Abstract
A series of novel tetracycline derivatives were synthesized with the goal of creating new antibiotics that would be unaffected by the known tetracycline resistance mechanisms. New C-9-position derivatives of minocycline (the aminomethylcyclines [AMCs]) were tested for in vitro activity against Gram-positive strains containing known tetracycline resistance mechanisms of ribosomal protection (Tet M in Staphylococcus aureus, Enterococcus faecalis, and Streptococcus pneumoniae) and efflux (Tet K in S. aureus and Tet L in E. faecalis). A number of aminomethylcyclines with potent in vitro activity (MIC range of ≤0.06 to 2.0 μg/ml) were identified. These novel tetracyclines were more active against one or more of the resistant strains than the reference antibiotics tested (MIC range, 16 to 64 μg/ml). The AMC derivatives were active against bacteria resistant to tetracycline by both efflux and ribosomal protection mechanisms. This study identified the AMCs as a novel class of antibiotics evolved from tetracycline that exhibit potent activity in vitro against tetracycline-resistant Gram-positive bacteria, including pathogenic strains of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE). One derivative, 9-neopentylaminomethylminocycline (generic name omadacycline), was identified and is currently in human trials for acute bacterial skin and skin structure infections (ABSSSI) and community-acquired bacterial pneumonia (CABP).
INTRODUCTION
First isolated in the 1940s, tetracyclines have been widely used for the treatment of a variety of infectious diseases, including those caused by susceptible Gram-positive and Gram-negative aerobic and anaerobic organisms, mycoplasmas, and intracellular pathogens (1). Initially produced from the fermentation of Streptomyces, the first tetracyclines (chlortetracycline and oxytetracycline) and the first semisynthetic compound tetracycline were later modified in the early 1970s through altered fermentation and semisynthesis to produce the clinically important broad-spectrum tetracyclines doxycycline 1 and minocycline 2 (Fig. 1). Although very effective for decades in clinical and agricultural use as broad-spectrum agents, widespread resistance development, difficulty in synthesis, and the emergence of the cephalosporins and the fluoroquinolones in the 1970s and 1980s relegated the use of the narrow- and expanded-spectrum tetracyclines to only a few indications.
FIG 1.
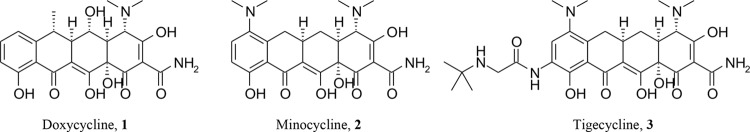
Structure of expanded-spectrum tetracyclines and tigecycline.
Acquired tetracycline resistance in bacteria is mediated by two major cellular phenotypes, drug efflux and ribosomal protection (1, 2). Tetracycline efflux is mediated by a family of structurally and mechanistically related proteins named the Tet proteins, which, as part of the major facilitator family of transport proteins, couple proton motive force to the exchange of a tetracycline-cation complex, thereby decreasing intracellular concentrations of tetracycline and allowing bacterial growth and survival. Ribosomal protection is mediated by GTP-dependent proteins structurally similar to ribosomal elongation factors EF-Tu and EF-G, blocking the ribosome binding of tetracyclines such as minocycline 2 and doxycycline 1 and thereby preventing the inhibition of ribosome function, allowing protein synthesis and bacterial survival. Ribosomal protection proteins are believed to bind to the ribosome, allosterically decreasing the affinity of tetracycline for its ribosomal binding site and allowing protein synthesis to occur. In all, over 35 different resistance genes derived from tet genetic determinants related to efflux and ribosomal protection are thought to be responsible for the decline in tetracycline effectiveness against community-acquired infections worldwide, especially those caused by Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae, and Enterobacteriaceae spp.
Advances in synthetic chemistry and the emergence of fluoroquinolone resistance renewed interest in the tetracycline class of antibiotics, and in the 1990s, Wyeth introduced the first of the tetracyclines known as the glycylcyclines (3). The glycylcycline tigecycline 3 (Fig. 1) was approved for the intravenous treatment of complicated intra-abdominal infections, complicated skin and skin structure infections, and community-acquired bacterial pneumonia (CABP). Tigecycline is a C-9-position, semisynthetic derivative of minocycline that shares a common mechanism of action with older tetracyclines but is not affected by the main tetracycline resistance mechanisms of efflux and ribosomal protection found in Gram-positive Staphylococcus species, Streptococcus species, and Enterococcus species, among others (4, 5).
The discovery of the glycylcyclines demonstrates the potential of novel tetracycline derivatives to combat resistant pathogens, and we sought to expand on the chemical diversity of semisynthetic modifications of the tetracyclines. By applying modern synthetic techniques to the tetracycline class, we generated a wide variety of diverse tetracycline derivatives through formation of novel carbon-carbon bonds at the C-7 and C-9 positions (6). In particular, we sought to identify novel reactive intermediates of the tetracycline class which were amenable to medicinal chemistry for optimization of activity against target bacteria. Here are described the results of this effort in identifying a novel class of C-9-substituted minocyclines, the aminomethylcyclines (AMCs), with potent activity against all major forms of tetracycline resistance in a variety of Gram-positive and Gram-negative pathogens, resulting in the discovery of omadacycline, the broad-spectrum antibiotic in development for the intravenous and oral treatment of acute bacterial skin and skin structure infections (ABSSSI) and community-acquired bacterial pneumonia (CABP) (7–9).
MATERIALS AND METHODS
Chemicals.
All novel tetracycline derivatives were prepared at Paratek Pharmaceuticals, Inc. (Boston, MA, USA). Tigecycline was obtained from Bosche Scientific, New Brunswick, NJ. Ciprofloxacin was obtained from Mediatech, Inc., Herndon, VA. Vancomycin and tetracycline were purchased from Sigma-Aldrich, Atlanta, GA. Minocycline HCl was obtained from Clariant, Holden, MA. Doxycycline was obtained from Hovione, East Windsor, NJ.
Synthesis of aminomethylcyclines.
