Abstract
Drug resistance is a major problem in Mycobacterium tuberculosis control, and it is critical to identify novel drug targets and new antimycobacterial compounds. We have previously identified an imidazo[1,2-a]pyridine-4-carbonitrile-based agent, MP-III-71, with strong activity against M. tuberculosis. In this study, we evaluated mechanisms of resistance to MP-III-71. We derived three independent M. tuberculosis mutants resistant to MP-III-71 and conducted whole-genome sequencing of these mutants. Loss-of-function mutations in Rv2887 were common to all three MP-III-71-resistant mutants, and we confirmed the role of Rv2887 as a gene required for MP-III-71 susceptibility using complementation. The Rv2887 protein was previously unannotated, but domain and homology analyses suggested it to be a transcriptional regulator in the MarR (multiple antibiotic resistance repressor) family, a group of proteins first identified in Escherichia coli to negatively regulate efflux pumps and other mechanisms of multidrug resistance. We found that two efflux pump inhibitors, verapamil and chlorpromazine, potentiate the action of MP-III-71 and that mutation of Rv2887 abrogates their activity. We also used transcriptome sequencing (RNA-seq) to identify genes which are differentially expressed in the presence and absence of a functional Rv2887 protein. We found that genes involved in benzoquinone and menaquinone biosynthesis were repressed by functional Rv2887. Thus, inactivating mutations of Rv2887, encoding a putative MarR-like transcriptional regulator, confer resistance to MP-III-71, an effective antimycobacterial compound that shows no cross-resistance to existing antituberculosis drugs. The mechanism of resistance of M. tuberculosis Rv2887 mutants may involve efflux pump upregulation and also drug methylation.
INTRODUCTION
Tuberculosis (TB) is a devastating disease that infects one-third of the world's population and killed 1.5 million people in 2013 (1). TB is caused by Mycobacterium tuberculosis and is challenging and time-consuming to treat. Standard TB treatment is currently 6 months long and involves a 2-month intensive phase consisting of treatment with four antibiotics (isoniazid, rifampin, ethambutol, and pyrazinamide) followed by a 4-month continuation phase (treatment with isoniazid and rifampin only). However, despite this combination therapy, drug resistance is on the rise, and in 2013, there were an estimated 480,000 cases of multidrug-resistant (MDR) TB, which is defined as TB caused by bacteria that are resistant to at least isoniazid and rifampin. These cases require up to 2 years of treatment, but drug resistance is developing even under these conditions, and in 2014, 9% of MDR TB cases were extensively drug resistant (XDR), meaning that they were also resistant to isoniazid, rifampin, a fluoroquinolone, and an injectable anti-TB drug (typically an aminoglycoside) (1). Thus, there is a great need both for new antibiotics to treat M. tuberculosis and for new drug targets that avoid cross-resistance to currently used therapies.
One such new compound is an imidazo[1,2-a]pyridine-4-carbonitrile-based agent, MP-III-71, a compound identified by Pieroni et al. as having an MIC of 0.5 μg/ml against both drug-susceptible and drug-resistant M. tuberculosis strains as well as a low Vero cell toxicity of >64 μg/ml (see Fig. S1a in the supplemental material) (2). Thus, this compound is a promising candidate for follow-up studies. We were particularly interested in its mechanism, since the susceptibility of MDR and XDR strains to MP-III-71 suggests that it acts against a novel target that can be used against drug-resistant strains. We addressed this question by identifying strains resistant to MP-III-71 and exploring the mechanism of resistance in these strains. In this study, we characterized these strains, with a focus on the effect of loss of function of Rv2887, a protein in the multiple antibiotic resistance repressor (MarR) family.
MATERIALS AND METHODS
MP-III-71.
MP-III-71 was synthesized as previously described (2). It was then dissolved in dimethyl sulfoxide (DMSO) at 2,048 μg/ml, and aliquots were stored in −80°C until use.
Synthesis of 2-(4-methoxybenzyl)-3,5-dimethyl-1-oxo-1,5-dihydrobenzo[4,5]imidazo[1,2-a]pyridine-4-carbonitrile (the N-Me derivative of MP III-71).
To a suspension of NaH (60% in mineral oil, 22 mg, 0.57 mmol) in dry dimethylformamide (DMF) (2 ml), 2-(4-methoxybenzyl)-3-methyl-1-oxo-1,5-dihydrobenzo[4,5]imidazo[1,2-a]pyridine-4-carbonitrile (60 mg, 0.19 mmol), prepared as previously reported, (2), was added at 0°C. After stirring for 15 min, iodomethane (0.023 ml, 0.38 mmol) was added portionwise, and the reaction mixture was refluxed for 4 h. The mixture was poured in ice-water and extracted with ethyl acetate (3 times, 10 ml each), and the organic layers were washed with water and brine, dried over anhydrous Na2SO4, and concentrated under reduced pressure. The crude material was purified through flash chromatography with elution with petroleum ether-ethyl acetate at 90:10 to 80:20, yielding the title compound as a white solid: yield, 76%; purity, 98%; 1H nuclear magnetic resonance (NMR) (400 MHz-d6-DMSO), δ 2.36 (s, 3H), 3.69 (s, 3H), 3.92 (s, 2H), 4.08 (s, 3H), 6.81 (d, J = 8 Hz, 2H), 7.15 (d, J = 8 Hz, 2H), 7.41 (t, J = 8 Hz, 1H), 7.59 (t, J = 8 Hz, 1H), 7.75 (d, J = 8 Hz, 1H), 8.69 (d, J = 8 Hz, 1H); 13C NMR (100.6 MHz-d6-DMSO) δ 18.8, 30.8, 31.4, 55.5, 70.0, 110.2, 114.2, 116.1, 116.7, 118.0, 123.1, 127.0, 127.3, 129.3, 132.6, 133.7, 144.8, 147.9, 158.0, 159.2; high-resolution mass spectrometry (HRMS) (electrospray ionization [ESI]) for C22H19N3O2 (M + H)+, 358.1477 (calculated) and 358.1482 (found).
