Abstract
A subset of bacterial pathogens, including the zoonotic Brucella species, are highly resistant against polymyxin antibiotics. Bacterial polymyxin resistance has been attributed primarily to the modification of lipopolysaccharide; however, it is unknown what additional mechanisms mediate high-level resistance against this class of drugs. This work identified a role for the Brucella melitensis gene bveA (BMEII0681), encoding a predicted esterase, in the resistance of B. melitensis to polymyxin B. Characterization of the enzymatic activity of BveA demonstrated that it is a phospholipase A1 with specificity for phosphatidylethanolamine (PE). Further, lipidomic analysis of B. melitensis revealed an excess of PE lipids in the bacterial membranes isolated from the bveA mutant. These results suggest that by lowering the PE content of the cell envelope, BveA increases the resistance of B. melitensis to polymyxin B. BveA was required for survival and replication of B. melitensis in macrophages and for persistent infection in mice. BveA family esterases are encoded in the genomes of the alphaproteobacterial species that coexist with the polymyxin-producing bacteria in the rhizosphere, suggesting that maintenance of a low PE content in the bacterial cell envelope may be a shared persistence strategy for association with plant and mammalian hosts.
INTRODUCTION
The polymyxin class of antibiotics, produced by soil bacteria such as Paenibacillus polymyxa (formerly Bacillus polymyxa), was abandoned for therapy in the 1970s in favor of newer drugs. However, with the emergence of multidrug-resistant bacteria, such as Acinetobacter baumannii and Klebsiella pneumoniae (1), rehabilitation of polymyxin B (PmB) and colistin (polymyxin E) has been proposed as a last-line treatment for infections with these organisms (2–4).
Polymyxins act at the cell envelope: their initial association with the outer membrane is dependent on displacing divalent cations (Mg2+ and Ca2+) from lipopolysaccharide (LPS). Their subsequent association with the cytoplasmic membrane results in the insertion of PmB, leading to the formation of pore-like structures and membrane permeabilization. The ensuing disruption of the cytoplasmic membrane leads to the inhibition of bacterial respiration via loss of the proton-motive force and, consequently, to growth inhibition (5).
Several bacterial species are inherently resistant to polymyxins, including Burkholderia, Proteus, Neisseria, and Brucella spp., and this characteristic is actually utilized for the primary isolation of Brucella spp. from clinical samples (6, 7). Understanding the basis for this resistance is important, as this knowledge may inform the design of novel therapies against drug-resistant organisms. Here, we identify a new mechanism for increased resistance to this class of drugs in the bacterial pathogen Brucella melitensis and show that this represents part of its adaptation in causing infections in mammalian hosts.
MATERIALS AND METHODS
Bacterial growth conditions.
Escherichia coli was grown in lysogeny broth (LB) and Brucella melitensis in tryptic soy broth (TSB). For mouse infections, B. melitensis was cultured on tryptic soy agar (TSA) plus 5% blood for 3 days (8). Salmonella enterica serovar Typhimurium strain IR715 was cultured under low-magnesium conditions (10 μM MgCl2) in chemically defined M9 minimal medium supplemented with 1% glucose following the method of Groisman et al. (9). When needed, nalidixic acid at 25 μg/ml, ampicillin at 250 μg/ml, kanamycin at 100 μg/ml, or gentamicin at 50 μg/ml was added to the medium. Work with B. melitensis wild-type and mutant strains was performed at biosafety level 3 and was approved by the Institutional Biosafety Committee at the University of California, Davis. For expression of proteins, an overnight culture was grown to an optical density of 0.4 in LB with glucose, and the expression was induced with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 6 h at 37°C with continuous shaking.
DNA manipulation, construction of mutants, and complementation.
The strains and plasmids used in this study are listed in Table 1. The bveA gene (BMEII0681) was identified in a bioinformatic screen for cell envelope and type IV secretion system-related functions (published in part in reference 10). It originally came to our attention because of its similarity to two proteins (AcvB and VirJ) in Agrobacterium tumefaciens, which are required for transfer of the Ti plasmid (11). However, a closer analysis showed that it had a lipase domain; therefore, we hypothesized a role for this protein related to membrane biogenesis. The bveA mutant was generated by allelic exchange. Regions up- and downstream of BMEII0681 (bveA) were amplified using the primer sets H1-For (5′-CTGCAGATAGCTGCGCTCCTGA-3′), H1-Rv (5′-TCTAGAGCTGCGCACTGTCTTCTATGG-3′), H2-Fw (5′-GTCGACCTTCCTGATCAGTGC-3′), and H2-Rv (5′-CTGCAGGCAAATCACATGCCGT-3′), and cloning of the amplicons into pCR2.1-TOPO (Invitrogen). The Tn903 kanamycin resistance cassette (KSAC from pUC4-KSAC) was introduced between the up- and downstream fragments to generate the plasmid pCR105 (Table 1). This plasmid was transferred to the B. melitensis wild-type strain 16M by electroporation, and the allelic exchange mutants of bveA were identified by screening for resistance to kanamycin and for loss of the plasmid-encoded ampicillin resistance. The correct position of the insertion was verified by PCR and Southern blotting of a HindIII-digested chromosomal preparation of bveA using a bveA-specific probe (data not shown). The correct strain was designated CMR27 (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype/description | Source |