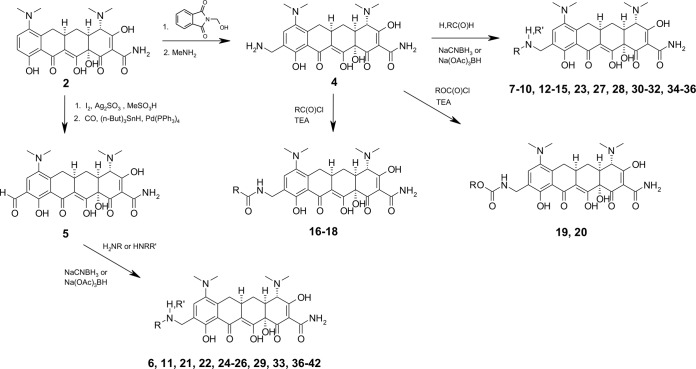
The initial goal of our synthetic effort was to identify novel reactive intermediates at the C-9 position of minocycline 2 which were amenable to synthetic medicinal chemistry. Through amidomethylation and deprotection of the resulting phthalimide using methylamine, 9-aminomethylminocycline 4 was produced (Fig. 2). The free amine moiety of this side chain provided a reactive moiety in which a variety of modifications were made to explore the effects of such substitutions on activity against resistant bacteria (Fig. 2). From 9-aminomethylminocycline, an initial chemical diversity set was prepared consisting of simple alkylamines, amides, carbamates, ureas, heterocycles, and aryl- or heteroarylamines. Desired modifications not accessible through the aminomethyl moiety were attained via direct reductive alkylation of the 9-formylminocycline intermediate 5. After screening and identification of hit compounds from the diversity set, lead optimization was carried out by targeted synthesis of analogs exploring the optimal modifications for activity against resistant organisms.
FIG 2.
Synthesis of novel aminomethylcyclines from minocycline.
Bacterial strains.
Clinical isolates of Gram-positive bacteria containing known tetracycline resistance genes were used, including isolates of Staphylococcus, Enterococcus, and Streptococcus (Table 1). All isolates were stored at −80°C in Trypticase soy broth plus 20% glycerol (Baltimore Biologics Labs [BBL]) or skim milk (Northeast Labs [NEL]). Horse or sheep blood supplementation was used for fastidious organisms. Isolates were subcultured twice onto appropriate solid media such as tryptic soy agar with 5% sheep blood (NEL), chocolate agar (NEL), or anaerobic blood agar (Anaerobe Systems) prior to MIC testing.
TABLE 1.
Bacterial strains and cell lines
| Strain or cell line | Species | Source | Relevant phenotype |
|---|---|---|---|
| RN450 | S. aureus | S. Levy, Tufts University Medical Center | Wild type, tetracycline susceptible |
| MRSA 5 | S. aureus | S. Levy, Tufts University Medical Center | MRSA, tetracycline resistant (ribosome protection, Tet M) |
| RN4250 | S. aureus | S. Levy, Tufts University Medical Center | Tetracycline resistant (tetracycline efflux, Tet K) |
| JH2-2 | Enterococcus faecalis | S. Levy, Tufts University Medical Center | Wild type, tetracycline susceptible |
| JH2-2(pAM211) | E. faecalis | S. Levy, Tufts University Medical Center | Tetracycline resistant (ribosome protection, Tet M) |
| JH2-2(pMV158) | E. faecalis | S. Levy, Tufts University Medical Center | Tetracycline resistant (tetracycline efflux, Tet L) |
| 494 | Enterococcus faecium | S. Levy, Tufts University Medical Center | Tetracycline resistant (ribosome protection, Tet M, plus tetracycline efflux, Tet L) |
| 157E | S. pneumoniae | GSK Laboratories, Verona, Italy | Wild type, tetracycline susceptible |
| ATCC 700905 | S. pneumoniae | American Type Culture Collection | Tetracycline resistant (ribosome protection, Tet M) |
| CHO-K1 | Chinese hamster ovary cell line | American Type Culture Collection | Mammalian cell, epithelial |
| COS-1 | African green monkey kidney cell line | American Type Culture Collection | Mammalian cell, fibroblast |
Susceptibility testing.
Microdilution MIC tests were performed according to Clinical and Laboratory Standards Institute (CLSI) standards (10). Media for omadacycline and tigecycline were freshly prepared. Microdilution MIC tests were modified to determine activity in the presence of pooled human serum (Atlanta Biologicals, Inc., Lawrenceville, GA) or sterile filtered mouse serum (Equitech-Bio Inc., Kerrville, TX). Serum was heat inactivated at 56°C for 45 min. Plates were prepared containing the final concentration of serum in cation-adjusted Mueller-Hinton broth of 50% for S. aureus or with the addition of 5% lysed horse blood of 25% for S. pneumoniae. Appropriate dilutions of novel tetracyclines and comparator compounds (20-mg/ml stock solutions in dimethyl sulfoxide [DMSO]) were made in cation-adjusted Mueller-Hinton broth (BBL) using the Tecan robotic workstation, followed by inoculation. Incubation was performed for 18 to 24 h at 35°C. Reading was done spectrophotometrically followed by manual checking for turbidity or other evidence of growth. Compounds were screened over the range of 0.06 to 64 μg/ml.
Cytotoxicity.
Cytotoxicity was determined using CHO and COS cell lines obtained from ATCC and grown in Dulbecco modified Eagle medium (DMEM) plus 10% fetal calf serum in 96-well microtiter plates. Drug was added, and plates were incubated for 20 to 24 h. Anhydrotetracycline was the standard positive control. Doxycycline was the comparator reference. Cell viability was determined by resazurin assay (11). Compounds were screened at 1 to 100 μg/ml, and the concentration that induced 50% cell toxicity (Tox50) was determined.
Systemic S. pneumoniae efficacy.
Six-week-old, specific-pathogen-free, male CD-1 mice, weighing 18 to 30 g (Charles River, Wilmington, MA), were used for all experiments. All animal protocols were reviewed and approved by the Paratek Pharmaceuticals Institutional Animal Care and Use Committee. Animals were acclimated for 1 week following delivery. Septicemia was induced by infecting mice intraperitoneally (i.p.) with 500 μl containing 1.02 × 105 ± 1.22 × 105 (mean ± standard deviation) CFU of S. pneumoniae 157E-2 in sterile bacteriological mucin (VWR Scientific, Pittsburgh, PA). At 1 h postinfection, mice were dosed subcutaneously with saline control, omadacycline, or comparator compounds of interest, dissolved in sterile saline for injection at a volume of 10 ml/kg of body weight. All drug doses were formulated fresh immediately prior to administration and adjusted to account for percent activity. Mice were housed in filter-topped cages in an isolated room and monitored for morbidity and mortality at least every 24 h for 7 days. All saline-treated control animals succumbed to infection within 48 h postinoculation.
S. aureus MRSA 5 thigh wound efficacy.
Animals were made neutropenic by i.p. administration of cyclophosphamide (150 mg/kg) 4 days before bacterial challenge and at a second dose of 100 mg/kg 1 day before challenge. Animals were infected with S. aureus MRSA 5 by direct injection into the thigh muscle. Two hours after inoculation, baseline colony counts were determined in a satellite group, and treatment groups received a single-dose treatment (or saline control) administered by subcutaneous injection. Animals were euthanized after 24 h, and the number of bacteria in the thigh was determined.
Pharmacokinetics.
