Abstract
Whole-genome sequences for Stenotrophomonas maltophilia serial isolates from a bacteremic patient before and after development of levofloxacin resistance were assembled de novo and differed by one single-nucleotide variant in smeT, a repressor for multidrug efflux operon smeDEF. Along with sequenced isolates from five contemporaneous cases, they displayed considerable diversity compared against all published complete genomes. Whole-genome sequencing and complete assembly can conclusively identify resistance mechanisms emerging in S. maltophilia strains during clinical therapy.
TEXT
Stenotrophomonas maltophilia is an aerobic, nonfermenting, and motile Gram-negative bacterium that is increasingly recognized as a cause of hospital-acquired infections, with crude mortality rates of 14 to 69% in cases of bacteremia (1). Treatment of S. maltophilia infections is challenging due to the pathogen's intrinsic resistance to many antibiotic classes via drug efflux pumps, beta-lactamase production, and decreased membrane permeability (1). Resistance phenotypes are known to change during the course of treatment, which complicates interpretation of automated drug susceptibility testing (DST) results (2). A mutant strain of S. maltophilia with emerging resistance to tetracycline, chloramphenicol, and quinolones was previously characterized following in vitro tetracycline selection (3, 4). However, little is known about the genetic and molecular mechanisms underlying acquired resistance in the clinical setting—particularly for quinolones, as, in contrast to other Gram-negative organisms, the quinolone-resistance determining region (QRDR) of topoisomerase genes is often unaltered (5). In this report, we describe the first reported use of whole-genome sequencing (WGS) in serial clinical isolates to definitively identify an acquired quinolone resistance mutation in S. maltophilia.
WGS was performed for the initial and subsequent S. maltophilia blood culture isolates from a patient for whom acquired quinolone resistance was observed (patient 1) and five other patients (patients 2 to 6) selected from a 2-month period in 2013 at The Mount Sinai Hospital. Patient 1 was a 56-year-old man requiring urgent placement of an intrahepatic shunt for complications of a Whipple procedure, which was followed by several episodes of bacteremia that were treated with multiple courses of antimicrobials, including levofloxacin. Two months later, another bacteremia developed, and blood cultures intermittently grew S. maltophilia despite antimicrobial therapy. Automated DST by Vitek2 (bioMérieux, Marcy l'Etoile, France) showed that the first S. maltophilia isolate acquired (ISMMS2) was susceptible to fluoroquinolones and trimethoprim-sulfamethoxazole (SXT). After treatment with ciprofloxacin the bacteremia initially cleared, but blood cultures 9 days later again grew S. maltophilia (isolated as ISMMS2R), now resistant to fluoroquinolones while still susceptible to SXT. Ciprofloxacin was stopped and intravenous SXT was given; subsequent cultures did not grow S. maltophilia.
Standard culturing and susceptibility testing for levofloxacin and SXT were performed by automated broth microdilution with Vitek2. Antimicrobial sensitivities were reported and interpreted according to the 2015 CLSI guidelines for S. maltophilia (6). Isolates were then stocked and frozen at −80°C. Levofloxacin and SXT susceptibilities for all isolates in this study were later confirmed by Etest (bioMérieux) at 24 h. To prepare for sequencing, isolates were grown from single colonies in tryptic soy broth, and DNA extraction was performed as previously described (7).
Sequencing was performed on the PacBio RSII platform (Pacific Biosciences, Menlo Park, CA), and reads were assembled de novo using PacBio's Hierarchical Genome Assembly Process (version 3) (8). Pairwise comparison and variant calling were performed with MUMmer 3.23 (9). Mugsy 2.2 (10) was used to align whole-genome sequences for phylogenetic reconstruction with RAxML-8.0.2 (11), and rearrangements were visualized with Mauve 2.4.0 (12). Additional details, including Sanger validation methods and sequence accession numbers, are in the supplemental methods.
Two complete whole-genome sequences were derived from patient 1's isolates before and after the change in levofloxacin MIC and compared to whole-genome sequences of five control S. maltophilia isolates (patients 2 to 6). All sequences were de novo assembled, i.e., without regard to reference assemblies. Table 1 summarizes the relative dates of collection, antimicrobial susceptibility results, and assembly statistics.
TABLE 1.
