Abstract
Qnr is a plasmid-encoded and chromosomally determined protein that protects DNA gyrase and topoisomerase IV from inhibition by quinolones. Despite its prevalence worldwide and existence prior to the discovery of quinolones, its native function is not known. Other synthetic compounds and natural products also target bacterial topoisomerases. A number were studied as molecular probes to gain insight into how Qnr acts. Qnr blocked inhibition by synthetic compounds with somewhat quinolone-like structure that target the GyrA subunit, such as the 2-pyridone ABT-719, the quinazoline-2,4-dione PD 0305970, and the spiropyrimidinetrione pyrazinyl-alkynyl-tetrahydroquinoline (PAT), indicating that Qnr is not strictly quinolone specific, but Qnr did not protect against GyrA-targeting simocyclinone D8 despite evidence that both simocyclinone D8 and Qnr affect DNA binding to gyrase. Qnr did not affect the activity of tricyclic pyrimidoindole or pyrazolopyridones, synthetic inhibitors of the GyrB subunit, or nonsynthetic GyrB inhibitors, such as coumermycin A1, novobiocin, gyramide A, or microcin B17.Thus, in this set of compounds the protective activity of Qnr was confined to those that, like quinolones, trap gyrase on DNA in cleaved complexes.
INTRODUCTION
Qnr was discovered as a plasmid-encoded protein that reduces susceptibility to quinolones (1). Quinolones are synthetic compounds that target the essential bacterial enzymes DNA gyrase and topoisomerase IV, homologous tetramers composed of GyrA and GyrB or ParC and ParE subunits, respectively, that introduce negative supercoils or unknot and decatenate the DNA helix with energy from ATP hydrolysis (2).Qnr is a pentapeptide repeat protein that blocks quinolone inhibition of both topoisomerases and binds to each of their subunits as well as to the holoenzymes (3–5). Many bacteria have qnr-like genes on the chromosome, some, especially in aquatic bacteria, closely related to plasmid-determined qnr varieties (6–8). The native function of these proteins, which clearly antedate the clinical use of quinolones, is not known.
A number of other agents target topoisomerases. Well-studied natural products include coumermycin A1 (9, 10), gyramide A (11), microcin B17 (12), novobiocin (9, 10), and simocyclinone D8 (13, 14). From mutational and other studies the sites of action of many of these agents are known. For example, the primary site of resistance mutations for quinolones in Gram-negative bacteria is a region on the GyrA subunit known as the quinolone resistance-determining region (QRDR) (15), while novobiocin targets ATPase activity of the GyrB subunit (9). Qnr does not protect against novobiocin inhibition of gyrase (16), but the protective effect of Qnr on other natural products is not yet known. Medicinal chemists have synthesized synthetic compounds of various structures intended to act on gyrase at sites different from those directly affected by quinolones, especially the GyrB subunit. Whether Qnr protects gyrase from such compounds has not been investigated. The aim of this study was to use such natural and synthetic inhibitors as molecular probes to gain insight into how Qnr protects DNA gyrase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used are shown in Table 1. Strains were routinely grown in Luria-Bertani broth at 37°C. Culture plates contained Mueller-Hinton agar (Becton, Dickinson and Co., Sparks, MD). Plasmids were transferred to Escherichia coli J53 Azir (azide resistant) by transformation or conjugation using 100 μg/ml of ampicillin, 25 μg/ml of ceftazidime, or 25 μg/ml of chloramphenicol for selection and, where necessary, 200 μg/ml of sodium azide for counterselection. Isopropyl-β-d-thiogalactopyranoside (IPTG) at 100 μM was used to maximize QnrB production with M15 pREP3 pQE-60-QnrB1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| J53 Azir | Plasmid recipient | 46 |
| BL21(DE3) | Expression host | Agilent Technologies |
| M15 pREP4 | Expression host | Qiagen |
| EW1b | ΔtolC | 47 |
| RYC1000 pMM39 | Microcin B17 producer | 48 |
| Plasmids | ||
| pMG252 | qnrA1 | 1 |
| pMG253 | qnrA1 cloned | 3 |
| pMG298 | qnrB1 | 27 |
| pET28a:QnrB1 | qnrB1 cloned, His6 tagged, IPTG inducible | 16 |
| pQE-60-qnrB1 | qnrB1 cloned, His6 tagged, IPTG inducible | 27 |
| pMG306 | qnrS1 | 49 |
Susceptibility testing.
Disk and agar dilution susceptibility testing was performed as described by the CLSI, using Mueller-Hinton agar, an inoculum of 104 CFU, and 16 to 20 h of incubation at 37°C (17). E. coli J53 Azir and ATCC 25922 were used for quality control. Blank disks were obtained from Becton, Dickinson, and Co.
Chemicals.