The title compound was characterized through 1H NMR and 13C NMR. The 1H NMR spectra were recorded on a Bruker 400 Avance spectrometer (400 MHz). The 13C NMR spectrum was recorded on a Bruker spectrometer at 100 MHz. Chemical shifts (δ scale) are reported in parts per million (ppm) relative to the central peak of the solvent. 1H NMR spectra are reported in the order multiplicity and number of protons; signals were characterized as s (singlet), dd (doublet of doublet), t (triplet), m (multiplet), or br s (broad signal). HRMS experiments were performed using an LTQ Orbitrap XL Thermo (Thermo Scientific) instrument coupled to high-pressure liquid chromatography (HPLC) with an Alltima C18 column (5 μm by 150 mm by 4.6 mm; Alltech Italia Srl) (flow rate, 1 ml/min; isocratic elution for 5 min with 70% CH3CN-H2O with 0.2% formic acid, gradient elution over 25 min from 95% CH3CN-H2O with 0.2% formic acid, and isocratic elution for 10 min with 95% CH3CN-H2O with 0.2% formic acid). Reactions were monitored by thin-layer chromatography (TLC) on a Kieselgel 60 F 254 instrument (DC-Alufolien, Merck).
Mutant isolation.
Wild-type M. tuberculosis H37Rv was grown in 7H9 broth to stationary phase (optical density at 600 nm [OD600], 1.98). A 500-μl portion was added to 7H10 plates containing 0.5, 1, 2, 4, or 8 μg/ml (2× to 32× the MIC) of MP-III-71 and incubated at 37°C for 1 month. Colonies grew on all plates, including three colonies on the 8-μg/ml plate. Each of these colonies was grown in 7H9 containing 8 μg/ml of MP-III-71. Of these, one colony did not grow. The other two were streaked onto 7H10 plates with 8 μg/ml MP-III-71, and single colonies were isolated. One colony was picked from each plate, and these became mutant 1 and mutant 2. Mutant 3 was selected using the same method but from a biological repeat. In this experiment, the plate with the highest concentration of drug with colonies contained two colonies growing on 1 μg/ml, one large and one small. The large colony became mutant 3.
Drug susceptibility testing.
All drug susceptibility testing was performed using the microplate alamarBlue assay (MABA) as previously described (3). In short, bacteria were added at an OD600 of 0.001 to drug dilutions in 7H9 without Tween in a 96-well plate. Plates were incubated at 37°C for 7 days, and then alamarBlue was added and left for 16 h before plates were read using a fluorescence microplate reader at 544-nm excitation and 590-nm emission wavelengths. Percent inhibition was calculated based on the relative fluorescence units, and the MIC was defined as minimum concentration that resulted in at least 90% inhibition. MP-III-71, N-methylated MP-III-71, rifampin (Sigma), and carbonyl cyanide 3-chlorophylhydrazone (CCCP) (Sigma) were dissolved in DMSO. Isoniazid (Fluka Analytical), ethambutol (Sigma), kanamycin (Sigma), verapamil hydrochloride (Sigma), chlorpromazine hydrochloride (Sigma), and sodium salicylate (Sigma) were dissolved in water.
DNA extraction.
Extraction of genomic DNA was performed on 10-ml cultures in 7H9 broth (mutants were grown in 7H9 with 8 μg/ml MP-III-71) using the cetyltrimethylammonium bromide (CTAB)-lysozyme method as previously described (4).
Whole-genome sequencing and analysis.
For samples run on the Ion Torrent Personal Genome Machine (PGM), 5 μg of genomic DNA was sheared using the Covaris S2 DNA system. The library was then prepared and enriched using the Ion Xpress Plus genomic DNA (gDNA) kit and Ion OneTouch Template kit on the Ion OneTouch, with sequencing performed using the Ion Sequencing kit v2.0 and 316 chip (Life Technologies). For samples run on the SOLiD, 10 μg of genomic DNA was submitted to the Johns Hopkins SKCC Next Generation Sequencing Center for sequencing. Libraries were constructed according to the protocols provided by Life Technologies (Fragment Library kit) and were run on a high-sensitivity chip using the Agilent Bioanalyzer to assess the size distribution and quality of the amplified library. Quantification of each library was performed by quantitative PCR (qPCR) using the TaqMan gene expression assay as outlined in the Applied Biosystems SOLiD Library Preparation Guide. Libraries were brought to a final concentration of 500 pM, and emulsion PCR was performed to generate 41,000,000 beads to deposit onto an Octet slide. Sequencing was performed on the Applied Biosystems SOLiD 3 Plus system using a single-read, 50-bp fragment run.
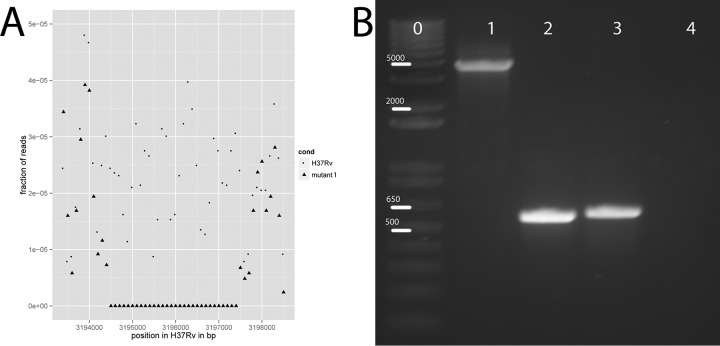
Reads were aligned to M. tuberculosis H37Rv (GI:41353971 from GenBank) using the Burrows-Wheeler alignment, and SNPs were called using the GATK toolkit (5–7). Coverage analysis to identify the deletion was performed by comparing the fraction of reads in the parent H37Rv sequence to the fraction of reads in the mutant sequence in each 100-bp window of the alignment and identifying the windows with >40% difference in coverage. From these data, the deletion was picked out as a region with no reads in mutant 1 and an average of 53.6 reads per 100-bp window in the Ion Torrent wild-type H37Rv sequence.
Mutation confirmation.