Sequenced clinical isolates and their antimicrobial susceptibilitiesa
| Patient | Time of collectionb (days) | Isolate name | Susceptibility (MIC [mg/liter]) |
Assembly quality | Depth of coverage | |||
|---|---|---|---|---|---|---|---|---|
| Levo |
SXT |
|||||||
| Vitek2 | Etest | Vitek2 | Etest | |||||
| 1 | 0 | ISMMS2 | S (0.5)c | S (1) | S (<20) | S (0.19) | 1 circular 4.51-Mbp chromosome | 160× |
| 1 | +10 | ISMMS2R | R (>32)c | R (16) | S (1) | S (0.38) | 1 circular 4.51-Mbp chromosome | 403× |
| 2 | −26 | ISMMS3 | S (0.25) | S (0.38) | U (80, <20)d | S (0.75) | 1 circular 4.80-Mbp chromosome | 153× |
| 3 | +14 | ISMMS4 | R (>8) | R (>12) | U (0.5, 80)d | S (0.75) | 3 contigs (4.73 Mbp, 6.5 kbp, 11.2 kbp) | 303× |
| 4 | −32 | ISMMS5 | S (1) | S (1) | S (<20) | S (0.25) | 18 contigs | 270× |
| 5 | 0 | ISMMS6 | S (<0.12) | S (0.125) | S (<20) | S (1.5) | 10 contigs | 262× |
| 6 | +2 | ISMMS7 | S (1) | S (0.75) | S (<20) | S (1.5) | 1 circular 4.69-Mbp chromosome, 1 additional 17.7-kbp contig | 318× |
Levo, levofloxacin; SXT, trimethoprim-sulfamethoxazole; S, susceptible; R, resistant; U, undetermined; Mbp, million base pairs; kbp, thousand base pairs.
Time of collection was defined in days relative to the date of collecting the initial S. maltophilia isolate in the case patient.
Change in levofloxacin susceptibility investigated in this study.
Inconsistent results were obtained in replicate.
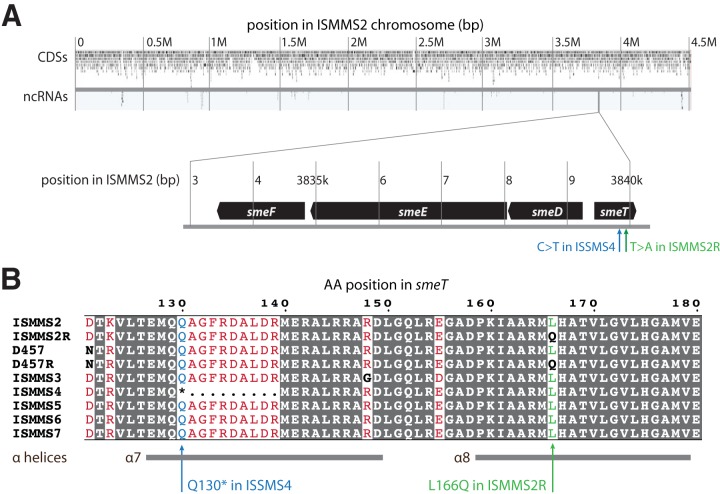
Assembled genome sequences for patient 1's isolates before (ISMMS2) and after (ISMMS2R) observation of levofloxacin resistance were compared directly and were identical except for one single-nucleotide variant (SNV) and five one-base indels. Sanger sequencing confirmed the presence of the SNV but identified the indels as homopolymer assembly errors. Coding domain sequence predictions for the surrounding locus (Fig. 1A) revealed that the SNV was inside smeT, a tetR-like repressor upstream of the structural operon for the smeDEF genes, which encode a multidrug efflux pump. The SNV is a T-to-A substitution at position 497 of smeT causing a nonsynonymous Leu-166→Gln mutation.
FIG 1.
Single-nucleotide variants (SNVs) observed in quinolone-resistant S. maltophilia clinical isolates. (A) Assembled circular chromosome for ISMMS2, including predicted coding domain sequence (CDS) and noncoding RNA (ncRNA) features drawn with ChromoZoom (23). Horizontal position corresponds to base pair location. The smeDEF operon is shown in the detail callout, which highlights both the smeT c.497T>A SNV that emerged in ISMMS2R and the aligned location of the smeT c.388C>T SNV (resulting in a premature stop codon) in ISMMS4. ISMMS2 and ISMMS2R are serial isolates from a single patient before and after development of quinolone resistance, while ISMMS4 was quinolone resistant at initial isolation from a different patient. (B) Multiple-sequence alignment of part of the predicted smeT product in each of the clinical isolates, the D457 reference assembly, and its quinolone-resistant counterpart D457R. Predicted α-helices (13) are depicted as gray bars below the sequence. Positions identical in all sequences are shaded with a dark gray background, equivalent substitutions are in red, and nonequivalent substitutions are in black and boldface. The L166Q and Q130* (*, stop codon) polymorphisms are highlighted.
The same nonsynonymous mutation has been previously observed in an in vitro strain of S. maltophilia, D457R, created by selecting single-step tetracycline-resistant mutants from the antibiotic-susceptible clinical strain D457 (3, 4). The mutation is in the eighth α-helix of the smeT protein (13), which homodimerizes to repress transcription of the smeDEF operon (3, 13). Although the mutation is not in the DNA-binding region, it has been shown to disable the repressor activity of SmeT (3), leading to upregulation of smeDEF and conferring a multidrug resistance (MDR) phenotype (14).
