Abstract
The mode of action of a group of glycosylated antimicrobial peptides known as glycocins remains to be elucidated. In the current study of one glycocin, sublancin, we identified the phosphoenolpyruvate:sugar phosphotransferase system (PTS) of Bacillus species as a key player in bacterial sensitivity. Sublancin kills several Gram-positive bacteria, such as Bacillus species and Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA). Unlike other classes of bacteriocins for which the PTS is involved in their mechanism of action, we show that the addition of PTS-requiring sugars leads to increased resistance rather than increased sensitivity, suggesting that sublancin has a distinct mechanism of action. Collectively, our present mutagenesis and genomic studies demonstrate that the histidine-containing phosphocarrier protein (HPr) and domain A of enzyme II (PtsG) in particular are critical determinants for bacterial sensitivity to sublancin.
INTRODUCTION
Bacteriocins are ribosomally synthesized peptides produced by a wide range of bacterial species. These bacteriocins endow the producing bacteria with a competitive advantage in their respective niche. Many bacteriocins are heavily posttranslationally processed during their biosynthesis, and these modifications are required for activity (1). Nisin is the best-studied bacteriocin and belongs to the lantibiotic family (2). The mode of action of nisin involves binding to lipid II, which prevents further cell wall synthesis, followed by the formation of pores within the membrane. The leakage of essential metabolites from these cells results in death of the bacteria. Targeting of lipid II by bacteriocins is a common mechanism of action (3–5). Other mechanisms include the targeting of phosphotransferase systems (6, 7), acting as Trojan horses (8, 9), parasitizing iron uptake pathways (10) and causing the collapse of membrane potential, together with leakage of ions and/or a decrease in intracellular ATP concentrations (11). There is much interest in bacteriocins for use in the control of bacterial infections and therefore in their mechanisms of action.
Sublancin is a bacteriocin produced by the Gram-positive soil bacterium Bacillus subtilis strain 168. It is capable of killing several species of Gram-positive bacteria, such as Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA) (12). Sublancin is encoded by the sunA gene on the SPβ prophage as a prepeptide (13). The genes necessary for the synthesis of sublancin are also included in this region and are expressed from two promoters. The biosynthetic operon is made up of five individual genes, which are responsible for producing active sublancin. The sunT gene is responsible for the export of sublancin and cleavage of its leader sequence. Two thiol-disulfide oxidoreductases, encoded by bdbA (the only gene of the operon that is dispensable for active sublancin production) and bdbB, are responsible for creating the two disulfide bonds of sublancin. These disulfide bonds involve four of the five cysteine residues that are present in the sublancin peptide (14). The fifth cysteine residue undergoes glucosylation by the glucosyltransferase encoded by the sunS gene (15). The second promoter drives the expression of a gene encoding the immunity protein SunI, which is also required for the production of active sublancin by protecting the producing organism from sublancin (16).
Sublancin is one of five bacteriocins that have been described as being S-glycosylated. Glycocin F is produced by Lactobacillus plantarum (17), ASM1 is produced by Lactobacillus plantarum strain A-1 and is an orthologue of glycocin F with five different residues in the C-terminal tail (18, 19), and the putative products of Bacillus thuringiensis, thurandacin A and B, have been produced in vitro (20). Sublancin is modified on Cys22, with a β-S-linked glucose (15, 21). For glycocin F, an S-linked N-acetylglucosamine (GlcNAc) is added to the C-terminal cysteine residue, and the other modification is on an internal serine residue with an O-linked GlcNAc (17, 22). Like glycocin F, thurandacin B is glucosylated on two residues: a cysteine residue undergoes S-linked glucosylation, and a serine is modified with an O-linked glucose moiety (20). Judged by homology searches to the bacteriocin sequence or the glucosyltransferase, many other Gram-positive bacteria appear to potentially encode such bacteriocins, with much variation in the sequences among them (15, 17).
The mechanisms by which glycosylated bacteriocins kill sensitive cells are currently unknown. Previous work has identified several genes in B. subtilis and S. aureus that alter sensitivity to sublancin. The mscL gene encodes the large mechanosensitive channel, and its deletion confers sublancin resistance in both S. aureus and B. subtilis (23). The addition of increased amounts of NaCl also results in increased resistance to sublancin, presumably due to the MscL channel being forced closed. This observation has led to speculation as to whether sublancin is able to enter the cell through this channel. Interestingly, since the connection between sublancin and MscL was reported, streptomycin has also been reported to use the MscL channel to enter the cell (24). In B. subtilis, the alternative sigma factor σW is known to play a role in the resistance to sublancin through its regulation of the yqeZ-yqfA-yqfB operon (25). The role these genes play in resistance to sublancin is unknown, but it is likely to be at the cell surface, due to their membrane localization (26).
In this study, we demonstrate that the phosphoenolpyruvate:sugar phosphotransferase system (PTS) of B. subtilis plays a major role in its sensitivity to sublancin. In the case of other bacteriocins for which the PTS was found to be involved, the addition of the PTS-requiring sugars resulted in increased sensitivity to the respective bacteriocin. However, for sublancin, the addition of PTS-requiring sugars leads to increased resistance, suggesting that sublancin has a distinct mechanism of action.
MATERIALS AND METHODS
Bacterial growth.
See Table 1 for the strains used in this study. B. subtilis 168, B. subtilis strain ATCC 6633, and Bacillus halodurans strain C-125 were grown in lysogeny broth (LB) at 37°C with vigorous shaking (250 rpm) on LB agar plates. B. subtilis was also grown on M9 agar plates (as described previously [27] but with the addition of 1.5% final concentration agar) with and without the addition of sugars at a final concentration of 0.3% glucose, 0.4% malate, or 0.4% citrate, as specified below. The LB agar used for sublancin inhibition plate assays did not include NaCl. Antibiotics were used for selection when necessary at the following concentrations: 100 μg/ml spectinomycin, 20 μg/ml kanamycin, 4 μg/ml phleomycin, 5 μg/ml chloramphenicol, and 2 μg/ml erythromycin. Stock sublancin solutions were prepared using phosphate-buffered saline (PBS).
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| 168 | Wild-type B. subtilis strain | Laboratory collection |
| ΔSPβ | Wild-type B. subtilis 168 strain lacking the entire SPβ prophage | 14 |
| C-125 | Wild-type B. halodurans strain | 56 |
| ATCC 6633 | Wild-type B. subtilis strain | 57 |
| 168 deletion collection | Collection of B. subtilis mutants lacking large genomic regions | 29 |
| ΔSPβ::cat | Chloramphenicol-selectable ΔSPβ prophage mutant of B. subtilis | 14 |
| ΔyvkS-yvkW | B. subtilis ΔyvkS-yvkW::phleo | This study |
| ΔykvY-glcT | B. subtilis ΔykvY-glcT::phleo | This study |
| ΔptsG-ptsI | B. subtilis ΔptsG-ptsI::phleo | This study |
| ΔSPβ-QB5435 | B. subtilis ΔpstG::cat; QB5435→SPβ | 48 |
| ΔSPβ-MZ303 | B. subtilis ΔptsH::cat; MZ303→SPβ | 58 |
| ΔSPβ-GP864 | B. subtilis ΔptsI::ermC; GP864→SPβ | 59 |
| ΔSPβ-QB5407 | B. subtilis ΔccpA::spec; QB5407→SPβ | 60 |
| ΔSPβ-GP202 | B. subtilis ΔhprK::spec; GP202→SPβ | 61 |
| ΔSPβ::cm-GP506 | B. subtilis ptsH H15A; SPβcm→GP506 | 62 |
| ΔSPβ::cm-GP507 | B. subtilis ptsH S46A; SPβcm→GP576 | 63 |
Production and isolation of sublancin 168.
The purification of sublancin from its natural producer, B. subtilis 168, was performed as previously reported (15).
Strain construction.
Chromosomal DNA was prepared from B. subtilis 168 using a standard procedure, as described previously (28). Deletion mutants in B. subtilis 168 were created as described by Tanaka et al. (29), and the oligonucleotides used are shown in Table S1 in the supplemental material. B. subtilis 168 was transformed using PCR products or chromosomal DNA, according to a standard procedure (30).
MIC determination.