Ciprofloxacin, coumermycin A1, and novobiocin came from Sigma-Aldrich Co., St. Louis, MO. ABT-719 was provided by Abbott Laboratories, Abbott Park, IL. PD 0305970 came from Pfizer Global Research and Development, Ann Arbor, MI. C3 and C4 came from Trius (subsequently acquired by Cubist). CB-220,404-AB-4 and CB-241,957-AD-2 came from Cubist Pharmaceuticals Inc., Lexington, MA. Simocyclinone D8 came from AdipoGen, San Diego, CA, and (R)-gyramide A came from Glixx Laboratories, Southborough, MA. Pyrazinyl-alkynyl-tetrahydroquinoline (PAT) was provided by AstraZeneca, Waltham, MA.
DNA gyrase supercoiling assay.
DNA supercoiling assays were performed with E. coli DNA gyrase and relaxed plasmid pUC19 in gyrase reaction buffer at pH 7.5 containing 35 mM Tris-HCl, 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol (DTT), 1.75 mM ATP, 5 mM spermidine, 0.1 mg/ml of bovine serum albumin (BSA), and 6.5% glycerol, all from New England BioLabs Inc., Ipswich, MA. The extent of supercoiling was gauged by the intensity of the lowest DNA band as measured with a Gel Doc EZ Imager (Bio-Rad, Hercules, CA). QnrB1 was purified from E. coli BL21(DE3) pET28a:QnrB1 as previously described (16).
Cross-streak assay.
The assay was adapted from a pyocin typing technique (18). The producer strain E. coli RYC1000 pMM39 was streaked diametrically across a glass petri dish containing Mueller-Hinton agar as a band about 1 cm wide. After overnight incubation, bacterial growth was removed with a glass slide, and 3 to 5 ml of CHCl3 was placed in the inverted lid and covered with the bottom of the dish for 15 min to kill residual organisms. CHCl3 in the lid was discarded, and the plate was opened for a few minutes to allow residual vapor to escape. Cultures of test organisms were then streaked at right angles to the original inoculum, and the plate was reincubated overnight.
RESULTS
Synthetic GyrA subunit inhibitors.
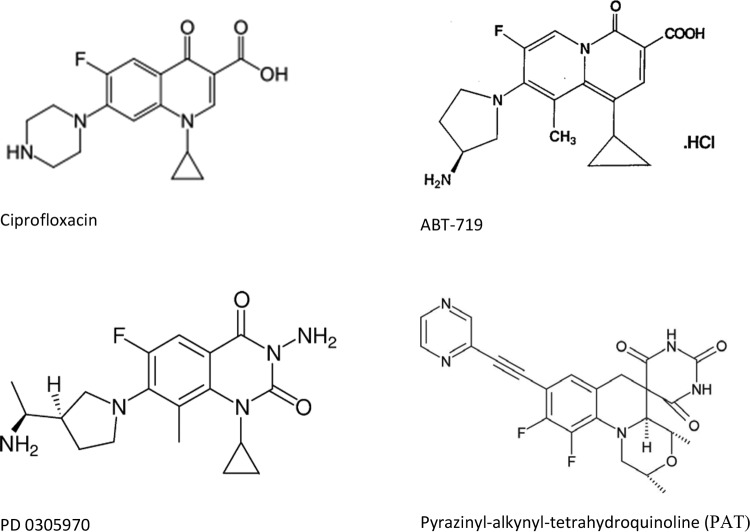
ABT-719 (Fig. 1) is a 2-pyridone inhibitor of bacterial DNA gyrase differing from a fluoroquinolone by placement of the nitrogen atom in the ring juncture (19, 20). Ten micrograms of ABT-719 added to a 6-mm blank disk produced an inhibitory zone of 13 mm on a lawn of E. coli J53 Azir but a 6-mm (no inhibition) zone with J53 Azir pMG252, indicating that qnrA1 protects against ABT-719.
FIG 1.
Chemical structures.
PD 0305970 is a quinazoline-2,4-dione (Fig. 1) that inhibits bacterial DNA gyrase and topoisomerase IV. It shares structural similarity with quinolones but remains effective against quinolone-resistant mutants of Streptococcus pneumoniae (21, 22). A 10-μg PD 0305970 disk produced an inhibitory zone of 29 mm on J53 Azir but only 18 mm with J53 Azir pMG252, indicating that qnrA1 also protects against PD0305970.
Pyrazinyl-alkynyl-tetrahydroquinoline (PAT) is a spiropyrimidinetrione (Fig. 1) that inhibits DNA gyrase and topoisomerase IV and retains activity against quinolone-resistant mutants of Staphylococcus aureus and S. pneumoniae (23). A 10-μg disk of PAT produced a 28-mm zone of inhibition with EW1b ΔtolC and a 24-mm zone with EW1b pMG253 containing the cloned qnrA1 gene. To confirm Qnr protection, the ability of purified QnrB1 to reverse PAT inhibition of gyrase supercoiling was studied in vitro. An 8 μM concentration of PAT inhibited gyrase activity by 78%, and QnrB reversed this inhibition with a 50% inhibitory concentration (IC50) of 14 nM (Fig. 2). Hence, Qnr also protects against compounds with a PAT-like structure.
FIG 2.
