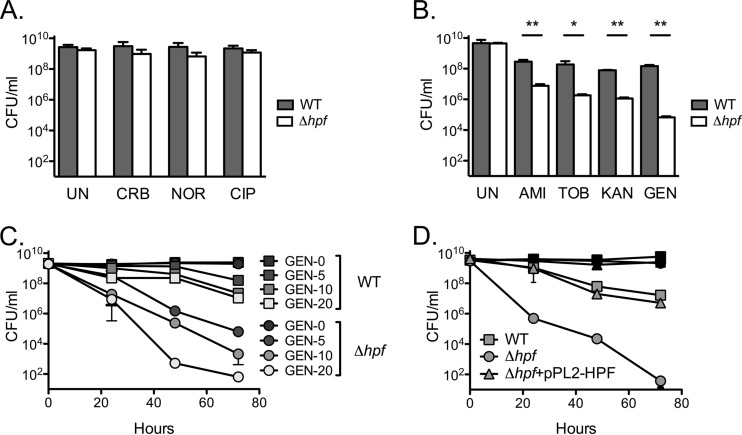
MICs were determined using cultures at a low density (105 to 106 CFU/ml). L. monocytogenes 100S ribosomes, however, are most prominent during entry to stationary phase, where cells are dense and growth has begun to slow (21). We therefore tested the sensitivity of stationary-phase L. monocytogenes cultures to prolonged antibiotic treatment. Neither the WT nor the Δhpf mutant in the untreated samples lost viability, even after an incubation of 3 days (Fig. 1). Both Δhpf mutant and WT stationary-phase bacteria were completely refractory to treatment with the beta-lactam carbenicillin or the quinolones ciprofloxacin and norfloxacin at levels greater than 10 times the MICs of these drugs (Fig. 1A). WT L. monocytogenes displayed a modest loss in the number of CFU when exposed to 10 times the MIC of gentamicin (10 μg/ml) for 72 h (Fig. 1B). Conversely, the Δhpf mutant exhibited a dramatic 5-log10-CFU/ml loss in viability. HPF mutant sensitivity was generalizable for all aminoglycosides tested, including amikacin, tobramycin, and kanamycin (Fig. 1B). Furthermore, killing by gentamicin occurred in a concentration-dependent manner. At the highest dose tested (20 μg/ml), the WT lost 2 log10 CFU/ml after 72 h, whereas the Δhpf mutant lost nearly 8 log10 CFU/ml and its growth approached the limit of detection (Fig. 1C). Finally, complementation of the Δhpf strain with pPL2-HPF fully rescued the gentamicin sensitivity phenotype (Fig. 1D). Taken together, these data indicate that stationary-phase Δhpf mutant cultures are specifically sensitive to aminoglycosides.
FIG 1.
Stationary-phase Δhpf mutant cultures are specifically susceptible to prolonged aminoglycoside exposure. (A) Stationary-phase Δhpf mutant cultures are insensitive to prolonged beta-lactam or quinolone exposure. UN, untreated culture; CRB, carbenicillin (200 μg/ml); NOR, norfloxacin (150 μg/ml); CIP, ciprofloxacin (30 μg/ml). Data are from 72 h. (B) Stationary-phase Δhpf mutant cultures are susceptible to prolonged aminoglycoside exposure. AMI, amikacin (160 μg/ml); TOB, tobramycin (10 μg/ml); KAN, kanamycin (150 μg/ml); GEN, gentamicin (10 μg/ml). Data are from 72 h. *, P < 0.05; **, P < 0.01. (C) Sensitivity to gentamicin is dose dependent. The gentamicin concentrations in the key are reported in micrograms per milliliter. (D) Sensitivity to gentamicin is fully complemented by the addition of pPL2-HPF. Means and SDs (n ≥ 3) are presented.
Inhibition of translation and transcription suppresses Δhpf mutant sensitivity.
