Abstract
The bacterial CRISPR endoribonuclease Csy4 has recently been described as a potential RNA processing tool. Csy4 recognizes substrate RNA through a specific 28-nt hairpin sequence and cleaves at the 3′ end of the stem. To further explore applicability in mammalian cells, we introduced this hairpin at various locations in mRNAs derived from reporter transgenes and systematically evaluated the effects of Csy4-mediated processing on transgene expression. Placing the hairpin in the 5′ UTR or immediately after the start codon resulted in efficient degradation of target mRNA by Csy4 and knockdown of transgene expression by 20- to 40-fold. When the hairpin was incorporated in the 3′ UTR prior to the poly(A) signal, the mRNA was cleaved, but only a modest decrease in transgene expression (∼2.5-fold) was observed. In the absence of a poly(A) tail, Csy4 rescued the target mRNA substrate from degradation, resulting in protein expression, which suggests that the cleaved mRNA was successfully translated. In contrast, neither catalytically inactive (H29A) nor binding-deficient (R115A/R119A) Csy4 mutants were able to exert any of the effects described above. Generation of a similar 3′ end by RNase P-mediated cleavage was unable to rescue transgene expression independent of Csy4. These results support the idea that the selective generation of the Csy4/hairpin complex resulting from cleavage of target mRNA might serve as a functional poly(A)/poly(A) binding protein (PABP) surrogate, stabilizing the mRNA and supporting translation. Although the exact mechanism(s) remain to be determined, our studies expand the potential utility of CRISPR nucleases as tools for controlling mRNA stability and translation.
Keywords: CRISPR, Csy4, mRNA stability, translation, gene regulation
INTRODUCTION
Endogenous control of gene expression is achieved by regulating transcription, processing, translation, and/or degradation of mRNA through a myriad of genetic elements. Artificial control of gene expression, on the other hand, requires the development of small molecule, protein or RNA-based tools and is essential for advancing synthetic biology and gene therapies. Post-transcriptional regulation of gene expression has been achieved by engineering RNA, for instance by using riboswitches that can not only be exploited to gain insight into endogenous RNA processing mechanisms, but also as programmable elements for building gene circuits (Chang et al. 2012). Another promising platform for developing RNA regulatory tools is the Pumilio/fem-3 binding factor or PUF proteins that recognize single-stranded RNA in a sequence-specific fashion (Wang et al. 2013). PUF domains can be exploited to engineer splicing factors, control translation, or develop artificial site-specific RNA endonucleases (Choudhury et al. 2012). Recent studies have suggested that bacterial CRISPR systems might be useful for RNA regulatory applications in comparison to the use of hammerhead ribozymes and RNase III (Qi et al. 2012). Regardless of the approach, controlling RNA stability and translation is a key aspect underlying these gene regulatory strategies.
The prokaryotic CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system provides adaptive immunity against invading genetic elements in ∼45% of bacteria and ∼83% of archaea (Grissa et al. 2007). Briefly, CRISPR loci encode a repeat-spacer array consisting of unique sequence elements (spacers), derived from previously encountered exogenous nucleic acid (Bolotin et al. 2005; Mojica et al. 2005; Pourcel et al. 2005; Barrangou et al. 2007). Each spacer is flanked by repeat elements and the entire array is transcribed as a single pre-crRNA (Tang et al. 2002, 2005). The pre-crRNA is processed into smaller crRNAs which serve as guides to target nucleic acid for neutralization by CRISPR-associated (Cas) proteins (Brouns et al. 2008; Garneau et al. 2010). The protein content of CRISPR loci is diverse and as such, three broad classes of CRISPR systems have been described based on the hallmark proteins Cas3, Cas9, and Cas10 (type I, type II, and type III, respectively) (Makarova et al. 2011). These groups are further divided into subclassifications denoted by alphabetical letters. The Pseudomonas aeruginosa type-IF CRISPR system uses Csy4 (Cas6f) to bind a 28-nucleotide (nt) repeat hairpin and cleave at the base of the 3′ end to generate crRNAs (Haurwitz et al. 2010; Cady and O'Toole 2011). The task of pre-crRNA processing in other systems falls to Cas5d, Cas6e (CasE/Cse3) and Cas6 in Bacillus halodurans, Escherichia coli, and Pyrococcus furiosus, respectively (Brouns et al. 2008; Carte et al. 2008; Nam et al. 2012).
