Abstract
Hepatitis C virus (HCV) genotype 4 (GT4) is genetically diverse, with 17 confirmed subtypes, and comprises approximately 13% of infections worldwide. In this study, we identified GT4 subtypes by phylogenetic analysis, assessed differences in patient demographics across GT4 subtypes, examined baseline sequence variability among subtypes and the potential impact on treatment outcome, and analyzed the development of viral resistance in patients who received a regimen of ombitasvir (nonstructural protein 5A [NS5A] inhibitor) plus ritonavir-boosted paritaprevir (NS3/4A inhibitor) with or without ribavirin (RBV) for the treatment of HCV GT4 infection. Phylogenetic analysis of HCV NS3/4A, NS5A, and NS5B nucleotide sequences identified 7 subtypes (4a, 4b, 4c, 4d, 4f, 4g/4k, and 4o) among 132 patient samples. Subtype prevalence varied by country, and the distributions of patient birth cohort and race were significantly different across GT4 subtypes 4a, 4d, and non-4a/4d. Baseline amino acid variability was detected in NS5A across GT4 subtypes but had no impact on treatment outcome. Three patients experienced virologic failure and were infected with subtype 4d, and the predominant resistance-associated variants at the time of failure were D168V in NS3 and L28V in NS5A. Overall, high response rates were observed among patients infected with 7 HCV GT4 subtypes, with no impact of baseline variants on treatment outcome. GT4 subtype distribution in this study differed based on patient demographics and geography.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major public health concern and a leading cause of chronic liver disease and hepatocellular carcinoma (1). Globally, 130 to 150 million people are chronically infected with HCV (2), which is genetically diverse and classified into 7 genotypes and 67 subtypes (3). HCV genotype 4 (GT4) encompasses 17 confirmed subtypes (3) and comprises approximately 13% of infections worldwide, with prevalence varying by geographic region (4). HCV GT4 is the most prevalent genotype in North Africa, the Middle East, and central and eastern sub-Saharan Africa, and its prevalence has been increasing in Europe (4–6). In Egypt, GT4 accounts for approximately 90% of infections, with subtype 4a predominating (5). Other geographic regions with high GT4 prevalence, such as Saudi Arabia and western Europe, have a higher incidence of subtypes 4c and 4d, or 4a and 4d, respectively (5–7).
Advancements have recently been made in the treatment of HCV infection through the use of interferon (IFN)-free regimens that combine direct-acting antiviral (DAA) agents, resulting in high efficacy rates with better safety and tolerability profiles than those of IFN-containing regimens (8–15). Ombitasvir (formerly ABT-267) is a nonstructural protein 5A (NS5A) inhibitor (16), and paritaprevir (formerly ABT-450) is an NS3/4A inhibitor that is coadministered with the pharmacokinetic enhancer ritonavir (paritaprevir/r) to increase the plasma concentration and half-life of paritaprevir (17). Ombitasvir demonstrated in vitro pangenotypic antiviral activity, with 50% effective concentration (EC50) values ranging from 0.82 to 19.3 pM against HCV GT1 to 5 and 366 pM against GT6, with an EC50 of 1.7 pM against GT4a (18). Paritaprevir demonstrated in vitro antiviral activity against HCV GT1 to -4 and GT6 (EC50 range, 0.09 to 19 nM), with an EC50 of 0.09 nM against GT4a (19). Based on the comparable in vitro potencies of ombitasvir and paritaprevir for GT1 and GT4a, this 2-DAA combination was evaluated in HCV GT4-infected patients.
The phase 2b PEARL-I study reported high sustained virologic response rates 12 weeks after the last dose of study drug (SVR12, 90.9 to 100%) in treatment-naive and treatment-experienced HCV GT4-infected patients without cirrhosis treated with ombitasvir plus paritaprevir/r with and without RBV (20). The present study was designed as a comprehensive clinical virology analysis to examine the frequency of GT4 subtypes among the PEARL-I study participants using phylogenetic analysis, demographic differences among patient populations infected with distinct GT4 subtypes, the impact of subtype and genetic variability at resistance-associated amino acid positions in NS3/4A and NS5A on SVR12rates, and the development and persistence of viral resistance mutations.
MATERIALS AND METHODS
Ethics statement.
The PEARL-I study was conducted in accordance with guidelines of the International Conference of Harmonisation, applicable regulations and guidelines governing clinical study conduct, and ethics principles expressed in the Declaration of Helsinki. The study protocol was approved by the relevant institutional review boards and regulatory agencies, and all patients provided written informed consent.
Study design and participants.
The PEARL-I study (ClinicalTrials.gov identifier NCT01685203) was a randomized, international, open-label, phase 2b study that evaluated the safety and efficacy of ombitasvir and paritaprevir/r with and without RBV in patients with HCV GT1b or GT4 infection. The study design, safety, and efficacy results through SVR12 from patients with HCV GT4 infection were previously reported (20). The GT4-infected patient population included noncirrhotic treatment-naive and pegylated-interferon (pegIFN)-RBV treatment-experienced patients in the United States and Europe (France, Italy, Poland, and Spain). The treatment duration was 12 weeks, followed by a 48-week posttreatment period when patients were monitored for durability of SVR. Virologic failure (VF) was defined as either on-treatment virologic failure (defined as a failure to achieve HCV RNA level of <25 IU/ml after 6 weeks of treatment, or two consecutive HCV RNA measurements of ≥25 IU/ml at any time point after achieving HCV RNA levels of <25 IU/ml) or posttreatment relapse (HCV RNA levels of ≥25 IU/ml between the end of treatment and posttreatment week 12 in patients who completed treatment with HCV RNA levels of <25 IU/ml).
Compounds.
Paritaprevir (2R,6S,12Z,13aS,14aR,16aS)-N-(cyclopropylsulfonyl)-6-{[(5-methylpyrazin-2-yl)carbonyl]amino}-5,16-dioxo-2-(phenanthridin-6-yloxy)-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamidehydrate and ombitasvir, dimethyl ([(2S,5S)-1-(4-tert-butylphenyl)pyrrolidine-2,5-diyl]bis{benzene-4,1-diylcarbamoyl(2S)pyrrolidine-2,1-diyl[(2S)-3-methyl-1-oxobutane-1,2-diyl]})biscarbamate hydrate were synthesized by AbbVie (16, 18, 19).
HCV genotype and subtype determination.