The pharmacokinetics of the 9-aminomethylcyclines was evaluated in Cynomolgus monkeys with sampling times of 0 (predose), 0.25 (end of infusion), 0.5, 1, 3, 5, 7, 16, and 24 h. Protein was precipitated from the plasma by addition of acetonitrile containing 0.1% formic acid. The samples were centrifuged, and an aliquot of the supernatant was evaporated to dryness. The samples were reconstituted in methanol-water (1:1), and were analyzed on a Sciex 3000 liquid chromatography-tandem mass spectrometry (LC-MS/MS) system. Concentration data were analyzed using a noncompartmental model in WinNonlin v.4.01.
RESULTS
Screening of initial AMC diversity set.
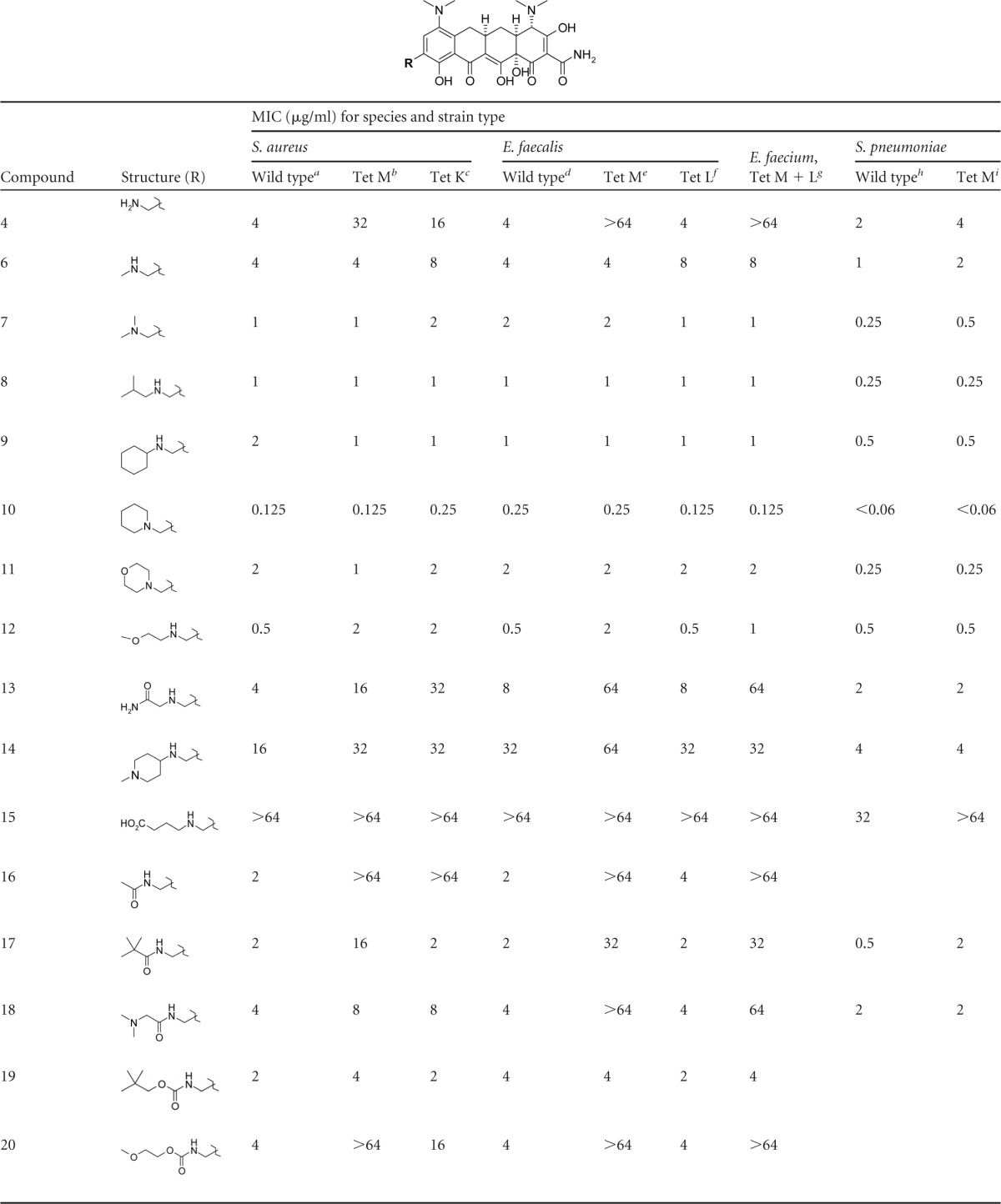
The initial goal was to generate a chemical diversity set based on modification of the 9-aminomethylminocycline intermediate 4 and identify hit compounds with activity against tetracycline-resistant Gram-positive organisms. Numerous alkyl, acyl, aryl, and heteroaryl derivatives were made to explore the effects of sterics, lipophilicity, polarity, and charge on the activity of the novel AMCs. The MIC values for representative compounds are shown in Table 2. Initial screening of the key intermediate AMC 4 demonstrated rather modest activity against wild-type (WT) Gram-positive organisms (MIC = 2 to 4 μg/ml) and even less activity against most resistant organisms (MIC, 16 to >64 μg/ml) with some moderate activity for the Tet L E. faecalis and Tet M S. pneumoniae mutants. However, a significant improvement in activity against all resistant organisms (MIC, 2 to 8 μg/ml) was observed by simple methyl substitution of the aminomethyl side chain compound 6, and potent activity (MIC, 0.25 to 2 μg/ml) was observed for the dimethyl-substituted compound 7 and compounds with larger, single-alkyl substitutions, such as compounds 8 and 9. The cyclic alkylamine AMC 10 displayed high potency (MIC, <0.06 to 0.25 μg/ml) against all resistant organisms in the screening panel with activity far superior to those of tetracycline and minocycline and comparable to that of tigecycline.
TABLE 2.
Initial structure-activity relationships of aminomethylcyclines against tetracycline-resistant Gram-positive bacteria
RN450.
MRSA 5.
RN4250.
JH2-2.
JH2-2(pAM211).
JH2-2(pMV158).
494.
157E.
ATCC 700905.
In general, AMCs with polar alkyl substitutions at the aminomethyl side chain were less active against the screening panel organisms than were lipophilic analogs, as shown by the slightly elevated MIC values determined for compounds 11 and 12, and particularly compound 13, which displayed very poor activity against most resistant strains (MIC, 16 to >64 μg/ml) despite being an isosteric analog of compound 8. The trend toward higher MIC values with increasing polarity was further confirmed by the very poor activity observed for compounds such as 14 and 15, which contain highly polar, charged substitutions on the aminomethyl side chain.