For mutant 1, primers were designed using Primer3 to span the putative deletion (8). The forward primer was 5′-GTAGCGTGCGAGGTTGAT-3′ and the reverse primer was 5′-GAAGCGTTCTTCAGTGGAGT-3′, with expected sizes of 3,753 bp in the parent strain and 663 bp in the mutant strain. In addition, primers to span Rv2887 were used for all three mutants. The forward primer was 5′-AATGCGATGTAGGCTTCAC-3′, and the reverse primer was 5′-ATCCACGCCCCAAATATC-3′. Primers were synthesized by Integrated DNA Technologies and diluted to 10 μM in nuclease water. PCR was performed using the 25-μl Taq 2× master mix (New England BioLabs), 1 μl forward primer, 1 μl reverse primer, 1 μl genomic DNA, and 22 μl nuclease-free water. The PCR program was 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 53°C for 30 s, and 1 min at 68°C. This was finished with 7 min at 68°C, and then products were held at 4°C. PCR products were run on a 1% agarose gel with a 1-kb Plus ladder (Life Technologies), and then the bands were purified using the QIAquick gel extraction kit (Qiagen). The purified product was diluted to 1.75 ng/μl and submitted with the forward and reverse primers to Genewiz for Sanger sequences. BLAST was used to align the resulting sequences against Rv2887 to confirm the presence of the mutation (9).
Complementation.
Primers were designed using Primer3 to include all of Rv2887 and 500 bp upstream of the start of Rv2887 (8). The forward primer was 5′-GGTATAGGTACCGGTCACGCCTACCACTTG-3′, which included a cut site for KpnI, and the reverse primer was 5′-ATCCTCTCTAGATGATGCTCTCGGCTGATAC-3′, which included a cut site for XbaI, with an expected size of 1,089 bp. The primers were dissolved in nuclease-free water and diluted to 10 μM. PCR was performed using 1 μl of each primer, 1,500 ng of genomic DNA from M. tuberculosis H37Rv, 25 μl Taq 2× master mix (New England BioLabs), and nuclease-free water to bring the volume to 50 μl. The resulting mix was thermocycled at 95°C for 3 min, followed by 30 rounds of 95°C for 30 s, 58°C for 30 s, and 60°C for 2 min. This was finished with 7 min at 68°C. The resulting product was purified using the QIAquick PCR purification kit (Qiagen).
Restriction digestion was performed on 2 μg of the pMH94h plasmid and 2 μg of the purified PCR product (10). The digest consisted of 0.2 μl KpnI-HF (New England BioLabs), 0.2 μl XbaI (New England BioLabs), 5 μl NEB buffer 4, and 44.6 μl DNA and nuclease-free water. The mix was incubated at 37°C for 4 h. The digested DNA was run on a 1% agarose gel and the bands extracted using the QIAquick gel extraction kit (Qiagen). The digested plasmid was then treated with calf intestinal alkaline phosphatase (CIP) (New England BioLabs). Fifty nanograms of CIP-treated digested plasmid was ligated with 150 ng of digested PCR product using the quick ligation kit from New England BioLabs. The ligated product was used to transform Escherichia coli One Shot TOP10 electrocompetent cells (Life Technologies), and the transformed bacteria were plated on LB agar containing 150 μg/ml hygromycin B (Roche). Plates were incubated overnight at 37°C, and four single colonies were picked the next day and grown in 5 ml LB broth with 150 μg/ml hygromycin B. The next day, the plasmid was isolated using the QIAprep spin miniprep kit (Qiagen).
To confirm the plasmid sequence, 600 ng of purified plasmid was digested as described above, and the digest was run on 1% agarose to confirm the presence of the insert. All four products were submitted to Genewiz for Sanger sequencing, along with the forward primer 5′-AGCGCATAGGAACGATTTAC-3′ and the reverse primer 5′-ACCCGGTAGAGCAGATAGC-3′. BLAST was used to confirm the correct sequence in all four plasmids; one was chosen randomly to continue (9).
Each strain was grown to an OD600 of around 0.8 in 50 ml of 7H9 and made electrocompetent. In brief, the culture was spun down for 10 min at 37°C at 4,500 rpm and was resuspended in 30 ml of 10% glycerol (Sigma). This was repeated 4 times, halving the volume used for resuspension each time, until the bacteria were finally resuspended in 2.5 ml 10% glycerol. One hundred microliters of cells was incubated at 55°C for 5 min, and then the bacteria were electroporated with 25 ng/μl plasmid in 50 μl water using 330 μF, 375 V, and 8 kΩ. The cells were then transferred into 2 ml of 7H9 and incubated at 37°C for 48 h. The transformed cells were then centrifuged at 10,000 rpm for 5 min and resuspended in 200 μl, and the entire volume was plated on 7H11 with 50 μg/ml hygromycin B. Plates were incubated 37°C for 4 weeks.
Transposon mutants.
Transposon mutants were available through the TARGET project (http://webhost.nts.jhu.edu/target/).
RNA extraction.
Total RNA was extracted from 50 ml of culture at an OD600 of around 1. M. tuberculosis cultures were centrifuged, and the bacterial pellet was resuspended in TRIzol (Invitrogen). This mixture was transferred to 1.8-ml O-ring tubes containing 0.5 ml of 0.1-mm glass beads (BioSpec Products). Cells were incubated at 25°C for 10 min and lysed by six cycles of bead beating for 30 s and cooling on ice for 1 min, using a mini-beadbeater at 4,800 rpm. Lysed cells were centrifuged for 5 min at 13,000 rpm, the supernatant was transferred to a fresh microcentrifuge tube, and RNA was then extracted as described previously (11, 12). The quality of RNA was assessed using a NanoDrop (ND-1000; Labtech) and an Agilent 2100 Bioanalyzer (Agilent Technologies).
qRT-PCR.
Fifteen micrograms of RNA was treated with DNase I (New England BioLabs) according to the manufacturer's instructions. Reverse transcription was performed using iScript reverse transcriptase (Bio-Rad) on 1 μl of DNase-treated RNA. The primers used for Rv2887 for quantitative reverse transcription-PCR (qRT-PCR) were 5′-GTTCGCTACCGGCTACATTG-3′ (forward) and 5′-CTAGTCGGACCCGAGCTTCT-3′ (reverse). The primers for our control housekeeping gene, sigA, were 5′-CCATCCCGAAAAGGAAGACC-3′ (forward) and 5′-TCGAGGTCTGGTTCAGCGTC-3′ (reverse). sigA is a housekeeping gene whose expression remains constant, and it is commonly used for M. tuberculosis (13). qRT-PCR was performed on 2 μl cDNA using the iQ SYBR green Supermix (Bio-Rad) on the Applied Biosystems StepOnePlus using 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 53°C for 1 min and then a melt curve from 55 to 95°C. The average fold changes compared to the wild type, normalized to sigA, were calculated using data from two technical replicates of three independent experiments.