|---|---|---|
| Brucella melitensis | ||
| B. melitensis 16M | Wild-type strain | Laboratory collection |
| CMR27 | ΔbveA::Kmr | This work |
| CMR28 | CMR27 complemented with pCR108 carrying bveA | This work |
| TOK09 | CMR27 carrying a chromosomal insertion of pTK19 in the tRNAtrp-secE intergenic region | This work |
| E. coli | ||
| Origami 2(DE3)pLysS | Host for expression of BveA fusion proteins | Novagen |
| Salmonella enterica serotype Typhimurium | ||
| IR715 | Spontaneous nalidixic acid-resistant derivative of ATCC 14028 | Laboratory collection |
| Plasmids | ||
| pBluescript KS | ColE1, Apr | Stratagene |
| pCR2.1-TOPO | TA cloning vector | Invitrogen |
| pUC4-KSAC | Source of Tn903 kanamycin resistance cassette | Pharmacia |
| pCR105 | Construct for allelic exchange to delete B. melitensis bveA, Kmr | This work |
| pCR108 | Carries the bveA gene in pBBR1MCS4 vector background, Apr | This work |
| pTK19 | Carries the bveA gene from B. melitensis 16M in exchange for the mCherry gene of pKSoriT-bla-PsecE-mCherry (13), Apr | This work |
| pTK07 | Overexpression construct of PelB-BveA-6×His fusion protein in E. coli; pET25b(+), Apr; pLac | This work |
| pTK25 | Overexpression construct of MBP-BveA fusion protein in E. coli; pMALp5x, Apr; pLac | This work |
To complement the bveA mutant (CMR27), plasmid pCR108 was constructed as follows. Primers CR-forward (5′-TCAGCGCGCAGGGCGCGGCGG-3′) and CR-reverse (5′-CGTATTCTTTATCGTCCTGGGGTTGCG-3′) were used to amplify bveA together with its promoter from the B. melitensis 16M genome. This PCR product was introduced into pCR2.1 by TA cloning (Invitrogen) and was subcloned into the pBBR1MCS4 vector (12) using EcoRI to obtain the plasmid pCR108. Plasmid pCR108 was introduced into CMR27 to yield strain CMR28. However, the expression of bveA in CMR28 was unstable during exposure to polymyxin B. We therefore utilized a previously described strategy for stable, single-copy chromosomal gene expression from the promoter of the secE gene encoding the preprotein translocase (13). To this end, we constructed a suicide plasmid (pTK19) by replacing the mCherry gene of pKSoriT-bla-PsecE-mCherry (kindly provided by X. DeBolle) (13) with an amplicon containing the bveA coding sequence (secE-bveA-Fr 5′-CCTGATCAGACAGAGTATGAAGAAAGAACGCGTATTCTTTATCGTCCTGGGGTTGG-3′ and secE-bveA-Rv 5′-CCCTGCAGGTCGAGGTCAGCGCGCAGGGCGCGG-3′) from B. melitensis 16M via Gibson assembly cloning (New England Biolabs [NEB]). All cloning steps were verified by sequencing, and following electroporation, the subsequent insertion of the fusion gene was confirmed by PCR.
For overexpression and purification of His-tagged BveA in E. coli, we amplified the gene using the primer combination His-681-SalI-For (5′-ATAGTCGACTCAGTGGTGGTGGTGGTGGTGGCGCGCAGGGCGCGGCGGC-3′) and 0681-BamHI-Rev (5′ ATAAGGATCCGAAGAAAGAACGCGTATTCTTTATCG-3′). This amplified the bveA gene with an additional in-frame 6×His tag and two restriction sites (SalI and BamHI) that were used to clone the fragment into the pET25b(+) vector (EMD Millipore), generating the plasmid pTK07. This step fused the BveA-His6 construct to the PelB leader that enables periplasmic localization of the final fusion protein. The construct was verified by sequencing. To express a fusion protein of BveA and the maltose-binding protein (MBP-BveA), we amplified bveA with the primers pMAL-Fr and pMAL-Rv (5′-GGGATCGAGGGAAGGAAGAAAGAACGCGTATTCTTTATCGTCCTGGGGTTGGC-3′ and 5′-catggacatatgtgaaatTCAGCGCGCAGGGCGCGG-3′) and cloned the resulting amplicon into pMALp5x (NEB), using the Gibson cloning method, to yield plasmid pTK25. Subsequent sequencing verified the correct sequence of the fusion construct.
Influence of polymyxin B on bacterial growth.
The bactericidal effect of polymyxin B (Difco) against B. melitensis strains and S. Typhimurium strain IR715 was tested by incubation of 1 × 106 CFU of each strain for 48 h at 37°C with 0 (control), 5, 10, 15, 50, and 150 μg/ml of PmB in TSB (pH 7.4) and subsequent enumeration of CFU on TSA.
Sensitivity of B. melitensis to antimicrobials.
The killing ability of several antimicrobial substances, peptides, and proteins against B. melitensis was investigated as described by Martinéz de Tejada et al. (14) with the exception of using 25 μg of polymyxin B sulfate (Difco), magainin 2, and cecropin P (Sigma-Aldrich).
Protein expression, periplasmic preparation, and purification of BveA.
C-terminal 6×His-tagged PelB-BveA protein was expressed in E. coli Origami B(DE3)pLysS (EMD Millipore), and a periplasmic fraction was obtained following the recommendations of Novagen's pET System Manual (11th edition). Periplasmic fractions were concentrated using spin concentrator units with a cutoff of 3 kDa (Millipore). The presence of the BveA fusion protein was verified by Western blotting detection using an anti-His antibody (Bio-Rad). An MBP-BveA fusion protein was purified from the periplasm of E. coli Origami B(DE3)pLysS (EMD Millipore) using amylose resin (NEB). The eluted protein was concentrated using a 50-kDa concentrator unit (Thermo Fisher Scientific), and the purity of the MBP-BveA protein sample was confirmed by SDS-PAGE.
Esterase activity assay.