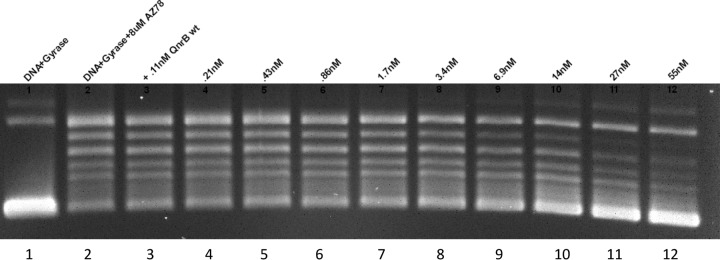
Protective effect of QnrB1 against gyrase inhibition by PAT. Lane 1, control reaction of relaxed pUC19 substrate and DNA gyrase. Lanes 2 to 12, DNA substrate, gyrase, and 8 μM PAT with no QnrB1 (lane 2) or with QnrB1 at concentrations of 0.11 nM (lane 3), 0.21 nM (lane 4), 0.43 nM (lane 5), 0.86 nM (lane 6),1.7 nM (lane 7), 3.4 nM (lane 8), 6.9 nM (lane 9), 14 nM (lane 10), 27 nM (lane 11), and 55 nM (lane 12).
Synthetic GyrB subunit inhibitors.
C3 and C4 are tricyclic pyrimidoindole inhibitors of the GyrB and ParE subunits (24). In E. coli J53 Azir or EW1b ΔtolC, QnrA, QnrB, or QnrS lacked a protective effect (Table 2).
TABLE 2.
MICs of test compounds with Qnr-containing strains
| E. coli strain | PMQRa | MIC (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cipro floxacin | Coumermycin A1 | Novobiocin | Gyramide A | Cubist CB-220 | Cubist CB-241 | Trius C3 | Trius C4 | ||
| J53 | 0.016 | 8 | 2,048 | >8 | >10 | >10 | 1.28 | 0.64 | |
| J53 pMG252 | qnrA1 | 0.512 | 16 | 512 | >8 | >10 | >10 | 1.28 | 0.64 |
| J53 pMG298 | qnrB1 aac(6′)Ib-cr | 0.512 | 16 | 2,048 | >8 | >10 | >10 | 0.64 | 0.64 |
| J53 pMG306 | qnrS1 | 2.048 | 16 | 512 | >8 | >10 | >10 | 1.28 | 0.64 |
| EW1b ΔtolC | 0.004 | 4 | 1 | 1.024 | 0.32 | 0.16 | 0.008 | 0.04 | |
| EW1b pMG253 | qnrA1 | 0.128 | 4 | 1 | 1.024 | 0.16 | 0.08 | 0.004 | 0.04 |
PMQR, plasmid-mediated quinolone resistance.
CB-220 and CB-241 are pyrazolopyridones that are also dual inhibitors of GyrB and ParE (25). QnrA, QnrB, or QnrS did not block their activity against E. coli with or without a tolC deletion (Table 2).
Natural product GyrA inhibitors.
Simocyclinone D8 is a chlorinated aminocoumarin linked to an angucyclic polyketide via a tetraene linker and a d-olivose sugar produced by Streptomyces antibioticus. Despite its aminocoumarin moiety, it does not inhibit GyrB ATPase activity but rather binds to the GyrA subunit near the QRDR, preventing DNA binding (14, 26). No inhibitory activity was evident with a 25-μmol simocyclinone D8 disk on a lawn of E. coli J53 Azir or EW1b, but in vitro, simocyclinone D8 was at least as potent as ciprofloxacin in inhibiting DNA gyrase supercoiling. QnrB1 did not block simocyclinone D8 inhibition of DNA gyrase, and QnrB1 at a high concentration (27) still inhibited the enzyme in the presence of simocyclinone (Fig. 3).
FIG 3.
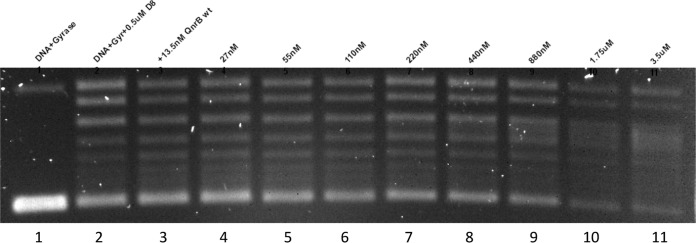
Lack of protection by QnrB1 against gyrase inhibition by simocyclinone D8. Lane 1, control reaction of pUC19 substrate and DNA gyrase. Lanes 2 to 11, DNA substrate, gyrase, and 0.5 μM simocyclinone D8 with no QnrB1 (lane 2) or with QnrB1 at concentrations of 13.5 nM (lane 3), 27 nM (lane 4), 55 nM (lane 5), 110 nM (lane 6), 220 nM (lane 7), 440 nM (lane 8), 880 nM (lane 9), 1.75 μM (lane 10), and 3.5 μM (lane 11).
Natural product GyrB inhibitors.
Coumermycin A1 is an aminocoumarin produced by a Streptomyces sp. Qnr had no protective effect (Table 2).