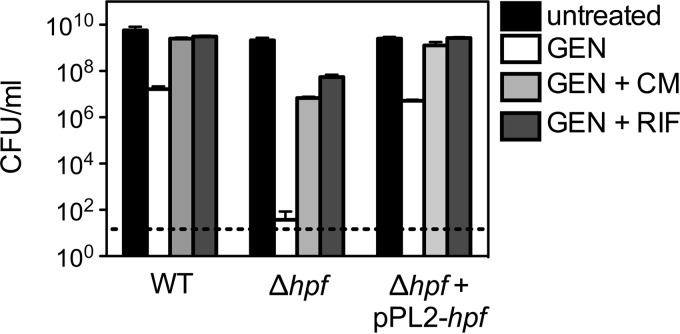
Translation is required for the bactericidal activity of aminoglycosides (44). Given the role of HPF in inactivating ribosomes during stationary phase, we hypothesized that inhibition of translation with a bacteriostatic translation inhibitor like chloramphenicol could suppress the sensitivity of the Δhpf mutant to gentamicin. To test this idea, translation was inhibited by pretreating stationary-phase cultures with a subinhibitory concentration of chloramphenicol. Chloramphenicol treatment alone did not affect the number of CFU for either the WT or the Δhpf mutant (data not shown). Chloramphenicol pretreatment significantly suppressed the sensitivity of the Δhpf mutant to gentamicin (Fig. 2). To determine whether this suppression was specific to inhibition of translation, transcription was inhibited with subinhibitory levels of rifampin, a bacteriostatic inhibitor of RNA polymerase. Rifampin treatment alone did not affect the number of CFU for the WT or the Δhpf mutant (data not shown). However, pretreatment with rifampin suppressed the sensitivity of the Δhpf mutant to prolonged gentamicin exposure (Fig. 2). These results confirm that active gene expression in the stationary-phase Δhpf mutant contributes to aminoglycoside susceptibility.
FIG 2.
Aminoglycoside susceptibility is suppressed by chemically arresting translation or transcription. Translation and transcription were inhibited by pretreating stationary-phase cultures for 30 min with subinhibitory levels of chloramphenicol and rifampin, respectively. Data are from 72 h. GEN, gentamicin (10 μg/ml); CM, chloramphenicol (20 μg/ml); RIF, rifampin (20 μg/ml). Means and SDs (n ≥ 3) are presented. Dashed line, limit of detection of 100 CFU/ml.
Disruption of the PMF suppresses Δhpf mutant sensitivity to gentamicin.
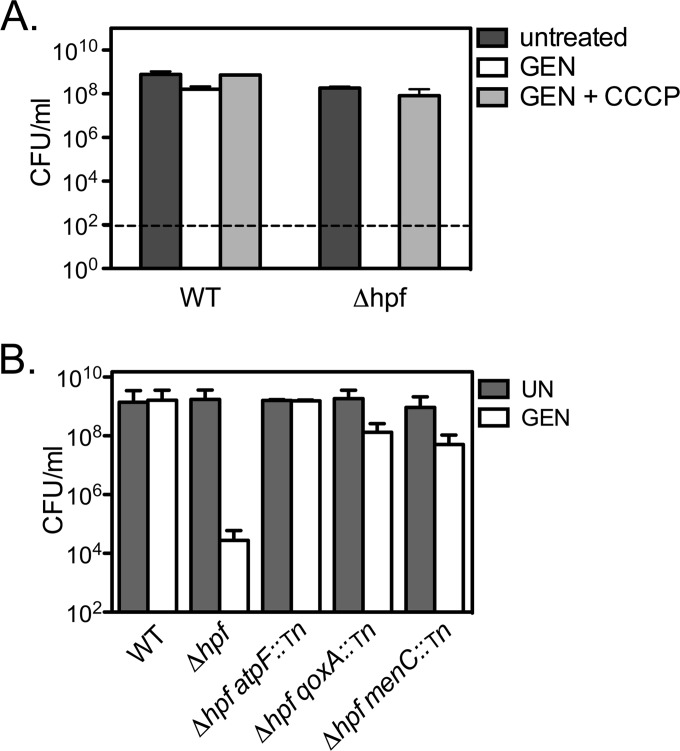
The uptake and accumulation of aminoglycosides are energy-dependent processes and require a threshold proton motive force (PMF) even in nondividing cells (42, 44). We hypothesized that the Δhpf strain remains metabolically active during stationary phase in a manner that facilitates aminoglycoside uptake and, consequently, enhances bacterial killing. To test this hypothesis, stationary-phase cultures were pretreated with the proton ionophore CCCP in order to disrupt the PMF and inhibit aminoglycoside uptake. The MIC of CCCP was determined to be 5 μM. Treatment with 0.5× the MIC of CCCP did not affect the number of CFU for either the WT or the Δhpf mutant (data not shown). Incubation with gentamicin for 72 h produced a modest reduction of 1.5 log10 CFU/ml for the WT, while the CFU counts for the Δhpf mutant were below the detection limit (102 CFU/ml) (Fig. 3A). Pretreatment with CCCP