Figure 1B shows an amino acid sequence alignment comparing smeT in D457 and D457R to aligned sequences from our seven isolates. Notably, while none of the remaining isolates shared the same Leu-166→Gln (c.497T>A) mutation, another isolate resistant to levofloxacin, ISMMS4, displayed a C-to-T mutation at position 388 of smeT, which creates a premature stop codon that likely disrupts smeT function (Fig. 1A and B).
The QRDRs are loci within genes encoding topoisomerase II and IV subunits known for mutations that confer quinolone resistance in Gram-negative bacteria, although they appear to play a secondary role to efflux systems for resistance emerging during treatment of S. maltophilia infection (5). An amino acid sequence alignment of the gyrA, gyrB, and parC genes of our seven isolates and the reference clinical isolates D457 and K279a revealed no differences in the QRDR. Some variants were observed within the QRDR of parE (see Fig. S1 in the supplemental material), all of which were consistent with past observations in clinical isolates (15) except for an Ile-599→Val variant observed in three of our isolates and the D457 reference sequence.
Significant genomic diversity was observed among the S. maltophilia isolates from all six patients. Figure S2 in the supplemental material shows a maximum-likelihood phylogeny with branch lengths scaled to SNV distances. Our isolates are distributed widely among all four reference assemblies for complete S. maltophilia genomes in GenBank. The distances of tens of thousands of SNVs seen in our phylogeny suggest that the natural diversity of pathogenic S. maltophilia is greater than that captured by the current set of reference assemblies, even within a single hospital setting.
Recombination is not an obvious source of diversity in our S. maltophilia isolates. Figure S3 in the supplemental material depicts whole-genome alignments between the four clinical isolates where assembly produced a circularized chromosome and the four GenBank references, showing small areas of nonhomology separating large regions of significant homology occurring generally in the same order for each genome. ISMMS2 and ISMMS2R are structurally identical, as expected for serial isolates, while recombination events among other strains are limited to small, 1- to 2-kb, segments. Epigenetics motif analysis also suggests that the isolates are not related. Table S1 in the supplemental material shows different motifs in isolates from separate patients, implicating differences in type II and III restriction modification systems between the isolates more likely to be caused by interstrain or interspecies horizontal transfer of methyltransferases than by intrastrain mutations (16). Together, these observations demonstrate that transmission did not occur among these six cases and that whole-genome sequencing can comprehensively capture genetic distances and structural variants among diverse clinical isolates of S. maltophilia.
This is the first report of WGS on serial isolates to characterize the emergence of a resistance mutation in S. maltophilia during antibiotic treatment of an active infection. In contrast to studies sequencing highly resistant strains of S. maltophilia to reveal various intrinsic and acquired antibiotic resistance genes (17, 18), where it remains difficult to assess their relative importance to the phenotype, performing WGS on serial isolates as resistance emerges in vivo allows the causative mutation(s) to be captured. In our patient, the mutation was a SNV that replicates a variant observed in an in vitro model strain created to study the MDR phenotype in 1997 (4). Using WGS and susceptibility testing, we can confirm that this SNV was the only variant to emerge and that it was sufficient to confer quinolone resistance in a clinical case. This underscores the need for clinicians to consider repeating DST during monotherapy if clinical signs suggest therapy failure.
smeT appears to play a central role in adaptive resistance to quinolones and other antibiotics effluxed by smeDEF, like tetracycline, chloramphenicol, erythromycin, and aminoglycosides. Since any mutation that inactivates this protein would be able to derepress smeDEF and confer resistance, smeT is under intense selective pressure in the presence of these drugs. In this study, we observed not only a deleterious SNV in the strain that displayed resistance (ISMMS2R) but also a premature stop codon in smeT in a strain that was already resistant at first isolation (ISMMS4). Certain nucleotide positions appear to be under greater selective pressure than others, as evidenced by our observation of the same mutation that occurred in D457R and a relative paucity of nonsynonymous coding mutations in smeT observed among clinical smeT isolates (19). Since sustained overexpression of smeDEF is physiologically unfavorable (20), it is possible that pathogenic strains of S. maltophilia rely on natural diversity of mutations in the smeT locus to activate or deactivate smeDEF expression, allowing for rapid adaptation to antibiotic stress, though further study is needed.
Since resistance from a single SNV emerged during a short course of ciprofloxacin, clinicians should be cautioned about using quinolone monotherapy for S. maltophilia bacteremia, as highlighted in recent retrospective studies (21, 22). The wide variety of MDR phenotypes and unreliability of DST results have created uncertainty about appropriate treatment for S. maltophilia, but SXT remains the most common choice for monotherapy (1, 21, 22). SXT resistance in S. maltophilia is not known to be caused by efflux systems but has been linked to class 1 integrons and ISCR elements (1). This suggests that spontaneous resistance is less likely to emerge with SXT monotherapy, although a clinical trial comparing the two antibiotics is warranted (21, 22).