Bioinformatics analysis.
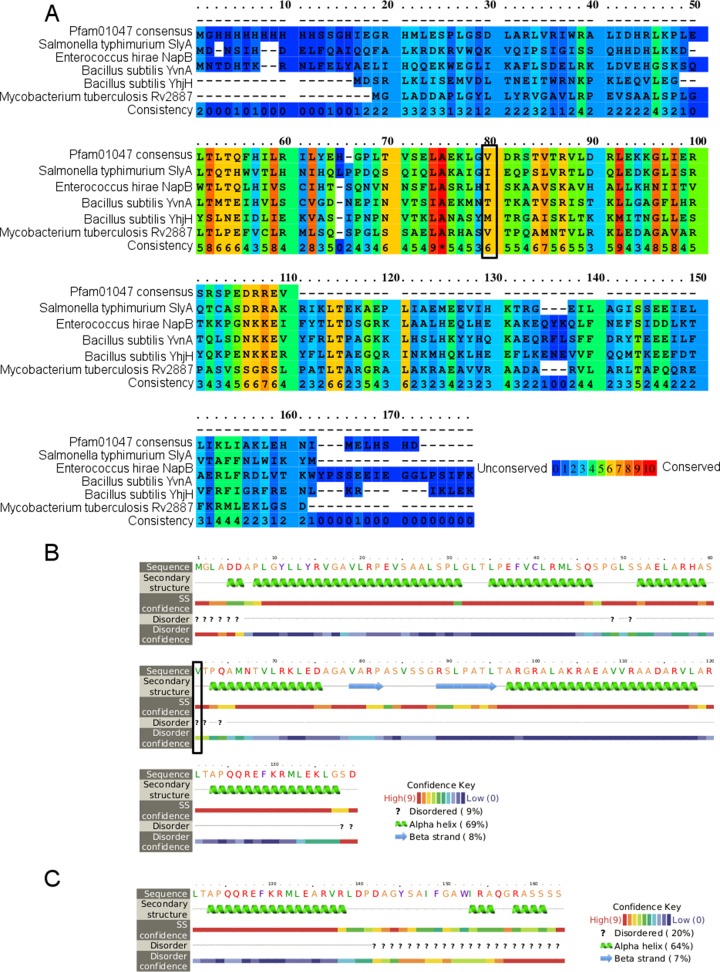
Protein BLAST (BLASTP) alignment was performed using the online tool (9). Consensus sequences for MarR and MarA were downloaded from the conserved domain database (14). Secondary structures were predicted using the default settings for the online versions of Phyre2, PROMALS3D, and PRALINE (15, 16). All tools made the same prediction; the results from Phyre2 are shown. Several alignment tools were used, including PRALINE, MUSCLE, and ClustalW, all on the default settings (17–19). The alignment from PRALINE is the one depicted in Fig. 2. The MarR protein sequences to align were selected from CDD, using the top five most diverse members, including the consensus sequence (14). The GI numbers for these sequences were 192988597 (Salmonella enterica serovar Typhimurium SlyA), 75341253 (Enterococcus hirae NapB), 81637589 (Bacillus subtilis YvnA), and 81703996 (Bacillus subtilis YhjH). The online tool for SIFT was used to assess the effect of nonsynonymous mutations (20).
FIG 2.
Rv2887 is in the MarR family. (A) Alignment of Rv2887 to the consensus sequence for Pfam01047 (MarR) and four other MarR family proteins (selected as the five most diverse members in the conserved domain database, including the consensus sequence) (14). The bottom row is the consistency, as estimated by PRALINE (17). (B) Secondary structure prediction from Phyre2 for Rv2887 (16). (C) Secondary structure prediction for the C terminus of mutant 3. The black boxes in panels A and B indicate V61, which is mutated to alanine in mutant 2.
Transcriptome sequencing (RNA-seq) and analysis.
The strains were grown to an OD600 of 0.6 in 150 ml, and MP-III-71 or an equivalent volume of DMSO was added at to a final concentration of 0.0625 μg/ml MP-III-71. Cultures were incubated with shaking at 37°C for 6 h, and then RNA extraction was performed as described above. rRNA was removed using the Ambion MICROBExpress bacterial mRNA enrichment kit (Life Technologies) and the results checked using the Agilent 2100 Bioanalyzer (Agilent Technologies). Library preparation was performed using the Ion Total RNA-Seq kit v2 (Life Technologies). Samples were barcoded using the Ion Xpress RNA barcodes, and each replicate was pooled into one library. Template preparation was performed with the Ion PGM template OT2 200 kit using the Ion OneTouch 2 and Ion OneTouch ES (Life Technologies). Sequencing was performed on the Ion Personal Genome Machine using the Ion 318 chip with the Ion PGM sequencing 300 kit with weighted buckets (Life Technologies). Reads were aligned to M. tuberculosis H37Rv (GI:41353971 from GenBank) using the Burrows-Wheeler alignment (6). Differential expression was assessed using DESeq, EdsgeR, and Cufflinks, all of which gave the same results (21–23). Results from DE-Seq are presented here.
Sequence accession numbers.
All sequencing data have been deposited in the GenBank Sequence Read Archive under BioProject PRJNA280011 with the following accession numbers: SRS889709, SRS891424, SRS891455, SRS891666, SRS893802, SRS891491, SRS891670, SRS891672, SRS891793, SRS891794, SRS891795, SRS891796, SRS891857, SRS891859, SRS891860, SRS891862, SRS891863.
RESULTS
Whole-genome sequencing reveals that Rv2887 mutations confer MP-III-71 resistance.
Spontaneous mutants resistant to MP-III-71 were selected by growing M. tuberculosis H37Rv on 7H9 agar plates containing up to 8 μg/ml of the compound, a concentration which is 32 times the MIC (see Materials and Methods). A total of three mutant colonies were identified during two separate selections, and their resistance to MP-III-71 was confirmed using the microplate alamarBlue assay (MABA), as shown in Table 1. Each mutant had an MIC of 1 to 2 μg/ml, which is 2 to 8 times the MIC of the parent strain, which showed an MIC of 0.25 to 0.5 μg/ml.