significantly rescued Δhpf mutant sensitivity. To further confirm that reduction of the PMF suppresses the susceptibility of the Δhpf mutant, genes involved in energy production were disrupted in the Δhpf mutant background. Transposon insertion into cytochrome quinol oxidase (qoxA), an ATP synthase subunit (atpF), or a member of the menaquinone biosynthetic pathway (menC) suppressed Δhpf mutant gentamicin sensitivity to levels near those for the WT (Fig. 3B). These data confirm that an active PMF is required for the sensitivity of the Δhpf mutant to gentamicin.
FIG 3.
Disruption of cellular energy production suppresses the sensitivity of the Δhpf mutant to gentamicin. Chemical inhibition of the proton motive force with 0.5× the MIC of CCCP (A) and disruption of genes involved in cellular energy production (B) suppress Δhpf mutant gentamicin sensitivity. Data are from 72-h cultures. Means and SDs (n ≥ 3) are presented. Tn, transposon.
Long-term stationary-phase Δhpf mutant cultures exhibit high levels of energy production.
In stationary phase, cells downregulate their levels of respiration and energy production to those commensurate with nutrient limitation and slow growth (45). Accordingly, overall ATP and NADH levels drop under nutrient starvation conditions (46–48). If ribosome hibernation facilitates the downregulation of energy production during stationary phase, then we predict that the Δhpf mutant will exhibit inappropriately high levels of ATP. To test this idea, overall ATP levels were quantified in a stationary-phase Δhpf mutant culture using the BacTiter-Glo microbial cell viability assay, which measures the amount of ATP present in a sample as a function of luminescence. Indeed, mutant cultures incubated for 72 h had 5-fold more ATP than the WT (Table 3). To measure the reducing potential of these strains, we employed an assay based on the reduction of the tetrazolium dye XTT into a formazan, which is directly related to the intracellular NADH pool. Similar to the measurements of ATP, the reducing power of the aged Δhpf mutant cultures was 5-fold higher than that of the WT (Table 3). Taken together, these analyses indicate that the Δhpf mutant remained in an inappropriately energized state during prolonged stationary phase.
TABLE 3.
Relative intracellular ATP and NADH measurements for L. monocytogenes WT and Δhpf mutant cultures at 72 ha
| Strain or P value | ||
|---|---|---|
| ATPb | XTT reduction (NADH)c | |
| WT | 1.1 ± 0.1 | 1.3 ± 0.2 |
| Δhpf mutant | 5.34 ± 1.7 | 5.04 ± 1.4 |
| P value | 0.022 | 0.011 |
Each value is the mean ± standard deviation from four biological replicates.
Relative luminescence units per CFU normalized to the value for the WT.
Absorbance at 450 nm per CFU normalized to the value for the WT.