The esterase activity of BveA was determined as published by Plou et al. (15) using A280-adjusted periplasmic samples, and the turbidity of the mixture was measured after 16 h of incubation in standard clear-bottom 96-well plates at 37°C using a FilterMax F3 reader (Molecular Devices) at 450 nm. To test the specificity of BveA for phosphatidylethanolamine (PE), phosphatidylcholine (PC), and phosphatidylglycerol (PG) (Sigma-Aldrich), we adopted the assay conditions but exchanged Tween 80 with 1 mM purified phospholipids. To investigate the specificity of BveA against crude Brucella envelope lipids, Tween 80 was substituted with 1% of a bacterial lipid extract from B. melitensis 16M. To this end, freshly extracted membrane lipids were resuspended by vortexing in 20 mM Tris (pH 7.4), and the reaction mixture was incubated at 37°C for 16 h and 120 h. The phospholipase A1 specificity was detected using the substrate PED-1 (EnzChek phospholipase A1 assay kit; Life Technologies).
Phospholipid extraction and thin-layer chromatography.
To obtain glycerophospholipids from the B. melitensis wild type and bveA mutant, we extracted membrane lipid from liquid cultures adjusted to an optical density at 600 nm (OD600) using methyl-tert-butyl ether (MTBE) (Sigma-Aldrich), according to the method of Matyash et al. (16). Two-dimensional thin-layer chromatography (2D-TLC) was performed on silica gel plates (Kieselgel 60; Merck) loaded with equal amounts of lipid preparations using the solvent chloroform-methanol-water (14:6:1) followed by chloroform-methanol-acetic acid (13:5:2). The phospholipids were identified with a molybdatophosphoric acid spray solution (Merck) and commercial standards (Avanti) (17). The relative amounts of the known membrane lipids were estimated by densitometry of stained 2D-TLC spots using a gray-scale, 8-bit transformed picture of the stained TLC plates with the BioSpectrum AC imaging system (UVP) and the densitometry tool of the Labworks software suite. The gray intensities for all known lipid spots were combined to calculate the percentage of the contribution of each single lipid species to the sum of all known lipid species.
Lipidomic analysis of the B. melitensis cell envelope.
The contributions of PE and PC to the total phospholipid contents of the B. melitensis cell envelope were analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) on a 6530 quadrupole time of flight (Q-TOF) mass spectrometer (Agilent Technologies) coupled with an automated high-performance liquid chromatograph (1200 series; Agilent Technologies) using a 2.1- by 100-mm C18 column from Waters. The MTBE extracts were redissolved in methanol and analyzed in the positive ionization mode. MS/MS spectra were analyzed manually with an in-house-developed MS/MS database (LipidBlast) that was used to identify single lipid species (18). To correct for machine drift and to normalize the data, each individual value (peak height) was divided against the sum of all peak intensities obtained in the corresponding chromatogram and multiplied by the average of all total intensities.
Cellular infection assays.
J774.A1 (ATCC TIB-67) macrophage-like cells were propagated in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies) and 1% nonessential amino acids (ATCC). For the macrophage survival assay, J774.A1 cells were infected with B. melitensis strains with a multiplicity of infection of 100 bacteria per cell, and the survival of intracellular bacteria was determined after 0, 4, 24, and 48 h as reported previously (19).
Mouse infections.
For mice inoculated intranasally (i.n.), groups (n = 4 to 5) of 8- to 10-week-old female C57BL/6J wild-type mice (The Jackson Laboratory) were inoculated with 1 × 107 CFU containing a 1:1 ratio of the B. melitensis wild type and bveA mutant in a total volume of 10 μl sterile phosphate-buffered saline (PBS). After 1 and 4 weeks postinfection (p.i.), cervical lymph nodes from infected mice were removed, weighed, and homogenized in 1 ml of PBS. For intraperitoneal (i.p.) infections, groups of 5 C57BL/6 mice were inoculated with 0.1 ml of PBS containing 1 × 105 CFU of either B. melitensis 16M or the bveA mutant (CMR27). For enumeration of the bacterial tissue loads, serial dilutions of tissue homogenates were spread on TSA with appropriate antibiotics. Mice were maintained in microisolator cages with sterile bedding and irradiated feed in a biosafety level 3 laboratory. All animal experiments were approved by the University of California Laboratory Animal Care and Use Committee and were conducted in accordance with institutional guidelines.
Statistical analysis.
All data in this work are presented as the mean and the standard error of the mean. To determine the statistical significance between experimental groups at a given time point, we used either Student's t test on log- or arc-sin-transformed data or the Mann-Whitney test on nonparametric values. A P value of <0.05 was considered significant.