MICs were determined by the broth dilution method (31). Serial dilutions of sublancin were prepared in sterile deionized water (SDW). Forty-eight-well microtiter plates (Costar; Corning) were utilized for both B. subtilis ATCC 6633 and B. halodurans C-125. The total volume of culture in each well was 300 μl; the experimental wells contained 30 μl of 10× stock sublancin at defined concentrations and 270 μl of a 1: 10 dilution (approximately 1 × 108 CFU ml−1) of a culture of an indicator strain diluted in fresh LB growth medium. In addition, each plate contained several blanks (270 μl of fresh growth medium and 30 μl of SDW) and control wells (270 μl of untreated culture diluted 1:10 and 30 μl of SDW). The optical density at 600 nm (OD600) was recorded at hourly intervals from 0 to 6 h, with an additional measurement at 18 h using a BioTek Synergy H4 plate reader. The plates were incubated under vigorous agitation at 37°C. The readings from triplicate experiments were averaged. Growth curves were developed using control (culture and SDW only) readings to ensure sufficient OD changes for accurate inhibition assessment. Curve fits for MIC determination were produced by fitting the data with the Origin8.5 software, using a dose-response curve with the equation y = A1 + (A2 − A1)/(1 + 10[logx0 − x]p), where p is the variable Hill slope.
Sublancin killing kinetics against sensitive Bacillus species.
Sensitive cultures were grown to mid-log phase in LB medium, as described above, transferred to 48-well microtiter plates (Costar; Corning), and exposed to sublancin at 1× and 4× the MIC. Immediately after the addition of sublancin, the OD600 was determined using a BioTek Synergy H4 plate reader. The cultures were incubated for 6 h, and the OD600 was recorded every 30 min. To verify that cells were killed, CFU counting was performed by serial dilution and plating.
Sublancin sensitivity screen of a gene deletion collection of B. subtilis.
Sublancin-induced growth inhibition assays were performed using the procedure described by Dorenbos et al. (14) but with a modification to enable the screening of large numbers of strains. Overnight cultures of B. subtilis mutants and a background control strain were grown in 96-well microtiter plates in a plate shaker at 37°C with shaking at 800 rpm. Bioassay dishes were prepared with LB agar without adding salt. The plates were thoroughly dried before being divided into 48 squares for inoculation. Cotton swabs were dipped into individual wells of the overnight culture before being spread on the appropriate square. The plates were allowed to dry before spotting 2 μl of an overnight culture of the B. subtilis 168 wild-type strain in the center of each inoculated square. The plates were incubated overnight, and visual analysis was used to determine zones of growth inhibition that were smaller or larger than that of the background strain. Strains with altered zones of inhibition were checked a further three times to ensure that the phenotype was reproducible.
Sublancin sensitivity assay in liquid medium.
Overnight cultures of B. subtilis grown in LB were diluted 1:100 in the same medium and grown to an OD600 of 0.5. The bacteria were then diluted 1:20 in a 96-well microtiter plate before growth was monitored every 10 min in a Synergy 4 BioTek plate reader (at 37°C with shaking). When the bacteria reached an OD600 of 0.185 (equivalent to 0.5 for a 1-cm path length), sublancin was added at the desired concentration before resuming the monitoring of growth. Sugars were added at the following final concentrations: 0.3% glucose, 0.4% malate, 0.3% sucrose, 0.3% fructose, 0.3% glycerol, 0.4% citrate, 0.4% galactose, and 0.4% succinate.
Membrane integrity assay.
B. subtilis was grown to an OD600 of 0.5 before purified sublancin was added at different concentrations (100 to 500 nM). As a positive control, nisin was added at 10 nM final concentration, and a negative-control sample contained no bacteriocin. Samples were taken at 30 and 90 min and prepared for LIVE/DEAD BacLight analysis (Molecular Probes, Life Technologies) (32). Samples were monitored by flow cytometry using an Accuri C6. The percentages of cells with intact or reduced membrane integrity were calculated.
Propidium iodide uptake.
Membrane integrity was also evaluated by measuring the uptake of propidium iodide (PI). B. subtilis ATCC 6633 and B. halodurans C-125 cultures were grown to a density of 4 × 106 cells ml−1 and then diluted with fresh LB medium to an OD600 of 0.1. Cells were transferred to tubes containing PI (final concentration, 25 μM; Molecular Probes, Inc., Leiden, the Netherlands), HEPES (1 mM), and sublancin (0, 0.2, 2.0, or 20 μM) or nisin (0, 0.2, 2.0, or 20 μM), incubated for 30 min at room temperature (RT), and analyzed. Data acquisition was performed with a BD Biosciences LSR II flow cytometer, using excitation at 488 nm with an argon laser and measurement of emission through a band-pass filter at 695/40 nm. A minimum of 50,000 events were detected for each sample, and the experiments were performed in triplicate. Data analysis to calculate the geometric mean fluorescence intensity (MFI) of gated cell populations was performed using FCS Express 3.00.0311 V Lite stand-alone software.
Generation of stable sublancin-resistant mutants and resistance frequency determination.
Genetically stable sublancin-resistant mutants were generated by growing the sublancin-susceptible strains B. halodurans C-125 and B. subtilis ATCC 6633 in LB, as described above (no additional sugars added), until an OD600 of 1.0 was reached (mid-log phase, 1-cm light path). The cultures were plated on agar plates containing 1× or 4× their respective sublancin MICs. Distinct colonies were observed by 24 h. Resistant colonies were picked, grown in LB, and plated on LB plates containing sublancin at 4× the MIC to confirm resistance. This procedure generated genetically stable sublancin-resistant mutants. The number of resistant mutants that emerged from each sublancin-susceptible culture was obtained by generating a serial dilution of each culture. Each dilution was plated on sublancin-containing plates. The total number of cells was determined by plating an appropriate (10−5, 10−6, 10−7, 10−8, or 10−9) dilution of the cultures on nonselective LB agar medium. Colonies from sublancin-containing and nonselective plates were counted after 24 h of incubation. The resistance frequency was determined as the mean number of resistant cells divided by the total number of viable cells per culture.
Single nucleotide polymorphisms detected by whole-genome sequencing.
Genomic DNA (gDNA) of the wild-type B. halodurans C-125 and four different sublancin-resistant isolates was extracted using an UltraClean microbial DNA isolation kit (Mo Bio). The gDNAs thus obtained were sequenced using a HiSeq 2000 Illumina sequencer, which generated close to 180 million single reads per lane, for an overall coverage of 360× for the 5-Mb genomes. All libraries were individually barcoded and constructed with the TruSeq sample prep kits (Illumina). The single nucleotide polymorphisms (SNPs) and the corresponding genes for resistant B. halodurans C-125 were identified. In addition, the wild-type B. halodurans C-125 strain was mapped to the published B. halodurans C-125 sequence (GenBank accession no. NC_002570.2) with CLC Genomics Workbench (CLC bio), using default parameters. A consensus sequence of the wild-type and reference genomes was obtained and used for SNP detection in sublancin-resistant mutants of B. halodurans.
PCR amplification and validation of SNPs in sublancin-resistant B. halodurans mutants.
PCR validation can serve as an iterative and informative process to modify and optimize the SNP filtering criteria to improve SNP calling (33). Primers flanking SNP-containing genes were synthesized and used for PCR amplification of the respective genes. The mutations reported here were all confirmed by PCR (see Table S2 in the supplemental material).
B. halodurans C-125 gene expression profile.
A culture of B. halodurans C-125 was grown in LB at 37°C with vigorous shaking until mid-log phase, at which point the culture was split into two 150-ml cultures, with one subjected to a subinhibitory concentration of sublancin (0.5× the MIC). RNA isolation was performed using the RNeasy minikit (Qiagen) and subsequent treatment with RNase-free DNase (Qiagen). The RNA was dissolved in RNase-free water and quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific). For each sample (i.e., with and without sublancin), 20 μg of total RNA was isolated from three biological replicates. cDNA synthesis was performed using the SuperScript double-stranded cDNA synthesis kit (Invitrogen), as per the manufacturer's instructions (NimbleGen arrays user's guide, version 5.1 [34]) and quantified with the NanoDrop instrument. Total cDNA was labeled overnight with the one-color DNA labeling kit (NimbleGen), as per the manufacturer's instructions. The arrays were scanned using an Axon 4000B array scanner.