A Δhpf mutant accumulates high levels of gentamicin.
Given that NADH drives the PMF required for aminoglycoside uptake, we hypothesized that enhanced uptake and enhanced accumulation of the drug underlie the mutant's sensitivity. Previous work has employed fluorescently labeled gentamicin to evaluate aminoglycoside uptake (42). Aminoglycoside uptake was quantified for the WT and the Δhpf mutant after an incubation of 24 h. The 24-h time point was chosen for analysis in order to avoid confounding of the results due to the dramatic loss in the number of CFU of the Δhpf mutant after 72 h of incubation. Texas Red-gentamicin killed the WT and Δhpf mutant at levels comparable to what we previously found for unlabeled gentamicin (Table 4). Quantification of the intracellular fluorescence for the Δhpf mutant showed that it was nearly 70% higher than that for the WT and correlated with a 1.5-log10-unit loss in the numbers of CFU per milliliter (Table 4). These data demonstrate that the high level of killing of the Δhpf mutant strain by gentamicin is related to increased intracellular concentrations of the drug.
TABLE 4.
Mean intracellular fluorescence and numbers of CFU of L. monocytogenes WT and Δhpf mutant cultures after incubation with Texas Red-gentamicin for 24 ha
| Strain or parameter | Relative fluorescenceb | Log10 CFU/ml |
|---|---|---|
| WT | 45.8 ± 5.4 | 9.0 ± 0.1 |
| Δhpf mutant | 76.1 ± 15.0 | 7.4 ± 0.2 |
| P value | 0.01 | 0.006 |
| Fold change | 1.7 | 0.8 |
Values are means ± standard deviations from five biological replicates.
Normalized to the value for untreated isolates.
An E. coli hibernation mutant exhibits sensitivity to gentamicin.
In order to kill bacteria, aminoglycosides must bind ribosomes and interfere with translation. The crystal structure of 100S ribosomes formed by E. coli RMF and HPF shows that short HPF binds ribosomes in a manner that occludes the aminoglycoside-binding site, while RMF binds near the peptide exit site (49). Given that short HPF and the N-terminal portion of long HPF are highly homologous, these data suggest the possibility that HPF homologs in general protect ribosomes from aminoglycosides by competing for binding. Because aminoglycosides bind to ribosomes very tightly (dissociation constant = 0.6 μM) (50), binding site competition could also explain why a Δhpf mutant harbors more intracellular gentamicin than the wild-type does. To test this hypothesis, E. coli Δrmf and Δhpf deletion mutants were evaluated for sensitivity to gentamicin (Fig. 4). Wild-type E. coli was much more sensitive to gentamicin than L. monocytogenes and required that the gentamicin concentration be reduced by half in order not to sterilize the cultures. At 5 μg/ml gentamicin, stationary-phase WT E. coli exhibited a 2-log10-unit loss in the numbers of CFU after 2 days. Contrary to the hypothesis of competitive binding, the Δrmf mutant but not the Δhpf mutant exhibited sensitivity to gentamicin. After 2 days, the numbers of CFU from the Δrmf mutant were at levels below the limit of detection. Conversely, the Δhpf mutant exhibited sensitivity similar to that of the WT. Given that RMF-bound ribosomes are translationally silent, these results suggest that ribosome hibernation protects bacteria by inactivating ribosomes rather than by blocking aminoglycoside binding.
FIG 4.
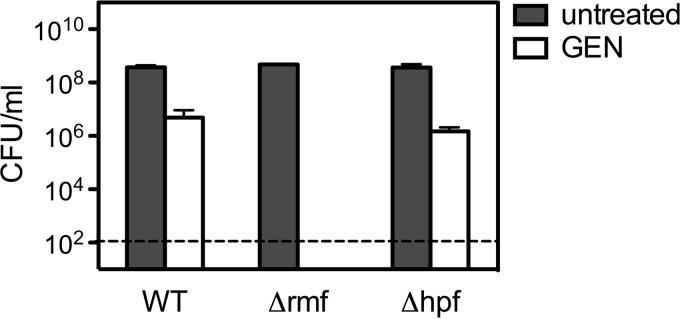
Stationary-phase E. coli Δrmf hibernation mutant cultures are susceptible to prolonged gentamicin exposure. The numbers of CFU per milliliter after a 48-h incubation with gentamicin at 5 μg/ml are provided. Means and SDs (n = 3) are presented.