RESULTS
B. melitensis BveA confers resistance against polymyxin B.
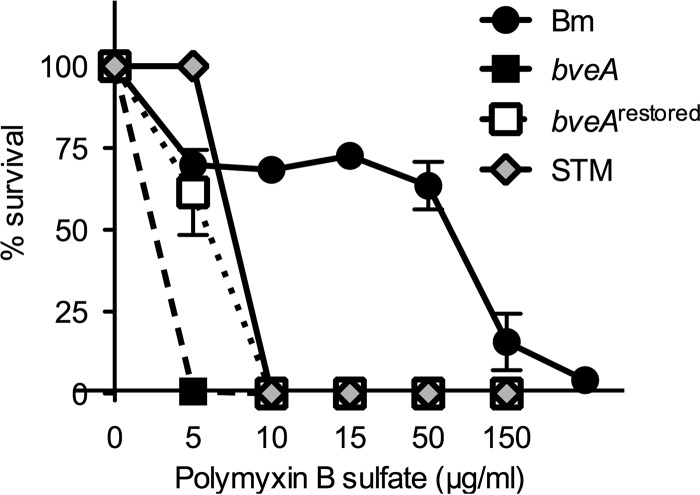
The remarkable resistance of Brucella spp. to cationic compounds such as polymyxins and host-derived antimicrobial peptides has been attributed to features of its cell envelope, including its unique lipopolysaccharide, and these properties contribute to the ability of Brucella spp. to infect their mammalian hosts (14, 20–22). However, since Brucella species with rough LPS, such as Brucella canis and Brucella ovis, are also resistant to cationic antimicrobials (23), it is possible that features of Brucella spp. in addition to LPS contribute to this phenotype. A gene with a predicted cell envelope-associated function, BMEII0681 (designated bveA for Brucella virulence-associated esterase A), was originally identified in a bioinformatic screen for Brucella genes with functions related to the cell envelope (partially published in reference 10). In silico analysis revealed that BMEII0681 was broadly conserved among Brucella spp., with paralogs in the soil-associated Alphaproteobacteria, including Ochrobactrum spp., Rhizobium spp., and Agrobacterium tumefaciens, as well as the more distantly related Burkholderia species associated with plant rhizospheres. To determine whether BveA functions in resistance against cationic antimicrobials, we constructed and characterized a B. melitensis strain deleted for bveA. The bveA mutant was indistinguishable from its wild-type parent with regard to growth in tryptic soy broth (see Fig. S1A in the supplemental material) and its ability to withstand short-term treatment (20 min) with cell envelope-disrupting agents, including the cationic lipopeptide polymyxin B (PmB), the antimicrobial peptides magainin 1 and cecropin P1, EDTA, lysozyme, and Tween 20 at concentrations that are bactericidal for smooth E. coli K-12 ATCC W1485 (14) (see Fig. S1B). Additionally, the mutant revealed no obvious defect in its lipopolysaccharide composition (see Fig. S1C). Considering that the antimicrobial action of PmB also induces bacteriostasis, we determined whether bveA contributes to resistance to the bacteriostatic effect of PmB. To this end, we incubated 1 × 106 CFU/ml of the wild-type B. melitensis 16M and an isogenic bveA mutant (CMR27) in TSB (pH 7.4) for 48 h with increasing concentrations of PmB to monitor the growth of the strain in the presence or absence of PmB (Fig. 1). As a reference, we used Salmonella enterica serotype Typhimurium strain IR715 grown under conditions that enhance its PmB resistance (9). As described previously (24), B. melitensis 16M exhibited a high level of PmB resistance compared to that of S. Typhimurium, with significant growth occurring at concentrations of PmB up to 150 μg/ml. In contrast, while S. Typhimurium grew in low (5 μg/ml) concentrations of PmB, at concentrations that exceeded 15 μg/ml PmB, a bactericidal effect of PmB was observed. The growth of the B. melitensis bveA mutant was inhibited at 5 μg/ml and a bactericidal effect was observed at 10 μg/ml, suggesting that BveA mediates resistance to polymyxin antibiotics. Partial restoration of the PmB resistance was achieved by chromosomal expression of the bveA gene under the control of the secE promoter (TOK09; bveArestored).
FIG 1.
BveA contributes to antibiotic resistance of B. melitensis. Impact of polymyxin B (PmB) on the growth of B. melitensis wild type (Bm), bveA mutant CMR27 (bveA), restored mutant TOK09 (bveArestored), and S. Typhimurium strain IR715 (STM) in TSB (pH 7.4) for 48 h at 37°C. The starting inoculum was 106 CFU/ml. S. Typhimurium IR715 was preincubated under conditions that induce the expression of the PhoP/Q regulon (9).
BveA is a phospholipase A1 that cleaves PE in the envelope of B. melitensis.
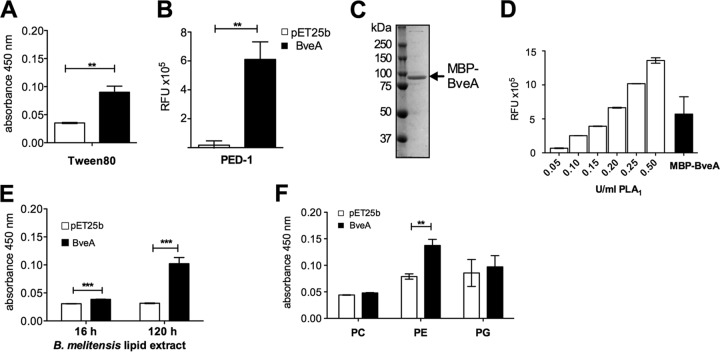
To predict the function of BveA in B. melitensis, we used the Phyre2.0 engine (25) to characterize orthologs in other organisms based on similarities to the solved protein structures. Table S1 in the supplemental material shows the five best hits based on similarity at the amino acid sequence and structural levels. All of these entries suggest that BveA is a putative hydrolase of the α/β2 superfamily, which includes lipases and esterases (26). Further, in silico prediction (SignalP) (27) indicated that BveA contains a signal peptide as well as two intramolecular disulfide bonds (DiANNA 1.1) (28), suggesting a periplasmic localization of the mature protein. To test for a potential esterase function for BveA, we expressed a 6×His-tagged fusion protein of BveA in E. coli (pTK07). To this end, we obtained periplasmic fractions from E. coli carrying pTK07 or the empty vector pET25b and determined the esterase activities in these fractions, using Tween 80 as a substrate. In this assay, cleavage of the ester bond linking the sorbitol backbone of Tween 80 to oleate leads to the release of free oleate, yielding a precipitate with CaCl2 that can be quantified spectrophotometrically (15). Cleavage of Tween 20 by periplasmic fractions containing BveA-6×His, but not control fractions from E. coli carrying the empty vector (Fig. 2A), demonstrated that BveA has esterase activity. To better characterize the specificity of BveA, we determined whether fractions containing BveA-6×His could cleave the fluorogenic phospholipase A1-selective substrate PED-1 (Fig. 2B). Both the periplasmic fraction containing BveA-6×His and a purified MBP-BveA fusion protein (Fig. 2C) were able to cleave PED-1, demonstrating that BveA is a phospholipase A1 (Fig. 2D).
FIG 2.