The Versant HCV Genotype Inno-LiPA assay version 2.0 (LiPA 2.0) was used to determine the HCV genotype for enrollment of patients with chronic HCV GT4 infection, but this assay was unable to identify the viral subtype for the majority of GT4-infected patients. Therefore, viral subtype was determined by phylogenetic analysis of a 329-nucleotide region of NS5B that was PCR amplified from baseline samples from HCV GT4-infected patients (21). The results from this analysis were used to assign a preliminary subtype to each GT4-infected patient sample, which then determined the subtype and gene-specific reverse transcriptase PCR (RT-PCR) and nested-PCR primer sets for the amplification of NS3/4A and NS5A genes from baseline samples. Phylogenetic analyses were subsequently conducted using full-length HCV NS3/4A and NS5A nucleotide sequences. The final HCV GT4 subtype assignment was determined by consensus between the NS3/4A, NS5A, and NS5B results.
Phylogenetic analysis.
The nucleotide sequences for NS3/4A, NS5A, and NS5B were aligned using the MAFFT sequence alignment method (22). Phylogenetic trees were constructed using the neighbor-joining tree-building method (23, 24) with the HKY85 nucleotide substitution model (25). Reliability of the tree topology was examined using bootstrap analysis, and 1,000 bootstrapping replicates were utilized to generate a consensus tree with a 50% threshold cutoff for each phylogenetic analysis. Nucleotide alignments and phylogenetic trees were generated using the Geneious (26) and MEGA5 (27) software packages.
Sequence analysis of patient samples.
Viral RNA isolation, RT-PCR, and nested-PCR were conducted on samples with HCV RNA levels of ≥1,000 IU/ml, as previously described (18, 19). Population nucleotide sequencing was conducted on full-length HCV NS3/4A and NS5A genes amplified from all available baseline samples (pretreatment) from patients with HCV GT4 infection. The baseline sequences were compared to HCV reference sequence 4a-ED43 (GenBank accession no. GU814265) for all GT4a and non-4a/4d sequences, and reference 4d-QC382 (GenBank accession no. FJ462437) was used for the GT4d sequences.
In patients who experienced VF, population nucleotide sequencing was conducted for the regions encoding full-length NS3/4A (amino acids 1 to 685) and NS5A (amino acids 1 to 444) at the time of failure, at posttreatment week 24, and at posttreatment week 48. Clonal sequencing was conducted on samples if only wild-type virus was detected by population sequencing. For clonal sequencing, the nested gel-purified NS3 or NS5A PCR products were inserted into pJET1.2/blunt cloning vector using the CloneJET PCR cloning kit (Thermo Scientific, Pittsburgh, PA), and >80 individual colonies were sequenced for each sample for the regions encoding NS3 amino acids 1 to 181 or NS5A amino acids 1 to 215. Treatment-emergent resistance-associated amino acid variants (RAVs) were identified by comparing translated postbaseline sequences to the respective baseline sequence. The following positions are considered to be signature resistance-associated amino acid positions in HCV GT4: 56, 155, 156, and 168 in NS3 for paritaprevir, and 28, 30, 31, 32, 58, and 93 in NS5A for ombitasvir.
HCV subgenomic replicons.
The generation and sequence of HCV GT1b strain Con1 (GenBank accession no. AJ238799) subgenomic chimeric transient replicons for NS3 genotypes 1, 3, 4, and 6 (19) and NS5A genotypes 1 to 6 (18) were previously described. The GT4a and 4d chimeric NS3 replicons were generated by insertion of the region encoding the first 251 amino acids of NS3 from a GT4a or GT4d clinical isolate. The GT4a chimeric NS5A replicon was generated by insertion of the region encoding the first 214 amino acids of NS5A from a GT4a clinical isolate. For NS5A GT4d, the chimeric replicon contained the region encoding the first 214 amino acids of an NS5A sequence that was generated as a synthetic consensus gene (IDT, Coralville, IA) from a GT4d clinical isolate. The wild-type GT4a or 4d chimeric replicon sequence for NS3 or NS5A matched the 4a-ED43 or 4d-QC382 reference amino acid sequence at resistance-associated positions.
HCV NS3 or NS5A variants identified either during in vitro selection experiments in cell culture, or in patients who experienced virologic failure in the PEARL-I study, were introduced into the wild-type NS3 or NS5A GT4a or 4d replicon constructs using the Change-IT multiple mutation site-directed mutagenesis kit (Affymetrix, Santa Clara, CA). For the transient replicon assay, subgenomic replicon RNA was generated by plasmid DNA linearization, followed by in vitro RNA transcription. The chimeric replicon RNA was transfected via electroporation into a Huh-7-derived cell line, and luciferase expression was measured 4 days posttransfection to determine the inhibitory effect of ombitasvir or paritaprevir on HCV chimeric replicon replication (28). The EC50 was calculated using a nonlinear regression curve fitting to the 4-parameter logistic equation in the Prism 4/5 software (GraphPad Software, Inc., La Jolla, CA). The mean EC50 and standard deviation were calculated from ≥3 independent experiments that were conducted in duplicate.
Statistical analysis.
Fisher's exact test with a two-sided significance level of 0.05 was used to compare the number and percentage of patients achieving SVR12 with or without an NS5A variant at baseline, as well as the distribution of patient demographics (birth cohort, race, and sex) across GT4 subtypes (4a, 4d, and non-4a/4d).
RESULTS
Determination of HCV GT4 subtype using phylogenetic analysis.