Structural studies have provided further mechanistic insight into pre-crRNA processing enzymes (Hochstrasser and Doudna 2015). Despite their functional similarity, these enzymes display minimal primary sequence homology. Likewise, the sequences which they process also differ both in sequence and structure, with Cas5d, Cas6e, and Csy4 associated repeat elements containing hairpin structures and Cas6 targeting a predicted unstructured sequence (Kunin et al. 2007). To carry out target neutralization, Csy4 remains bound to the processed crRNA and associates with additional Cas proteins, Csy1, Csy2, and Csy3 for target recognition (Wiedenheft et al. 2011). This complex is guided to target DNA based on sequence complementarity provided by the crRNA. In type-I CRISPR systems, Cas3 is then recruited to cleave and degrade the target DNA, neutralizing the invader (Westra et al. 2012; Sinkunas et al. 2013).
Recent studies have adapted the type-IF CRISPR endoribonuclease, Csy4, for processing of RNA encoding multigene operons and gene regulation in E. coli, Bacillus subtilis, and Saccharomyces cerevisiae (Qi et al. 2012). In particular, this study demonstrated that RNA processing by Csy4 is an effective strategy to maintain the endogenous function of different promoters, genes and regulatory elements without interference when engineered into complex genetic circuits. More recently, Csy4 has been applied for processing guide RNA (gRNA) for mammalian Cas9 technology and the programming of complex genetic circuits (Nissim et al. 2014; Tsai et al. 2014). These advancements have led to the expression of multiple gRNAs from a single promoter, removed the restriction of a 5′ guanosine imposed by the U6 promoter, and permitted gRNA expression from RNA polymerase II promoters. Furthermore, Csy4 has been utilized in the isolation of RNA-interacting proteins and has the potential to help analyze the protein associations of diverse transcripts (Lee et al. 2013). In the current study, we systematically evaluated the ability of Csy4 to exercise post-transcriptional control of transgene expression in mammalian cells. Specifically, we investigate the positional effects of Csy4-mediated cleavage in the 5′ UTR, coding sequence, and 3′ UTR of transcripts. Surprisingly, we find that the Csy4 processing of the 3′ ends of mRNA supports translation and stabilizes the mRNA in lieu of a poly(A) signal.
RESULTS AND DISCUSSION
Csy4-mediated knockdown of 5′ UTR-hairpin (HP) and ATG-HP constructs
We first evaluated the effect of Csy4-mediated cleavage of the substrate HP incorporated within the 5′ untranslated region (UTR) or HP inserted in-frame immediately following the start codon of reporter transcripts. To this end, we cloned the Csy4 target hairpin (Fig. 1A) derived from the P. aeruginosa strain UCBPP-PA14 CRISPR locus, upstream (5′ UTR-HP) or immediately downstream (ATG-HP) of the initiator AUG in two separate reporter genes, GFP and Gaussia Luciferase (GLuc), and compared the activity of these constructs to the control cassettes lacking the Csy4 hairpin (Fig. 1B). The reporters were transfected into HEK293 cells with or without a plasmid expressing Csy4. While no change in mRNA levels was observed, as indicated by random primed RT-PCR in the case of the control plasmid lacking the Csy4 HP substrate, we observed a marked decrease in the case of the 5′ UTR-HP and the ATG-HP templates (Fig. 1C). This observation was supported by a decrease in GFP signal from the 5′ UTR-HP and ATG-HP templates, but not the control substrate as assessed by fluorescence microscopy (Fig. 1D). Moreover, there was an ∼24-fold decrease in GLuc reporter transgene expression mediated by Csy4 for the 5′ UTR-HP construct and an ∼37-fold decrease in signal for the ATG-HP construct (Fig. 1E). In contrast, the control cassette displayed a minimal change in gene expression in the presence of Csy4. It is important to consider that the ATG-HP approach results in the incorporation of a 10 amino acid residue peptide tag with the sequence SSLPYRQLRN into the GFP or luciferase proteins. Further, it should be noted that we also observed a significant decrease in GLuc reporter activity of the 5′ UTR-HP reporter construct in comparison with the unmodified or ATG-HP GLuc construct, even in the absence of Csy4. This is likely due to decreased efficiency of translational initiation by incorporation of a RNA hairpin structure in the 5′ UTR region (Kozak 1986, 1989; Babendure et al. 2006). These results are consistent in part with studies in S. cerevisiae, B. subtilis, and E. coli (Qi et al. 2012). In summary, our results confirm that Csy4 is functional in mammalian cells and can process RNA in a highly selective fashion mediated by the cognate hairpin substrate.
FIGURE 1.