BveA is a phospholipase A1 with specificity for PE lipase activity against Tween 80 (A) and the phospholipase A1 substrate PED-1 (B) using periplasmic fractions containing BveA and the empty vector control. Purification (C) and lipase activity (D) of the MBP-BveA fusion protein (97-kDa) compared to those of increasing units of gelatinase, a positive control for phospholipase A1 activity (PLA1) (Life Technologies). Lipase activity of BveA against lipid extractions of the B. melitensis cell envelope (E) and against phospholipid standards (Sigma-Aldrich) PC, PE, and PG (F). Significant differences between BveA-containing samples and the empty vector control: **, P < 0.01; ***, P < 0.001. RFU, relative fluorescence units.
Since both the in silico analysis and the expression in E. coli indicated a periplasmic localization of BveA, we tested whether BveA would act on the cell envelope lipids of B. melitensis. To this end, periplasmic fractions containing BveA-6×His were incubated with phospholipids extracted from B. melitensis 16M (Fig. 2E). We observed a low activity of the enzyme after 16 h of incubation, which increased with prolonged incubation times. Assuming a small amount of Brucella lipids in the assay, we extended the assay duration to 120 h of incubation, and this gave precipitate accumulations comparable to those observed for Tween 80. Since the major lipid components of the Brucella envelope are the glycerolipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), we tested the activities of the BveA-6×His-containing periplasmic fractions against those of purified PC, PE, and PG (1 mM). This assay detected phospholipase activity against PE, but when PC or PG was provided as the substrate, no esterase activity was detected (Fig. 2F).
BveA fine-tunes the PE content of the B. melitensis cell envelope.
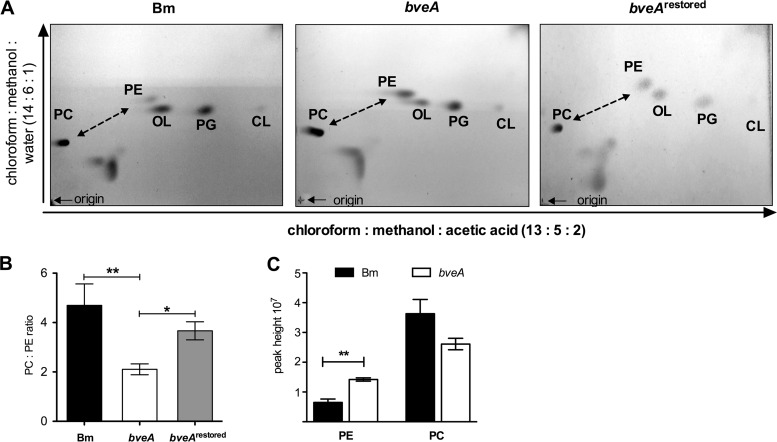
The apparent specificity of BveA for PE suggested that it targets PE in the B. melitensis cell envelope. To test this idea, we compared the relative amounts of the major membrane lipid species in the methyl-tert-butyl ether (MTBE) extracts of the B. melitensis wild type (16M), the bveA mutant (CMR27), and the chromosomally restored bveA mutant (TOK09; bveArestored) by two-dimensional thin-layer chromatography (2D-TLC), using the conditions reported previously (17). The abundances of the main membrane lipids PC, PE, ornithine lipids (OL), PG, and cardiolipin (CL) were determined. Individual spots were assigned according to the position of the corresponding lipid standards from E. coli (Avanti-Polar; data not shown). While PC, PE, OL, PG, and CL were detected in the cell envelope of the wild type, the bveA mutant (CMR27), and the bveArestored strain (TOK09), the signal intensity of the PE spot compared to that of the PC spot was higher in the bveA mutant (CMR27) than in the wild type and in the bveArestored strain (TOK09) (Fig. 3A, dashed arrows). Based on three biological replicates for each strain, we quantified the contribution of each lipid species to the known lipid content based on the densitometry of the spot intensities (see Fig. S2 in the supplemental material) and calculated the ratio of PC to PE for each strain (Fig. 3B). Significantly more PE was observed in the bveA mutant (CMR27) (P = 0.018) than in the wild type. When expressed as a PC/PE ratio, the values were 4.6 for the B. melitensis wild type, 2.1 for the bveA mutant, and 3.7 for the complemented mutant (TOK09). No significant differences between strains were observed for the other major cell envelope lipid species (see Table S2 in the supplemental material). These results supported the specificity of BveA for PE. Compared to that of the wild type, the lipid profile of the bveArestored strain (TOK09) was only altered in the PG levels. The significance of this result is not clear, but it may be related to the expression of bveA from the strong secE promoter at its chromosomal insertion site rather than from its own promoter. This slight alteration in lipid composition in the restored bveA mutant (TOK09) compared to that of the wild type may underlie the partial restoration of resistance against PmB in this strain (Fig. 1).
FIG 3.
BveA fine-tunes the PE content of the B. melitensis cell envelope, Membrane lipid composition of wild-type B. melitensis (Bm) compared to an isogenic bveA mutant CMR27 (bveA) and the chromosomally restored strain TOK09 (bveArestored). (A) 2D-TLC of lipid extracts of the cell envelope of B. melitensis 16M, bveA mutant (CMR27), and restored strain TOK09 extracted with MTBE. Phospholipids were detected with molybdatophosphoric acid spray solution (Merck). The dashed line highlights the PC/PE ratio. (B) Ratio of phospholipids PC/PE in the cell envelope of B. melitensis 16M, bveA mutant CMR27, and chromosomally restored bveA mutant TOK09. Ratios were calculated based on the relative densitometric abundance (percentage) of individual MBTE-extracted membrane lipids from 2D-TLC of PC, PE, OL, PG, and CL (see Fig. S2 in the supplemental material). (C) Q-TOF analysis to estimate the relative quantities of PE and PC in B. melitensis 16M and the bveA mutant CMR27. Significant differences between the wild-type 16M and bveA mutant CMR27: *, P < 0.05; **, P < 0.01.