Novobiocin is another aminocoumarin produced by Streptomyces niveus. Qnr was not protective in whole cells (Table 2), just as QnrB1 failed to block novobiocin inhibition of DNA gyrase in vitro (16).
Gyramide A is an N-benzyl-3-sulfonamidopyrrolidine produced by an Streptomyces sp. It inhibits GyrB ATPase activity and produces chromosome condensation halting DNA replication and segregation (11). Qnr did not protect against gyramide A inhibition (Table 2).
Microcin B17 is a plasmid encoded, peptide-derived antibiotic containing oxazoles and thiazoles. It was tested by cross-streaking a microcin B17 producer with test strains. Figure 4 shows that E. coli producing microcin B17 is protected from its action, but E. coli J53 Azir with qnrA, qnrB, or qnrS was just as susceptible as the plasmid-free strain, and addition of IPTG to stimulate QnrB production when the gene was cloned in an IPTG-inducible expression vector failed to disclose a protective effect.
FIG 4.
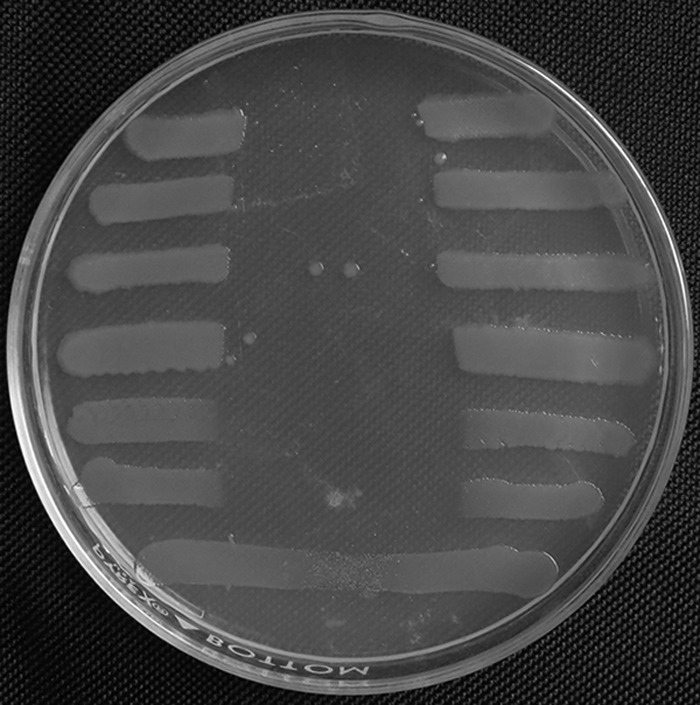
Cross-streak test for microcin B17 inhibition. The producer strain RYC1000 pMM39 was grown as a vertical band and removed, leaving microcin B17 in the agar, which was cross-streaked at right angles with (from the top) E. coli J53 Azir, J53 pMG298 (qnrB1), J53 pMG252 (qnrA1), J53 pMG306 (qnrS1), M15 pREP4 pQE-60, M15 pREP4 pQE-60-qnrB1, and RYC1000 pMM39 and reincubated. The Mueller-Hinton agar medium contained 100 μM IPTG to maximize QnrB1 production from expression plasmid pQE-60-qnrB1.
DISCUSSION
Qnr protects against all fluoroquinolones tested (28, 29) and at a lower level against nalidixic acid, technically a naphthyridone. Reduced susceptibility to these compounds maps to the QRDR of GyrA, where in E. coli amino acid substitutions at specific residues between amino acids 51 and 106 affect susceptibility. This is the cleavage-ligation region of the enzyme where gyrase forms a covalent bond with DNA and quinolone intercalation into DNA increases the concentration of cleavage complexes and facilitates formation of lethal double-strand DNA breaks (30, 31). In Gram-negative bacteria, the GyrB subunit and topoisomerase IV are more resistant to quinolones than GyrA, but once GyrA becomes less susceptible by mutation, mutations in GyrB at residues 426 and 447, in ParC at positions 78, 80, and 84, and in ParE at residue 445 can further reduce susceptibility (32). Qnr is a pentapeptide repeat protein that dimerizes and folds into a rod-like molecule with a size and surface charge similar to those of B-form DNA (16). It competes with quinolone for gyrase in vitro (3) and in cells lowers susceptibility to the level of a GyrA mutation, suggesting that it may interact with GyrA as a DNA mimic in the QRDR region or DNA gate. However, in a gel displacement assay (4, 5) or bacterial two-hybrid system (33), Qnr binds to GyrB as well as to GyrA and to both subunits of topoisomerase IV as well as to the holoenzymes, suggesting that a broader range of potential interactions is possible.