DISCUSSION
The 100S ribosomes were first observed over 50 years ago (51), yet the precise function of ribosome hibernation remains unclear. This study demonstrated that ribosome hibernation is critical for the high level of tolerance to aminoglycosides exhibited by stationary-phase bacteria. Stationary-phase Δhpf mutant bacteria remained inappropriately energized and, accordingly, exhibited enhanced gentamicin uptake. Bacterial killing was suppressed by genetically or chemically disrupting processes like energy production. Taken together, these data imply that during stationary phase the Δhpf mutant fails to adequately downregulate cellular activities that contribute to the bactericidal activity of aminoglycosides.
On the basis of these data, we propose that ribosome hibernation is an important mechanism for energy conservation under conditions where nutrients are limiting. Exactly how ribosome hibernation contributes to aminoglycoside tolerance remains unclear. A simple model is that by inactivating a significant portion of ribosomes available for translation, ribosome hibernation reduces overall protein production, which has downstream effects on growth, metabolism, and energy production. The net result is a reduction in PMF, which blocks aminoglycoside import and subsequent killing.
The data presented here indicate that ribosome hibernation influences the two requirements for the bactericidal activity of aminoglycosides: energy-dependent uptake and active translation. While aminoglycoside uptake has been extensively studied (44), the lethal mode of action for aminoglycosides remains a matter of debate. It has been proposed that aminoglycosides reduce translation fidelity and result in the production of toxic peptides that cause lethal membrane damage (52). It is possible that, in the absence of 100S ribosomes, translation fidelity is further reduced and potentiates the effect of these mistranslated proteins. Although the role of ribosome hibernation in translation fidelity has not been studied, there are many other examples of cellular mechanisms for ensuring translation fidelity, for instance, the stringent response mediated by (p)ppGpp, which is crucial for tuning the translational capacity so that it is commensurate with the amounts of nutrients available during periods of amino acid deprivation (53). The stringent response contributes to translation fidelity, survival, and antibiotic tolerance (53–56). Ribosome hibernation factors are regulated in response to (p)ppGpp (24, 57, 58), and the connection between 100S ribosomes, the stringent response, translation fidelity, and antibiotic tolerance remains to be explored.
The recently described crystal structure of heterologous 100S ribosomes comprising Thermus thermophilus 70S ribosomes and E. coli RMF and HPF demonstrated that the short HPF homolog binds the ribosome at a site directly overlapping the binding site of aminoglycosides (49). We therefore hypothesized that ribosomes bound by HPF would naturally be protected from aminoglycoside binding and subsequent death. Long HPF is thought to perform the activities of both short HPF and RMF and likely binds ribosomes in a similar manner. We therefore attempted to test our hypothesis by generating constructs that split the L. monocytogenes long HPF into the N-terminal portion, which corresponds to the short HPF, and the C-terminal portion, which is thought to serve the function of RMF. However, coexpression of these constructs failed to reconstitute the activity of HPF, and bacteria expressing these constructs did not form 100S ribosomes (data not shown). So, we tested ribosome hibernation mutants of E. coli, which provide a nice system for uncoupling the dimerization activity of RMF (which is required for 90S ribosome formation) and the binding activity of HPF. Contrary to our hypothesis of competitive binding, the E. coli Δrmf mutant but not the Δhpf mutant exhibited sensitivity to gentamicin. These data suggest that ribosome inactivation rather than blocking of ribosome binding protects bacteria from the bactericidal activity of aminoglycosides. Although short HPF does not contribute to aminoglycoside tolerance per se, it is possible that, by protecting ribosomes from irreversible aminoglycoside binding, HPF aids in recovery once the antibiotic stress is removed. Further studies on outgrowth after antibiotic treatment will be required to examine any additional roles that HPF may play in recovery from stress.