While densitometric quantification of 2D-TLC signals provided a first clue to changes in the abundance of PE, a more quantitative measure of lipid abundance requires specific measurement of the lipid species. We then determined the levels of PE and PC species in MTBE extracts of the wild type and bveA mutant by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS). The lipid species were identified using the LipidBlast in silico tandem mass spectrometry database search (18), with a focus on PE and PC species. All individual identifiable PE and PC species were grouped and analyzed to determine the overall changes in these phospholipids. We chose this approach because PE is used by Brucella spp. for the synthesis of PC species (29), and, thus, a non-PE-specific membrane defect might be consequently reflected in changes in the PC pool. As presented in Fig. 3C, we again detected a significant increase in the total amount of PE species in the mutant over that in the wild type. In contrast, no significant difference in the amount of PC species between the wild type and mutant was observed. Taken together, these results suggest that BveA modulates the phospholipid content of B. melitensis by degrading PE and that loss of BveA results in an increased abundance of PE in the cell envelope.
BveA is important for interaction of B. melitensis with mammalian hosts.
To determine whether modulation of PE in the cell envelope is important for the ability of B. melitensis to interact with host cells, we compared the abilities of the bveA mutant (CMR27) and the wild type to survive intracellularly within macrophages, using a gentamicin protection assay. To this end, we infected the murine macrophage-like cell line J774.A1 with the B. melitensis wild-type strain 16M, the bveA mutant (CMR27), and the bveA mutant complemented with a plasmid-carried copy of bveA (CMR28). As presented in Fig. 4A, the survival of the bveA mutant was reduced beginning at 4 h postinfection compared to that of the wild-type strain 16M, and this defect was restored by complementation.
FIG 4.
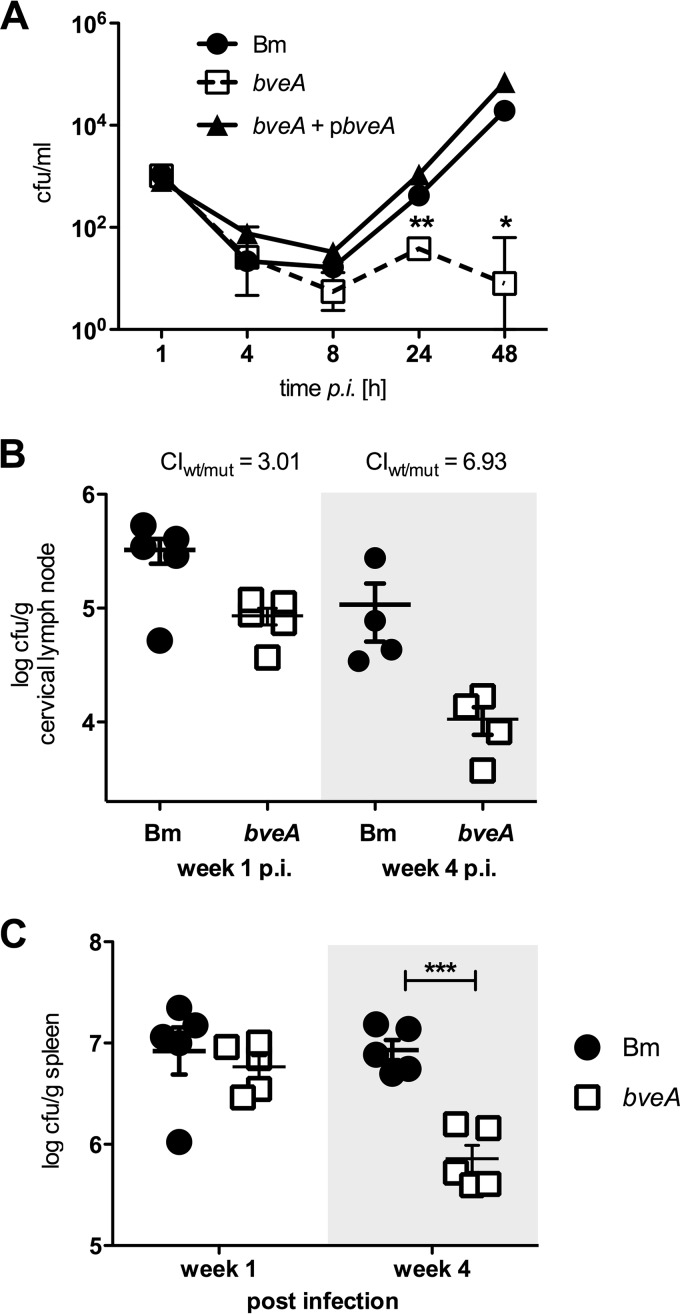
BveA contributes to intracellular survival of B. melitensis in macrophages and to persistent infection in vivo. (A) Intracellular replication of wild-type B. melitensis (Bm), bveA mutant CMR27 (bveA), and complemented strain CMR28 (pbveA) as recovered CFU/ml at specific time points postinfection in J774.A1 macrophage-like cells. (B) Colonization levels of wild-type and bveA mutant CMR27 in cervical lymph nodes of C57BL/6J mice at 1 week (n = 5) and 4 weeks (n = 4) after inoculation via the i.n. route with a 1:1 mixture of the wild type (WT) and CMR27 (bveA). The competitive index (ratio of WT to CMR27) was calculated based on recovered CFU/g tissue. (C) Colonization levels of B. melitensis WT and CMR27 (bveA) after inoculation of individual strains via the i.p. route. Symbols represent the values from individual mice, and horizontal bars represent the geometric means of each group. Significant differences between the wild type and CMR27 (bveA): *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next determined whether modulation of PE abundance by BveA was required for colonization and persistence of B. melitensis in a mammalian host. To this end, we used a competitive infection assay, in which C57BL/6J mice were inoculated via the intranasal route with an inoculum containing a 1:1 ratio of the B. melitensis 16M and bveA mutant. At 1 and 4 weeks after inoculation, colonization of the draining cervical lymph nodes was assessed (Fig. 4B). At 1 week postinfection, 3-fold-higher levels of the B. melitensis wild type than the bveA mutant (CMR27) were recovered. This competitive advantage was increased at 4 weeks, suggesting that modulation of the membrane lipid contents by BveA contributes to the fitness of B. melitensis during persistent infection.