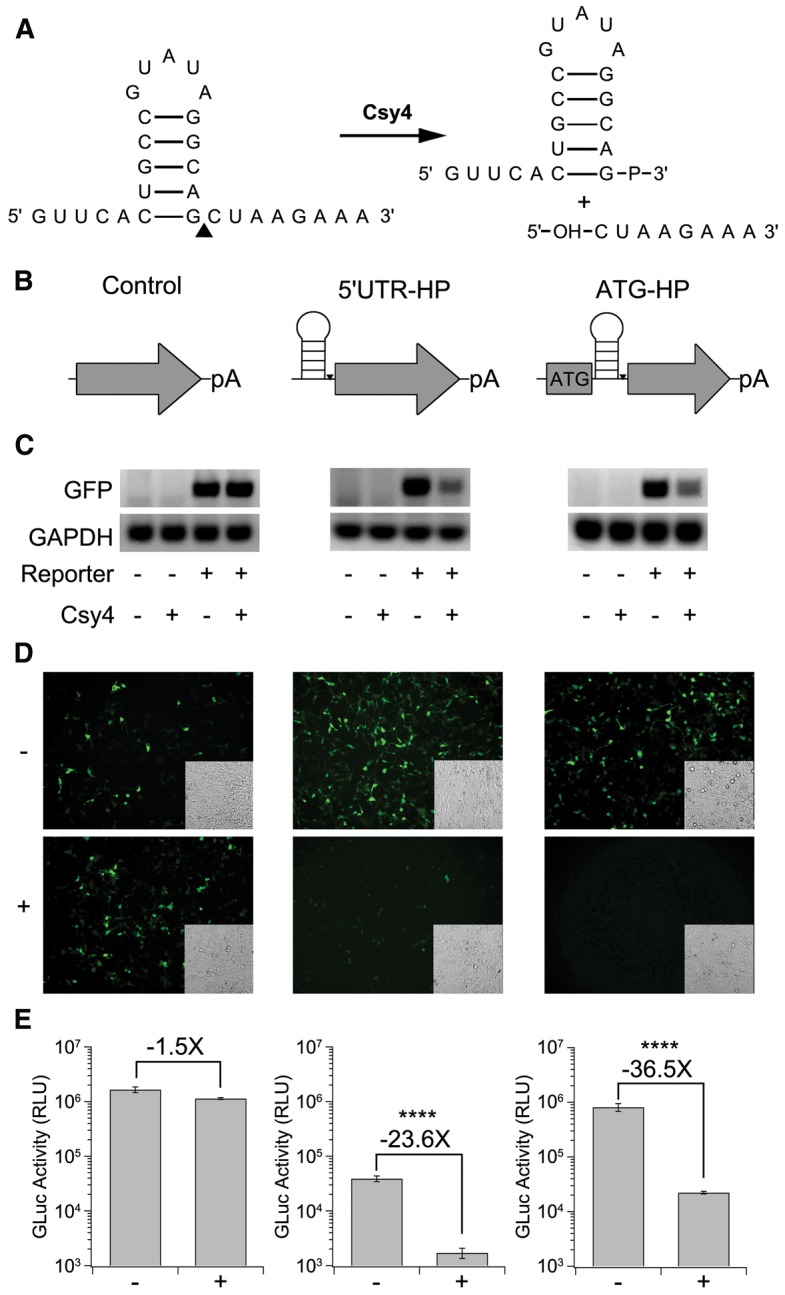
Csy4-mediated knockdown of 5′ UTR-hairpin (HP) and ATG-HP constructs. (A) Sequence of the Csy4 hairpin substrate and fragments after enzymatic cleavage. The cleavage site is indicated by a black triangle on the unprocessed hairpin. (B) Schematics of the unmodified control (left), 5′ UTR-HP (middle), and ATG-HP (right) reporters. (C) PCR products of randomly primed RNA isolated from HEK293 cells transfected as indicated. (Left) unmodified control, (middle) 5′ UTR-HP-GFP, and (right) ATG-HP-GFP. (D) Fluorescent images of HEK293 cells expressing unmodified control (left), 5′ UTR-HP (middle), and ATG-HP GFP (right) reporters in the absence (−) or presence (+) of Csy4. Corresponding transmitted light images are shown as insets in each fluorescent image. (E) Quantitation of GLuc activity by luminometric analysis at 24 h post-transfection for unmodified control (left), 5′ UTR-HP (middle), and ATG-HP (right) GLuc reporters. Error bars indicate standard deviation of four replicates. Statistical significance was calculated using a two-tailed Student's t-test ([****] P ≤ 0.0001).