A B. halodurans C-125 NimbleGen custom array, containing a probe set of 22 unique 45-mer to 60-mer oligonucleotide probes for each of the 4,066 genes of this bacterium, was used. The NimbleScan software (version 2.6.0.0; Roche NimbleGen) was used to generate one normalized value per probe set using the robust multiarray average (RMA) algorithm (background correction, normalization, and summarization; data not logged). The data were then imported into R (35) using the limma package (36) and log2 transformed. Statistical analysis for differential expression between the mutant and wild-type groups was performed, taking into account the correlation due to processing batch (37, 38). Raw P values were corrected for multiple hypothesis testing using the false discovery rate method (39). Annotation for the probe sets was primarily provided by NimbleGen and included BH identification numbers (IDs) (e.g., BH0001), gene names, descriptions, genomic locations, and URL links to NCBI. Entrez Gene IDs and official gene symbols were downloaded from the B. halodurans genome record in NCBI (GenBank accession no. NC_002570). For analysis, we filtered genes to identify those that were altered by a ≥1.5-fold change in transcription (upregulation and downregulation). For data mining, we used DAVID bioinformatics resources, consisting of an integrated biological knowledge base and analytical tools that use the results from the statistical analysis to explore and interpret gene regulation data (40).
RESULTS
Sublancin displays submicromolar MICs against Bacillus strains.
The antibacterial activity of sublancin against selected Bacillus strains was first determined by solid agar diffusion assays with sublancin at a range of concentrations (0.097 μM to 50 μM). After confirmation that B. subtilis ATCC 6633 and B. halodurans C-125 were sensitive, the MICs were determined by the broth dilution method (41, 42). A series of dilutions of sublancin (0.097 μM to 50 μM) were made and incubated with a defined number of bacterial cells in LB medium. Plates were incubated for 18 to 24 h at 37°C, and growth was assessed by measuring the optical density of each well at 600 nm. The MICs were determined by fitting the data to a dose-response curve. In liquid cultures, the MICs of sublancin against B. halodurans C-125 and B. subtilis ATCC 6633 were 0.312 μM and 0.625 μM, respectively (see Fig. S1 in the supplemental material).
Bactericidal activity of sublancin.
One element for consideration, when trying to understand how an antimicrobial compound functions, is whether it is bactericidal or bacteriostatic. Furthermore, some bacteria are lysed after being killed, others are lysed immediately, and yet others undergo nonlytic death (43, 44). The ability of sublancin to kill or arrest the sensitive Bacillus strains was therefore evaluated. B. subtilis ATCC 6633 and B. halodurans C-125 cultures were grown to mid-log phase, transferred to a 48-well plate, and exposed to sublancin (at 1× and 4× the MIC). After the addition of sublancin, the OD600 was monitored periodically. After a 6-h incubation period, the B. halodurans C-125 and B. subtilis ATCC 6633 cultures showed a decrease in optical density, suggesting that sublancin has bactericidal activity (Fig. 1). To verify whether the activity of sublancin was bactericidal, CFU were determined by plating, which confirmed the bactericidal activity observed by the OD readings. The decrease in optical density was not nearly as great as the decrease in CFU, which implies that sublancin kills without immediate lysis.
FIG 1.
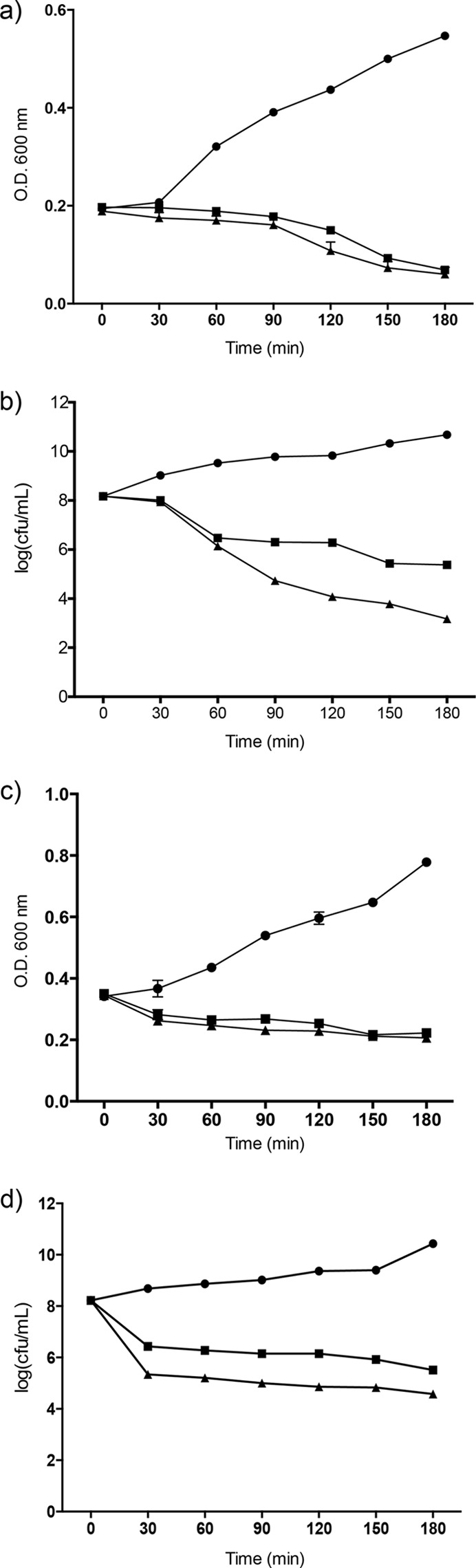
Growth inhibition of B. subtilis ATCC 6633 and B. halodurans C-125 by sublancin 168. (a) Time-dependent changes in OD600 of cultures of B. subtilis ATCC 6633 in the absence (circles) or presence of sublancin 168 at 1× MIC (squares) and 4× MIC (triangles). (b) Aliquots of the cultures in panel a were analyzed for CFU. (c) Time-dependent changes in OD600 of cultures of B. halodurans C-125. The same symbols are used as in panel a. (d) Aliquots of the cultures in panel c were analyzed for CFU. The mean values of the data from one experiment conducted in triplicate are shown. The data are representative of the results from three independent experiments. The error bars indicate standard deviations. When error bars are not visible, they are smaller than the size of the symbol used.
Sublancin does not affect the integrity of the cell membrane.
Some bacteriocins destabilize the membrane or form pores (4, 45). Nisin is the prototypical pore-forming bacteriocin, which binds to lipid II within the membrane (2). To determine whether sublancin affects membrane integrity, we challenged cultures of B. subtilis strain ΔSPβ, B. subtilis ATCC 6633, and B. halodurans C-125 with several different concentrations of sublancin. We monitored the membrane integrity of B. subtilis ATCC 6633 and B. halodurans C-125 by flow cytometric analysis, using the cell-impermeable propidium iodide (PI) dye, after a 30-min exposure to sublancin. Our nisin control showed an increase in fluorescence due to membrane permeabilization, but sublancin-treated cells did not, even at concentrations as high as 32× the MIC for B. subtilis ATCC 6633 and 64× the MIC for B. halodurans C-125 (Fig. 2). We monitored the membrane integrity of B. subtilis ΔSPβ with the LIVE/DEAD BacLight bacterial cell viability assay at 30 and 90 min after the addition of sublancin (see Fig. S2 in the supplemental material). At both time points, we found no change in membrane integrity. When the same strain was exposed to nisin as a positive control, a dramatic loss of membrane integrity was seen already after 30 min of incubation. Collectively, these experiments show that sublancin does not affect membrane integrity and likely acts through an alternative mechanism.
FIG 2.