The intranasal route of inoculation, while mimicking the natural mucosal infection route, is very inefficient in establishing infection, as dissemination to the draining cervical lymph nodes requires passage through a narrow infection bottleneck. We therefore performed an additional experiment in which we inoculated mice via the i.p. route with individual B. melitensis strains. The results of this experiment, shown in Fig. 4C, revealed that the bveA mutant disseminated to the spleen initially at wild-type levels, but at 4 weeks, the mutant was recovered at an order of magnitude less than the wild type. Thus, the bveA mutant is deficient for persistence in both draining lymph nodes and systemically in the spleen.
DISCUSSION
It is known that the cell envelope of Brucella species has a phospholipid composition that differs markedly from that of other Gram-negative mammalian pathogens (30). Two major differences between Brucella spp. and other Gram-negative bacteria, such as S. Typhimurium or E. coli, are the presence of PC and a remarkably small PE content: the PE content is 80% in E. coli, whereas it is only 25% in B. melitensis (30–33). Interestingly, a link between the PE abundance and antimicrobial susceptibility has been noted previously in studies with model biomembranes. The preferential binding of the amphiphilic cationic peptide NK2 to PE-rich model membranes (34) was related to a higher affinity of NK2 for PE than for other lipids and correlated with the degree of accessibility of the lipid phosphate groups to the cationic side chains of the NK2 peptide (35).
Previous work on Brucella abortus demonstrated that disruption of the individual synthesis pathways for either PC (pcs and pmtA) (29) or PE (pssA) (17) resulted in reduced fitness of B. abortus strains in vitro and in vivo (17), demonstrating that the phospholipid composition of the envelope is critical to the biology of this organism. Our results detail a mechanism by which B. melitensis maintains a low abundance of PE in the cell envelope via expression of the BveA phospholipase A1 enzyme. This property of the cell envelope contributes to both polymyxin resistance and persistence in an infected host.
Although inactivation of bveA and the consequent increase in PE abundance did not compromise the integrity of the B. melitensis cell envelope as evidenced by an unchanged sensitivity to membrane-disrupting agents, it did affect its ability to survive in a mammalian host. BveA contributed to host-pathogen interactions of B. melitensis either directly, by enhancing resistance to host antimicrobials, or indirectly. Since phospholipids are determinants of membrane topology (36), a change in the balance between the phospholipid species might affect the insertion or proper function of membrane proteins involved in interaction with host cells. For example, Agrobacterium tumefaciens encodes two proteins of this family, VirJ (encoded on the octopine-type plasmid pTiA6) and AcvB, that play overlapping roles in tumorigenesis, by promoting transfer DNA (T-DNA) transfer to plant cells (11). Thus, it is possible that the esterase activity of these proteins modulates the phospholipid composition of the cytoplasmic membrane to enable the proper assembly of the type IV secretion system that mediates the transfer of T-DNA and effector proteins.
The best-characterized mechanism of resistance to polymyxins in bacteria is modification of the lipid A moiety of LPS (37). The regulation and biochemistry of these processes have been described in detail for S. Typhimurium and Pseudomonas aeruginosa, which respond to the presence of antimicrobial peptides by modification of lipid A to reduce its net negative charge, a strategy that prevents PmB binding to the bacterial surface (37–39). Recently, part of the S. Typhimurium response to cationic peptides was shown to be palmitoylation of PG by the lipid A-modifying enzyme PagP, suggesting that modifications of other membrane lipids may also play a role in defense against cationic peptides such as PmB (40).
The B. abortus LPS is known to have several distinctive features that mediate its resistance to PmB, including a lack of divalent cations (14), a low degree of acyl chain fluidity (41), and the glycosylation pattern of its LPS core (22, 42). These features of the LPS might be considered a first line of defense against PmB, since the B. melitensis mutants with rough LPS are killed rapidly, in contrast to the bveA mutant, which is killed after a prolonged incubation with higher concentrations of PmB. Thus, it is likely that the LPS of Brucella spp. mediates resistance to PmB, but during prolonged incubation small amounts can enter the cell envelope, and at this point the reduction in PE composition in the cell envelope by BveA is important for resisting the formation of pore-like structures and the permeabilization of the cytoplasmic membrane by PmB that result in the loss of the proton-motive force and the inhibition of growth (5). Considering the presence of bveA orthologs (e.g., AtvA from Rhizobium tropici CIAT899 [43]) in the phylogenetically related alphaproteobacterial species inhabiting the rhizosphere, a reduction in PE abundance may be a conserved mechanism among this group of bacteria with relevance for bacterial-plant interactions as well as interactions with mammalian hosts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by PHS grants AI050553 and AI109799 to R.M.T. V.L.A. was supported by T32AI060555.
We thank Andreas Bäumler, Sebastian Winter, Chuck Bevins, Ignacio Moriyón, Maite Iriarte, and Raquel Conde-Álvarez for helpful discussions during the course of this work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00792-15.