Table 3 summarizes information about the compounds tested to date for interaction with Qnr. Those that Qnr fails to protect against include coumermycin A1, novobiocin, and gyramide A, which act at the ATPase site of GyrB or ParE. Resistance to the aminocoumarin compounds in both E. coli (34) and S. aureus (35) map in GyrB at sites distinct from those contributing to quinolone resistance. Similarly, resistance to gyramide A, another competitive inhibitor of GyrB ATPase activity, occurs at sites separate from those for both quinolones and aminocoumarins. Qnr also failed to protect against synthetic GyrB ATPase inhibitors, such as pyrazolopyridone or tricyclic pyrimidoindoles compounds. Lack of protection against the GyrB targeting microcin B17 is interesting because organisms that produce this toxin protect themselves in part by coproduction of McbG, a pentapeptide repeat protein with antiquinolone activity (D. Hooper and J.-L. Yu, personal communication) that blocks gyrase inhibition by the microcin. Evidently the pentapeptide repeat Qnr proteins lack this microcin protective effect.
TABLE 3.
Summary of gyrase inhibitors tested for Qnr protection
| Compound(s) | Target | Stabilization of covalent complex | ATPase inhibition | Inhibition of topoisomerase IV | Resistance mutations (in E. coli unless otherwise specified) | Qnr Protection | Reference(s) |
|---|---|---|---|---|---|---|---|
| CcdB | Binds as a dimer to the dimerization domain of GyrA while strand passage takes place | Yes | No | GyrA mutation at amino acid 462 with no cross-resistance with quinolone | No | 37–39, 50 | |
| Coumermycin A1 | Competitve inhibitor of GyrB and ParE ATPase activity | No | Yes | Yes | GyrB mutations at sites other than those associated with quinolone resistance: amino acids 73, 77, 136, 164, and 165 | No | 34, 35, 51, 52 |
| Gyramide A | Competitive inhibitor of GyrB ATPase activity | No | Yes | No | At sites in GyrA (amino acids 34, 35, 45, 96, 97, 98, 169, 170, 172, 173, 267, and 335) and GyrB (amino acid 508) without effect on quinolone or novobiocin susceptibility | No | 11, 53 |
| Microcin B17 | Binds to C-terminal domain of GyrB, slowing DNA strand passage | Yes | No | GyrB mutation at amino acid 751 confers resistance, but some quinolone resistance mutations at amino acid 83 in GyrA or amino acid 447 in GyrB provide partial resistance | No | 12, 54, 55 | |
| Novobiocin | Competitve inhibitor of GyrB and ParE ATPase activity | No | Yes | Yes | GyrB mutations at sites other than those associated with quinolone resistance: amino acids 73, 77, 136, 164, and 165 | No | 16, 34, 35, 51, 52 |
| ParE toxin | DNA gyrase | Yes | No | 37, 56 | |||
| Simocyclinone D8 | N-terminal domain of GyrA, preventing DNA binding. Second weak binding site on C terminus of GyrB. | No | No | No | GyrA mutations (amino acids 42, 44, 45, 80, 81, 83, 84, 87, 91, and 120), some of which confer quinolone resistance | No | 13, 14, 26, 57–59 |
| ABT-719 | DNA gyrase | Yes | Yes | 19 | |||
| PD 0305970 | DNA gyrase | Yes | Yes | In S. pneumoniae mutations in toprim domain of GyrB at amino acids 456 and 474 or in ParE at 435 and 475. No cross-resistance with several quinolone resistance mutations. | Yes | 21, 22, 30 | |
| PAT | DNA gyrase | Yes | No cross-resistance with quinolones in S. aureus or S. pneumoniae | Yes | 23, 60, 61 | ||
| Pyrazolopyridones | GyrB | No | 25 | ||||
| Quinolones | GyrA, ParC, GyrB, ParE with gyrase more susceptible than topoisomerase IV for E. coli | Yes | No | Yes | QRDR GyrA: amino acids 51, 67, 81, 82, 83, 84, 87, and 106. QRDR GyrB: amino acids 426 and 447. QRDR ParC: amino acids 78, 80, and 84. QRDR ParE: amino acid 445. | Yes | 3–5, 32 |
| Tricyclic pyrimidoindoles | GyrB and ParE at the ATP binding site | Yes | Yes | No | 24 |
Qnr does protect against the synthetic compounds shown in Fig. 1, which have some structural similarity to a quinolone. Detailed structural analysis is available for the interaction of the dione PD 0305970 with a partially reconstructed topoisomerase IV of Streptococcus pneumoniae (30) and a similar quinazoline dione with topoisomerase IV of Bacillus anthracis (36). Quinolone and dione form similar cleavage complexes at the DNA gate but differ in their binding to neighboring amino acid residues, thus explaining the lack of cross-resistance between the two inhibitors. Evidently Qnr does not recognize such differences and blocks gyrase inhibition by both agents. ABT-719 and PAT probably have binding sites sufficiently near the quinolone binding site so that Qnr can block their action as well. The CcdB and ParE toxins of plasmid addiction systems have recently been shown to act without inhibition by Qnr (37). The mechanism of CcdB toxicity has been studied in greater detail than that of ParE (38, 39). CcdB binding requires gyrase to be in an open confirmation. The toxin also inhibits gyrase by stabilizing the cleavage complex but differs from quinolones in the site of resistance mutations. Lack of protection against simocyclinone D8 is intriguing because this agent binds to the N-terminal domain of GyrA, like quinolones, with mutations at such GyrA residues as 81, 83, 84, and 87 providing resistance to both agents (14). Furthermore, simocyclinone blocks DNA binding such that if Qnr functions as a DNA mimic, competition between Qnr and simocylinone would be expected. Nevertheless, none was seen. Not only did QnrB not prevent simocyclinone inhibition of gyrase supercoiling, but also it failed to prevent gyrase inhibition seen with QnrB at high concentrations (27, 40). Thus, no nonsynthetic agent has been found for which Qnr provides protection, although several potential candidates have not been available for testing, including albicidin (41, 42), clerocidin (43, 44), and cystobactamid (45).