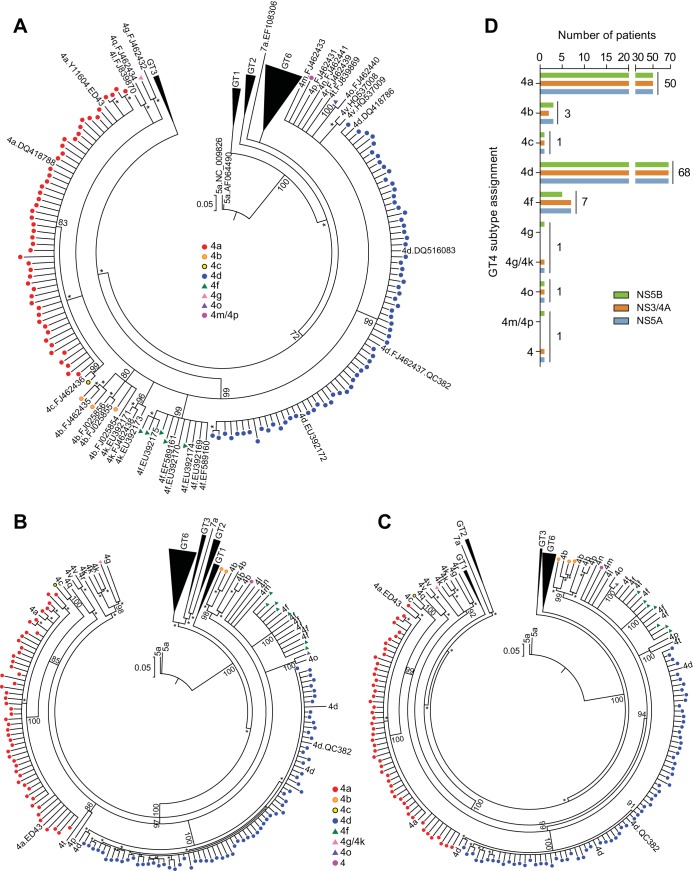
Phylogenetic analyses were conducted to ensure proper assignment of HCV GT4 subtype using NS5B (a 329-nucleotide region [21]) and full-length NS3/4A and NS5A gene sequences from the baseline samples of HCV GT4-infected patients. A total of 135 GT4-infected patients were enrolled in the PEARL-I study, and baseline samples from 132 patients were available for analysis. The NS5B phylogenetic analysis (Fig. 1A) identified 8 GT4 subtypes (4a, 4b, 4c, 4d, 4f, 4g, 4m/4p, and 4o) among the 132 patients. The preliminary subtype results were used to determine the gene-specific primer sets for subsequent amplification of the NS3/4A and NS5A genes from baseline samples; therefore, ambiguous subtypes were identified by the closest evolutionary taxa in the NS5B phylogenetic tree. Subsequent NS3/4A and NS5A phylogenetic analyses (Fig. 1B and C) identified 7 subtypes among 131 patient samples (4a, 4b, 4c, 4d, 4f, 4g/4k, and 4o) and 1 sample for which the GT4 subtype could not be determined.
FIG 1.
Phylogenetic analysis of HCV GT4 baseline sequences. Neighbor-joining phylogenetic trees displayed in circular format for NS5B (A), NS3/4A (B), and NS5A (C). HCV patient isolates are represented by colored circles or triangles indicating the subtype of the sequence. Reference sequences are labeled by subtype and GenBank accession number (A), or by subtype (B and C). Bootstrap values of ≥70 are listed at key nodes, and bootstrap values of ≥50 are indicated with an asterisk. The genetic distance scale bar indicates the number of nucleotide substitutions per site. (D) Subtype assignment for virus from HCV GT4-infected patients was determined by phylogenetic analysis.
Overall, there was high subtype concordance between the three target sequences, with most patients infected with subtype 4a or 4d (Fig. 1D). Among the 135 patients, 37.0% were infected with GT4a, 2.2% were infected with GT4b, 50.4% were infected with GT4d, and 5.2% were infected with GT4f. Subtypes 4c, 4g/4k, and 4o were each identified in one patient (0.7%), while the subtype was not determined for four patients (3.0%) by either phylogenetic analysis or the LiPA 2.0 assay. NS3/4A could not be amplified from one sample identified as GT4b, and NS5B could not be amplified from 2 samples that were subsequently identified as GT4f.
Discrepancies in the phylogenetic subtype assignment between targets occurred for two patient samples. The NS5B sequence from one patient sample sorted as GT4g but could not be distinguished between subtypes 4g or 4k by NS3/4A and NS5A phylogenetic analyses, so the final consensus subtype was labeled 4g/4k. For a second patient sample, the NS5B sequence sorted ambiguously and was assigned a preliminary subtype of 4m/4p, but further analysis of NS3/4A and NS5A did not confirm a specific GT4 subtype. A comparison between the phylogenetic and LiPA 2.0 subtyping results (Table 1) revealed that while the phylogenetic analysis was able to assign a subtype to 131 out of 132 patient samples, the LiPA 2.0 assay was unable to subtype 47 samples and classified 79 samples as subtype 4a/c/d, but it was able to correctly identify 6 out of 7 subtype 4f samples.
TABLE 1.
HCV GT4 subtype comparison between the LiPA 2.0 assay and phylogenetic analysis
| Phylogenetic analysis subtype | No. of samples by LiPA 2.0 subtype |
||||
|---|---|---|---|---|---|
| 4 | 4a/c/d | 4e | 4f | 4h | |
| 4a | 26 | 22 | 2 | ||
| 4b | 3 | ||||
| 4c | 1 | ||||
| 4d | 13 | 55 | |||
| 4f | 1 | 6 | |||
| 4g/4k | 1 | ||||
| 4o | 1 | ||||
| 4a | 1 | ||||
| No sampleb | 1 | 2 | |||
Subtype not determined; non-4a/4d sample.
A baseline plasma sample was not available for sequencing and phylogenetic analysis.
Prevalence of HCV GT4 subtype and birth cohort distribution by geographic region.
Since HCV subtype prevalence varies by geographic region, an assessment of HCV subtype by the country of enrollment was conducted (Table 2). HCV subtype 4a was predominant in patients enrolled in the United States, while subtypes 4a and 4d were both common in patients enrolled in Europe. Subtype 4d was predominant in patients from Italy, Poland, and Spain, while both GT4a and 4d infections were observed in France. The majority of HCV non-4a/4d subtypes were observed in patients enrolled in France (4b, 4f, 4g/4k, 4o, 4 [unidentified non-4a/4d subtype]). Birth cohort distribution by country (Table 2) revealed that the majority (74 to 89%) of GT4-infected patients from France, Italy, Spain, and the United States were born prior to 1970. The opposite was true for patients enrolled in Poland, where only 31% of the GT4-infected patient population was born prior to 1970, and 50% of the patients were born after 1980. All seven of the patients born during or after 1990 were from Poland.
TABLE 2.