TABLE 1.
Summary of mutants
| Strain | MIC (μg/ml) |
Sequencing platform | Rv2887 mutation | |
|---|---|---|---|---|
| MP-III-71 | Complement MP-III-71 | |||
| H37Rv | 0.25–0.5 | SOLiD and PGM | ||
| Mutant 1 | 2 | 0.25–0.5 | PGM | Deletion at positions 3194362–3197545 (3,183 bp), which includes Rv2885c, Rv2886c, Rv2887, and Rv2888c |
| Mutant 2 | 1–2 | 0.25–0.5 | PGM | 2-bp deletion at positions 3196830–3196831, which is nucleotide 399 in Rv2887 (amino acid 133) |
| Mutant 3 | 1 | 0.25–0.5 | SOLiD | V61A nonsynonymous SNP in Rv2887 |
Genomic DNA from each of the three mutants and the wild-type parent H37Rv strain was submitted for deep sequencing. Mutants 1 and 2 and wild-type H37Rv were sequenced on an Ion Torrent Personal Genome Machine (PGM), with an average of 1.4 million reads and an average read length of 111 bp (see Table S1 in the supplemental material). The parent H37Rv strain was resequenced using an ABI SOLiD instrument, along with mutant 3, with an average of 25.2 million 50-bp reads (see Table S1 in the supplemental material). Reads were aligned to the M. tuberculosis H37Rv reference genome, and mutations identified in either of the two parental H37Rv sequences were removed from further analysis. After this analysis, mutant 1 had 7 SNPs and 11 indels, mutant 2 had 9 SNPs and 4 indels, and mutant 3 had 1 SNP (see Table S2 in the supplemental material). Using the TARGET collection of mutants, we tested the MP-III-71 susceptibility of M. tuberculosis CDC1551 strains with a transposon inserted in Rv2658c or Rv3668c, the only two genes mutated in our study that had an available transposon mutant (24). However, neither of these mutants had an altered susceptibility to MP-III-71 (see Table S3 in the supplemental material).
Interestingly, Rv2887 was mutated in both mutants 2 and 3, with a 2-bp deletion in mutant 2 and a nonsynonymous mutation in mutant 3, both of which were confirmed with Sanger sequencing of PCR-amplified DNA fragments (Table 1). In addition, we analyzed the coverage of each strain and found a 3,183-bp deletion in mutant 1, which includes Rv2887 (Table 1; Fig. 1A). The deletion was confirmed by PCR with primers inside and outside the deleted area (Fig. 1B), and the outside PCR product was submitted for Sanger sequencing to identify the exact boundaries of the deletion. From this, we determined that the deletion was from position 3194362 to position 3197545 in the H37Rv reference nucleotide sequence, resulting in partial deletion of Rv2885c and Rv2888c and full deletion of Rv2886c and Rv2887. Thus, we hypothesized that loss of function of Rv2887 confers resistance to MP-III-71.
FIG 1.
The 3,183-bp deletion in mutant 1, which includes Rv2887. (A) Coverage analysis of sequencing results for M. tuberculosis H37Rv and mutant 1, showing a lack of reads in this region for the mutant but not the parent H37Rv strain. (B) Confirmation of the deletion mutation. Lane 0, Invitrogen 1-kb Plus DNA ladder. Numbers over the bars indicate the size in base pairs of the ladder. Lane 1, PCR of genomic DNA from the parent H37Rv strain with primers outside the region of the deletion. Lane 2, PCR of genomic DNA from mutant 1 with the same primers as for lane 1. Lane 3, PCR of genomic DNA from H37Rv with primers inside the deletion (in Rv2887). Lane 4, PCR of mutant 1 genomic DNA with the same primers as for lane 3.
Complementation of Rv2887 confirms that mutation of this gene confers resistance to MP-III-71.
Wild-type Rv2887 was cloned into an integrating pMH94-derived plasmid, along with 500 bp upstream to capture its native promoter (10). The plasmid was introduced into each of the three mutants, and MABA was performed with MP-III-71. In addition, in mutant 1, which had a full deletion of Rv2887, real-time PCR was performed to confirm that expression of Rv2887 from the plasmid is the same as wild-type levels (see Fig. S2 in the supplemental material). Introduction of the wild-type version of Rv2887 was sufficient to restore susceptibility to MP-III-71 to wild-type levels in all three mutants (Table 1), confirming that loss of function of Rv2887 results in resistance to MP-III-71.
Comparative analysis of Rv2887 with known MarR transcriptional regulators.
BLAST analysis revealed that Rv2887 belongs to the MarR Pfam family, Pfam01047, with an Expect (E) value of 6.38 × 10−13 (9, 25, 26). MarR homologs contain a winged helix-turn-helix motif, and they have been shown to play a role as either transcriptional repressors or activators of several different pathways, including response to antibiotics and oxidative stress (27, 28). The E. coli mar (multiple antibiotic resistance) locus, which has been well characterized, consists of two transcriptional units, marC and marRAB, which are under the control of a centrally located promoter region between these two divergent operons. MarR, which has been shown to function as a repressor in E. coli, binds to repeats within the operator and prevents transcription until bound by certain chemical compounds, such as tetracycline, chloramphenicol, and sodium salicylate (29). Binding induces a conformational change in MarR, which reduces its affinity for the repressor DNA sequence, allowing transcription of marC and marRAB. MarA, which functions as a transcriptional activator in E. coli, is then expressed, leading to elevated transcription of a diverse regulon of genes (29). In E. coli, this regulon governs numerous functions, including upregulation of efflux pumps, and is associated with the multiple antibiotic resistance phenotype.
Alignment of Rv2887 and the mutants against the consensus Pfam01047 sequence and other members of this family showed that valine 61, mutated to alanine in mutant 3, is a conserved amino acid in MarR-like proteins in multiple species, and thus this small amino acid change may have a significant impact on protein function (Fig. 2) (14). Several alignment tools were used, including PRALINE, MUSCLE, and ClustalW, to confirm this finding; Fig. 2 shows the results from PRALINE (17–19). The importance of this amino acid was confirmed by SIFT, which predicted that the V61A mutation will affect protein function, based on the conservation of amino acid residues (20). In addition, the 2-bp deletion in mutant 2 changes the four C-terminal residues into 29 residues, increasing the length of the protein and adding an additional disordered region including two new alpha helices (Fig. 2C). Thus, the changes in both mutant 2 and mutant 3 are predicted to affect Rv2887 function.