REFERENCES
- 1.Storm DR, Rosenthal KS, Swanson PE. 1977. Polymyxin and related peptide antibiotics. Annu Rev Biochem 46:723–763. doi: 10.1146/annurev.bi.46.070177.003451. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos A, Falagas ME. 2008. Colistin and polymyxin B in critical care. Crit Care Clin 24:377–391. doi: 10.1016/j.ccc.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 4.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaPorte DC, Rosenthal KS, Storm DR. 1977. Inhibition of Escherichia coli growth and respiration by polymyxin B covalently attached to agarose beads. Biochemistry 16:1642–1648. doi: 10.1021/bi00627a019. [DOI] [PubMed] [Google Scholar]
- 6.Farrell ID. 1974. The development of a new selective medium for the isolation of Brucella abortus from contaminated sources. Res Vet Sci 16:280–286. [PubMed] [Google Scholar]
- 7.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat 13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Alton GG, Jones LM, Pietz DE. 1975. Laboratory techniques in brucellosis. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 9.Groisman EA, Kayser J, Soncini FC. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol 179:7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolan HG, Tsolis RM. 2012. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol 2:47. doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan SQ, Jin S, Boulton MI, Hawes M, Gordon MP, Nester EW. 1995. An Agrobacterium virulence factor encoded by a Ti plasmid gene or a chromosomal gene is required for T-DNA transfer into plants. Mol Microbiol 17:259–269. doi: 10.1111/j.1365-2958.1995.mmi_17020259.x. [DOI] [PubMed] [Google Scholar]
- 12.Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop Ii RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 13.Copin R, Vitry MA, Hanot Mambres D, Machelart A, De Trez C, Vanderwinden JM, Magez S, Akira S, Ryffel B, Carlier Y, Letesson JJ, Muraille E. 2012. In situ microscopy analysis reveals local innate immune response developed around Brucella infected cells in resistant and susceptible mice. PLoS Pathog 8:e1002575. doi: 10.1371/journal.ppat.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez de Tejada G, Pizarro-Cerda J, Moreno E, Moriyón I. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect Immun 63:3054–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plou FJ, Ferrer M, Nuero OM, Calvo MV, Alcalde M, Reyes F, Ballesteros A. 1998. Analysis of Tween 80 as an esterase/lipase substrate for lipolytic activity assay. Biotechnol Tech 12:183–186. doi: 10.1023/A:1008809105270. [DOI] [Google Scholar]
- 16.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukata L, Altabe S, de Mendoza D, Ugalde RA, Comerci DJ. 2008. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J Bacteriol 190:8197–8203. doi: 10.1128/JB.01069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. 2013. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.den Hartigh AB, Rolan HG, De Jong MF, Tsolis RM. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J Bacteriol 190:4427–4436. doi: 10.1128/JB.00406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freer E, Pizarro-Cerdá J, Weintraub A, Bengoechea J-A, Moriyón I, Hultenby K, Gorvel J-P, Moreno E. 1999. The outer membrane of Brucella ovis shows increased permeability to hydrophobic probes and is more susceptible to cationic peptides than are the outer membranes of mutant rough Brucella abortus strains. Infect Immun 67:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paixão TA, Roux CM, Den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, Tsolis RM. 2009. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect Immun 77:4197. doi: 10.1128/IAI.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conde-Álvarez R, Arce-Gorvel V, Iriarte M, Manček-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C, Chaves-Olarte E, Martirosyan A, von Bargen K, Grilló M-J, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyón I, Gorvel J-P. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog 8:e1002675. doi: 10.1371/journal.ppat.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen AE, Halling SM. 2010. Effect of polymyxin B and environmental conditions on isolation of Brucella species and the vaccine strain RB51. Comp Immunol Microbiol Infect Dis 33:121–131. doi: 10.1016/j.cimid.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Moreno E, Moriyón I. 2001. The genus Brucella. In Dworkin M. (ed), The prokaryotes: an evolving electronic resource for the microbiological community, Springer Verlag, New York, NY. [Google Scholar]
- 25.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Nardini M, Dijkstra BW. 1999. α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 27.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Ferre F, Clote P. 2006. DiANNA 1.1: an extension of the DiANNA Web server for ternary cysteine classification. Nucleic Acids Res 34(Web Server issue):W182−W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conde-Alvarez R, Grilló MJ, Salcedo SP, De Miguel MJ, Fugier E, Gorvel JP, Moriyón I, Iriarte M. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell Microbiol 8:1322–1335. doi: 10.1111/j.1462-5822.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- 30.Gamazo C, Moriyón I. 1987. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect Immun 55:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ames GF. 1968. Lipids of Salmonella Typhimurium and Escherichia coli: structure and metabolism. J Bacteriol 95:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pramanik J, Keasling JD. 1997. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng 56:398–421. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Thiele OW, Schwinn G. 1973. The free lipids of Brucella melitensis and Bordetella pertussis. Eur J Biochem 34:333–344. doi: 10.1111/j.1432-1033.1973.tb02764.x. [DOI] [PubMed] [Google Scholar]
- 34.von Deuster CI, Knecht V. 2012. Antimicrobial selectivity based on zwitterionic lipids and underlying balance of interactions. Biochim Biophys Acta 1818:2192–2201. doi: 10.1016/j.bbamem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Schröder-Borm H, Willumeit R, Brandenburg K, Andrä J. 2003. Molecular basis for membrane selectivity of NK-2, a potent peptide antibiotic derived from NK-lysin. Biochim Biophys Acta 1612:164–171. doi: 10.1016/S0005-2736(03)00115-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 37.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA/PmrB regulated genes necessary for 4 aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 39.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manterola L, Moriyón I, Moreno E, Sola-Landa A, Weiss DS, Koch MH, Howe J, Brandenburg K, Lopez-Goni I. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J Bacteriol 187:5631–5639. doi: 10.1128/JB.187.16.5631-5639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gil-Ramírez Y, Conde-Álvarez R, Palacios-Chaves L, Zuniga-Ripa A, Grillo MJ, Arce-Gorvel V, Hanniffy S, Moriyón I, Iriarte M. 2014. The identification of wadB, a new glycosyltransferase gene, confirms the branched structure and the role in virulence of the lipopolysaccharide core of Brucella abortus. Microb Pathog 73:53–59. doi: 10.1016/j.micpath.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Vinuesa P, Neumann-Silkow F, Pacios-Bras C, Spaink HP, Martínez-Romero E, Werner D. 2003. Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol Plant Microbe Interact 16:159–168. doi: 10.1094/MPMI.2003.16.2.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.