HCV GT4 subtype and birth cohort distribution by country
| Characteristic | No. of patients by country |
||||
|---|---|---|---|---|---|
| France | Italy | Poland | Spain | USA | |
| Total | 54 | 16 | 16 | 30 | 19 |
| Subtypea | |||||
| 4a | 26 | 1 | 7 | 16 | |
| 4b | 3 | ||||
| 4c | 1 | ||||
| 4d | 16 | 15 | 16 | 20 | 1 |
| 4f | 6 | 1 | |||
| 4g/4k | 1 | ||||
| 4o | 1 | ||||
| 4b | 1 | ||||
| No samplec | 2 | 1 | |||
| Birth cohort | |||||
| Pre-1960 | 24 | 4 | 2 | 7 | 10 |
| 1960–1969 | 24 | 9 | 3 | 15 | 4 |
| 1970–1979 | 5 | 1 | 3 | 6 | 1 |
| 1980–1989 | 1 | 2 | 1 | 2 | 4 |
| 1990 and after | 0 | 0 | 7 | 0 | 0 |
Determined by phylogenetic analysis.
Subtype not determined; non-4a/4d sample.
A baseline plasma sample was not available for sequencing and phylogenetic analysis.
HCV GT4-infected patient demographic characteristics by subtype.
Patient demographic characteristics, including birth cohort, race, and sex, were compared across GT4 subtypes (Table 3). The GT4 subtypes were divided into three categories for this analysis: 4a, 4d, and non-4a/4d (4b, 4c, 4f, 4g/4k, 4o, and 4). The distributions of patient birth cohort and race were significantly different across GT4 subtypes (P, 0.012 and <0.001, respectively). The percentages of patients born prior to 1960 were different across the subtypes and encompassed 42%, 24%, and 71% of the 4a, 4d, and non-4a/4d-infected populations, respectively. Similarly, the proportions of patients born after 1980 were different across subtypes and included 6%, 16%, and 7% of the 4a, 4d, and non-4a/4d-infected populations, respectively. The distribution of patient race was also significantly different across GT4 subtypes, with those of white race comprising 92%, 100%, and 29% and those of black race comprising 4%, 0%, and 71% of the 4a, 4d, and non-4a/4d-infected populations, respectively. Notably, 10 out of the 12 patients of black race enrolled in the study were determined to be infected with an HCV non-4a/4d subtype. Of these 10 patients, 8 were from France (subtypes 4b, 4f, 4g/4k, 4o, and 4), 1 was from Spain (subtype 4f), and 1 was from the United States (subtype 4c). Patient sex distribution was similar across GT4 subtypes (P = 0.809). However, an analysis of the 17 patients born during or after 1980 across all subtypes revealed a higher male-to-female ratio of approximately 5:1 compared to a 2:1 ratio in the overall study population, with male sex representing 65% (88/135) of the total patient population and 82% (14/17) of the patients in the 1980-and-after birth cohorts.
TABLE 3.
HCV GT4-infected patient demographic characteristics by subtype
| Characteristic | No. (% of subtype) of patients by subtypea |
P valueb | ||||
|---|---|---|---|---|---|---|
| 4a | 4d | Non-4a/4d | No samplec | Total | ||
| Patients | 50 | 68 | 14 | 3 | 135 | |
| Birth cohort | 0.012 | |||||
| Pre-1960 | 21 (42) | 16 (24) | 10 (71) | 0 | 47 | |
| 1960–1969 | 19 (38) | 33 (49) | 2 (14) | 1 (33) | 55 | |
| 1970–1979 | 7 (14) | 8 (12) | 1 (7) | 0 | 16 | |
| 1980–1989 | 3 (6) | 4 (6) | 1 (7) | 2 (67) | 10 | |
| 1990 and after | 0 | 7 (10) | 0 | 0 | 7 | |
| Race | <0.001 | |||||
| White | 46 (92) | 68 (100) | 4 (29) | 2 (67) | 120 | |
| Black | 2 (4) | 0 | 10 (71)d | 0 | 12 | |
| Egyptian | 2 (4) | 0 | 0 | 1 (33) | 3 | |
| Sex | 0.809 | |||||
| Male | 32 (64) | 45 (66) | 8 (57) | 3 (100) | 88 | |
| Female | 18 (36) | 23 (34) | 6 (43) | 0 | 47 | |
Subtype determined by phylogenetic analysis.
P value from the comparison of proportions across subtypes (4a, 4d, and non-4a/4d) using Fisher's exact test.
A baseline plasma sample was not available for sequencing and phylogenetic analysis. Patients with no sample were not included in the statistical analysis.
Of these 10 patients, 8 were from France, 1 was from Spain, and 1 was from the United States.
SVR12 rates by subtype for GT4-infected patients.
In this analysis, the SVR12 rates were examined by treatment regimen and viral subtype using the subtype assignment determined by phylogenetic analysis (Table 4). The GT4 subtypes were divided into three categories for the determination of SVR12 rates: 4a, 4d, and non-4a/4d (4b, 4c, 4f, 4g/4k, 4o, and 4). HCV GT4-infected patients with an undetermined subtype were grouped in the non-4a/4d category, including three patients who did not have an available baseline sample for phylogenetic analysis. All HCV GT4a- and non-4a/4d-infected patients achieved SVR12 regardless of treatment regimen, with the exception of 1 GT4b-infected treatment-naive patient who prematurely discontinued treatment for nonvirologic reasons. The SVR12 rate for GT4d-infected patients was 81.3% (13/16) for treatment-naive patients receiving ombitasvir plus paritaprevir/r without RBV and 100% for treatment-naive (22/22) and treatment-experienced (30/30) patients receiving ombitasvir plus paritaprevir/r with RBV.
TABLE 4.
SVR12 rates by treatment regimen and viral subtype
| GT4 subtype | SVR12 rates (% [n/N]) in HCV GT4-infected patientsa |
||
|---|---|---|---|
| Treatment naive |
Treatment experienced |
||
| Ombitasvir + paritaprevir/r regimen (N = 44) | Ombitasvir + paritaprevir/r + RBV regimen (N = 42) | Ombitasvir + paritaprevir/r + RBV regimen (N = 49) | |
| All GT4 | 90.9 | 100 | 100 |
| 4a | 100 (21/21) | 100 (13/13) | 100 (16/16) |
| 4d | 81.3 (13/16) | 100 (22/22) | 100 (30/30) |
| 4b, 4c, 4f, 4g/4k, 4o, 4b | 85.7 (6/7)c | 100 (7/7) | 100 (3/3) |
n/N = (number of SVR12-achieving patients)/(total number of treated patients).
4 = GT4 subtype could not be determined by phylogenetic analysis or the LiPA 2.0 assay.
One GT 4b-infected treatment-naive patient prematurely discontinued treatment.
