Abstract
We report here the emergence of seven IMP-4-producing Raoultella ornithinolytica isolates obtained from one patient. All isolates carried the blaIMP-4 carbapenemase gene, five isolates also carried blaSHV-12, four contained blaTEM-1, and one contained blaOXA-1. Notably, the R. ornithinolytica isolate Ro25724 also expressed Klebsiella pneumoniae carbapenemase (KPC)-2. The blaKPC-2 gene was located on a Tn3-Tn4401 integration structure on a plasmid of ∼450 kb. This is the first description of the coexistence of blaKPC-2 and blaIMP-4 from the genus Raoultella.
TEXT
Raoultella ornithinolytica is mostly recovered from the environment and rarely causes severe infections in humans; so far, only a few invasive R. ornithinolytica infections have been reported (1, 2). Carbapenem resistance in Enterobacteriaceae represents one of the greatest problems in the realm of antibiotic resistance, and resistance is most often mediated by carbapenemases, such as Klebsiella pneumoniae carbapenemase (KPC), IMP, NDM, and OXA-48 (3). Plasmid-mediated carbapenem resistance may further aggravate the problem (4). However, the prevalence and molecular diversity of carbapenemases in Raoultella are largely unknown. Until now, the emergence of carbapenemases in the genus Raoultella has scarcely been described (1, 5–7).
In June 2014, a 13-year-old boy was admitted to our hospital. He had suffered a sport-related injury and required an urgent operation. The patient underwent an open reduction and internal fixation surgery. Two weeks later, his clinical status worsened, with episodes of obvious wound infection. Urgent debridement and drainage were carried out for four times in the following period of 80 days, and seven samples obtained from wound fluid and necrotic tissue were sequentially collected at different time points for routine clinical microbiology investigation. The general information and phenotypic characteristics of the seven isolates are summarized in Table 1.
TABLE 1.
General features and phenotypic characteristics of the seven R. ornithinolytica isolates
| Isolate no. | Isolation date (yr-mo-day) | Source | Antibiotic resistance gene(s) | MIC (μg/ml) fora: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMK | CIP | CTRX | IPM | ETP | AZT | GEN | FEP | TZP | TOB | SXT | SAM | ||||
| Ro23980 | 2014-7-18 | Exudate | blaIMP-4 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 8 | ≥16 | ≤20 | ≥32 |
| Ro24005 | 2014-7-19 | Exudate | blaIMP-4, blaSHV-12, blaTEM-1 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 8 | ≥16 | ≤20 | ≥32 |
| Ro24362 | 2014-8-4 | Necrotic tissue | blaIMP-4, blaSHV-12, blaTEM-1 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 8 | ≥16 | ≤20 | ≥32 |
| Ro24522 | 2014-8-11 | Exudate | blaIMP-4, blaSHV-12, blaTEM-1, blaOXA-1 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 64 | ≥16 | ≤20 | ≥32 |
| Ro24724 | 2014-8-21 | Necrotic tissue | blaIMP-4, blaKPC-2, blaSHV-12, | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 64 | ≥16 | ≤20 | ≥32 |
| Ro25277 | 2014-9-18 | Exudate | blaIMP-4, blaSHV-12, blaTEM-1 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 8 | ≥16 | ≤20 | ≥32 |
| Ro25687 | 2014-10-9 | Exudate | blaIMP-4 | ≥32 | ≥64 | 1 | ≥64 | 8 | ≥8 | ≥64 | ≥16 | ≥64 | 8 | ≥16 | ≥320 | ≥32 |
AMP, ampicillin; AMK, amikacin; CIP, ciprofloxacin; CTRX, ceftriaxone; IPM, imipenem; ETP, ertapenem; AZT, aztreonam; GEN, gentamicin; FEP, cefepime; TZP, piperacillin-tazobactam; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole; SAM, ampicillin-sulbactam.