Qnr reduces quinolone susceptibility but not to the CLSI-defined breakpoint. It does, however, facilitate the selection of more quinolone-resistant mutants (1). Lack of protection by Qnr is thus a desirable property for a new therapeutic agent, and the results of this study suggest which compounds that target DNA gyrase are likely to escape this effect.
ACKNOWLEDGMENTS
We thank the suppliers of the proprietary compounds used in this study.
This work was supported by grant R01 AI057576 from the U.S. Public Health Service, National Institutes of Health, to D.C.H. and G.A.J.
REFERENCES
- 1.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 2.Drlica K, Hooper DC. 2003. Mechanisms of quinolone action, p 19–40. In Hooper DC, Rubinstein E (ed), Quinolone antimicrobial agents, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 3.Tran JH, Jacoby GA. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A 99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother 49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother 49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Liard A, Rodriguez-Martinez JM, Nordmann P. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother 56:1118–1121. doi: 10.1093/jac/dki371. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Martínez JM, Velasco C, Briales A, García I, Conejo MC, Pascual A. 2008. Qnr-like pentapeptide repeat proteins in gram-positive bacteria. J Antimicrob Chemother 61:1240–1243. doi: 10.1093/jac/dkn115. [DOI] [PubMed] [Google Scholar]
- 8.Jacoby GA, Hooper DC. 2013. Phylogenetic analysis of chromosomally determined Qnr and related proteins. Antimicrob Agents Chemother 57:1930–1934. doi: 10.1128/AAC.02080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gellert M, O'Dea MH, Itoh T, Tomizawa J. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A 73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper DC, Wolfson JS, McHugh GL, Winters MB, Swartz MN. 1982. Effects of novobiocin, coumermycin A1, clorobiocin, and their analogs on Escherichia coli DNA gyrase and bacterial growth. Antimicrob Agents Chemother 22:662–671. doi: 10.1128/AAC.22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajendram M, Hurley KA, Foss MH, Thornton KM, Moore JT, Shaw JT, Weibel DB. 2014. Gyramides prevent bacterial growth by inhibiting DNA gyrase and altering chromosome topology. ACS Chem Biol 9:1312–1319. doi: 10.1021/cb500154m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks WM, Bottrill AR, Pierrat OA, Durrant MC, Maxwell A. 2007. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie 89:500–507. doi: 10.1016/j.biochi.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Sissi C, Vazquez E, Chemello A, Mitchenall LA, Maxwell A, Palumbo M. 2010. Mapping simocyclinone D8 interaction with DNA gyrase: evidence for a new binding site on GyrB. Antimicrob Agents Chemother 54:213–220. doi: 10.1128/AAC.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearnshaw SJ, Edwards MJ, Stevenson CE, Lawson DM, Maxwell A. 2014. A new crystal structure of the bifunctional antibiotic simocyclinone D8 bound to DNA gyrase gives fresh insight into the mechanism of inhibition. J Mol Biol 426:2023–2033. doi: 10.1016/j.jmb.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Bogaki M, Nakamura M, Nakamura S. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother 34:1271–1272. doi: 10.1128/AAC.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS. 2011. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J Biol Chem 286:25265–25273. doi: 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed CLSI document M07-A9. CLSI, Wayne, PA. [Google Scholar]
- 18.Gillies RR, Govan JR. 1966. Typing of Pseudomonas pyocyanea by pyocine production. J Pathol Bacteriol 91:339–345. doi: 10.1002/path.1700910207. [DOI] [PubMed] [Google Scholar]
- 19.Flamm RK, Vojtko C, Chu DT, Li Q, Beyer J, Hensey D, Ramer N, Clement JJ, Tanaka SK. 1995. In vitro evaluation of ABT-719, a novel DNA gyrase inhibitor. Antimicrob Agents Chemother 39:964–970. doi: 10.1128/AAC.39.4.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alder J, Clement J, Meulbroek J, Shipkowitz N, Mitten M, Jarvis K, Oleksijew A, Hutch T Sr, Paige L, Flamm B. 1995. Efficacies of ABT-719 and related 2-pyridones, members of a new class of antibacterial agents, against experimental bacterial infections. Antimicrob Agents Chemother 39:971–975. doi: 10.1128/AAC.39.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huband MD, Cohen MA, Zurack M, Hanna DL, Skerlos LA, Sulavik MC, Gibson GW, Gage JW, Ellsworth E, Stier MA, Gracheck SJ. 2007. In vitro and in vivo activities of PD 0305970 and PD 0326448, new bacterial gyrase/topoisomerase inhibitors with potent antibacterial activities versus multidrug-resistant gram-positive and fastidious organism groups. Antimicrob Agents Chemother 51:1191–1201. doi: 10.1128/AAC.01321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan XS, Gould KA, Fisher LM. 2009. Probing the differential interactions of quinazolinedione PD 0305970 and quinolones with gyrase and topoisomerase IV. Antimicrob Agents Chemother 53:3822–3831. doi: 10.1128/AAC.