Variability at signature amino acid positions in NS3/4A and NS5A across HCV GT4 subtypes.
The prevalence of baseline polymorphisms at resistance-associated amino acid positions in NS3/4A and NS5A was determined using sequences from 132/135 HCV GT4-infected patients with available baseline samples. The amino acids Y56, R155, A156, and D168 in NS3/4A were 100% conserved across all of the HCV GT4 isolates. Baseline polymorphisms at signature resistance-associated amino acid positions (28, 30, 31, 32, 58, and 93) in NS5A were present in 57.6% (76/132) of the GT4 samples relative to the respective prototypic reference sequences (Fig. 2A). The majority of these polymorphisms occurred at position 58 in NS5A GT4d sequences, and the T58P polymorphism was the most prevalent, being present in 70.6% (48/68) of the GT4d samples. In the GT4d chimeric replicon, NS5A variants A58, P58, S58, and T58 do not confer resistance to ombitasvir (Table 5). Variability at NS5A amino acid positions 28, 30, 31, 32, and 93 was detected in 1.5 to 12% of the samples. Overall, the SVR12 rates were 96.1% and 98.2% for HCV GT4-infected patients with or without an NS5A variant at baseline, respectively (Fig. 2B) (P = 0.64, two-sided Fisher's exact test). Overall, the variability of amino acid polymorphisms at signature resistance-associated positions in NS5A had no apparent impact on treatment outcome.
FIG 2.
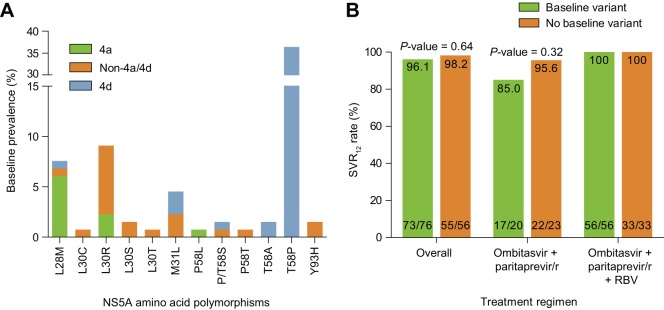
Variability at signature NS5A amino acid positions across HCV GT4 subtypes. (A) Baseline prevalence of NS5A polymorphisms at signature resistance-associated amino acid positions. NS5A sequences were divided into subtypes 4a, 4d, and non-4a/4d for analysis. Baseline sequences were compared to reference sequence 4a-ED43 for all GT4a and non-4a/4d sequences, and reference 4d-QC382 was used for the GT4d sequences. (B) Lack of impact of NS5A baseline variants on SVR12 rates by treatment regimen. P values were calculated using a two-sided Fisher's exact test.
TABLE 5.
Activity of paritaprevir or ombitasvir against chimeric GT4 transient subgenomic replicons containing amino acid variants in NS3 or NS5A
| HCV GT4 replicon subtype by targeta | Varianta | EC50 (mean ± SD) (nM)b | Fold resistancec |
|---|---|---|---|
| NS3 | |||
| 4a | Wild typed | 0.048 ± 0.0066 | |
| D168V | 15 ± 1.6 | 313 | |
| 4d | Wild type | 0.015 ± 0.001 | |
| Y56H | 0.12 ± 0.028 | 8.0 | |
| D168V | 4.7 ± 0.9 | 313 | |
| Y56H+D168V | 188 ± 50 | 12,533 | |
| NS5A | |||
| 4a | Wild type | 0.00035 ± 0.00007 | |
| L28V | 0.008 ± 0.0028 | 23 | |
| 4d | Wild type | 0.00038 ± 0.00006 | |
| L28S | NDe | ||
| L28V | 0.118 ± 0.016 | 310 | |
| M31I | 0.00096 ± 0.00012 | 2.5 | |
| M31L | 0.00039 ± 0.00008 | 1.0 | |
| T58A | 0.00053 ± 0.00002 | 1.4 | |
| T58P | 0.00042 ± 0.00004 | 1.1 | |
| T58S | 0.00052 ± 0.00009 | 1.4 | |
| L28S+M31I | ND | ||
| L28V+T58S | 0.289 ± 0.029 | 760 |
Chimeric replicons containing the NS3 or NS5A gene cloned from a GT4a or GT4d treatment-naive clinical isolate.
The mean ± SD EC50 values were derived from the results from at least three independent experiments.
Calculated as variant EC50/wild-type EC50.
Wild type indicates that amino acids at resistance-associated positions match those of reference sequence 4a-ED43 or 4d-QC382.
ND, EC50 could not be determined due to the poor replication capacity of the chimeric replicon.
Resistance-associated variants in patients not achieving SVR12.
The three patients who experienced VF in this study were infected with HCV GT4d, the most prevalent subtype among trial participants. All three were treatment-naive patients who received ombitasvir plus paritaprevir/r without RBV, including one patient who experienced on-treatment virologic failure at treatment week 8 and two patients who experienced virologic relapse before posttreatment week 12 (Table 6). Resistance-associated variants were not present in NS3 or NS5A at baseline prior to treatment. The predominant resistance-associated treatment-emergent variants at the time of VF were D168V in NS3 and L28V in NS5A. In NS3, D168V confers 313-fold resistance to paritaprevir in GT4 subtype 4a or 4d, and the double mutant Y56H+D168V, which was observed in 1 of the 3 patients who experienced VF, confers 12,533-fold resistance to paritaprevir in GT4d (Table 5). The NS3 D168V variant persisted in 2 of the 3 patients at posttreatment week 24 but was not detectable by clonal sequencing at posttreatment week 48. The NS5A variant L28V confers 23-fold resistance to ombitasvir in GT4a and 310-fold resistance in GT4d. The L28V+T58S double variant in GT4d confers 760-fold resistance to ombitasvir (Table 5). The EC50 for the L28S variant in GT4d NS5A could not be determined due to the poor replication capacity of the chimeric replicon containing this variant. Treatment-emergent NS5A variants at resistance-associated amino acid positions persisted to posttreatment week 48 in 2 of the 3 patients.
TABLE 6.