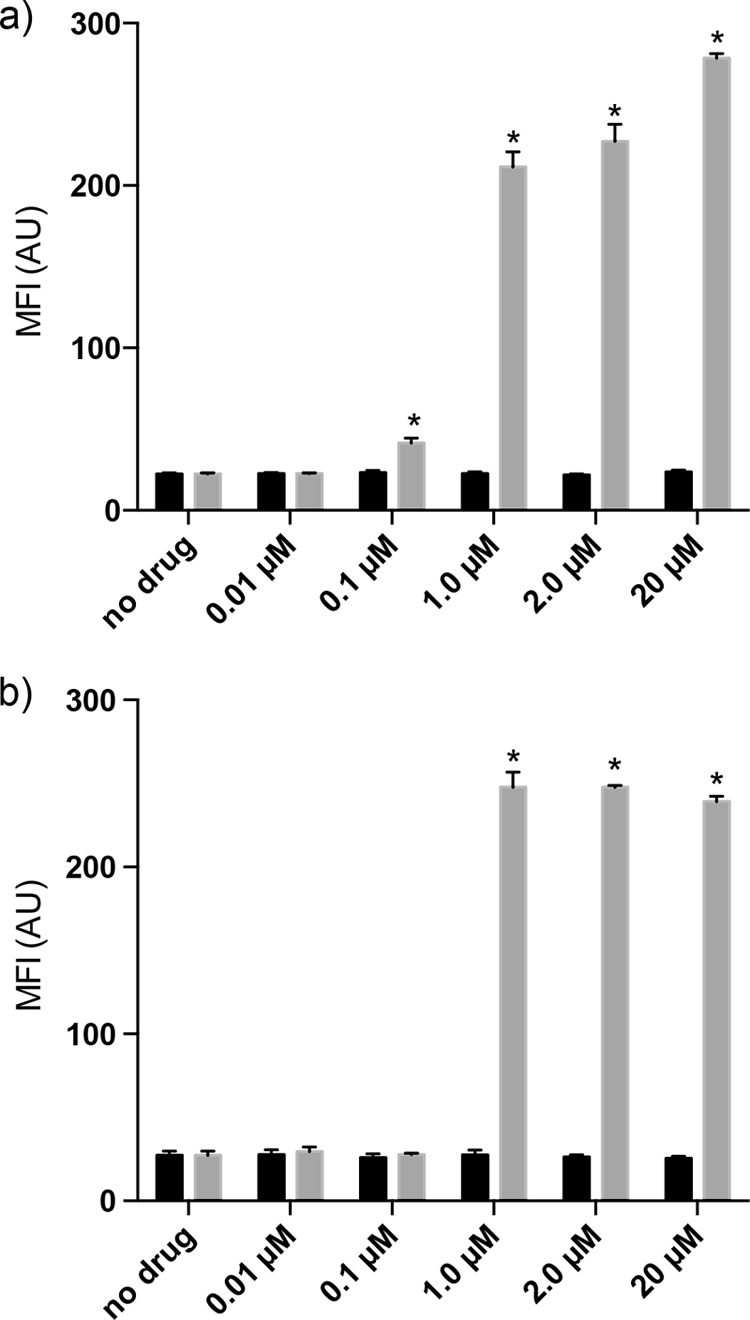
Membrane integrity assays by measuring propidium iodide (PI) uptake. The addition of sublancin at the indicated concentrations (black bars) does not alter the membrane permeability of B. halodurans C-125 (a) and B. subtilis ATCC 6633 (b). Nisin was used as the positive control (gray bars). *, P < 0.05 between nisin (0.1 μM to 20 μM)-treated cells relative to no drug. In all experiments in which the cells were exposed to sublancin, the increase in MFI relative to the control was not statistically significant (P > 0.05). The means of the data from one experiment conducted in triplicate are shown. The data are representative of the results from three independent experiments. The error bars indicate standard deviations.
Resistance frequency.
The manifestation of antibiotic resistance to clinically used antibiotics suggests that resistance is likely to develop against any antibacterial compound. It is useful, however, to analyze the frequency at which resistance to novel antibacterial compounds arises (46). The spontaneous resistance frequency is defined as the number of resistant mutants per total number of viable cells that grow after an established period of time. The resistance frequency of sublancin was determined by plating aliquots of bacterial culture onto agar containing the antibacterial compound at 4× the MIC. Aliquots were also plated onto agar plates with no antibiotic to determine the number of viable bacterial cells in the liquid culture. The resistance frequencies determined were relatively high, with resistance frequencies of 10−5 for B. halodurans C-125 and 10−6 for B. subtilis ATCC 6633. To verify that the observed colonies were indeed resistant to the antibiotic, they were subcultured in sublancin-free LB medium and plated on LB agar containing the antibacterial compound at a concentration of 4× the MIC. For both strains, the plated resistant strains grew a full lawn.
Identification of B. subtilis chromosomal regions that affect sensitivity to sublancin.
We aimed at finding genetic factors that affect sensitivity to sublancin. To do this, we first employed the set of deletion mutants described by Tanaka et al. (29). These mutants were created with a strain in which the prophages of B. subtilis, including SPβ, had been deleted. Therefore, all mutant strains lack the gene encoding the immunity protein for sublancin, sunI (47), making it an ideal collection of mutants for identifying interesting genomic regions with respect to sublancin sensitivity. During the screening, we used LB agar without NaCl, as it was previously shown that B. subtilis is more sensitive to sublancin under low-osmosis conditions (23). The strains were plated in duplicate on the LB agar and spotted with 2 μl of an overnight culture of the sublancin-producing strain B. subtilis 168. We found B. subtilis strain JJS-DIn010, in which rsiW and sigW are deleted, to have increased sensitivity (i.e., a larger zone of clearing around the producing colony) (Fig. 3a). This finding is in concordance with previously reported observations (25), suggesting that our assay was able to identify strains with altered sensitivity. Another strain was identified (B. subtilis JJS-DIn042), in which the genes ykvS, ykvT, ykvU, stoA, zosA, ykvY, ykvZ, glcT, ptsG, ptsH, and ptsI were deleted. JJS-DIn042 was resistant to the effects of sublancin (Fig. 3a) under conditions in which the ΔSPβ strain did not survive. Because of this interesting observation, we investigated this region further by constructing several different individual gene deletion mutants. This approach revealed that only the deletion of the pts operon (ptsGHI) results in resistance to sublancin (Fig. 3b and Table 2). In contrast, a deletion of glcT, which plays a regulatory role in the pts operon (48), did not result in sublancin resistance (Fig. 3b). PtsG is the major glucose transporter of the phosphotransferase system (49), and PtsH and PtsI are general components of the sugar transport system that phosphorylates the incoming sugar (50). PtsH is more commonly known as HPr (as referred to here), and PtsI is also called EI. In B. subtilis, the PTS transfers a phosphate group from phosphorylated PtsI to HPr, which in turn transfers the phosphate to a variety of different PTS permeases. To utilize glucose, HPr transfers the phosphate to the IIA domain of the sugar permease PtsG. The IIA domain then phosphorylates the IIB domain of PtsG, which in turn transfers the phosphate to the incoming sugar. Last, the phosphorylated sugar moves into glycolysis. It is intriguing that the PTS was identified in our screen for sublancin sensitivity, as the most common PTS substrate is glucose, whereas sublancin is glucosylated. A functional homologue of HPr is present in B. subtilis, i.e., Crh. We therefore tested a crh deletion mutant in the presence of sublancin, but no change in sensitivity was observed compared to the wild type (data not shown), suggesting that the sensitivity to sublancin is specifically dependent on HPr.
FIG 3.
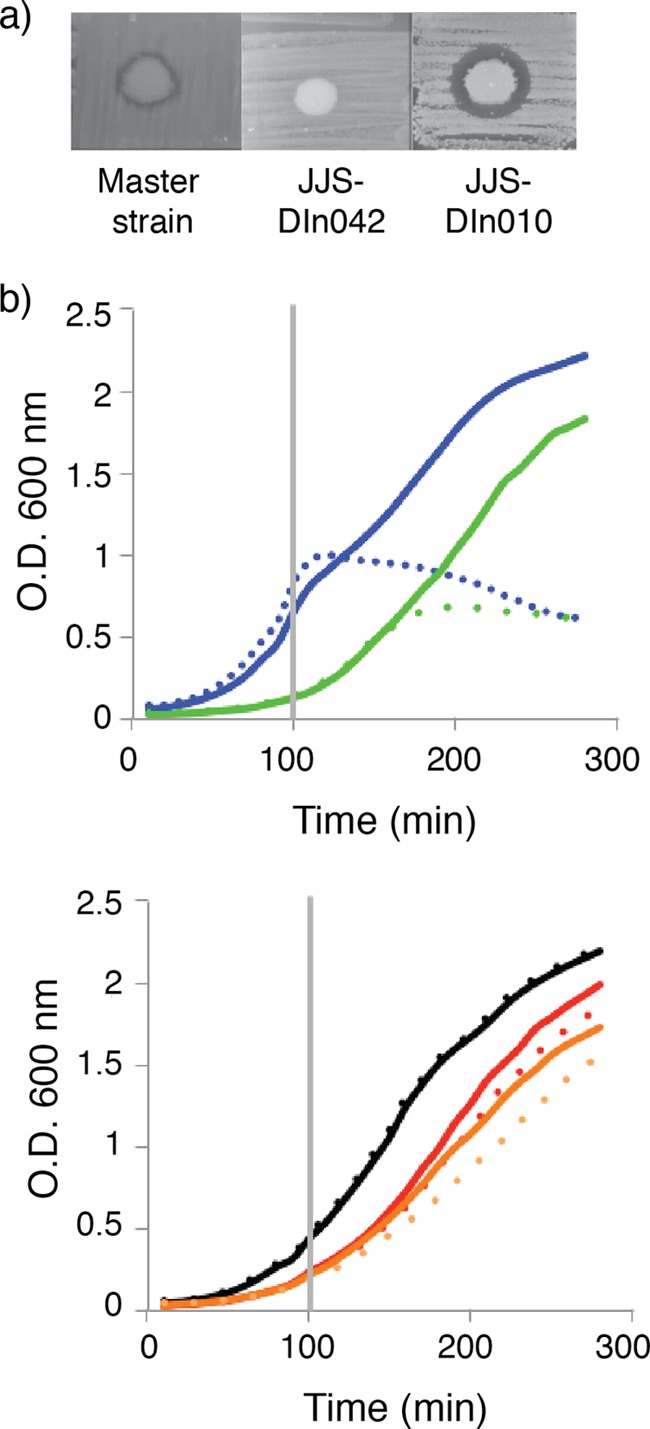
Deletion of the ptsGHI operon results in resistance to sublancin. (a) B. subtilis strains described by Tanaka et al. (29) were screened for increased and reduced sensitivity to sublancin. The parental strain of the collection is labeled “Master strain.” In JJS-DIn042, the region from ykvS to ptsI is deleted, and in JJS-DIn010, rsiW and sigW are deleted. (b) Growth curves of mutant strains with individual deletions of the genes that are responsible for the resistance in strain JJS-DIn042. In the top graph, blue line, B. subtilis ΔSPβ; green line, B. subtilis ΔSPβ-glcT. In the bottom graph, black line, B. subtilis ΔSPβ-ptsG; red line, B. subtilis ΔSPβ-ptsH; orange line, B. subtilis ΔSPβ-ptsI; solid lines, no sublancin; dotted lines, 200 nM sublancin added at 100 min, as indicated by the vertical gray line. Deletion of any of the genes within the ptsGHI operon results in resistance to sublancin. Deletion of the gene encoding the transcriptional antiterminator glcT has no effect on the sensitivity of B. subtilis to sublancin. The means of the data from one experiment conducted in triplicate are shown. The data are representative of the results from three independent experiments.