Effect of placing the HP in the 3′ UTR on Csy4-mediated knockdown
In contrast to insertions near the 5′ UTR or the start codon, placement of the Csy4 target HP following the stop codon and prior to the poly(A) signal (3′ UTR-HP, Fig. 2A, left column) resulted in only a modest decrease in mRNA levels (as indicated by random primed RT-PCR) and GFP expression, when coexpressed with Csy4 (Fig. 2B,C, left columns). Further, quantitation of GLuc reporter activity indicated only an ∼2.5-fold reduction in transgene expression upon treatment with Csy4 (Fig. 2D, left column). However, there was a substantial reduction in poly-adenylated reporter RNA levels as measured by oligo-dT20 primed RT-PCR, consistent with mRNA cleavage by Csy4 (Fig. 2E, left column). These results are particularly intriguing, since removal of the poly(A) signal is expected to destabilize mRNA.
FIGURE 2.

Effect of placing the HP in the 3′ UTR on Csy4-mediated knockdown. (A) Schematics of the 3′ UTR-HP reporter (left) and 3′ UTR-HP-Δp(A) reporter (right). (B) PCR products of randomly primed RNA isolated from HEK293 cells transfected with the GFP-3′ UTR-HP reporter (left) or GFP-3′ UTR-HP-Δp(A) reporter (right) and Csy4 as indicated. (C) Fluorescent images of HEK293 cells expressing GFP-3′ UTR-HP (left) or GFP-3′ UTR-HP-Δp(A) (right) in the absence (−) or presence (+) of Csy4. Corresponding transmitted light images are shown as insets in each fluorescent image. (D) Quantitation of Gluc activity by luminometric analysis at 24 h post-transfection for GLuc-3′ UTR-HP (left) or GLuc-3′ UTR-HP-Δp(A) (right) reporters. Error bars indicate standard deviation of four replicates. Statistical significance was calculated using a two-tailed Student's t-test ([****] P ≤ 0.0001). (E) PCR products of oligo-d(T)20 primed RNA isolated from HEK293 cells expressing GLuc-3′ UTR-HP (left) or Gluc-3′ UTR-HP-Δp(A) (right) with or without Csy4.
The latter observations might arise from the fact that following cleavage, Csy4 remains bound to the cognate HP substrate (Haurwitz et al. 2010), which in turn could protect the 3′ end of the transcript from degradation, despite removal of the poly(A) tail. Since translation is only slightly reduced, the Csy4/RNA complex must also be compatible with translation. To test this hypothesis, we generated a second set of reporters identical to the 3′ UTR-HP constructs, but lacking a poly(A) signal [3′ UTR-HP-Δp(A)] (Fig. 2A, right column). As seen in Figure 2B (right column), due to the lack of an efficient 3′ processing signal, the 3′ UTR-HP-Δp(A) constructs accumulate only a small amount of RNA. However, when co-expressed with Csy4, we observed an increase in mRNA levels by random primed RT-PCR, as well as an increase in GFP transgene expression (Fig. 2B,C, right columns). As described earlier, we then assessed the poly-adenylation status of this reporter mRNA by generating reverse transcribed cDNA using an oligo-dT20 primer. Regardless of the presence or absence of Csy4, 3′ UTR-HP-Δp(A) reporter mRNA levels primed by oligo-dT20 appeared low relative to 3′ UTR-HP (Fig. 2E, right column). This is consistent with the 3′ UTR-HP-Δp(A) RNA lacking a poly(A) signal. Importantly, Csy4 coexpression does not alter the amount of poly-adenylated RNA detected. Furthermore, quantitation of GLuc activity revealed a ninefold increase in signal with Csy4 coexpression relative to a control lacking Csy4 (Fig. 2D). This corroborates the notion that Csy4 can potentially stabilize mRNA containing the HP substrate in the 3′ UTR and support translation.