Efflux inhibitors and resistance to MP-III-71.
Based on the role of MarR in regulating efflux pumps in E. coli, we tested the efflux pump inhibitors verapamil, chlorpromazine, and carbonyl cyanide 3-chlorophenyl hydrazone (CCCP) (30–33). Verapamil and chlorpromazine compounds had modest potentiating effects on the MIC of MP-III-71 to wild-type H37Rv and complemented mutant 1; however, they did not alter the MIC of the resistant strain, mutant 1 (see Table S4 in the supplemental material). On the other hand, CCCP, which is an oxidative phosphorylation inhibitor that uncouples the membrane proton gradient required for some efflux pumps to act, actually inhibited the activity of MP-III-71 for wild-type H37Rv and complemented mutant 1. This may be related to the highly pleiotropic effects of uncoupling the membrane proton gradient rather than to a specific effect on small-molecule efflux. Thus, MP-III-71 does appear to be subject to drug efflux by pumps that are susceptible to verapamil and chlorpromazine, and mutation of Rv2887 abrogates the efflux processes targeted by verapamil or chlorpromazine. M. tuberculosis H37Rv has an estimated 148 efflux pumps, and so mutation of Rv2887 may have additional effects on efflux pumps other than those blocked by verapamil and chlorpromazine (34).
Given the role of MarR in drug resistance in E. coli and other organisms, we also tested whether any of the mutants had altered susceptibility to the anti-TB antibiotics isoniazid, rifampin, ethambutol, and kanamycin. None of the mutants showed MIC changes to these drugs compared to the wild type (see Table S4 in the supplemental material; data not shown for mutants 2 and 3). This correlates with our previous findings that MP-III-71 is effective against drug-resistant clinical isolates and inhibits a unique pathway not targeted by existing anti-TB drugs.
Transcriptional profiling of an MP-III-71-resistant mutant.
Since Rv2887 is a putative transcriptional regulator, we performed RNA-seq on the Ion Torrent PGM to determine whether it governs the transcription of other genes. We chose to focus on mutant 1 and complemented mutant 1 because mutant 1 had the full deletion of Rv2887 and because it had the highest MP-III-71 MIC. We harvested the RNA after 6 h of incubation with 0.0625 μg/ml MP-III-71 (1/4 of the wild-type MIC) or with an equivalent volume of DMSO only. Triplicate samples were run on the Ion Torrent PGM, with an average of 1.2 million reads and a 115-bp read length (see Table S1 in the supplemental material).
After performing differential expression analysis comparing the mutant and its complemented strain, we identified six genes that were significantly differentially expressed between the mutant and the wild type in the presence of MP-III-71, including Rv2887 (Table 2). All genes identified as significant were upregulated in the MP-III-71-resistant mutant compared to the complemented strain, with the exceptions of Rv2887 and Rv2886c. Rv2886c downregulation in the mutant is probably due to artifactual overexpression of this gene in the comparator strain, since the end of this gene is at the start of Rv2887, and the complementation plasmid contained 500 bp upstream of Rv2887. The fact that the other genes are upregulated with the deletion of Rv2887 in the presence of MP-III-71 is consistent with the hypothesis that Rv2887 serves as a transcriptional repressor when the drug is present. Furthermore, the fact that these genes have a small change in expression when treated only with DMSO suggests that Rv2887 does not constitutively repress these genes but rather prevents their expression in the presence of MP-III-71.
TABLE 2.
Significant hits from RNA-seq analysis
| Gene | Mutant 1 + MP-III-71 vs complement + MP-III-71 |
Mutant 1 + DMSO vs complement + DMSO |
TubercuList annotation | ||
|---|---|---|---|---|---|
| Log2 fold change | Adjusted P value | Log2 fold change | Adjusted P value | ||
| Rv0560c | −7.67 | 6.98E−58 | 0.00 | 3.29E−53 | Possible benzoquinone methyltransferase (methylase) |
| Rv2887 | 7.16 | 4.38E−20 | 75.40 | 1.38E−12 | |
| Rv0558 | −3.06 | 1.16E−12 | 0.08 | 1.96E−14 | MenH (probable ubiquinone/menaquinone biosynthesis methyltransferase MenH) |
| Rv0559c | −3.63 | 4.70E−11 | 0.05 | 3.38E−13 | Possible conserved secreted protein |
| Rv2463 | −1.59 | 0.01 | 0.35 | 0.02 | LipP (probable esterase/lipase) |
| Rv2886c | 2.34 | 0.03 | 3.28 | 1.00 | Probable resolvase |
The regulon of Rv2887 includes genes involved in menaquinone biosynthesis and a potential MarA protein.
Three of the significant genes identified in our RNA-seq analysis as being upregulated upon Rv2887 deletion were Rv0558, Rv0559c, and Rv0560c. These three genes form a cluster in the H37Rv genome, although Rv0558 is convergently transcribed compared with the Rv0560c and Rv0559c genes and thus is unlikely to be in the same operon. Rv0559c encodes a nonessential exported protein found in culture filtrate, membrane, and whole-cell lysate and, along with Rv0560c, is upregulated in rifampin-resistant strains and induced by salicylate (35–39). Rv0560c is a benzoquinone methyltransferase involved in the biosynthesis of isoprenoid compounds, and it is also upregulated under iron-limited and anaerobic conditions (35, 40–42). Rv0558 (menH) encodes an S-adenosylmethionine-dependent methyltransferase found in the membrane that catalyzes the final step in menaquinone biosynthesis (35, 43, 44). Menaquinone (vitamin K) is an essential electron carrier in the respiratory chain and is particularly important in M. tuberculosis survival under low-oxygen conditions (44). The other gene upregulated by Rv2887 deletion was Rv2463. This gene has been annotated as lipP, a probable esterase/lipase gene (35). LipP has a low level of long-chain triacylglycerol hydrolase activity and is induced after 6 h of nutrient starvation (45).