TABLE 2.
Resistance phenotype of B. subtilis ΔSPβ single-gene deletion strains upon exposure to sublancin
| Deleted gene | Sublancin resistant or sensitive |
|---|---|
| ykvS | Sensitive |
| ykvT | Sensitive |
| ykvU | Sensitive |
| stoA | Sensitive |
| zosA | Inconclusive |
| ykvY | Sensitive |
| ykvZ | Sensitive |
| glcT | Sensitive |
| ptsG | Resistant |
| ptsH | Resistant |
| ptsI | Resistant |
Comparative genomics.
Bacteria often acquire stable resistance to antibiotics due to gene mutations. A comparative genomics analysis was therefore performed to identify the mutations that conferred resistance to B. halodurans C-125 after exposure to sublancin. The gDNA of the sensitive strain B. halodurans C-125 and of four of the spontaneous resistant mutants obtained as described above were extracted and sequenced using a HiSeq 2000 Illumina sequencer. The wild-type B. halodurans strain was mapped to the published B. halodurans C-125 genome sequence (GenBank accession no. NC_002570.2) to generate a consensus sequence that was used for SNP detection in sublancin-resistant mutants of B. halodurans. A comparison of the gDNA of the wild-type sensitive strain with the four sublancin-resistant mutants revealed several mutations. One strain contained three mutations in the intergenic region between the transcriptional antiterminator (locus tag BH0845) and ptsG (locus tag BH0844), another strain contained a missense mutation in the gene for mannitol-1-phosphate 5-dehydrogenase (locus tag BH3851), and most interestingly, the three strains that did not have a mutation in the intergenic region mentioned above all had nonsense mutations in the gene for the glucose-specific transporter subunit IIC, which is part of the multidomain protein PtsG (see Table S2 in the supplemental material). The missense mutation prevents the production of PtsG, and the three mutations in the intergenic region between the antiterminator and ptsG are predicted to considerably stabilize the structure of the terminator (see Fig. S3 in the supplemental material), thus potentially also preventing ptsG transcription. Once more, these findings point to the PTS being important for sensitivity to sublancin.
Gene expression profile by microarray analysis of B. halodurans C-125.
Antimicrobial-resistant mutants provide valuable insights, but the information obtained from a resistance phenotype is not always representative of the identity of the target. We therefore also monitored changes in global gene expression upon exposure of B. halodurans C-125 to sublancin.
The expression profiles revealed four genes that are part of sulfur metabolism that are highly upregulated (changes from 9- to 14-fold; see Table S3 in the supplemental material). The analysis also revealed upregulated genes involved in transmembrane transporter activities, whereas genes involved in amino sugar and nucleotide sugar metabolism were up- and downregulated. Interestingly, the genes for HPr and for PtsG that were also identified in the set of deletion mutants and in the comparative genomics analysis were downregulated (see Table S3), as was another PTS protein that is homologous to YpqE in B. subtilis, a putative phosphotransferase enzyme IIA component (51). These data again suggest that, like in B. subtilis, the PTS is involved in the sensitivity of B. halodurans toward sublancin.
Addition of PTS sugars to the growth medium results in increased resistance to sublancin.
Several bacteriocins have been shown to target PTS proteins as part of their mode of action. In these reported cases, the addition of the relevant sugar resulted in an increased sensitivity to the bacteriocin (6, 7, 11). This effect is due to the elevated uptake of the respective bacteriocins via the PTS. We therefore investigated whether this was also true for sublancin. The PTS is able to use many sugars, employing a different transporter for each sugar, together with the HPr and PtsI proteins. Once the sugar is phosphorylated, it moves into glycolysis at the relevant metabolic junction. To investigate the influence of added sugars, B. subtilis ΔSPβ cultures were diluted in LB medium (without NaCl) containing different sugars and grown in 96-well microtiter plates with shaking to an OD600 of 0.5 before the addition of sublancin at the MIC of 200 nM, as measured for this strain under these conditions. The presence of the PTS sugars glucose, sucrose, and fructose prevented the growth inhibition imposed by sublancin (Fig. 4a), since no significant reduction in the OD was observed. In contrast, the non-PTS sugars citrate, galactose, and succinate had no influence on the activity of sublancin (Fig. 4b). The two exceptions were the non-PTS sugars glycerol and malate. In this respect, it is noteworthy that the glycerol kinase GlpK requires phosphorylation by HPr for glycerol utilization (48, 52). Malate is a preferred carbon source for B. subtilis and is known to influence the carbon catabolite repression response (53). In this context, it is noteworthy that a decrease in antimicrobial activity was also reported for glycocin F upon supplementation of the medium with GlcNAc, which is the sugar that is attached to glycocin F at two positions (17).
FIG 4.