Csy4 binding and cleavage are necessary for regulation of transgene expression
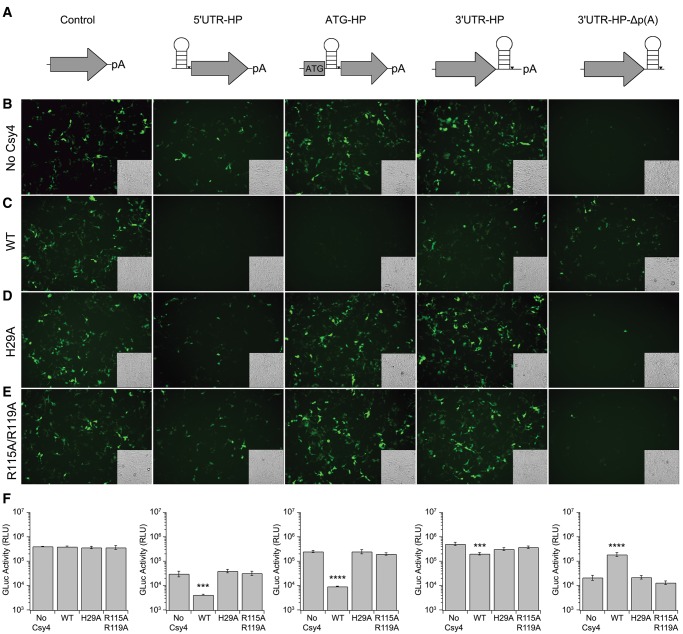
We next sought to determine whether Csy4 binding to the substrate HP alone was sufficient or if binding followed by enzymatic cleavage was essential for regulation of transgene expression. Specifically, we used two mutants of Csy4; Csy4-R115A/R119A, a binding-deficient mutant that still retains catalytic activity, and Csy4-H29A, a catalytically inactive, but binding competent mutant (Haurwitz et al. 2010; Sternberg et al. 2012). Each construct including the unmodified control, 5′ UTR-HP, ATG-HP, 3′ UTR-HP, and the 3′ UTR-HP-Δp(A) reporter (Fig. 3A) was expressed in the presence or absence of native Csy4, Csy4-H29A, or Csy4-R115A/R119A. Expression from unmodified GFP and GLuc constructs was unaffected by either mutant. The 5′ UTR-HP GFP construct was also unaffected by coexpressing either the catalytically inactive or the binding-deficient mutant (Fig. 3B,D,E). Similar observations were made in the case of the ATG-HP GFP reporter expressed with the catalytically inactive mutant, suggesting that steric hindrance from binding alone was insufficient to block translation initiation (Fig. 3B,D). Further, the Csy4-R115A/R119A mutant also did not seem to substantially affect ATG-HP reporter expression (Fig. 3B,E). Treatment with Csy4-H29A and Csy4-R115A/R119A did not dramatically affect expression of the corresponding GLuc reporters, supporting the previous data (Fig. 3F). Taken together, these data suggest that both binding and cleavage are essential for Csy4-mediated knockdown of transgene expression. In addition, neither mutant was able to rescue reporter signal in the case of the 3′ UTR-HP and the 3′ UTR-HP-Δp(A) reporters (Fig. 3B–F). These results support the notion that rescue of transgene expression from reporter constructs lacking a poly(A) tail requires both Csy4 binding to and cleavage of target mRNA. Thus, Csy4 bound to the cleaved hairpin at the 3′ end of an mRNA appears to have the remarkable property of stabilizing the mRNA, translocating to the cytoplasm, and supporting translation.
FIGURE 3.
Csy4 binding and cleavage are necessary for regulation of transgene expression. (A) Schematics of reporters (left to right): unmodified control, 5′ UTR-HP, ATG-HP, 3′ UTR-HP, and 3′ UTR-HP-Δp(A). Fluorescent images of HEK293 cells expressing each reporter in the absence of Csy4 (B), or presence of either native Csy4 (C), Csy4 H29A (D), or Csy4 R115A/R119A (E). Corresponding transmitted light images are shown as insets in each fluorescent image. (F) Quantitation of GLuc activity at 24 h post-transfection by luminometric analysis for each reporter. Error bars indicate standard deviation of four replicates. Statistical significance was expressed relative to the “No Csy4” control and calculated using a two-tailed Student's t-test ([****] P ≤ 0.0001, [***] P ≤ 0.001).