Given that Rv2887 is in the MarR family but M. tuberculosis lacks the adjacent marRAB locus identified in E. coli, we used BLAST to seek E. coli MarA (accession number EDV65186.1) homologs which may be situated elsewhere in the H37Rv genome (9). Only 6 genes were identified, one of which was Rv2463, with an E value of 0.23 (see Table S5 in the supplemental material). Although a function for Rv2463 as a MarA-like transcriptional activator seems unlikely for a protein with lipase activity, especially given the relatively high E value, we used this same method to look for other MarR-like proteins in H37Rv with E. coli MarR (accession number AAK21292.1) as the query, and picked up Rv2887, in addition to 7 other potential MarR-like proteins (see Table S5 in the supplemental material). Thus, M. tuberculosis H37Rv possesses several putative MarR- and MarA-like proteins. However, further studies will be needed to assess function.
Rv2887-dependent susceptibility to MP-III-71 may involve drug methylation.
We hypothesized that mutation of Rv2887 may be indirectly causing resistance to MP-III-71 by altering expression of its true target. We had access to Rv0559c and Rv2463 transposon mutants (24). Given that loss of Rv2887 caused these genes to be upregulated, we hypothesized that deletion of these genes might result in MP-III-71 hypersusceptibility. We tested the MIC of MP-III-71 for the Rv0559c and Rv2463 mutants. However, neither of these mutants had an altered susceptibility to MP-III-71, suggesting that their upregulation with loss of Rv2887 is not the cause of MP-III-71 resistance (see Table S3 in the supplemental material). However, the transposon in Rv2463 was inserted 2 amino acids from the C terminus, so this mutant may retain some degree of intact function (see Table S3 in the supplemental material).
This left altered expression of Rv0558 and Rv0560c as potential reasons for MP-III-71 resistance. Rv0560c is upregulated in the presence of salicylate, which is of particular interest since salicylate is one of the compounds that interferes with the repressor activity of MarR in E. coli (46–48). As a result, we hypothesized that salicylate may turn off the repressor activity of Rv2887 and thus may affect the susceptibility of the wild-type strains to MP-III-71. Thus, we tested the susceptibility of mutant 1 to salicylate, but we found that deletion of Rv2887 had no effect on the MIC of salicylate (all strains had MICs of 250 to 500 μg/ml). Sub-MIC levels of salicylate did not affect the susceptibility of either the wild-type or complemented strain to MP-III-71, although it did slightly reduce the MIC of the mutant strain (see Table S4 in the supplemental material). These findings suggest that our hypothesis that salicylate inhibits repressor activity of Rv2887 is incorrect and that, in contrast to the case for E. coli MarR, the M. tuberculosis MarR-like regulator Rv2887 may not be susceptible to salicylate inhibition.
Since Rv0558 and Rv0560c are methylases, we next hypothesized that MP-III-71 may be methylated, leading to inactivation of the compound. Upregulation of these genes by altering Rv2887 function would increase the amount of methylated, inactivated drug. We tested our strains for susceptibility to N-methylated MP-III-71 (see Fig. S1b in the supplemental material). Interestingly, this compound had no effect on the growth of any strain, even at the highest concentrations tested (32 μg/ml) (see Table S4 in the supplemental material). Thus, methylation of MP-III-71 abrogates its antimicrobial activity. Although we have not demonstrated that methylation occurs in vivo or that the purified Rv0558 or Rv0560c is capable of methylating MP-III-71, the inactivity of N-methyl-MP-III-71 suggests that one potential mechanism of resistance to MP-III-71 may be through drug methylation.
DISCUSSION
The development of new drug targets and new drugs to combat M. tuberculosis is critical as rates of drug resistance increase. Here, we explored the function of one potential new antimycobacterial compound, MP-III-71, and we showed that loss-of-function mutations in Rv2887 confer resistance to this compound.
Rv2887 is a transcriptional regulator in the MarR family. Very little is known about the role of the mar operon in mycobacteria. McDermott et al. showed that expression of E. coli MarA in M. smegmatis increases resistance to several antibiotics, including rifampin, isoniazid, ethambutol, tetracycline, and chloramphenicol, suggesting that a mar-like system is present in mycobacteria (49). Following this, Zhang et al. showed that in M. smegmatis, Ms6508 is a MarR-like protein whose corresponding marRAB operon confers rifampin resistance (50). However, both of these studies were performed in the nonpathogenic organism M. smegmatis. To our knowledge, only one other study has focused on the mar operon in M. tuberculosis. Radhakrishnan et al. showed that Rv0678 is a MarR-like regulator that controls transcription of the MmpS5-MmpL5 transporter, providing direct evidence of a mar-like operon in M. tuberculosis (51).
Despite the function of its homologs in drug resistance, loss-of-function mutations in Rv2887 do not confer resistance to rifampin or isoniazid. We did observe that efflux pump inhibitors potentiate the action of MP-III-71 and that this potentiation is abrogated by Rv2887 mutation. Hence, loss of Rv2887 appears to increase drug efflux in M. tuberculosis, and this may be one mechanism of drug resistance in Rv2887 mutants. We also found that intact Rv2887 negatively regulates the transcription of Rv0558, Rv0559c, Rv0560c, and Rv2463. Rv2463, also known as lipP, encodes a lipase that has weak homology to E. coli MarA (BLAST E value of 0.23) (see Table S5 in the supplemental material); MarA serves as the activator in the E. coli mar system, which in E. coli is negatively regulated by MarR. While the only known function of Rv0559c is that it is secreted, Rv0560c methylates benzoquinone (coenzyme Q) and Rv0558 methylates menaquinone, both of which play an important role in electron transport. Thus, mutation of Rv2887 may confer resistance to MP-III-71 by upregulating expression of these two genes, resulting in altered cellular energy levels and transport. This may lead to enhanced methylation of MP-III-71, causing resistance through drug inactivation.
This study revealed that M. tuberculosis susceptibility to MP-III-71 is Rv2887 dependent. The efflux pump inhibitors verapamil and chlorpromazine potentiated the action of MP-III-71, but this potentiation was absent in the Rv2887 mutant. Rv2887 encodes a MarR-like protein which inhibits the expression of at least six genes, including Rv0558, encoding a menaquinone methyltransferase, and Rv0560c, encoding a benzoquinone methyltransferase. A methylated derivative of MP-III-71 is inactive. These results suggest that loss of Rv2887, leading to elevated efflux pump activity and methylase expression, accounts for MP-III-71 resistance in M. tuberculosis Rv2887 mutants.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge National Institutes of Health grants AI39856, AI36973, and AI097138 and the Howard Hughes Medical Institute for funding this work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01341-15.