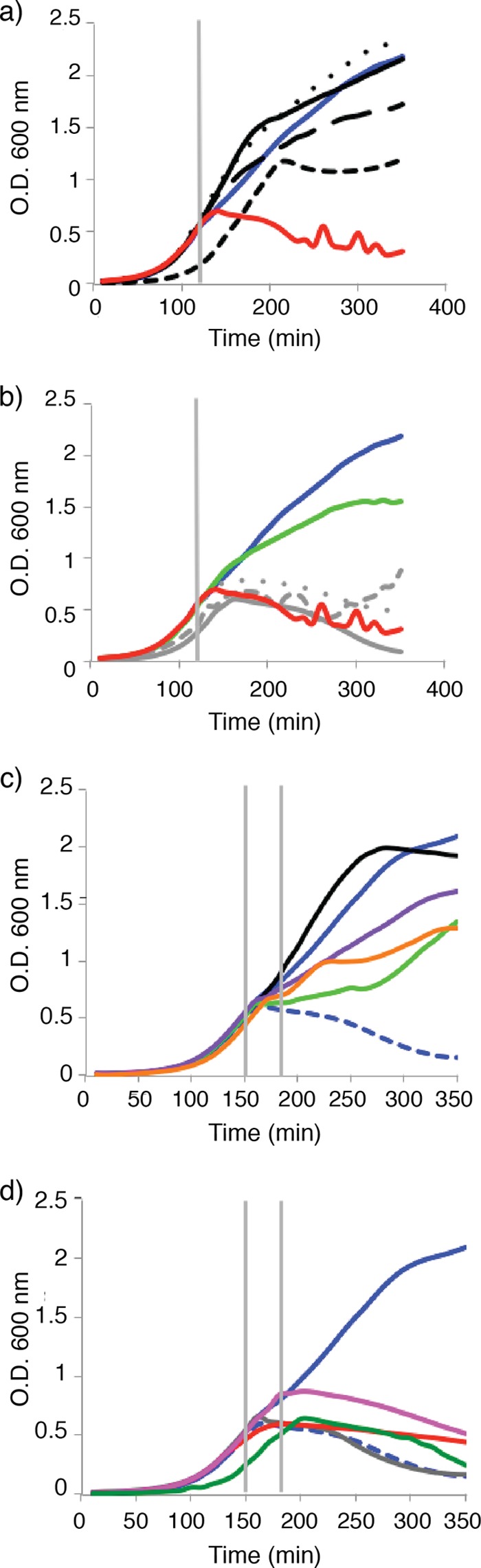
Addition of PTS sugars to LB blocks growth inhibition by sublancin. (a) Growth curves of the B. subtilis ΔSPβ strain in LB medium with salt with added PTS sugars. Blue line, no addition of sublancin; red line, addition of sublancin; black solid line, addition of 0.3% glucose; black dotted line, addition of 0.3% fructose; black long-dashed line, addition of 0.3% sucrose; black short-dashed line, addition of 0.3% glycerol. Sublancin was added at 120 min, as indicated by the vertical gray line. (b) Growth curve of the ΔSPβ strain in LB medium with addition of non-PTS sugars. Blue line, no addition of sublancin; red line, addition of sublancin; green line, addition of 0.4% malate; gray short-dashed line, addition of 0.4% citrate; gray dotted line, addition of 0.4% galactose; gray solid line, addition of 0.4% succinate. (c) Growth curve of the ΔSPβ strain with sublancin added at 150 min, followed by the addition of PTS and non-PTS sugars 30 min later, as depicted by the left and right vertical lines, respectively. Blue line, no sublancin; blue dashed line, addition of sublancin; black line, 0.3% glucose; purple line, 0.3% fructose; orange line, 0.3% glycerol; green line, 0.4% malate (final concentration of sugars shown). (d) Growth curve of the ΔSPβ strain with sublancin added at 150 min, followed by the addition of PTS and non-PTS sugars 30 min later, as depicted by the left and right vertical lines, respectively. Blue line, no sublancin; blue dashed line, addition of sublancin; gray line, 0.4% citrate; red line, 0.3% sucrose; pink line, 0.4% galactose; green line, 0.4% succinate (final concentration of sugars shown). The means of the data from one experiment conducted in triplicate are shown. The data are representative of the results from three independent experiments.
To further delineate the effects of sugars on sublancin sensitivity, purified sublancin was spotted onto lawns of B. subtilis ΔSPβ, which were grown on agar plates containing the defined M9 minimal medium supplemented with glucose, malate, or citrate. When glucose was present, the cells were always resistant to sublancin. In contrast, with citrate, a large zone of clearing was observed. In the presence of malate, an intermediate-sized zone of inhibition was observed. This observation underpins the view that the carbon source affects the sensitivity to sublancin (see Fig. S4 in the supplemental material).
Since glucose was found to prevent the effect of sublancin, we wondered whether it would be possible to rescue sublancin-treated cells by adding glucose. We therefore grew the bacteria on LB medium (with NaCl) and added sublancin at an OD600 of 0.5. The cells were then incubated for 30 min before adding the same PTS and non-PTS sugars as in the previous experiment (Fig. 4c and d). Glucose almost instantaneously rescued the cells from the growth inhibition that sublancin imposed on the cells. Fructose also rescued the cells, but to a smaller extent than glucose. The non-PTS sugar glycerol rescued the cells in a manner similar to that with fructose. Malate was also able to rescue the cells, but this took approximately 100 min following the addition of the sugar, whereas the effect for glycerol and fructose was observed immediately after the addition of the respective sugar. In contrast, the addition of the other non-PTS sugars or sucrose had no effect on the survival of the bacteria.
The observed rescue of growth by addition of the different PTS sugars and glycerol suggests that the PTS transporter is not irreversibly inactivated by sublancin; perhaps instead, sublancin affects the pathway that leads to the phosphorylation of the sugar. The addition of sublancin and the PTS sugar at the same time might result in competition for phosphorylation of the sugar or the glucose on sublancin (or its metabolite). With this in mind, we looked at the phosphorylation sites on the HPr protein.
Phosphorylation of HPr is responsible for sensitivity to sublancin.
The HPr protein is phosphorylated on two sites. The first, His15, is phosphorylated by PtsI. HPr then transfers the phosphate group to PtsG, and the phosphate is subsequently used to phosphorylate the incoming sugar. The second, Ser46, is phosphorylated by HPr kinase (HPrK) under conditions of large glycolytic flux. This phosphorylation subsequently allows HPr to form a complex with the catabolite control protein A (CcpA). This HPr-CcpA complex mediates carbon catabolite repression by binding to its cognate operator regions. To link sublancin sensitivity to one of these HPr-mediated processes, we tested two B. subtilis ΔSPβ strains with point mutations at one of the two HPr phosphorylation sites. As shown in Fig. 5a, B. subtilis ΔSPβ carrying the S46A point mutation in HPr was fully sensitive to sublancin. In contrast, the strain carrying the H15A point mutation in HPr displayed an increased level of resistance to sublancin. This observation suggests that hprK and ccpA deletion mutants would remain sensitive to sublancin, since the carbon catabolite-repressing function of HPr is not affected. This prediction was indeed confirmed, as the deletion of either of these two genes had no effect on sublancin sensitivity (Fig. 5b). Also, the addition of glucose to the ΔccpA mutant conferred resistance to sublancin (not shown), providing further evidence that it is not the carbon catabolite-repressing branch of HPr regulation that leads to sublancin sensitivity. Instead, it seems to be the PTS branch involving the H15 phosphorylation site that is responsible for the effects on sublancin sensitivity. However, how phosphorylation of His15 of HPr is exactly tied to sublancin sensitivity is presently unknown.
FIG 5.
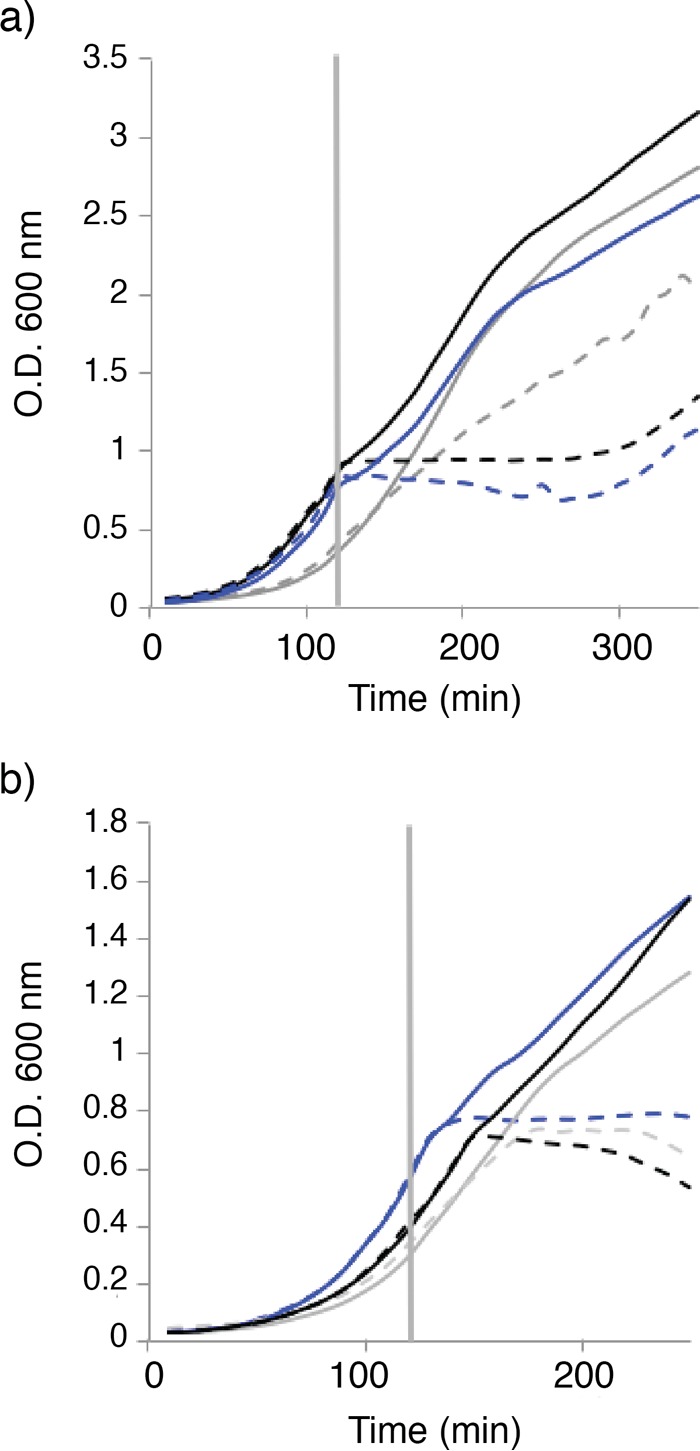
The H15A mutation in HPr results in increased resistance to sublancin. (a) The two phosphorylation sites in the HPr protein were mutated individually to alanine residues. The growth curves of the resulting strains are shown, with 200 nM sublancin added at 120 min, as depicted by the vertical line. Blue line, B. subtilis ΔSPβ; gray line, B. subtilis ΔSPβ-H15A; black line, B. subtilis ΔSPβ-S46A; solid line, no sublancin; dashed line, with sublancin. (b) Blue line, B. subtilis ΔSPβ; gray line, B. subtilis ΔSPβ-hprK; black line, B. subtilis ΔSPβ-ccpA; solid line, no sublancin; dashed line, with sublancin. The ΔccpA and ΔhprK mutations have no effect on the sensitivity of the cells to sublancin. Sublancin at 200 nM was added at 120 min. The means of the data from one experiment conducted in triplicate are shown. The data are representative of the results from three independent experiments.