The bacterial growth rate is correlated with the translational capacity of bacteria (59) as well as ATP levels (60). Given that a Δhpf mutant is unable to inactivate ribosomes into 100S particles (21), we hypothesized that the Δhpf mutant may continue to divide even during stationary phase. However, we have been unable to demonstrate a significant effect of ribosome hibernation on growth. We previously reported that the doubling time of the Δhpf mutant does not significantly differ from that of the WT (43 and 40 min, respectively) (21), and time-lapse microscopy studies failed to provide evidence that the Δhpf mutant continues to divide during stationary phase (data not shown). Previous work has demonstrated that bacteria can remain metabolically active without dividing (42, 61, 62). Taken together, these data suggest that hibernating ribosomes do not influence cell division but probably do have effects on other cellular activities.
The reduction of ATP levels and the subsequent homeostasis of bacteria in no-growth states are important adaptive responses to nutrient limitation. In E. coli, ATP levels decrease 5-fold upon entry into stationary phase, which triggers an important homeostatic loop for regulating ribosome biogenesis (47). Furthermore, this drop in ATP levels is required for the stability and activation of RpoS and σB, which are the major regulators of adaptation to stationary phase in E. coli and Bacillus subtilis, respectively (63, 64). Finally, like E. coli, when Mycobacterium tuberculosis shifts to a nonreplicative state, ATP levels are reduced by 5-fold and maintained at this reduced level (46, 48). This ATP reduction is important for survival and correlated with dramatically enhanced antibiotic tolerance (46, 65). It will be interesting to determine whether ribosome hibernation contributes to maintaining the 5-fold drop in ATP levels and the notorious drug recalcitrance of nonreplicating M. tuberculosis cells.
This work supports the idea proposed by others that ribosome hibernation adjusts the cell's translational capacity according to the available nutrients (15, 18). Although in vitro studies have conclusively shown that 100S ribosomes are translationally silent (13, 17, 49), the effect of ribosome hibernation on protein production in vivo has not yet been determined and should be the focus of future work. While global translation is reduced during stationary phase, translation continues at low levels and is critical for survival and subsequent recovery (61, 62, 66). It is possible that, in addition to reducing the overall translation level, ribosome hibernation is required to facilitate the efficient translation of the proteins required for stationary-phase survival. Future studies of the role of ribosome hibernation in translation regulation and adaptation are critical to obtain a complete understanding of this fundamental bacterial process.
In the broader sense, this work suggests that ribosome hibernation represents an Achilles heel for populations of slow-growing or latent bacteria. Pathogens like M. tuberculosis and Pseudomonas aeruginosa within biofilms that cause recurrent infections may employ ribosome hibernation to avoid being killed by antibiotics. The development of strategies that disrupt 100S ribosomes may vastly improve the efficacy of conventional antimicrobial agents.
ACKNOWLEDGMENTS
We thank Gabriel Mitchell for helpful discussions and careful review of the manuscript, Raymond Yin for help constructing the DP-L6182 strain, Carolyn Huangci for help with the MIC determinations, and Jonathan Portman for the menC mutant strain. We also thank Xiaoxue Zhou, Leigh Harris, and Julie Theriot for their generous help with time-lapse microscopy analysis of the Δhpf mutant.
This project was supported by Agriculture and Food Research Initiative competitive grant no. 2013-67012-21274 from the USDA National Institute of Food and Agriculture to S.L.M. and by National Institutes of Health grant 1PO1 AI63302 and National Institutes of Health (http://www.nih.gov) grant 1R01 AI27655 to D.A.P.