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kern G, Basarab GS, Andrews B, Schuck V, Stone G, Kutschke A, Beaudoin M-E, San Martin M, Brassil P, Fan JH, Mills SD, Doig P. 2011. A DNA gyrase inhibitor with a novel mode of inhibition and in vivo efficacy, abstr F1-1840. Abstr 51st Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 24.Tari LW, Li X, Trzoss M, Bensen DC, Chen Z, Lam T, Zhang J, Lee SJ, Hough G, Phillipson D, Akers-Rodriguez S, Cunningham ML, Kwan BP, Nelson KJ, Castellano A, Locke JB, Brown-Driver V, Murphy TM, Ong VS, Pillar CM, Shinabarger DL, Nix J, Lightstone FC, Wong SE, Nguyen TB, Shaw KJ, Finn J. 2013. Tricyclic GyrB/ParE (TriBE) inhibitors: a new class of broad-spectrum dual-targeting antibacterial agents. PLoS One 8:e84409. doi: 10.1371/journal.pone.0084409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Cross J, Yang Q, Mesleh M, Romero J, Wang B, Bevan D, Hall K, Epie F, Moy T, Daniel A, Shotwell J, Chamberlain B, Carter N, Ryan D, Metcalf C, Silverman J, Nguyen K, Lippa B, Dolle R. 2013. The discovery of pyrazolopyridones as a novel class of gyrase B inhibitors through fragment- and structure-based drug discovery, abstr F-1224. Abstr 53rd Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 26.Edwards MJ, Flatman RH, Mitchenall LA, Stevenson CE, Le TB, Clarke TA, McKay AR, Fiedler HP, Buttner MJ, Lawson DM, Maxwell A. 2009. A crystal structure of the bifunctional antibiotic simocyclinone D8, bound to DNA gyrase. Science 326:1415–1418. doi: 10.1126/science.1179123. [DOI] [PubMed] [Google Scholar]
- 27.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Martínez L, Pascual A, García I, Tran J, Jacoby GA. 2003. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother 51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Sahm DF, Jacoby GA, Zhang Y, Hooper DC. 2004. Activities of newer quinolones against Escherichia coli and Klebsiella pneumoniae containing the plasmid-mediated quinolone resistance determinant qnr. Antimicrob Agents Chemother 48:1400–1401. doi: 10.1128/AAC.48.4.1400-1401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. 2010. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper DC. 2003. Mechanisms of quinolone resistance, p 41–67. In Hooper DC, Rubinstein E (ed), Quinolone antimicrobial agents, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 33.Kim ES, Chen C, Braun M, Kim HY, Okumura R, Wang Y, Jacoby GA, Hooper DC. 22 June 2015. Interactions between QnrB, QnrB mutants, and DNA gyrase. Antimicrob Agents Chemother doi: 10.1128/AAC.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross CH, Parsons JD, Grossman TH, Charifson PS, Bellon S, Jernee J, Dwyer M, Chambers SP, Markland W, Botfield M, Raybuck SA. 2003. Active-site residues of Escherichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob Agents Chemother 47:1037–1046. doi: 10.1128/AAC.47.3.1037-1046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto-Nakamura M, Ito H, Oyamada Y, Nishino T, Yamagishi J. 2005. Accumulation of mutations in both gyrB and parE genes is associated with high-level resistance to novobiocin in Staphylococcus aureus. Antimicrob Agents Chemother 49:3810–3815. doi: 10.1128/AAC.49.9.3810-3815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aldred KJ, McPherson SA, Wang P, Kerns RJ, Graves DE, Turnbough CL Jr, Osheroff N. 2012. Drug interactions with Bacillus anthracis topoisomerase IV: biochemical basis for quinolone action and resistance. Biochemistry 51:370–381. doi: 10.1021/bi2013905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak YG, Jacoby GA, Hooper DC. 2015. Effect of Qnr on plasmid gyrase toxins CcdB and ParE. Antimicrob Agents Chemother 59:5078–5079. doi: 10.1128/AAC.00524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dao-Thi MH, Van Melderen L, De Genst E, Afif H, Buts L, Wyns L, Loris R. 2005. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 348:1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 39.Smith AB, Maxwell A. 2006. A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res 34:4667–4676. doi: 10.1093/nar/gkl636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mérens A, Matrat S, Aubry A, Lascols C, Jarlier V, Soussy CJ, Cavallo JD, Cambau E. 2009. The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J Bacteriol 191:1587–1594. doi: 10.1128/JB.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimi SM, Wall MK, Smith AB, Maxwell A, Birch RG. 2007. The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob Agents Chemother 51:181–187. doi: 10.1128/AAC.00918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cociancich S, Pesic A, Petras D, Uhlmann S, Kretz J, Schubert V, Vieweg L, Duplan S, Marguerettaz M, Noell J, Pieretti I, Hugelland M, Kemper S, Mainz A, Rott P, Royer M, Sussmuth RD. 19 January 2015. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat Chem Biol doi: 10.1038/nchembio.1734. [DOI] [PubMed] [Google Scholar]