DISCUSSION
Sublancin is a bacteriocin that was recently found to be glucosylated as part of its maturation process, and this glucosylation is required for its activity (15). We showed in this study that sublancin is bactericidal and that it does not kill by pore formation or disruption of membrane integrity. Four different lines of evidence point toward the phosphoenolpyruvate:sugar phosphotransferase system as a factor affecting the activity of the bacteriocin. Experiments with deletion mutants of B. subtilis identified PtsG, HPr, and PtsI, but not GlcT and Crh, as being important for sensitivity to sublancin. In addition, three of four B. halodurans sublancin-resistant mutants contained stop codon mutations within the ptsG gene, with the fourth resistant strain having three mutations that potentially interfere with ptsG expression. The transcriptional profile also indicated a strong downregulation of PTS genes upon exposure to sublancin and, last, the addition of PTS sugars decreased sensitivity to sublancin.
The PTS has been described as affecting sensitivity to other bacteriocins, including dysgalacticin and lactococcin A (6, 7, 54). Dysgalacticin appears to bind to the glucose and mannose transporters of the PTS (7). Dysgalacticin was shown to block the uptake of glucose and the nonmetabolizable analog 2-deoxyglucose; it also perturbs the membrane of the cell, causing the dissipation of membrane potential (7). This activity appears to be different from the mechanism used by sublancin, as the addition of glucose or any other PTS sugar blocked the killing activity of sublancin, and membrane disruption was not observed. Lactococcin A also uses components of the mannose PTS in its mode of action. Lactococcin A binds to the membrane-located complex of the IIC and IID subunits of the mannose transporter (6), resulting in dissipation of the membrane potential (45). Like the observations with dysgalacticin, decreased growth rates were observed for cells grown with mannose or glucose as the sole carbon source in the presence of lactococcin A (6). The current study suggests that sublancin is also functioning in a manner different from that of lactococcin A, since the studies with gene deletion strains and spontaneous resistance mutants both point to the phosphorylation components of the PTS, rather than the membrane components, as being key to sublancin sensitivity. When we monitored sublancin susceptibility using M9 minimal medium supplemented with glucose as the single carbon source, cells were completely immune to the effects of sublancin. A third bacteriocin, the circular molecule garvicin ML, requires the maltose-binding protein for activity. Growth in medium in which the sole carbon source is either maltose or galactose again resulted in increased sensitivity to this bacteriocin (55).
Two regions of the B. subtilis chromosome that result in resistance to sublancin have now been identified. The first is mscL, which encodes a mechanosensitive channel, as described by Kouwen et al. (23) and, in this work, the proteins encoded by the ptsGHI operon. Several scenarios may describe the mechanism by which sublancin interacts with the PTS. First, it is intriguing that it is the glucose transporter that was identified, given that sublancin is modified with an S-linked glucose moiety. The glucose moiety on sublancin might be recognized by the transporter to facilitate entry into the cell or potentially to kill it through its interaction with this system. Competition between sublancin and glucose might explain the observed decrease in sensitivity upon the addition of glucose. We note that HPr and domains IIA and IIB of PtsG are located in the cytoplasm (see Table S4 in the supplemental material); hence, for this mechanism to be correct, the glucose on sublancin would have to traverse the transporter or bypass the glucose transporter via the MscL channel. In this respect, it is noteworthy that the other sugars that were able to protect the bacteria from sublancin are either gluconeogenic or feed into glycolysis elsewhere in the pathway, therefore possibly bypassing the need for the glucose transporter. When we tested strains that express variants of the HPr protein with point mutations that remove the phosphorylation sites, the mutation that led to increased resistance to sublancin was H15A. The phosphorylation of His15 is responsible for transferring a phosphate group to the incoming PTS sugar. This points to a critical role of phosphorylation in the growth inhibition by sublancin and seems to suggest that sublancin may need to be phosphorylated upon its entry into the cell to exert its bactericidal effect. Interestingly, bacterial growth was rescued when PTS sugars were added to the growth medium 30 min after the challenge with sublancin. This finding implies that either the specific growth-inhibiting mechanism employed by sublancin is reversible or that the addition of the PTS sugars results in the cell using a different biological process that allows survival.
In conclusion, we have shown here that sublancin exerts its bactericidal effects in a novel manner. At present, it is not clear exactly how sublancin interacts with the PTS, and several questions remain for future research. Is there a physical interaction between sublancin and the PTS in the inhibited cells? Is there a link between the PTS and the MscL channel? How does sublancin actually inhibit the growth of the cell? How do the strong structural similarities of glycocin F and sublancin fit into the mechanism, and what role does the three-dimensional structure of the peptide components of these glycocins play (21, 22)? In a time in which bacteria are becoming resistant to the antimicrobial compounds that are used in clinical practice, research to understand how infections can be fought in alternative manners is essential. The apparently novel mechanism by which sublancin kills sensitive species of bacteria may therefore offer biological insights for the development of new antimicrobial strategies. Whether the mechanism identified in Bacillus is also operational in sensitive pathogens, such as S. aureus and Listeria monocytogenes, requires further in-depth analyses.
Supplementary Material
ACKNOWLEDGMENTS
E.L.D., R.A.T.M., and J.M.V.D. were supported by the Commission of the European Union (projects LSHG-CT-2006-037469 and 244093) and the transnational Systems Biology of Microorganisms (SysMO) organization (project BACELL SysMO2) through the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research. W.A.V.D.D. received support from the Howard Hughes Medical Institute. C.V.G.D.G was supported by an NIGMS-NIH Chemistry-Biology Interface Training grant (5T32-GM070421).
We thank Jenny Drnevich and Mike Band at the Roy J. Carver Biotechnology Center and W. M. Keck Center for Comparative and Functional Genomics for assistance with the microarray data and Alvaro Hernandez for assistance with the comparative genomics. Barbara Pilas assisted with the flow cytometry.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01519-15.