Acyl derivatives of the aminomethyl side chain, including amides and carbamates, generally had poorer activity against resistant organisms than did the alkyl derivatives as demonstrated by the MIC values for compounds 16 to 20. In particular, amide derivatives tended to have poor activity against Tet M resistant strains, although derivatives with larger alkyl substitutions, such as compound 17, retained some modest activity (MIC, 2 μg/ml) against the Tet K and Tet L efflux mutants. Carbamates with lipophilic groups, such as compound 19, displayed modest activity (MIC, 4 μg/ml) against all resistant strains, but as with alkyl derivatives, polar substitutions significantly reduced activity against Tet M resistant strains (compound 20).
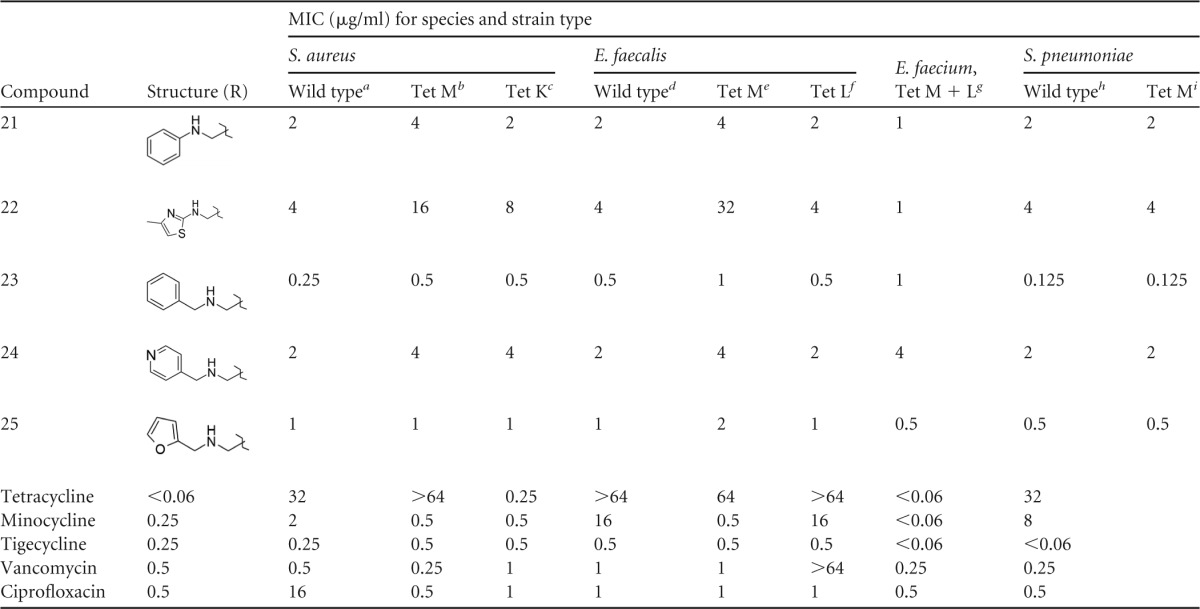
The unsubstituted phenyl AMC derivative compound 21 displayed modest activity (MIC, 2 to 4 μg/ml) against resistant strains, but heteroaryl derivatives such as compound 22 demonstrated poor activity against resistant organisms, particularly against Tet M mutants. However, a significant improvement in activity was observed when the phenyl group of compound 21 was extended a single carbon away from the amine of the aminomethyl side chain as in compound 23. Compound 23 displayed very good activity (MIC, 0.125 to 0.5 μg/ml) against all forms of resistance and approached the activity of the cyclic alkyl compound 10. As observed with other derivatives, more polar heterocyclic analogs of compound 23, such as the pyridinyl derivative compound 24 and the furanyl derivative compound 25, had less activity against all organisms than did the nonpolar benzylic group of compound 23 but still retained modest activity (MIC, 1 to 4 μg/ml).
Screening of the initial diversity set identified several AMCs with potent activity against all forms of tetracycline resistance. In particular, compounds with lipophilic alkyl or benzylic substituents off the aminomethyl side chain (compounds 7 to 10 and 23) displayed the lowest MIC values against ribosomal protection and efflux mutants of S. aureus, Enterococcus spp., and S. pneumoniae. One derivative, compound 10, had an activity profile comparable to that of tigecycline in our screening panel.
Optimization of the AMCs for activity against tetracycline resistance.
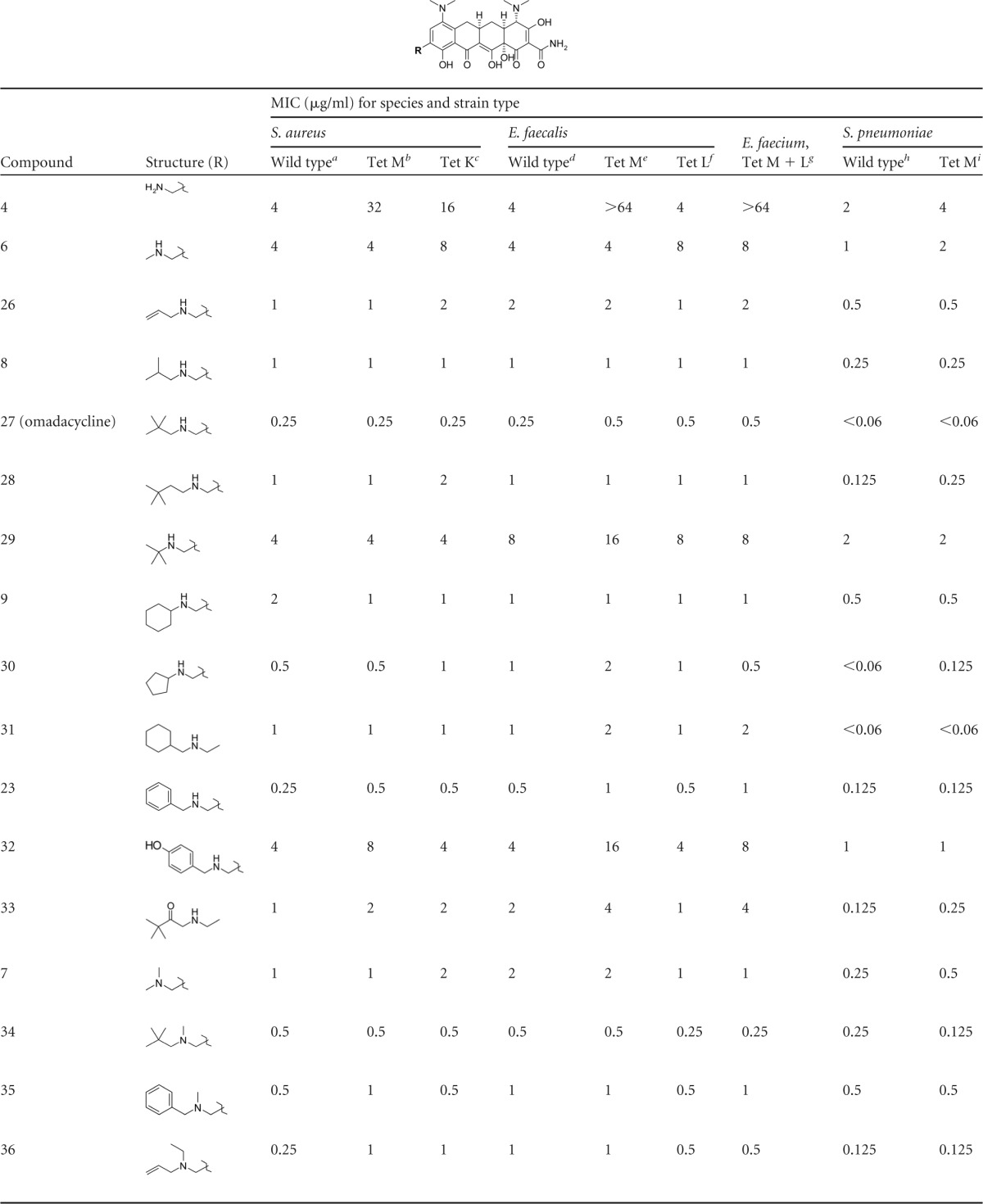
Compound series identified from the initial diversity set were optimized for activity against Gram-positive organisms with both major types of tetracycline resistance. Compounds 8, 9, 10, and 23 were chosen as the most promising candidates from the diversity set screening, and focused analogs of each were prepared to identify the most potent derivatives against Gram-positive pathogens with both ribosomal protection and efflux mechanisms of resistance. The results of this optimization are summarized in Table 3. The initial optimization focused on exploring the effects of alkyl group structure on activity. In agreement with the diversity set screening results, activity against tetracycline-resistant organisms increased with increasing alkyl group size as evidenced by the significant increase in activity from the unsubstituted compound 4 to the three-carbon alkenyl group of compound 26. Introduction of a single methyl branch at the 2-position of the side chain as in compound 8 slightly improved activity over that of the unbranched compound 26, and significant improvement in potency was observed with the addition of a second methyl group at this position as in the neopentyl group of compound 27 (omadacycline). Compound 27 (omadacycline) was very potent against resistant organisms (MIC, ≤0.06 to 0.50 μg/ml) and was comparable in activity to the piperidine compound 10 and tigecycline. Further extension of the side chain of compound 27 (omadacycline) by a single carbon as in compound 28 had a negative impact on activity, as MIC values against all organisms increased by 2 to 3 dilutions and yet were still comparable to those of the hit compound 8. Likewise, shortening of the side chain of compound 27 (omadacycline) by a single carbon had a negative effect on activity, as shown by the rather poor MIC profile of compound 29.
TABLE 3.
Activities of aminomethylcyclines against tetracycline-resistant Gram-positive bacteria