All seven strains were identified as R. ornithinolytica by 16S rRNA sequencing analysis. In vitro antimicrobial susceptibility testing was conducted by Vitek 2 and Etest (bioMérieux, France) and interpreted as in previous studies (5, 8). The MIC results demonstrated that all isolates were resistant to ampicillin, amikacin, ceftriaxone, imipenem, ertapenem, aztreonam, gentamicin, cefepime, tobramycin, ceftazidime, and ampicillin-sulbactam but susceptible to ciprofloxacin and piperacillin-tazobactam (Table 1). It is worth noting that all isolates were susceptible to trimethoprim-sulfamethoxazole (SXT) except R. ornithinolytica strain Ro25687, which indicates that the patient was not infected by the same strain or that the use of antibiotics led to SXT resistance under the selective pressure of SXT usage, given that the patient received 2 weeks of SXT therapy before the isolation of Ro25687. To date, carbapenemase-resistant R. ornithinolytica strains have been reported only in North America (1, 9).
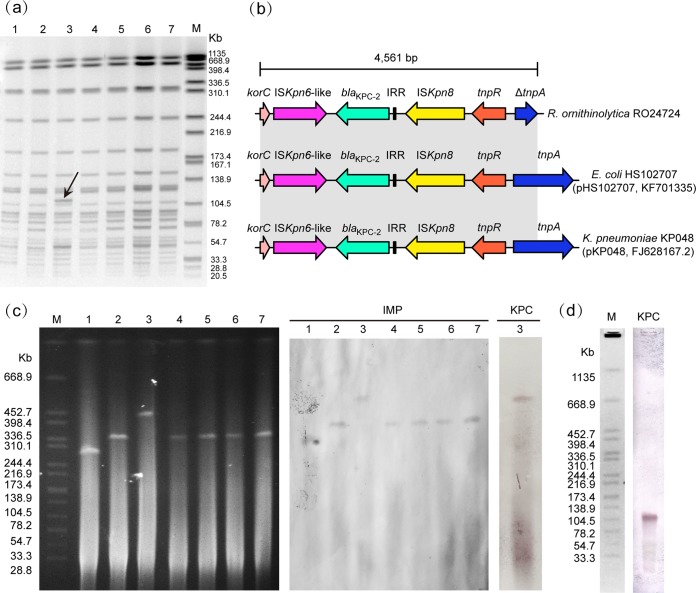
To confirm the clonality of the isolates, pulsed-field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) were therefore conducted according to published protocols (8, 10). All isolates shared identical or highly similar PFGE patterns (Fig. 1a) and ERIC-PCR profiles (see Fig. S1 in the supplemental material), indicating that all isolates were closely related. Notably, R. ornithinolytica strain Ro24724 yielded a different PFGE and ERIC-PCR genotype, with a difference of only one band.
FIG 1.
Molecular characterization of seven IMP-4-producing R. ornithinolytica strains. (a) PFGE patterns generated by XbaI restriction enzyme treatment of seven isolates. Lane 1, Ro25687; lane 2, Ro25277; lane 3, Ro24724; lane 4, Ro24522; lane 5, Ro24362; lane 6, Ro24005; lane 7, Ro23980A; M, Salmonella enterica subsp. enterica serotype Braenderup strain H9812 molecular marker. The arrow shows the band present only in strain Ro24724. (b) Schematic representation of the genetic structures surrounding the blaKPC-2 gene in Ro24724. Various genes and their directions of transcription are depicted as broad arrows. Regions with identical sequences are shown in gray between the different plasmids. On the right are the plasmids carried in the various strains, followed by their accession numbers. IRR, inverted repeat region. (c) S1-PFGE and Southern hybridization results with the blaIMP- and blaKPC-specific probes. Lane 1, Ro25687; lane 2, Ro25277; lane 3, Ro24724; lane 4, Ro24522; lane 5, Ro24362; lane 6, Ro24005; lane 7, Ro23980A; M, molecular mark. (d) PFGE followed by Southern hybridization using blaKPC-2 probe.