Selective processing by Csy4 is essential for rescue of 3′ UTR-HP-Δp(A) constructs
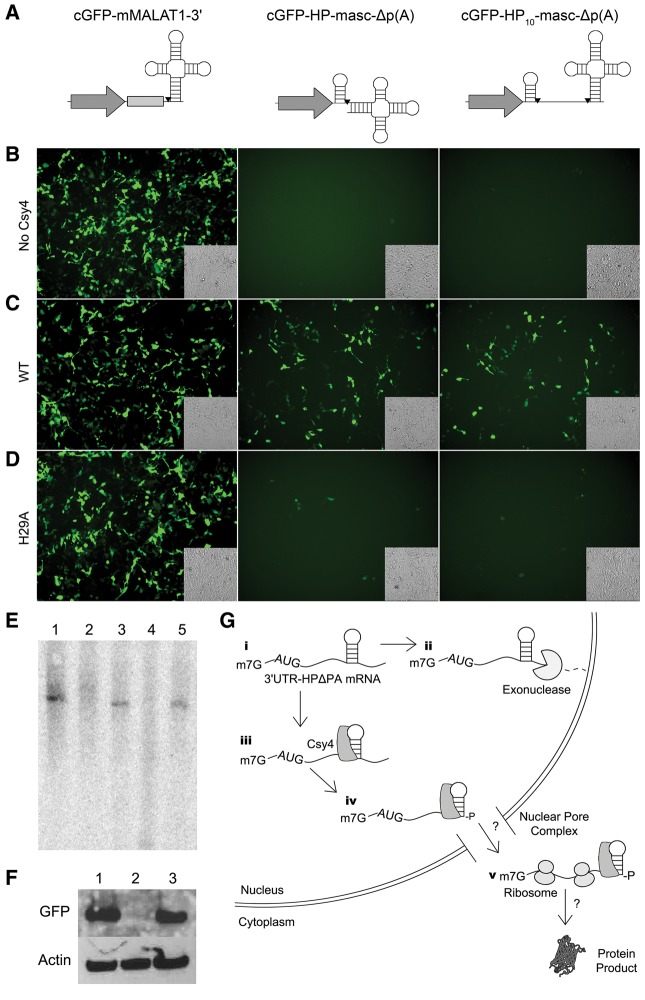
To further confirm the role of Csy4-mediated recognition and cleavage in rescuing 3′ UTR-HP-Δp(A) reporters, we attempted to generate a similar cleaved 3′ end product using a different endoribonuclease. Specifically, we engineered a novel reporter cassette [cGFP-HP-masc-Δp(A)], wherein a tRNA-like structure (mascRNA) was placed adjacent to the Csy4 HP (Fig. 4A). This mascRNA motif is derived from the 3′ end of a previously described MALAT1 RNA and is selectively cleaved by RNase P (Wilusz et al. 2008), releasing the capped 5′ region of the transcript. In the endogenous MALAT1 RNA, the 3′ end generated by RNase P processing folds into a triple helix structure that is capable of stabilizing the RNA and supporting translation in the absence of a poly(A) signal (Brown et al. 2012; Wilusz et al. 2012). The cGFP-HP-masc-Δp(A) construct was engineered in such a manner that RNase P would cleave at the same nucleotide as Csy4, at the base of the Csy4 HP stem, releasing the mascRNA motif, but generating the same RNA product as Csy4 would generate. In addition, we also constructed a cassette wherein the Csy4 HP and the mascRNA motif were separated by 10 nt to allow RNA processing at two different sites [Fig. 4A, cGFP-cHP10-masc-Δp(A)]. In this construct, the initial RNaseP product can be subsequently cleaved by Csy4. A construct containing the full MALAT1 3′ end, including the 3′ end stabilizing triple helix element in place of the Csy4 hairpin, was used as a control (Fig. 4A, cGFP-mMALAT1-3′).
FIGURE 4.
Selective processing by Csy4 is essential for rescue of 3′ UTR-HP-Δp(A) constructs. (A) Schematics of the unmodified cGFP-mMALAT1-3′ reporter (left), cGFP-HP-masc-Δp(A) (middle), and cGFP-HP10-masc-Δp(A) (right) reporters. Cleavage sites are indicated by black inverted triangles. Fluorescent images of HEK293 cells expressing either cGFP-mMALAT3′ (left), cGFP-HP-masc-Δp(A) (middle), or cGFP-HP10-masc-Δp(A) (right) in the absence of Csy4 (B), with Csy4 wild type (C), or Csy4-H29A (D). Corresponding transmitted light images are shown as insets in each fluorescent image. (E) Northern blot of cGFP-based reporters in the presence or absence of Csy4. cGFP-mMALAT1-3′ (lane 1), cGFP-HP-mascΔp(A) (lane 2), cGFP-HP-mascΔp(A) with Csy4 (lane 3), cGFP-HP10-masc-Δp(A) (lane 4), and cGFP-HP10-masc-Δp(A) with Csy4 (lane 5). (F) A Western blot probing for cGFP using cell lysates of HEK293s expressing either cGFP-mMALAT1-3′ (lane 1), cGFP-HP-mascΔp(A) (lane 2), or cGFP-HP-mascΔp(A) with Csy4 (lane 3). A Western blot for Actin is provided as a loading control. (G) Potential events outlining Csy4-mediated processing of 3′ UTR-HP-Δp(A) mRNA. First, within the nucleus, poly(A)-deficient constructs are likely degraded by cellular exonucleases in the absence of Csy4 [steps (i) and (ii)]. However, Csy4 binding (iii) followed by cleavage of the substrate (iv) appears to stabilize the mRNA and enable cytosolic transport as well as translation (v) by unknown mechanisms.