RN450.
MRSA 5.
RN4250.
JH2-2.
JH2-2(pAM211).
JH2-2(pMV158).
494.
157E.
ATCC 700905.
The active compound 9 contains a cyclic cyclohexyl side chain with a branch point at the 1-position and displayed potent activity against all organisms (MIC, 0.5 to 2 μg/ml) but did not approach the activity of compound 27 (omadacycline). Compound 30, an analog of compound 9 with a smaller cyclopentyl side chain, displayed a MIC profile comparable to that of compound 9 against Staphylococcus and Enterococcus spp. but was particularly active against S. pneumoniae, approaching the MIC values observed for compound 27 (omadacycline). Compound 31 extends the cyclohexyl group of compound 9 by one carbon atom from the aminomethyl group, thus moving the branch position to the 2-position of the side chain. The MIC profile of compound 31 was comparable to that of compound 9 against all organisms, except that activity against S. pneumoniae improved by more than 3 dilutions to the potency observed for compound 27 (omadacycline). Furthermore, compound 23 is the benzylic analog of compound 31 and displayed comparable but consistently more potent activity against all organisms and approached the MIC profile of compound 27 (omadacycline). The above results indicate a clear preference for alkyl groups which extend to at least 3 carbons from the aminomethyl side chain and contain branching at the 2-position. Branching at the 1-position of the alkyl side chain appears to have a negative steric impact on activity, particularly for S. pneumoniae.
To confirm the observations of the initial diversity set screening, several focused analogs of hit compounds with polar substitutions were prepared. Compounds 32 (analog of compound 23) and 33 (analog of compound 28) had poorer MIC profiles than did their lipophilic counterparts, which again confirms the observation that polar groups, particularly groups with hydrogen bond donors such as compound 32, tend to decrease the potency of the AMCs against resistant organisms (Table 3).
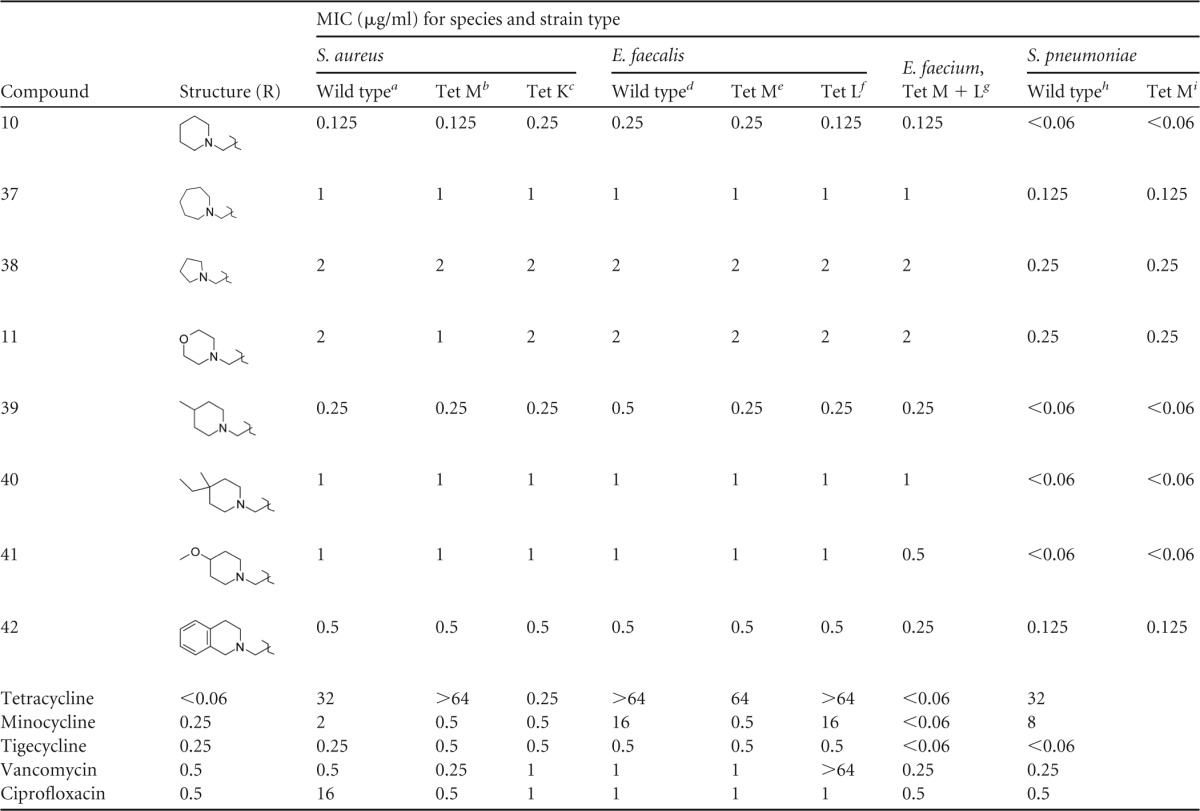
Despite the absence of an alkyl side chain extending at least 3 carbons, the dimethyl-substituted AMC 7 had potent activity comparable to those of other singly alkylated active compounds (8 and 9). Combination of the neopentyl side chain of compound 27 (omadacycline) along with a second methyl alkylation of the amine yielded a compound, 34, with very potent activity against all resistant organisms, approaching the activity of compound 27 (omadacycline), but with slightly less activity against S. pneumoniae. Similarly, amine methylation of compound 23 as in compound 35 yielded a compound with activity comparable to that of compound 23, but with less activity against S. pneumoniae. Compound 36 contains the 3-carbon alkenyl group of compound 26 and an ethyl group off the amine side chain. As observed for the methyl addition, the addition of the ethyl group did not significantly improve activity against most organisms; however, a slight but consistent 2-dilution improvement was observed in the activity against S. pneumoniae, which suggests an advantage to having combinations of alkyl groups more than 2 carbons. A large improvement in activity against all organisms for the bis-alkylated series was observed for cyclic compounds such as 10, which was very potent against all organisms and comparable in activity to compound 27 (omadacycline) and tigecycline. Analogs designed to explore the effect of ring size on activity (compounds 37 and 38) demonstrated a preference for the six-membered ring of compound 10, and polar analogs such as compound 11 displayed poorer activity. Methyl substitution of the 3-position of compound 10 as in compound 39 had no effect on activity relative to that of compound 10, but further substitution at this position as in compound 40 had a negative effect on MICs against Staphylococcus and Enterococcus spp. Polar substitutions at this position as in compound 41 also had a negative influence on potency relative to that of compound 10. Compound 42 represents a structural combination of the AMC side chains of compounds 23 and 10. No advantage in activity was observed for the combination, but compound 42 was quite potent against all panel organisms and was comparable in activity to compound 23.