Screening for the presence of carbapenemase and extended-spectrum β-lactamase genes was carried out as described previously (11–15). The primers are detailed in Table S1 in the supplemental material. PCR screening revealed the presence of blaIMP-4 in all strains. All isolates were negative for other metallo-β-lactamase (MBL) or KPC-type β-lactamase genes, except strain Ro24724, which also carried blaKPC-2 (Table 1). In addition, there were 5 isolates positive for blaSHV-12, 4 isolates contained blaTEM-1, and one isolate contained blaOXA-1. Of note, the blaOXA-1 gene was first identified in the genus Raoultella. Interestingly, in China, KPC-2 was first reported in our hospital and is now rapidly disseminated throughout China (8, 16). Additionally, blaKPC-2 genes are largely disseminated worldwide among K. pneumoniae isolates but less frequently among other Enterobacteriaceae (8). Until now, the coexistence of blaKPC-2 and blaIMP-4 had been described in K. pneumoniae and in China only (17–19). The clinical isolate Ro24724 reported here is the first documented KPC- and IMP-producing R. ornithinolytica.
The genetic environment of blaKPC-2 was sought by long-PCR mapping based on previously described primers (8). PCR amplification generated a 4,561-bp segment, which suggested the blaKPC-2-flanking region in Ro24724 shared 100% identity with blaKPC-2-carrying plasmid K. pneumoniae pKP048 and Escherichia coli pHS102707 (Fig. 1b). The blaKPC-2 gene in R. ornithinolytica is also located on a Tn3-Tn4401 integration structure, as previously described from K. pneumoniae and E. coli (8, 20). Of note, the blaKPC-2 gene has been detected in K. pneumoniae, E. coli, Enterobacter cloacae, Morganella morganii, and even Pseudomonas aeruginosa isolates from Zhejiang province (16, 21–23). Our observation further documents the transmission of this structure in different Enterobacteriaceae in China.
The in vitro conjugation experiments were unsuccessful. We also tried to transfer plasmids extracted from seven isolates by transformation experiments. However, repeated transformation methods failed to move the plasmids to recipient E. coli DH5α cells. This suggests that plasmids were likely to be nonconjugative, and this may primarily be due to the large size of the plasmids, which may limit the efficiency of transformation.
Plasmid sizes were then determined using an S1-nuclease PFGE (S1-PFGE) method (24). S1-PFGE revealed that all isolates contained a single plasmid, with sizes of ∼300 to 450 kb (Fig. 1c). Of note, strain Ro24724 contained a larger plasmid than that of other isolates, while isolate Ro25687 carried the smallest plasmid, which is in agreement with their phenotypic and molecular differences. The hybridization results showed that the blaKPC-2 and blaIMP-4 genes were located on a plasmid of ∼450 kb in strain Ro24724. The blaIMP-4 gene was present on a plasmid of ∼300 kb in strain Ro25687 and on plasmids of ∼340 kb in R. ornithinolytica strains Ro23980, Ro24005, Ro24362, Ro24522, and Ro25277. PFGE followed by Southern blotting revealed that blaKPC-2 was located on the additional band observed in PFGE, which suggested that this band is part of a large plasmid (Fig. 1a and d). Previous studies showed that blaKPC was localized on an 11-kb plasmid in R. ornithinolytica and Raoultella planticola, and blaIMP-8 was carried on a 249-kb plasmid in R. planticola (1, 6). Our observations further exhibited that the blaKPC-2 and blaIMP genes have the potential to spread widely among members of the genus Raoultella.
In conclusion, we provide here the first report of the sequential isolation of plasmid-medicated IMP-4-producing R. ornithinolytica and the first description of the coexistence of blaKPC-2 and blaIMP-4 in Raoultella. This study reinforces the idea of a rapid dissemination of the blaIMP-4 and blaKPC-2 genes in clinical isolates of Enterobacteriaceae in China. Despite being only one case, our study is of great concern for its epidemic potential, since the emergence of KPC-2 is a severe threat in China. Further research to explore the detailed genetic features of these plasmids and their evolution in different clinical isolates is now under way.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 program, grant 2015CB554201), the National Natural Science Foundation of China (grants 81361138021 and 81301461), and the Zhejiang Provincial Natural Science Foundation of China (grant LQ13H190002).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01363-15.