REFERENCES
- 1.World Health Organization. 2014. Tuberculosis. http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf?ua=1 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Pieroni M, Tipparaju SK, Lun S, Song Y, Sturm AW, Bishai WR, Kozikowski AP. 2011. Pyrido[1,2-a]benzimidazole-based agents active against tuberculosis (TB), multidrug-resistant (MDR) TB and extensively drug-resistant (XDR) TB. Chemmedchem 6:334–342. doi: 10.1002/cmdc.201000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins L, Franzblau SG. 1997. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother 41:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR Jr. 2007. Genetic manipulation of Mycobacterium tuberculosis. Curr Protoc Microbiol Chapter 10:Unit 10A12. [DOI] [PubMed] [Google Scholar]
- 5.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 11:11–10-11–11-10-33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee MH, Pascopella L, Jacobs WR Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A 88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatfull GF, Jacobs WR Jr (ed). 2000. Molecular genetics of mycobacteria. ASM Press, Washington, DC. [Google Scholar]
- 12.Larsen MH. 2000. Appendix 1. Some common methods in mycobacterial genetics. RNA preparation—Trizol, p 317 In Hatfull GF, Jacobs WR Jr (ed), Molecular genetics of mycobacteria. ASM Press, Washington, DC. [Google Scholar]
- 13.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol 31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 14.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei J, Tang M, Grishin NV. 2008. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res 36:W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 17.Simossis VA, Heringa J. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 21.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulavik MC, Dazer M, Miller PF. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol 179:1857–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol 8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol 8:51–62. [PubMed] [Google Scholar]
- 28.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol 2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 29.Alekshun MN, Levy SB. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 7:410–413. doi: 10.1016/S0966-842X(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 30.Szumowski JD, Adams KN, Edelstein PH, Ramakrishnan L. 2013. Antimicrobial efflux pumps and Mycobacterium tuberculosis drug tolerance: evolutionary considerations. Curr Top Microbiol Immunol 374:81–108. doi: 10.1007/82_2012_300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawkins MJ, Judah JD, Rees KR. 1959. The mechanism of action of chlorpromazine. Reduced diphosphopyridine nucleotidecytochrome c reductase and coupled phosphorylation. Biochem J 73:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawkins MJ, Judah JD, Rees KR. 1959. The effect of chlorpromazine on the respiratory chain; cytochrome oxidase. Biochem J 72:204–209. doi: 10.1042/bj0720204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkelstein A. 1970. Weak-acid uncouplers of oxidative phosphorylation. Mechanism of action on thin lipid membranes. Biochim Biophys Acta 205:1–6. [DOI] [PubMed] [Google Scholar]
- 34.Ren Q, Chen K, Paulsen IT. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res 35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList—10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Denkin S, Byrne S, Jie C, Zhang Y. 2005. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch Microbiol 184:152–157. doi: 10.1007/s00203-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 37.de Knegt GJ, Bruning O, ten Kate MT, de Jong M, van Belkum A, Endtz HP, Breit TM, Bakker-Woudenberg IA, de Steenwinkel JE. 2013. Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis (Edinb) 93:96–101. doi: 10.1016/j.tube.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 38.de Souza GA, Leversen NA, Malen H, Wiker HG. 2011. Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J Proteomics 75:502–510. doi: 10.1016/j.jprot.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Malen H, Berven FS, Fladmark KE, Wiker HG. 2007. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7:1702–1718. doi: 10.1002/pmic.200600853. [DOI] [PubMed] [Google Scholar]
- 40.Garbe TR. 2004. Co-induction of methyltransferase Rv0560c by naphthoquinones and fibric acids suggests attenuation of isoprenoid quinone action in Mycobacterium tuberculosis. Can J Microbiol 50:771–778. doi: 10.1139/w04-067. [DOI] [PubMed] [Google Scholar]
- 41.Starck J, Kallenius G, Marklund BI, Andersson DI, Akerlund T. 2004. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150:3821–3829. doi: 10.1099/mic.0.27284-0. [DOI] [PubMed] [Google Scholar]
- 42.Bacon J, Dover LG, Hatch KA, Zhang Y, Gomes JM, Kendall S, Wernisch L, Stoker NG, Butcher PD, Besra GS, Marsh PD. 2007. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology 153:1435–1444. doi: 10.1099/mic.0.2006/004317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu S, Chen J, Dobos KM, Bradbury EM, Belisle JT, Chen X. 2003. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics 2:1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Johnston JM, Arcus VL, Morton CJ, Parker MW, Baker EN. 2003. Crystal structure of a putative methyltransferase from Mycobacterium tuberculosis: misannotation of a genome clarified by protein structural analysis. J Bacteriol 185:4057–4065. doi: 10.1128/JB.185.14.4057-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. 2006. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem 281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol 181:4669–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, Cheng SJ, Zhang H, Zhang Y. 2001. Salicylate uniquely induces a 27-kDa protein in tubercle bacillus. FEMS Microbiol Lett 203:211–216. doi: 10.1111/j.1574-6968.2001.tb10843.x. [DOI] [PubMed] [Google Scholar]
- 48.Schuessler DL, Parish T. 2012. The promoter of Rv0560c is induced by salicylate and structurally-related compounds in Mycobacterium tuberculosis. PLoS One 7:e34471. doi: 10.1371/journal.pone.0034471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDermott PF, White DG, Podglajen I, Alekshun MN, Levy SB. 1998. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J Bacteriol 180:2995–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Gao L, Zhang J, Li W, Yang M, Zhang H, Gao C, He ZG. 2014. A novel marRAB operon contributes to the rifampicin resistance in Mycobacterium smegmatis. PLoS One 9:e106016. doi: 10.1371/journal.pone.0106016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radhakrishnan A, Kumar N, Wright CC, Chou TH, Tringides ML, Bolla JR, Lei HT, Rajashankar KR, Su CC, Purdy GE, Yu EW. 2014. Crystal structure of the transcriptional regulator Rv0678 of Mycobacterium tuberculosis. J Biol Chem 289:16526–16540. doi: 10.1074/jbc.M113.538959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.