The results of our optimization effort identified several AMCs with potent activity against resistant Gram-positive organisms comparable to that of tigecycline. The most potent compounds fell into two subtly distinct structural classes which included the singly alkylated series, including compounds 27 (omadacycline), 8, 23, 30, and 31 and the bis-alkylated series, including the cyclic compounds 10, 40, and 42 and the acyclic compounds 34 and 36.
Initial efficacy and safety screening of lead compounds in an S. pneumoniae infection model in mice.
Lead AMCs identified from susceptibility testing above were further characterized by cytotoxicity testing against animal cells grown in culture, serum MICs against a sensitive S. pneumoniae strain, and efficacy screening in an S. pneumoniae i.p. challenge infection model in mice. Most active AMC compounds had no measurable cytotoxicity up to 100 μg/ml (Table 4). Also, no measurable cytotoxicity was observed for compound 31 or 42 up to the concentrations at which solubility limited the ability to test them. Only one active AMC compound, 23, displayed measurable cytotoxicity at high concentrations, displaying a Tox50 of 54 μg/ml for CHO cells and 70 μg/ml for COS cells. These concentrations are 10-fold in excess of the measured or predicted Tox50 values observed for the AMCs and several orders of magnitude greater than the observed MIC values for sensitive S. pneumoniae wild type (WT) (Table 5). This relative selectivity of activity indicates their inherent specificity for bacteria compared to mammalian cells, typical of the tetracycline antibiotic family. This result demonstrates the selective nature of the antibacterial activity of the AMCs, typical of the tetracycline class.
TABLE 4.
Animal cell cytotoxicity
| Compound | Tox50a (μg/ml) for cell line: |
|
|---|---|---|
| COS | CHO | |
| 8 | >100 | >100 |
| 27 (omadacycline) | >100 | >100 |
| 28 | >100 | >100 |
| 30 | >100 | >100 |
| 31 | >36 ppt | >36 ppt |
| 23 | 70 | 54 |
| 34 | >100 | >100 |
| 10 | >100 | >100 |
| 40 | >100 | >100 |
| 42 | >40 ppt | >40 ppt |
| Minocycline | >100 | >100 |
| Tigecycline | >100 | >100 |
Concentration which decreases viable cell numbers by 50%. ppt, compound precipitated in culture medium above concentration shown; COS, African green money kidney fibroblast; CHO, Chinese hamster ovary fibroblast.
TABLE 5.
Animal efficacy of lead compounds in an S. pneumoniae intraperitoneal challenge infection model in mice
| Compound |
S. pneumoniae WT serum MIC (μg/ml)a |
% survival for i.p. challenge model in miceb | |
|---|---|---|---|
| None | Mouse | ||
| 8 | 0.25 | 0.25 | 100 |
| 27 (omadacycline) | <0.06 | <0.06 | 100 |
| 28 | 0.125 | 0.5 | 100 |
| 30 | <0.06 | <0.06 | 100 |
| 31 | <0.06 | 100 | |
| 23 | 0.125 | 0.25 | 100 |
| 34 | 0.25 | 0.25 | 100 |
| 10 | <0.06 | 0.125 | 100 |
| 40 | <0.06 | 0.25 | 80 |
| 42 | 0.125 | 4 | 100 |
| Minocycline | <0.06 | 0.125 | 100 |
| Tigecycline | <0.06 | <0.06 | 100 |
MIC against S. pneumoniae 157E in the absence or presence of 50% mouse serum.
Efficacy in S. pneumoniae 157E intraperitoneal challenge infection model in CD-1 mice. Compounds were screened at 5 mg/kg via single subcutaneous injection. Percent survival was measured versus untreated animal group at 7 days postinfection.
Prior to in vivo screening, the susceptibility of each lead compound in the presence of 25% (S. pneumoniae) or 50% (S. aureus) mouse plasma was assessed to determine the effects of plasma protein on activity. As shown in Table 5, the presence of mouse serum had little effect on the MIC values for most AMCs against the sensitive S. pneumoniae strain, and most leads retained potent activity (MIC of <1 μg/ml). One notable exception was the serum MIC value observed for compound 42, which increased 5 dilutions, from 0.125 μg/ml to 4 μg/ml, in the presence of mouse serum. AMC 42 represents one of the most lipophilic derivatives tested, which may explain the significant effect of mouse serum on susceptibility. Although not as dramatic, a similar trend of increased MICs in the presence of serum (at least a 2-dilution increase) is observed for compounds 40 and 28, which contain lipophilic side chains of 6 or more carbons.