- 43.McCullough JE, Muller MT, Howells AJ, Maxwell A, O'Sullivan J, Summerill RS, Parker WL, Wells JS, Bonner DP, Fernandes PB. 1993. Clerocidin, a terpenoid antibiotic, inhibits bacterial DNA gyrase. J Antibiot (Tokyo) 46:526–530. doi: 10.7164/antibiotics.46.526. [DOI] [PubMed] [Google Scholar]
- 44.Pan XS, Dias M, Palumbo M, Fisher LM. 2008. Clerocidin selectively modifies the gyrase-DNA gate to induce irreversible and reversible DNA damage. Nucleic Acids Res 36:5516–5529. doi: 10.1093/nar/gkn539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann S, Herrmann J, Raju R, Steinmetz H, Mohr KI, Huttel S, Harmrolfs K, Stadler M, Muller R. 2014. Cystobactamids: myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew Chem Int Ed Engl 53:14605–14609. doi: 10.1002/anie.201409964. [DOI] [PubMed] [Google Scholar]
- 46.Jacoby GA, Han P. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitney EN. 1971. The tolC locus in Escherichia coli K12. Genetics 67:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrido MC, Herrero M, Kolter R, Moreno F. 1988. The export of the DNA replication inhibitor Microcin B17 provides immunity for the host cell. EMBO J 7:1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, Medalla F, Chiller TM, Hooper DC. 2006. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis 43:297–304. doi: 10.1086/505397. [DOI] [PubMed] [Google Scholar]
- 50.Bernard P, Couturier M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226:735–745. doi: 10.1016/0022-2836(92)90629-X. [DOI] [PubMed] [Google Scholar]
- 51.Contreras A, Maxwell A. 1992. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol Microbiol 6:1617–1624. doi: 10.1111/j.1365-2958.1992.tb00886.x. [DOI] [PubMed] [Google Scholar]
- 52.Fournier B, Hooper DC. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother 42:121–128. doi: 10.1093/jac/42.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foss MH, Hurley KA, Sorto N, Lackner LL, Thornton KM, Shaw JT, Weibel DB. 2011. N-Benzyl-3-sulfonamidopyrrolidines are a new class of bacterial DNA gyrase inhibitors. ACS Med Chem Lett 2:289–292. doi: 10.1021/ml1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vizán JL, Hernández-Chico C, del Castillo I, Moreno F. 1991. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J 10:467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heddle JG, Blance SJ, Zamble DB, Hollfelder F, Miller DA, Wentzell LM, Walsh CT, Maxwell A. 2001. The antibiotic microcin B17 is a DNA gyrase poison: characterisation of the mode of inhibition. J Mol Biol 307:1223–1234. doi: 10.1006/jmbi.2001.4562. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Y, Pogliano J, Helinski DR, Konieczny I. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 57.Schimana J, Fiedler HP, Groth I, Sussmuth R, Beil W, Walker M, Zeeck A. 2000. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tu 6040. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot (Tokyo) 53:779–787. [DOI] [PubMed] [Google Scholar]
- 58.Oppegard LM, Hamann BL, Streck KR, Ellis KC, Fiedler HP, Khodursky AB, Hiasa H. 2009. In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob Agents Chemother 53:2110–2119. doi: 10.1128/AAC.01440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flatman RH, Howells AJ, Heide L, Fiedler HP, Maxwell A. 2005. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob Agents Chemother 49:1093–1100. doi: 10.1128/AAC.49.3.1093-1100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller AA, Bundy GL, Mott JE, Skepner JE, Boyle TP, Harris DW, Hromockyj AE, Marotti KR, Zurenko GE, Munzner JB, Sweeney MT, Bammert GF, Hamel JC, Ford CW, Zhong WZ, Graber DR, Martin GE, Han F, Dolak LA, Seest EP, Ruble JC, Kamilar GM, Palmer JR, Banitt LS, Hurd AR, Barbachyn MR. 2008. Discovery and characterization of QPT-1, the progenitor of a new class of bacterial topoisomerase inhibitors. Antimicrob Agents Chemother 52:2806–2812. doi: 10.1128/AAC.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basarab GS, Brassil P, Doig P, Galullo V, Haimes HB, Kern G, Kutschke A, McNulty J, Schuck VJ, Stone G, Gowravaram M. 2014. Novel DNA gyrase inhibiting spiropyrimidinetriones with a benzisoxazole scaffold: SAR and in vivo characterization. J Med Chem 57:9078–9095. doi: 10.1021/jm501174m. [DOI] [PubMed] [Google Scholar]