Variants in NS3/4A and NS5A at resistance-associated amino acid positions in patients experiencing virologic failure
| Patienta | Failure typeb | NS3/4A |
NS5A |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | Virologic failure | PTW24 | PTW48 | Baseline | Virologic failure | PTW24 | PTW48 | ||
| 1 | OTVF at wk 8 | Nonec | Y56H+ D168V | Noned | NDe | None | L28V | L28V/L | Noned |
| 2 | Relapse at PTW4 | None | D168V | D168Vd | Noned | T58P | L28S, M31I/M, T58P | L28S+M31I+T58P | L28S/L, M31I/M, T58P |
| 3 | Relapse at PTW8 | None | D168V | D168V | Noned | T58T/S | L28V+T58S | L28V, T58T/S | L28V+T58S |
All three patients were infected with HCV genotype 4d.
OTVF, on-treatment virologic failure; PTW, posttreatment week.
None, variants at resistance-associated amino acid positions were not detected.
Determined by clonal sequencing analysis; includes variants detected in ≥2 clones.
ND, sequence not determined because resistance-associated variants were not detected at the previous time point.
DISCUSSION
The consensus classification and nomenclature system for HCV genotype and subtype assignment was established using phylogenetic analysis (3). In the current study, GT4 subtype classification was determined by phylogenetic analysis for HCV GT4-infected patients, which identified 7 GT4 subtypes among 132 patient samples (4a, 4b, 4c, 4d, 4f, 4g/4k, and 4o). The LiPA 2.0 assay is a commercially available HCV genotyping assay that is based on reverse hybridization of the 5′-nontranslated region (NTR) and core regions of the HCV genome to immobilized oligonucleotide probes (29). Although it is widely used as a diagnostic HCV genotyping assay, several studies have reported HCV subtype misclassification using the LiPA method (30–32). In the current study, a comparison between the two subtyping methods revealed that the LiPA 2.0 HCV genotyping assay was not able to accurately identify GT4 subtypes, while the sequence-based phylogenetic method demonstrated high concordance among the three targets analyzed. These data suggest that a phylogenetic methodology encompassing a small region of NS5B is sufficient to accurately determine GT4 subtypes and should be used in future clinical trials that aim to subtype HCV GT4-infected patients.
Substantial variation exists in the predominant circulating subtypes of HCV GT4 in northern and sub-Saharan Africa, the Middle East, and Europe. Our analysis of GT4 subtype prevalence by geographic region revealed differences in the circulating GT4 subtypes in France, Italy, Spain, Poland, and the United States. The most common subtypes identified were 4a and 4d, representing 37.0% and 50.4% of the study population, respectively. The HCV isolates from Italy, Spain, and Poland were predominantly subtype 4d, while the most subtype diversity was seen in France (4a, 4b, 4d, 4f, 4g/4k, and 4o). These observations are consistent with previous reports of higher GT4a and 4d prevalence in France, Italy, and Spain (4–7). Epidemiological studies of HCV GT4 in France have identified the emergence of GT4a and 4d infections associated with blood transfusion or intravenous drug use, while infection with more heterogeneous GT4 subtypes was identified in patients of African origin with undetermined risk factors (33). In this study, we identified a high proportion (83%) of patients of black race who were infected with an HCV non-4a/4d subtype. Although the sample size is small, eight of the 10 patients of black race and a heterogeneous HCV subtype were from France (subtypes 4b, 4f, 4g/4k, 4o, and 4).
In the analysis of birth cohort distribution by country, we found that 82% of the HCV-infected patients from France, Italy, Spain, and the United States were ≥45 years of age, while 44% of the patients enrolled in Poland were ≤25 years of age. The prevalence of HCV GT4 infection in Poland varies by province and is higher in patients <20 years of age and in HIV-coinfected patient populations (34). Additional risk factors associated with HCV infection in Poland include male sex, blood transfusions before 1992, and intravenous drug use (35). Notably, all patients from Poland were infected with GT4d, the spread of which has been associated with intravenous drug use in other European countries (33, 36). In addition, 75% of the 8 Polish patients born after 1980 were male, perhaps indicating a population of young males at a higher risk of acquiring HCV GT4 infection in Poland. In general, we found a higher male-to-female ratio of approximately 5:1 in the younger patient population (born during or after 1980) across all GT4 subtypes and countries, although due to the small sample size, it is difficult to draw conclusions regarding overall HCV GT4 infection risk in young males. Across Europe, intravenous drug users are at the highest risk of HCV GT4 infection, particularly for subtypes 4a and 4d (5, 6, 33, 35, 36). Given the recent approval of treatment regimens for HCV GT4 infection containing sofosbuvir (14, 37, 38), daclatasvir (13, 39), or ombitasvir plus paritaprevir/r (20), it will be important to identify patients at a high risk for potential HCV reinfection in order to apply targeted prevention and patient education programs and ultimately reduce the occurrence and spread of drug-resistant HCV.
HCV reinfection did not occur in GT4-infected patients through posttreatment week 48 in this study. SVR12 rates of 100% were observed among GT4-infected treatment-naive and treatment-experienced patients receiving ombitasvir plus paritaprevir/r with RBV. None of the GT4a- or non-4a/4d-infected patients receiving the regimen without RBV experienced virologic failure. Three out of 135 patients (2%) experienced virologic failure; all three were treatment naive, received ombitasvir plus paritaprevir/r without RBV, and were infected with HCV GT4d. The predominant resistance-associated variants at the time of VF were D168V in NS3, which conferred 313-fold resistance to paritaprevir, and L28V in NS5A, which conferred 310-fold resistance to ombitasvir in a GT4d chimeric replicon. The degree of resistance conferred by D168V in NS3 was similar between the 4a and 4d subtypes, whereas in NS5A, the L28V variant conferred approximately 15-fold higher resistance to ombitasvir in an NS5A subtype 4d background compared to that in subtype 4a. This finding may partially explain the lower SVR12 rates in GT4d-infected patients who received an RBV-free regimen.
Genetic variability between and among HCV GT4 subtypes, as measured by nucleotide pairwise distance, ranges between 12.7 and 15.3%, which is similar to the variability of 12.9 to 17.0% seen in GT1 (3). In this study, an assessment of amino acid variability in NS3 and NS5A at baseline revealed that amino acids at signature resistance-associated positions in NS3 were conserved across GT4 subtypes. Although substantial amino acid variability (57.6%) was observed at signature resistance-associated amino acid positions in NS5A across GT4 subtypes, most variants remained susceptible to ombitasvir, indicating that genetic variability between GT4 subtypes did not impact treatment outcome.