REFERENCES
- 1.Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. 2009. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J Clin Microbiol 47:4129–4130. doi: 10.1128/JCM.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haruki Y, Hagiya H, Sakuma A, Murase T, Sugiyama T, Kondo S. 2014. Clinical characteristics of Raoultella ornithinolytica bacteremia: a case series and literature review. J Infect Chemother 20:589–591. doi: 10.1016/j.jiac.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen Ø, Seifert H, Woodford N, Nordmann P, European Network on Carbapenemases. 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 5.Khajuria A, Praharaj AK, Grover N, Kumar M. 2013. First report of blaNDM-1 in Raoultella ornithinolytica. Antimicrob Agents Chemother 57:1092–1093. doi: 10.1128/AAC.02147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng SP, Wang JT, Liang CY, Lee PS, Chen YC, Lu PL. 2014. First report of bla(IMP-8) in Raoultella planticola. Antimicrob Agents Chemother 58:593–595. doi: 10.1128/AAC.00231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou G, Guo S, Luo Y, Ye L, Song Y, Sun G, Guo L, Chen Y, Han L, Yang J. 2014. NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg Infect Dis 20:340–342. doi: 10.3201/eid2002.121263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tijet N, Sheth PM, Lastovetska O, Chung C, Patel SN, Melano RG. 2014. Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008–2011. PLoS One 9:e116421. doi: 10.1371/journal.pone.0116421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endimiani A, Carias LL, Hujer AM, Bethel CR, Hujer KM, Perez F, Hutton RA, Fox WR, Hall GS, Jacobs MR, Paterson DL, Rice LB, Jenkins SG, Tenover FC, Bonomo RA. 2008. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob Agents Chemother 52:2680–2682. doi: 10.1128/AAC.00158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother 57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 15.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 16.Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. 2007. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother 51:763–765. doi: 10.1128/AAC.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Z, Yu T, Qi Y, Ji S, Shen P, Yu Y, Chen Y. 2011. Coexistence of plasmid-mediated KPC-2 and IMP-4 carbapenemases in isolates of Klebsiella pneumoniae from China. J Antimicrob Chemother 66:2670–2671. doi: 10.1093/jac/dkr330. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Cao W, Zhu X, Chen Z, Li L, Zhang B, Wang B, Tian L, Wang F, Liu C, Sun Z. 2012. Characterization of a novel Klebsiella pneumoniae sequence type 476 carrying both bla KPC-2 and bla IMP-4. Eur J Clin Microbiol Infect Dis 31:1867–1872. doi: 10.1007/s10096-011-1512-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Wan LG, Deng Q, Cao XW, Yu Y, Xu QF. 2015. First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol Infect 143:376–384. doi: 10.1017/S0950268814000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G, Zhang Y, Bi D, Shen P, Ai F, Liu H, Tian Y, Ma Y, Wang B, Rajakumar K, Ou HY, Jiang X. 2015. First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob Agents Chemother 59:338–343. doi: 10.1128/AAC.03061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai JC, Zhou HW, Zhang R, Chen GX. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi Y, Wei Z, Li L, Ji S, Du X, Shen P, Yu Y. 2010. Detection of a common plasmid carrying blaKPC-2 in Enterobacteriaceae isolates from distinct cities in China. Microb Drug Resist 16:297–301. doi: 10.1089/mdr.2010.0023. [DOI] [PubMed] [Google Scholar]
- 23.Hu YY, Gu DX, Cai JC, Zhou HW, Zhang R. 2015. Emergence of KPC-2-producing Pseudomonas aeruginosa sequence type 463 isolates in Hangzhou, China. Antimicrob Agents Chemother 59:2914–2917. doi: 10.1128/AAC.04903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. 2009. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother 53:4240–4246. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.