When transfected into HEK293 cells, cGFP-mMALAT1-3′ reporter expression was not affected by coexpression with Csy4 wild type (Fig. 4B,C). This is expected due to the absence of a Csy4 target hairpin in this construct. In contrast, neither of the hairpin-containing reporters, cGFP-HP-masc-Δp(A) or cGFP-HP10-masc-Δp(A), alone was able to support transgene expression due to the lack of a poly(A) signal or the MALAT1 triple helical motif (Fig. 4B). However, coexpression of native Csy4 with these reporters rescued GFP expression, consistent with earlier results demonstrating the ability of Csy4 to stabilize the mRNA 3′ end (Fig. 4C). Northern blot using a cGFP probe confirmed these observations (Fig. 4E). When expressed independently of Csy4, no cGFP RNA is detected for cGFP-HP-masc-Δp(A) or cGFP-HP10-masc-Δp(A) (Fig. 4E, lanes 2,4). However, when coexpressed with Csy4 (lanes 3,5), a band is detected for each reporter that runs slightly below that of cGFP-mMALAT1-3′ (Fig. 4E, lane 1). This is expected as the cleavage products of cGFP-HP-masc-Δp(A) and cGFP-HP10-masc-Δp(A) are expected at 754 nt while the processed cGFP-mMALAT1-3′ transcript runs at 844 nt. Our observations were further confirmed by Western blot for cGFP on lysates expressing cGFP-mMALAT1-3′ (lane 1), cGFP-HP-masc-Δp(A) (lane 2), or cGFP-HP-masc-Δp(A) with Csy4 (lane 3) (Fig. 4F). Further, we expressed the cGFP-mMALAT1-3′ construct, cGFP-HP-masc-Δp(A) or cGFP-HP10-masc-Δp(A) construct with the catalytically inactive Csy4-H29A mutant. In this setting, the reporter mRNA would only be processed by RNase P to generate a 3′ end, however, Csy4-H29A is still expected to bind the HP substrate. As seen in Figure 4D, while cGFP-mMALAT1-3′ expression is unaffected, rescue of GFP transgene expression from both Csy4 HP-containing constructs using the catalytically inactive Csy4 mutant was markedly reduced. Interestingly, a few cells expressing GFP were still detectable when Csy4-H29A was expressed with these reporter constructs. This observation suggests that generation of a similar 3′ end using RNase P in conjunction with binding by the Csy4-H29A mutant can at least partially rescue expression as seen with wild-type Csy4. It should be noted that Csy4-mediated cleavage of the hairpin results in a 3′-phosphate at the base of the HP stem, while RNase P processing generates a 3′-hydroxyl group at the same position (Wilusz et al. 2008; Wiedenheft et al. 2011). A specific role for the 3′-phosphate group, if any, on Csy4-mediated rescue of nonpolyadenylated RNAs is the subject of further exploration. Nevertheless, when taken together, these results affirm that both Csy4 binding and 3′ end processing at the base of the HP stem are essential for stabilizing mRNA.
Implications for studying and manipulating mRNA
Our studies suggest that Csy4 is a robust tool for knockdown of transgene-derived mRNA. Potential applications for this versatile system include (i) post-transcriptional control of transgenes, e.g., the development of “safety” switches that can turn off gene expression (Di Stasi et al. 2011; Ketzer et al. 2014); (ii) spatio-temporal knockdown of overexpressed transgenes in animal models, where RNAi-mediated approaches to knockdown endogenous transgenes might not be effective; and (iii) multiplex processing of noncoding RNAs such as microRNAs without risking saturation of endogenous nucleus-to-cytosol transport machinery (Grimm et al. 2006). Another intriguing possibility is the potential to utilize different combinations of Type I CRISPR endoribonucleases such as Cas6, Cas5d, Cas6e, and their corresponding substrates in multiplexed RNA processing applications (Brouns et al. 2008; Carte et al. 2008; Nam et al. 2012). In addition to providing tools for controlling transgene expression, the current studies might also help expand the application of CRISPR-based tools for understanding RNA processing.