Molecular and phylogenetic analyses in this study revealed that regardless of the identified GT4 subtype or the presence of NS5A baseline polymorphisms at resistance-associated amino acid positions, HCV GT4-infected patients who received a regimen containing ombitasvir plus paritaprevir/r achieved high SVR12 rates. The regimen of ombitasvir plus paritaprevir/r with RBV was recently approved in the United States and Europe for the treatment of HCV GT4 infection, which expands treatment options for GT4-infected patients. Finally, GT4 subtype distribution in this study varied by geography and patient demographics of birth cohort and race.
ACKNOWLEDGMENTS
We acknowledge the clinical providers and patients for their study participation and the study coordinators for assistance provided in the preparation and operation of the study. We also thank Barbara McGovern for critical review of the manuscript.
All authors are employees of AbbVie and may hold stock or stock options. The design, study conduct, and financial support for this research were provided by AbbVie. AbbVie participated in the interpretation of data, review, and approval of the publication.
REFERENCES
- 1.Abdel-Hamid M, El-Daly M, Molnegren V, El-Kafrawy S, Abdel-Latif S, Esmat G, Strickland GT, Loffredo C, Albert J, Widell A. 2007. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol 88:1526–1531. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2014. Hepatitis C: fact sheet no. 164. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs164/en/. [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment Web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. 2014. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Kamal SM. 2007. Improving outcome in patients with hepatitis C virus genotype 4. Am J Gastroenterol 102:2582–2588. doi: 10.1111/j.1572-0241.2007.01538.x. [DOI] [PubMed] [Google Scholar]
- 6.Kamal SM, Nasser IA. 2008. Hepatitis C genotype 4: what we know and what we don't yet know. Hepatology 47:1371–1383. doi: 10.1002/hep.22127. [DOI] [PubMed] [Google Scholar]
- 7.Di Lello FA, Neukam K, Parra-Sanchez M, Plaza Z, Soriano V, Cifuentes C, Mira JA, Poveda E, Pineda JA. 2013. Hepatitis C virus genotype 4 in southern and central Spain does not originate from recent foreign migration waves. J Med Virol 85:1734–1740. doi: 10.1002/jmv.23657. [DOI] [PubMed] [Google Scholar]
- 8.Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, Ghalib R, Gitlin N, Herring R, Lalezari J, Younes ZH, Pockros PJ, Di Bisceglie AM, Arora S, Subramanian GM, Zhu Y, Dvory-Sobol H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Sulkowski M, Kwo P, ION-2 Investigators. 2014. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med 370:1483–1493. doi: 10.1056/NEJMoa1316366. [DOI] [PubMed] [Google Scholar]
- 9.Afdhal N, Zeuzem S, Kwo P, Chojkier M, Gitlin N, Puoti M, Romero-Gomez M, Zarski JP, Agarwal K, Buggisch P, Foster GR, Brau N, Buti M, Jacobson IM, Subramanian GM, Ding X, Mo H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Mangia A, Marcellin P, ION-1 Investigators. 2014. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med 370:1889–1898. doi: 10.1056/NEJMoa1402454. [DOI] [PubMed] [Google Scholar]
- 10.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 11.Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M, Rustgi V, Chojkier M, Herring R, Di Bisceglie AM, Pockros PJ, Subramanian GM, An D, Svarovskaia E, Hyland RH, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Pound D, Fried MW, ION-3 Investigators. 2014. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 370:1879–1888. doi: 10.1056/NEJMoa1402355. [DOI] [PubMed] [Google Scholar]
- 12.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, Sulkowski MS, Wedemeyer H, Tam E, Desmond P, Jensen DM, Di Bisceglie AM, Varunok P, Hassanein T, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
- 13.Hézode C, Hirschfield GM, Ghesquiere W, Sievert W, Rodriguez-Torres M, Shafran SD, Thuluvath PJ, Tatum HA, Waked I, Esmat G, Lawitz EJ, Rustgi VK, Pol S, Weis N, Pockros PJ, Bourliere M, Serfaty L, Vierling JM, Fried MW, Weiland O, Brunetto MR, Everson GT, Zeuzem S, Kwo PY, Sulkowski M, Brau N, Hernandez D, McPhee F, Wind-Rotolo M, Liu Z, Noviello S, Hughes EA, Yin PD, Schnittman S. 2014. Daclatasvir plus peginterferon alfa and ribavirin for treatment-naive chronic hepatitis C genotype 1 or 4 infection: a randomised study. Gut doi: 10.1136/gutjnl-2014-307498. [DOI] [PubMed] [Google Scholar]
- 14.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 15.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT, Subramanian GM, McHutchison JG, Weiland O, Reesink HW, Ferenci P, Hezode C, Esteban R, VALENCE Investigators . 2014. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med 370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 16.DeGoey DA, Randolph JT, Liu D, Pratt J, Hutchins C, Donner P, Krueger AC, Matulenko M, Patel S, Motter CE, Nelson L, Keddy R, Tufano M, Caspi DD, Krishnan P, Mistry N, Koev G, Reisch TJ, Mondal R, Pilot-Matias T, Gao Y, Beno DW, Maring CJ, Molla A, Dumas E, Campbell A, Williams L, Collins C, Wagner R, Kati WM. 2014. Discovery of ABT-267, a pan-genotypic inhibitor of HCV NS5A. J Med Chem 57:2047–2057. doi: 10.1021/jm401398x. [DOI] [PubMed] [Google Scholar]
- 17.Menon RM, Klein CE, Lawal AA, Chiu YL, Awni WM, Podsadecki TJ, Nada A, Bernstein BM. 2009. Pharmacokinetics and tolerability of the HCV protease inhibitor ABT-450 following single ascending doses in healthy adult volunteers with and without ritonavir. Global Antiviral J 5:53. [Google Scholar]