In conclusion, characterizing the full extent of genetic alterations that S. maltophilia utilizes to develop antibiotic resistance in vivo and improving genomic surveillance of clinical strains will help refine antibiotic selection criteria available to clinicians. Furthermore, this study highlights the utility of WGS for profiling the precise mutations underlying emerging antibiotic resistance in clinical cases of bacteremia.
Nucleotide sequence accession numbers.
Complete genome sequences for ISMMS2, ISMMS2R, and ISMMS3 were deposited in GenBank under accession numbers CP011305, CP011306, and CP011010, respectively. Deposited sequences for ISMMS2 and ISMMS2R incorporate the Sanger corrected regions described above. Sequences for ISMMS4, ISMMS5, ISMMS6, and ISMMS7 were deposited as whole-genome shotgun projects at DDBJ/EMBL/GenBank under accession numbers JZIU00000000, JZIV00000000, JZIW00000000, and JZTX00000000, respectively, with the versions described in this paper at JZIU01000000, JZIV01000000, JZIW01000000, and JZTX01000000, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Icahn Institute for Genomics and Multiscale Biology at Mount Sinai, in part by the NIAID-supported NRSA Institutional Research Training Grant (5 T32 AI 7647-13) for Global Health Research (D.R.A.), and in part by the resources and expertise of the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
We thank Timothy O'Donnell, Tavi Nathanson, Jose Clemente, Flora Samaroo, Angelo Rendo, and members of the clinical microbiology laboratory at Mount Sinai for their contributions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01723-15.
REFERENCES
- 1.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrison MW, Anderson DE, Campbell DM, Carroll KC, Malone CL, Anderson JD, Hollis RJ, Pfaller MA. 1996. Stenotrophomonas maltophilia: emergence of multidrug-resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob Agents Chemother 40:2859–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sánchez P, Alonso A, Martinez JL. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob Agents Chemother 46:3386–3393. doi: 10.1128/AAC.46.11.3386-3393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso A, Martínez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 41:1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valdezate S, Vindel A, Saéz-Nieto JA, Baquero F, Cantón R. 2005. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J Antimicrob Chemother 56:220–223. doi: 10.1093/jac/dki182. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Altman DR, Sebra R, Hand J, Attie O, Deikus G, Carpini KWD, Patel G, Rana M, Arvelakis A, Grewal P, Dutta J, Rose H, Shopsin B, Daefler S, Schadt EE, Kasarskis A, van Bakel H, Bashir A, Huprikar S. 2014. Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am J Transplant 14:2640–2644. doi: 10.1111/ajt.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 9.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 10.Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández A, Maté MJ, Sánchez-Díaz PC, Romero A, Rojo F, Martínez JL. 2009. Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J Biol Chem 284:14428–14438. doi: 10.1074/jbc.M809221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso A, Martinez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:1879–1881. doi: 10.1128/AAC.45.6.1879-1881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdezate S, Vindel A, Echeita A, Baquero F, Cantón R. 2002. Topoisomerase II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob Agents Chemother 46:665–671. doi: 10.1128/AAC.46.3.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikhanta YN, Fox KL, Jennings MP. 2010. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol 8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 17.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream M-A, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Niu W, Sun Y, Hao H, Yu D, Xu G, Shang X, Tang X, Lu S, Yue J, Li Y. 2015. Identification and characterization of a serious multidrug resistant Stenotrophomonas maltophilia strain in China. Biomed Res Int 2015:580240. doi: 10.1155/2015/580240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez P, Alonso A, Martinez JL. 2004. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob Agents Chemother 48:2274–2276. doi: 10.1128/AAC.48.6.2274-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso A, Morales G, Escalante R, Campanario E, Sastre L, Martinez JL. 2004. Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J Antimicrob Chemother 53:432–434. doi: 10.1093/jac/dkh074. [DOI] [PubMed] [Google Scholar]
- 21.Cho SY, Kang CI, Kim J, Ha YE, Chung DR, Lee NY, Peck KR, Song JH. 2014. Can levofloxacin be a useful alternative to trimethoprim-sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob Agents Chemother 58:581–583. doi: 10.1128/AAC.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YL, Scipione MR, Dubrovskaya Y, Papadopoulos J. 2014. Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for treatment of Stenotrophomonas maltophilia infections. Antimicrob Agents Chemother 58:176–182. doi: 10.1128/AAC.01324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pak TR, Roth FP. 2013. ChromoZoom: a flexible, fluid, Web-based genome browser. Bioinformatics 29:384–386. doi: 10.1093/bioinformatics/bts695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.