Daniel A. Portnoy has a consulting relationship with and a financial interest in Aduro Biotech, and both he and the company stand to benefit from the commercialization of the results of this research.
REFERENCES
- 1.Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A 108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlech WF., III 2000. Foodborne listeriosis. Clin Infect Dis 31:770–775. doi: 10.1086/314008. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2011. Multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado; CDC, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html. [Google Scholar]
- 4.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moors MA, Portnoy DA. 1995. Identification of bacterial genes that contribute to survival and growth in an intracellular environment. Trends Microbiol 3:83–85. doi: 10.1016/S0966-842X(00)88885-X. [DOI] [PubMed] [Google Scholar]
- 6.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 7.Borucki MK, Peppin JD, White D, Loge F, Call DR. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69:7336–7342. doi: 10.1128/AEM.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65:1811–1829. [DOI] [PubMed] [Google Scholar]
- 9.Conly J, Johnston B. 2008. Listeria: a persistent food-borne pathogen. Can J Infect Dis Med Microbiol 19:327–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly CW. 1990. Concerns of microbial pathogens in association with dairy foods. J Dairy Sci 73:1656–1661. doi: 10.3168/jds.S0022-0302(90)78838-8. [DOI] [PubMed] [Google Scholar]
- 11.Ringus DL, Ivy RA, Wiedmann M, Boor KJ. 2012. Salt stress-induced transcription of sigmaB- and CtsR-regulated genes in persistent and non-persistent Listeria monocytogenes strains from food processing plants. Foodborne Pathog Dis 9:198–206. doi: 10.1089/fpd.2011.1000. [DOI] [PubMed] [Google Scholar]
- 12.Kato T, Yoshida H, Miyata T, Maki Y, Wada A, Namba K. 2010. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. 1995. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun 214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 14.Wada A, Yamazaki Y, Fujita N, Ishihama A. 1990. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A 87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 16.Maki Y, Yoshida H, Wada A. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 17.Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. 2008. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem 143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 18.Puri P, Eckhardt TH, Franken LE, Fusetti F, Stuart MC, Boekema EJ, Kuipers OP, Kok J, Poolman B. 2014. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol Microbiol 91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- 19.Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H, Kato T, Miyata T, Namba K, Wada A. 2013. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 20.Wada A, Mikkola R, Kurland CG, Ishihama A. 2000. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J Bacteriol 182:2893–2899. doi: 10.1128/JB.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kline BC, McKay SL, Tang WW, Portnoy DA. 2015. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol 197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. 2012. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol 194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. 1993. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J 12:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tagami K, Nanamiya H, Kazo Y, Maehashi M, Suzuki S, Namba E, Hoshiya M, Hanai R, Tozawa Y, Morimoto T, Ogasawara N, Kageyama Y, Ara K, Ozaki K, Yoshida M, Kuroiwa H, Kuroiwa T, Ohashi Y, Kawamura F. 2012. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. Microbiologyopen 1:115–134. doi: 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terui Y, Tabei Y, Akiyama M, Higashi K, Tomitori H, Yamamoto K, Ishihama A, Igarashi K, Kashiwagi K. 2010. Ribosome modulation factor, an important protein for cell viability encoded by the polyamine modulon. J Biol Chem 285:28698–28707. doi: 10.1074/jbc.M110.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Sharoud WM, Niven GW. 2007. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology 153:247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 27.Niven GW. 2004. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch Microbiol 182:60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida H, Wada A. 2014. The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip Rev RNA 5:723–732. doi: 10.1002/wrna.1242. [DOI] [PubMed] [Google Scholar]