- 18.Krishnan P, Beyer J, Mistry N, Koev G, Reisch T, DeGoey D, Kati W, Campbell A, Williams L, Xie W, Setze C, Molla A, Collins C, Pilot-Matias T. 2015. In vitro and in vivo antiviral activity and resistance profile of ombitasvir, an inhibitor of hepatitis C Virus NS5A. Antimicrob Agents Chemother 59:979–987. doi: 10.1128/AAC.04226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilot-Matias T, Tripathi R, Cohen D, Gaultier I, Dekhtyar T, Lu L, Reisch T, Irvin M, Hopkins T, Pithawalla R, Middleton T, Ng T, McDaniel K, Or YS, Menon R, Kempf D, Molla A, Collins C. 2015. In vitro and in vivo antiviral activity and resistance profile of the hepatitis C virus NS3/4A protease inhibitor ABT-450. Antimicrob Agents Chemother 59:988–997. doi: 10.1128/AAC.04227-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hézode C, Asselah T, Reddy KR, Hassanein T, Berenguer M, Fleischer-Stepniewska K, Marcellin P, Hall C, Schnell G, Pilot-Matias T, Mobashery N, Redman R, Vilchez RA, Pol S. 2015. Ombitasvir plus paritaprevir plus ritonavir with or without ribavirin in treatment-naive and treatment-experienced patients with genotype 4 chronic hepatitis C virus infection (PEARL-I): a randomized, open-label trial. Lancet 385:2502–2509. doi: 10.1016/S0140-6736(15)60159-3. [DOI] [PubMed] [Google Scholar]
- 21.Koletzki D, Dumont S, Vermeiren H, Fevery B, De Smet P, Stuyver LJ. 2010. Development and evaluation of an automated hepatitis C virus NS5B sequence-based subtyping assay. Clin Chem Lab Med 48:1095–1102. [DOI] [PubMed] [Google Scholar]
- 22.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 24.Studier JA, Keppler KJ. 1988. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol 5:729–731. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 26.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi RL, Krishnan P, He Y, Middleton T, Pilot-Matias T, Chen CM, Lau DT, Lemon SM, Mo H, Kati W, Molla A. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antiviral Res 73:40–49. doi: 10.1016/j.antiviral.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Bouchardeau F, Cantaloube JF, Chevaliez S, Portal C, Razer A, Lefrere JJ, Pawlotsky JM, De Micco P, Laperche S. 2007. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol 45:1140–1145. doi: 10.1128/JCM.01982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avó AP, Agua-Doce I, Andrade A, Pádua E. 2013. Hepatitis C virus subtyping based on sequencing of the C/E1 and NS5B genomic regions in comparison to a commercially available line probe assay. J Med Virol 85:815–822. doi: 10.1002/jmv.23545. [DOI] [PubMed] [Google Scholar]
- 31.Cai Q, Zhao Z, Liu Y, Shao X, Gao Z. 2013. Comparison of three different HCV genotyping methods: core, NS5B sequence analysis and line probe assay. Int J Mol Med 31:347–352. [DOI] [PubMed] [Google Scholar]
- 32.Guelfo JR, Macias J, Neukam K, Di Lello FA, Mira JA, Merchante N, Mancebo M, Nunez-Torres R, Pineda JA, Real LM. 2014. Reassessment of genotype 1 hepatitis C virus subtype misclassification by LiPA 2.0: implications for direct-acting antiviral treatment. J Clin Microbiol 52:4027–4029. doi: 10.1128/JCM.02209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morice Y, Roulot D, Grando V, Stirnemann J, Gault E, Jeantils V, Bentata M, Jarrousse B, Lortholary O, Pallier C, Deny P. 2001. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J Gen Virol 82:1001–1012. [DOI] [PubMed] [Google Scholar]
- 34.Panasiuk A, Flisiak R, Mozer-Lisewska I, Adamek A, Tyczyno M, Halota W, Pawlowska M, Stanczak J, Berak H, Wawrzynowicz-Syczewska M, Boron-Kaczmarska A, Lapinski TW, Grzeszczuk A, Piekarska A, Tomasiewicz K, Jablkowski M, Kryczka W, Zarebska-Michaluk D, Stepien P, Garlicki AM, Kozlowska J, Wiercinska-Drapalo A, Zasik E, Mazur W, Dobracka B, Dobracki W, Simon K, Ryzko J, Pawlowska J, Dzierzanowska-Fangrat K, Januszkiewicz-Lewandowska D, Szenborn L, Zaleska I, Rokitka M, Strawinska E, Balinowska K, Smiatacz T, Stalke P, Sikorska K, Lakomy A, Zdrojewski M, Lachowicz A. 2013. Distribution of HCV genotypes in Poland. Przegl Epidemiol 67:11–16, 99–103. [PubMed] [Google Scholar]
- 35.Flisiak R, Halota W, Horban A, Juszczyk J, Pawlowska M, Simon K. 2011. Prevalence and risk factors of HCV infection in Poland. Eur J Gastroenterol Hepatol 23:1213–1217. doi: 10.1097/MEG.0b013e32834d173c. [DOI] [PubMed] [Google Scholar]
- 36.Cantaloube JF, Gallian P, Laperche S, Elghouzzi MH, Piquet Y, Bouchardeau F, Jordier F, Biagini P, Attoui H, de Micco P. 2008. Molecular characterization of genotype 2 and 4 hepatitis C virus isolates in French blood donors. J Med Virol 80:1732–1739. doi: 10.1002/jmv.21285. [DOI] [PubMed] [Google Scholar]
- 37.Esmat GE, Shiha G, Omar RF, Hassany M, Hammad R, Khairy M, Samir W, Soliman R, Brainard DM, Jiang D, Kersey K, Knox SJ, Massetto B, McHutchison JG, Doss WH. 2014. Sofosbuvir plus ribavirin in the treatment of Egyptian patients with chronic genotype 4 HCV infection. Hepatology 60:662A–663A. [Google Scholar]
- 38.Ruane PJ, Ain D, Stryker R, Meshrekey R, Soliman M, Wolfe PR, Riad J, Mikhail S, Kersey K, Jiang D, Massetto B, Doehle B, Kirby BJ, Knox SJ, McHutchison JG, Symonds WT. 2014. Sofosbuvir plus ribavirin for the treatment of chronic genotype 4 hepatitis C virus infection in patients of Egyptian ancestry. J Hepatol doi: 10.1016/j.jhep.2014.10.044. [DOI] [PubMed] [Google Scholar]
- 39.Hassanein T, Sims KD, Bennett M, Gitlin N, Lawitz E, Nguyen T, Webster L, Younossi Z, Schwartz H, Thuluvath PJ, Zhou H, Rege B, McPhee F, Zhou N, Wind-Rotolo M, Chung E, Griffies A, Grasela DM, Gardiner DF. 2015. A randomized trial of daclatasvir in combination with asunaprevir and beclabuvir in patients with chronic hepatitis C virus genotype 4 infection. J Hepatol doi: 10.1016/j.jhep.2014.12.025. [DOI] [PubMed] [Google Scholar]