REFERENCES
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 3.Schneider T, Sahl HG. 2010. Lipid II and other bactoprenol-bound cell wall precursors as drug targets. Curr Opin Investig Drugs 11:157–164. [PubMed] [Google Scholar]
- 4.Breukink E, de Kruijff B. 2006. Lipid II as a target for antibiotics. Nat Rev Drug Discov 5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 5.Islam MR, Nagao J, Zendo T, Sonomoto K. 2012. Antimicrobial mechanism of lantibiotics. Biochem Soc Trans 40:1528–1533. doi: 10.1042/BST20120190. [DOI] [PubMed] [Google Scholar]
- 6.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A 104:2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swe PM, Cook GM, Tagg JR, Jack RW. 2009. Mode of action of dysgalacticin: a large heat-labile bacteriocin. J Antimicrob Chemother 63:679–686. doi: 10.1093/jac/dkn552. [DOI] [PubMed] [Google Scholar]
- 8.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J Biol Chem 281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 9.Nolan EM, Walsh CT. 2008. Investigations of the MceIJ-catalyzed posttranslational modification of the microcin E492 C-terminus: linkage of ribosomal and nonribosomal peptides to form “Trojan horse” antibiotics. Biochemistry 47:9289–9299. doi: 10.1021/bi800826j. [DOI] [PubMed] [Google Scholar]
- 10.Grinter R, Milner J, Walker D. 2012. Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One 7:e33033. doi: 10.1371/journal.pone.0033033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau S, Martin NI, Vederas JC. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577–592. doi: 10.1016/S0300-9084(02)01414-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Zeng X, Wang S, Hou C, Yang F, Ma X, Thacker P, Qiao S. 2014. The bacteriocin sublancin attenuates intestinal injury in young mice infected with Staphylococcus aureus. Anat Rec (Hoboken) 297:1454–1461. doi: 10.1002/ar.22941. [DOI] [PubMed] [Google Scholar]
- 13.Paik SH, Chakicherla A, Hansen JN. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem 273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 14.Dorenbos R, Stein T, Kabel J, Bruand C, Bolhuis A, Bron S, Quax WJ, Van Dijl JM. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem 277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- 15.Oman TJ, Boettcher JM, Wang H, Okalibe XN, van der Donk WA. 2011. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat Chem Biol 7:78–80. doi: 10.1038/nchembio.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois JY, Kouwen TR, Schurich AK, Reis CR, Ensing HT, Trip EN, Zweers JC, van Dijl JM. 2009. Immunity to the bacteriocin sublancin 168 is determined by the SunI (YolF) protein of Bacillus subtilis. Antimicrob Agents Chemother 53:651–661. doi: 10.1128/AAC.01189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepper J, Shastri S, Loo TS, Preston JC, Novak P, Man P, Moore CH, Havlicek V, Patchett ML, Norris GE. 2011. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett 585:645–650. doi: 10.1016/j.febslet.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Norris GE, Patchett ML. 2014. Glycosylated ribosomally synthesized peptide toxins, p 507–543. In Havlíček V, Spí̌zek J (ed), Natural products analysis: instrumentation, methods, and applications. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 19.Hata T, Tanaka R, Ohmomo S. 2010. Isolation and characterization of plantaricin ASM1: a new bacteriocin produced by Lactobacillus plantarum A-1. Int J Food Microbiol 137:94–99. doi: 10.1016/j.ijfoodmicro.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Oman TJ, Zhang R, Garcia De Gonzalo CV, Zhang Q, van der Donk WA. 2014. The glycosyltransferase involved in thurandacin biosynthesis catalyzes both O- and S-glycosylation. J Am Chem Soc 136:84–87. doi: 10.1021/ja411159k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia De Gonzalo CV, Zhu L, Oman TJ, van der Donk WA. 2014. NMR structure of the S-linked glycopeptide sublancin 168. ACS Chem Biol 9:796–801. doi: 10.1021/cb4008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venugopal H, Edwards PJ, Schwalbe M, Claridge JK, Libich DS, Stepper J, Loo T, Patchett ML, Norris GE, Pascal SM. 2011. Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry 50:2748–2755. doi: 10.1021/bi200217u. [DOI] [PubMed] [Google Scholar]
- 23.Kouwen TR, Trip EN, Denham EL, Sibbald MJ, Dubois JY, van Dijl JM. 2009. The large mechanosensitive channel MscL determines bacterial susceptibility to the bacteriocin sublancin 168. Antimicrob Agents Chemother 53:4702–4711. doi: 10.1128/AAC.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iscla I, Wray R, Wei S, Posner B, Blount P. 2014. Streptomycin potency is dependent on MscL channel expression. Nat Commun 5:4891. doi: 10.1038/ncomms5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butcher BG, Helmann JD. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol Microbiol 60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 26.Dempwolff F, Möller HM, Graumann PL. 2012. Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J Bacteriol 194:4652–4661. doi: 10.1128/JB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mäder U, Nicolas P, Piersma S, Rugheimer F, Becher D, Bessières P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hübner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, et al. 2012. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 28.Bron S, Venema G. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat Res 15:1–10. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Henry CS, Zinner JF, Jolivet E, Cohoon MP, Xia F, Bidnenko V, Ehrlich SD, Stevens RL, Noirot P. 2013. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res 41:687–699. doi: 10.1093/nar/gks963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 32.Goosens VJ, Mars RA, Akeroyd M, Vente A, Dreisbach A, Denham EL, Kouwen TR, van Rij T, Olsthoorn M, van Dijl JM. 2013. Is proteomics a reliable tool to probe the oxidative folding of bacterial membrane proteins? Antioxid Redox Signal 18:1159–1164. doi: 10.1089/ars.2012.4664. [DOI] [PubMed] [Google Scholar]
- 33.Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche NimbleGen. 2010. NimbleGen arrays user's guide, version 5.1. Roche NimbleGen, Inc., Madison, WI. [Google Scholar]
- 35.R Development Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 36.Smyth GK. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 37.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK, Michaud J, Scott HS. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57:289–300. [Google Scholar]
- 40.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. [DOI] [PubMed] [Google Scholar]
- 41.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):S5–S16. [DOI] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, vol 26 CLSI document M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob Agents Chemother 52:2223–2225. doi: 10.1128/AAC.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDowell TD, Lemanski CL. 1988. Absence of autolytic activity (peptidoglycan nicking) in penicillin-induced nonlytic death in a group A streptococcus. J Bacteriol 170:1783–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Belkum MJ, Kok J, Venema G, Holo H, Nes IF, Konings WN, Abee T. 1991. The bacteriocin lactococcin A specifically increases permeability of lactococcal cytoplasmic membranes in a voltage-independent, protein-mediated manner. J Bacteriol 173:7934–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol Biol Evol 20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 48.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol Microbiol 25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 49.Gonzy-Tréboul G, de Waard JH, Zagorec M, Postma PW. 1991. The glucose permease of the phosphotransferase system of Bacillus subtilis: evidence for IIGlc and IIIGlc domains. Mol Microbiol 5:1241–1249. doi: 10.1111/j.1365-2958.1991.tb01898.x. [DOI] [PubMed] [Google Scholar]
- 50.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reizer J, Bachem S, Reizer A, Arnaud M, Saier MH Jr, Stülke J. 1999. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145(Pt 12):3419–3429. [DOI] [PubMed] [Google Scholar]
- 52.Darbon E, Galinier A, Le Coq D, Deutscher J. 2001. Phosphotransfer functions mutated Bacillus subtilis HPr-like protein Crh carrying a histidine in the active site. J Mol Microbiol Biotechnol 3:439–444. [PubMed] [Google Scholar]
- 53.Meyer FM, Jules M, Mehne FM, Le Coq D, Landmann JJ, Görke B, Aymerich S, Stülke J. 2011. Malate-mediated carbon catabolite repression in Bacillus subtilis involves the HPrK/CcpA pathway. J Bacteriol 193:6939–6949. doi: 10.1128/JB.06197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol 77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabrielsen C, Brede DA, Hernandez PE, Nes IF, Diep DB. 2012. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob Agents Chemother 56:2908–2915. doi: 10.1128/AAC.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res 28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishio C, Komura S, Kurahashi K. 1983. Peptide antibiotic subtilin is synthesized via precursor proteins. Biochem Biophys Res Commun 116:751–758. doi: 10.1016/0006-291X(83)90588-0. [DOI] [PubMed] [Google Scholar]
- 58.Arnaud M, Vary P, Zagorec M, Klier A, Débarbouillé M, Postma P, Rapoport G. 1992. Regulation of the sacPA operon of Bacillus subtilis: identification of phosphotransferase system components involved in SacT activity. J Bacteriol 174:3161–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh KD, Schmalisch MH, Stülke J, Görke B. 2008. Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources. J Bacteriol 190:7275–7284. doi: 10.1128/JB.00848-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faires N, Tobisch S, Bachem S, Martin-Verstraete I, Hecker M, Stülke J. 1999. The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J Mol Microbiol Biotechnol 1:141–148. [PubMed] [Google Scholar]
- 61.Hanson KG, Steinhauer K, Reizer J, Hillen W, Stülke J. 2002. HPr kinase/phosphatase of Bacillus subtilis: expression of the gene and effects of mutations on enzyme activity, growth and carbon catabolite repression. Microbiology 148:1805–1811. [DOI] [PubMed] [Google Scholar]
- 62.Reizer J, Bergstedt U, Galinier A, Küster E, Saier MH Jr, Hillen W, Steinmetz M, Deutscher J. 1996. Catabolite repression resistance of gnt operon expression in Bacillus subtilis conferred by mutation of His-15, the site of phosphoenolpyruvate-dependent phosphorylation of the phosphocarrier protein HPr. J Bacteriol 178:5480–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deutscher J, Reizer J, Fischer C, Galinier A, Saier MH Jr, Steinmetz M. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol 176:3336–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