- 29.Weis J, Seeliger HP. 1975. Incidence of Listeria monocytogenes in nature. Appl Microbiol 30:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narisawa N, Haruta S, Arai H, Ishii M, Igarashi Y. 2008. Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl Environ Microbiol 74:3887–3894. doi: 10.1128/AEM.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelsic ED, Zhao J, Vetsigian K, Kishony R. 2015. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG. 1999. Microbial antagonism: a neglected avenue of natural products research. J Biotechnol 70:27–32. doi: 10.1016/S0168-1656(99)00054-1. [DOI] [PubMed] [Google Scholar]
- 33.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. [DOI] [PubMed] [Google Scholar]
- 35.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 36.Lewis K. 2012. Persister cells: molecular mechanisms related to antibiotic tolerance. Handb Exp Pharmacol 8:121–133. doi: 10.1007/978-3-642-28951-4_810.1007/978-3-642-28951-4_8. [DOI] [PubMed] [Google Scholar]
- 37.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wikler MA, Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard, 8th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Tsukatani T, Suenaga H, Higuchi T, Akao T, Ishiyama M, Ezoe K, Matsumoto K. 2008. Colorimetric cell proliferation assay for microorganisms in microtiter plate using water-soluble tetrazolium salts. J Microbiol Methods 75:109–116. doi: 10.1016/j.mimet.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Sandoval R, Leiser J, Molitoris BA. 1998. Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. J Am Soc Nephrol 9:167–174. [DOI] [PubMed] [Google Scholar]
- 42.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 44.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starosta AL, Lassak J, Jung K, Wilson DN. 2014. The bacterial translation stress response. FEMS Microbiol Rev 38:1172–1201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gengenbacher M, Rao SP, Pethe K, Dick T. 2010. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 47.Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12:125–134. doi: 10.1016/S1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 48.Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tangy F, Moukkadem M, Vindimian E, Capmau ML, Le Goffic F. 1985. Mechanism of action of gentamicin components. Characteristics of their binding to Escherichia coli ribosomes. Eur J Biochem 147:381–386. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy BJ. 1960. Variations in bacterial ribosomes. Biochim Biophys Acta 39:563–564. doi: 10.1016/0006-3002(60)90221-3. [DOI] [Google Scholar]
- 52.Davis BD. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaca AO, Colomer-Winter C, Lemos JA. 2015. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Farrell PH. 1978. The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14:545–557. doi: 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 56.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 57.Eymann C, Homuth G, Scharf C, Hecker M. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol 184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Izutsu K, Wada A, Wada C. 2001. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells 6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- 59.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. 2010. Interdependence of cell growth and gene expression: origins and consequences. Science 330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 60.Schneider DA, Gourse RL. 2003. Changes in Escherichia coli rRNA promoter activity correlate with changes in initiating nucleoside triphosphate and guanosine 5′ diphosphate 3′-diphosphate concentrations after induction of feedback control of ribosome synthesis. J Bacteriol 185:6185–6191. doi: 10.1128/JB.185.20.6185-6191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gefen O, Fridman O, Ronin I, Balaban NQ. 2014. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci U S A 111:556–561. doi: 10.1073/pnas.1314114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinai L, Rosenberg A, Smith Y, Segev E, Ben-Yehuda S. 2015. The molecular timeline of a reviving bacterial spore. Mol Cell 57:695–707. doi: 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peterson CN, Levchenko I, Rabinowitz JD, Baker TA, Silhavy TJ. 2012. RpoS proteolysis is controlled directly by ATP levels in Escherichia coli. Genes Dev 26:548–553. doi: 10.1101/gad.183517.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang S, Haldenwang WG. 2005. Contributions of ATP, GTP, and redox state to nutritional stress activation of the Bacillus subtilis sigmaB transcription factor. J Bacteriol 187:7554–7560. doi: 10.1128/JB.187.22.7554-7560.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarathy J, Dartois V, Dick T, Gengenbacher M. 2013. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segev E, Smith Y, Ben-Yehuda S. 2012. RNA dynamics in aging bacterial spores. Cell 148:139–149. doi: 10.1016/j.cell.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 67.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J Immunol 139:2005–2009. [PubMed] [Google Scholar]
- 68.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]