Despite the effects of mouse serum on MIC values for some AMCs, all active compounds were very effective in the S. pneumoniae (WT) i.p. challenge infection model in mice, including compound 42. After a single subcutaneous dose of 5 mg/kg, all of the compounds prevented the death of all animals with the exception of compound 40, which was effective in 4 out of 5 animals in the treated group (Table 5). In this model, all animals in the untreated group succumbed to disease at 48 h. These results demonstrate that all AMCs were well tolerated in mice and achieved therapeutic plasma exposure after a single injection. In addition, the efficacy of all but one lead compound was comparable to those of minocycline and tigecycline against the sensitive S. pneumoniae strain.
Efficacy of AMCs against resistant S. aureus in a thigh wound infection model in mice.
Based on the positive results from the initial efficacy screening against sensitive S. pneumoniae, the AMCs were further tested for efficacy against a resistant S. aureus strain (MRSA 5, Tet M) in a thigh wound infection model in mice. As with the systemic infection model, AMCs were initially screened for antibacterial activity in vitro against the resistant S. aureus strain in the presence of 50% mouse serum. Unlike the results against the sensitive S. pneumoniae strain, dramatic serum effects on activity were observed against the Tet M S. aureus strain (Table 6). Six of the 10 AMCs tested displayed very poor MIC values (8 to >64 μg/ml) against strain MRSA 5 in the presence of mouse serum despite showing potent activity in the absence of serum. As before, compound 42 displayed the highest differential between the absence and presence of plasma, showing an increase in MIC value of more than 7 dilutions. This result again supports the hypothesis that highly lipophilic side chains increase plasma protein binding and negatively impact activity. In fact, all AMCs with more than 5 total carbons off the amine side chain displayed serum MIC values at least 4 dilutions higher than their respective MICs in the absence of mouse serum. This observation was further confirmed by MIC values attained in the presence of 50% human serum (Table 6). AMC derivatives with 5 or fewer carbons off the amine side chain (compounds 8, 27 [omadacycline], 30, and 10) were little affected by the presence of plasma and retained potent activity against MRSA 5.
TABLE 6.
Efficacy of leads against resistant S. aureus MRSA 5 (Tet M)
| Compound |
S. aureus MRSA 5 (Tet M) |
|||
|---|---|---|---|---|
| Serum MIC (μg/ml)a |
Log CFU change at 24 h for thigh wound infection model in miceb | |||
| None | Mouse | Human | ||
| 8 | 1 | 2 | 2 | −3.11 |
| 27 (omadacycline) | 0.25 | 1 | 0.5 | −2.49 |
| 28 | 1 | 16 | 16 | Not tested |
| 30 | 0.5 | 2 | 4 | −2.8 |
| 31 | 1 | 16 | 8 | −1.09 |
| 23 | 0.25 | 32 | 32 | −1.8 |
| 34 | 1 | 32 | >64 | Not tested |
| 10 | 0.125 | 1 | 1 | −2.82 |
| 40 | 1 | 8 | 32 | Not tested |
| 42 | 0.5 | >64 | >64 | Not tested |
| Minocycline | 2 | 64 | 16 | −0.98 |
| Tigecycline | 0.25 | 1 | 1 | −3.74 |
| Vancomycin | 0.5 | 2 | 1 | −1.71 |
MIC values in the absence or presence of 50% mouse or human serum.
Log CFU change relative to untreated control group at 24 h after single subcutaneous dose of 20 mg/kg.
The potency of AMCs in the presence of serum translated well to efficacy in the S. aureus MRSA 5 thigh wound infection model. AMCs with fewer than 5 side chain carbons were very effective at reducing CFU of S. aureus MRSA 5 at 24 h after a single subcutaneous injection of 20 mg/kg. As shown in Table 6, AMCs 8, 27 [omadacycline], 30, and 10 reduced colony counts by log10 2.49 to 3.11 relative to the untreated control animals and approached the activity observed for tigecycline (log10 CFU reduction, −3.74). In comparison, minocycline displayed poor efficacy in this model, reducing the colony count by only log10 0.98, as this strain is resistant to minocycline. Representative AMCs with poor activity against MRSA 5 in the presence of serum (compounds 23 and 31) failed to achieve a 2-log reduction in colony count after subcutaneous injection of 20 mg/kg despite having potent MIC values in the initial screening panel.
Pharmacokinetics of AMCs.
The most favorable AMC compounds, 27 (omadacycline) and 10, were chosen for pharmacokinetic studies in Cynomolgus monkeys to further characterize the potential of these novel tetracyclines as human therapeutics. Both AMCs were well tolerated after intravenous infusion and displayed pharmacokinetics comparable to those of both minocycline and tigecycline when corrected for dose with somewhat longer terminal half-lives (Table 7). After oral administration, AMC 27 (omadacycline) achieved significant plasma exposure in monkeys with a determined bioavailability of 31% (Table 7). Although lower than the bioavailability observed for minocycline (%F = 80), compound 27 (omadacycline) achieved oral plasma exposures 10-fold higher than those of tigecycline, which was poorly absorbed (%F = 3). AMC 10 demonstrated less oral bioavailability (%F = 7) than did compound 27 (omadacycline) but still achieved a plasma exposure 3-fold higher than that of tigecycline at the same dose.
TABLE 7.
Pharmacokinetics of AMCs and comparators in nonhuman primatesb
| Compound | Route | Dose (mg/kg) | t1/2 (h)a | Cmax (μg/ml)a | Tmax (h)a | AUC (μg · h/ml) | %F |
|---|---|---|---|---|---|---|---|
| 27 (omadacycline) | i.v. | 2.6 | 14.4 | 1.88 | 0.2 | 9.0 | |
| p.o. | 5 | 11.1 | 0.41 | 2.3 | 5.3 | 31 | |
| 10 | i.v. | 5 | 15.1 | 10.10 | 0.2 | 19.5 | |
| p.o. | 5 | 4.7 | 0.21 | 4.0 | 1.5 | 7.8 | |
| Minocycline | i.v. | 5 | 10.4 | 14.8 | 0.1 | 27.6 | |
| p.o. | 5 | 7.0 | 2.20 | 3.0 | 22.1 | 80 | |
| Tigecycline | i.v. | 5 | 10.9 | 23.2 | 0.2 | 16.3 | |
| p.o. | 5 | 3.4 | 0.04 | 4.0 | 0.5 | 3.1 |
Kinetic parameters determined from plasma concentration-versus-time curve.
Abbreviations: t1/2, half-life; Cmax, maximum concentration of drug in serum; Tmax, time to maximum concentration of drug in serum; AUC, area under the concentration-time curve; %F, oral bioavailability; i.v., intravenous; p.o., oral.
DISCUSSION
The discovery and development of tigecycline highlight the promise of novel tetracycline derivatives for combating antibiotic-resistant bacteria. Although effective clinically, tigecycline is reported to have very low oral bioavailability (5), which limits its use to intravenous administration in a hospital setting. In addition, nausea and vomiting have been prominent in the clinical use of tigecycline and have been reported for another glycylcycline, eravacycline (12). Since minocycline is highly orally available and well tolerated, the addition of the glycyl side chain moiety limits the oral availability and imparts the as-yet-undefined mechanism of nausea and vomiting of the resulting derivatives. It was our goal to identify novel C-9-position derivatives of minocycline that could overcome tetracycline resistance and have sufficient in vitro activity, spectrum, and in vivo efficacy to progress to clinical development, with oral bioavailability to address an important medical need and avoid the poor tolerability of the glycylcyclines.
In this study, we demonstrate the successful discovery of potent, orally available tetracycline derivatives known as aminomethylcyclines (AMCs) that have activity against resistant pathogens and have successfully created a well-tolerated broad-spectrum tetracycline that has avoided the nausea and vomiting described for the glycylcyclines (9). The key step in this process was the development of the novel reactive intermediate 9-aminomethylminocycline 4, which can be readily prepared from commercially available minocycline in two synthetic steps.
The amine side chain of compound 4 is highly amenable to a wide variety of chemical modifications and allowed the generation of a chemically diverse set of aminomethylcyclines to explore the effects of various substitutions on antibacterial activity. A second intermediate, 9-formylminocycline 5, further expanded the accessible chemical diversity of the AMCs and, together with compound 4, provided the necessary starting materials to generate a large number of diverse AMCs in good yield to support optimization of the activity, efficacy, and pharmacokinetics of the AMCs.
Utilizing this chemistry, an extensive discovery and optimization effort identified the alkyl AMCs as top lead candidates, exemplified by AMC 27 (omadacycline). This novel tetracycline derivative displays potent in vitro activity against resistant Gram-positive bacteria, low serum binding effects on activity, and potent efficacy against sensitive and resistant Gram-positive organisms, including MRSA in mouse models of infection. The antibacterial profile and efficacy of compound 27 (omadacycline) are superior to those of the early tetracyclines such as minocycline and are comparable to those of tigecycline. However, the unique structural modification of the 9-position of compound 27 (omadacycline) greatly improves the oral bioavailability of the drug versus tigecycline, making it very attractive for development as an oral agent for community-acquired infections.
Based on this work, compound 27 (omadacycline) was identified for clinical development, with the generic designation omadacycline, and is now entering phase 3 human trials as an intravenous and oral therapy for ABSSSI and CABP. Omadacycline has been shown to inhibit bacterial protein synthesis by targeting the classic tetracycline ribosomal binding site of the 30S subunit, in either the presence or the absence of ribosome protection-mediated tetracycline resistance (7). Furthermore, the unique 9-alkylaminomethyl side chain of omadacycline allows the drug to thwart efflux-mediated resistance mechanisms. Omadacycline has demonstrated positive efficacy and safety in phase 1 and 2 human trials with an oral bioavailability of approximately 35% in human patients (9). Importantly, no significant emesis or nausea has been observed in patients receiving omadacycline, which is a common and dose-limiting side effect of the glycylcyclines tigecycline and eravacycline (12). These results demonstrate the promise of omadacycline as a novel and orally available antibiotic therapy to address the increasing threat of multidrug-resistant bacteria.
ACKNOWLEDGMENTS
We acknowledge the editorial assistance of Richard S. Perry in the preparation of the manuscript.
All authors are or were full-time, paid employees of Paratek Pharmaceuticals, Boston, MA.
This work, including the editorial assistance of Richard S. Perry in the preparation of the manuscript, was entirely supported by Paratek Pharmaceuticals, Boston, MA.
REFERENCES
- 1.Nelson ML, Levy SB. 2011. The history of the tetracyclines. Ann N Y Acad Sci 1241:17–32. doi: 10.1111/j.1749-6632.2011.06354.x. [DOI] [PubMed] [Google Scholar]
- 2.Thaker M, Spanogiannopoulos P, Wright GD. 2010. The tetracycline resistome. Cell Mol Life Sci 67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra I. 2001. Glycylcyclines: third-generation tetracycline antibiotics. Curr Opin Pharmacol 1:464–469. doi: 10.1016/S1471-4892(01)00081-9. [DOI] [PubMed] [Google Scholar]
- 4.Pankey GA. 2005. Tigecycline. J Antimicrob Chemother 56:470–480. doi: 10.1093/jac/dki248. [DOI] [PubMed] [Google Scholar]
- 5.Rubinstein E, Vaughan D. 2005. Tigecycline: a novel glycylcycline. Drugs 65:1317–1336. doi: 10.2165/00003495-200565100-00002. [DOI] [PubMed] [Google Scholar]
- 6.Nelson ML, Ismail MY, McIntyre L, Bhatia B, Viski P, Hawkins P, Rennie G, Andorsky D, Messersmith D, Stapleton K, Dumornay J, Sheahan P, Verma AK, Warchol T, Levy SB. 2003. Versatile and facile synthesis of diverse semisynthetic tetracycline derivatives via Pd-catalyzed reactions. J Org Chem 68:5838–5851. doi: 10.1021/jo030047d. [DOI] [PubMed] [Google Scholar]
- 7.Draper MP, Weir S, Macone A, Donatelli J, Trieber CA, Tanaka SK, Levy SB. 2014. Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob Agents Chemother 58:1279–1283. doi: 10.1128/AAC.01066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macone AB, Caruso BK, Leahy RG, Donatelli J, Weir S, Draper MP, Tanaka SK, Levy SB. 2014. In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob Agents Chemother 58:1127–1135. doi: 10.1128/AAC.01242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noel GJ, Draper MP, Hait H, Tanaka SK, Arbeit RD. 2012. A randomized, evaluator-blind, phase 2 study comparing the safety and efficacy of omadacycline to those of linezolid for treatment of complicated skin and skin structure infections. Antimicrob Agents Chemother 56:5650–5654. doi: 10.1128/AAC.00948-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—7th ed. Clinical and Laboratory Standards Institute document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Ahmed SA, Gogal RM Jr, Walsh JE. 1994. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods 170:211–224. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 12.Connors KP, Housman ST, Pope JS, Russomanno J, Salerno E, Shore E, Redican S, Nicolau DP. 2014. Phase I, open-label, safety and pharmacokinetic study to assess bronchopulmonary disposition of intravenous eravacycline in healthy men and women. Antimicrob Agents Chemother 58:2113–2118. doi: 10.1128/AAC.02036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]