Potential cellular processing events for 3′ UTR constructs containing the Csy4 HP substrate and lacking a poly(A) signal [3′ UTR-HP-Δp(A)] are shown (Fig. 4G). First, within the nucleus, poly(A)-deficient constructs are likely degraded by cellular exonucleases in the absence of Csy4 [steps (i) and (ii)]. However, Csy4 binding (iii) followed by cleavage of the substrate (iv) appears to stabilize the mRNA and enable cytosolic transport as well as translation (v) by unknown mechanisms. It is well known that the poly(A) tail plays a multifunctional role in protecting mRNA from degradation, transcriptional termination, nuclear-to-cytosolic transport, and translation. Thus, the absence of a poly(A) tail is generally associated with rapid mRNA degradation and a lack (or exceedingly low levels) of transgene product. In the current study, we clearly demonstrate the ability of Csy4 to stabilize such poly(A) deleted mRNA constructs subsequent to binding and processing of the substrate hairpin at the 3′ end. Secondly, cytoplasmic export of the Csy4-bound transcript from the nucleus and translation are likely essential for rescuing transgene expression. The mechanisms underlying these critical events are currently under investigation. Similar paradigms have been proposed in relation to 3′ UTR processing for two nuclear noncoding RNAs—MALAT1 and MEN β, which form a triple helical structure capable of stabilizing transgene-derived mRNA, supporting nuclear export and translation (Wilusz et al. 2012). Other examples of such tertiary RNA structures have also been reported in the case of histone and viral-derived RNAs (Marzluff 2012). For instance, Kaposi's sarcoma-associated herpesvirus (KSHV) is one such virus that uses an expression and nuclear retention element (ENE) which also forms a triple helical structure (Mitton-Fry et al. 2010). Together, our studies and other examples in the literature strongly support the notion that the Csy4–hairpin complex can potentially function as a surrogate poly(A) tail. From a broader perspective, our studies expand the potential utility of CRISPR nucleases as tools for controlling mRNA stability and translation.
MATERIALS AND METHODS
Plasmids
The Csy4 gene was amplified from P. aerugionsa strain UCBPP-PA14 genomic DNA and cloned under the control of the chicken β-actin (CBA) promoter using the following primers: Csy4wt Forward 5′-ATCGTCTAGAATGGACCACTACCTCGACATTCGCTTGC-3′ and Csy4wt Reverse 5′-CGATGCGGCCGCTCAGAACCAGGGAACGAAACCTCCTTTGC-3′ (IDT DNA Technologies). P. aeruginosa UCBPP-PA14 genomic DNA was kindly provided by Dr. Matthew Wolfgang (University of North Carolina at Chapel Hill). Csy4-H29A was amplified from pHMGWA-Pa14Csy4H29A (Addgene plasmid #41092), which was provided as a gift from Jennifer Doudna (Haurwitz et al. 2010). Csy4-H29A was cloned under the control of the CBA reporter. Csy4 R115A/R119A was synthesized as a gBlock (IDT DNA Technologies) and cloned under the control of the CBA promoter.
The reporter plasmid constructs containing enhanced green fluorescent protein (GFP) and Gaussia Luciferase (GLuc) were modified to incorporate the Csy4 hairpin (HP) in different locations as follows. (i) Reporter cassettes containing the HP after the start codon (ATG-HP) were generated using overlap extension PCR with primers that generated two fragments containing an extended Csy4 HP (5′-AGTTCACTGCCGTATAGGCAGCTAAGAAAT-3′) in the 3′ or 5′ end. The two fragments were then gel purified and combined in equimolar quantities (40 ng each) in consecutive PCR reactions without primers (35 cycles) and with flanking primers (30 additional cycles) to obtain the ATG-HP construct. The latter PCR products were then cloned under the control of the CBA promoter. (ii) The 5′ UTR-HP-GFP was constructed by overlap extension PCR in a similar fashion, while the 5′ UTR-HP-GLuc was synthesized as a gBlock (IDT DNA Technologies) and cloned under the control of the CBA promoter. (iii) 3′ UTR-HP reporters and poly(A) deleted 3′ UTR-HP [3′ UTR-HP-Δp(A)] reporters were synthesized from gBlocks and cloned as described earlier. The cGFP-mMALAT1-3′ reporter was generously provided by Dr. Jeremy Wilusz (University of Pennsylvania). A partial Csy4 hairpin (5′-GTTCACTGCCGTATAGGCAG-3′) and mascRNA were synthesized as a gblock (IDT DNA Technologies) and cloned in place of the mMALAT1 3′ UTR in the cGFP-mMALAT1-3′ reporter to generate cGFP-HP-masc-Δp(A). Similarly, a second gblock (IDT DNA Technologies) was synthesized containing the partial Csy4 hairpin and mascRNA separated by 10 nt (CTAAACGCGT) and cloned as described earlier, to generate cGFP-HP10-masc-Δp(A).
Cell culture
HEK293 cells were cultured in Dulbecco's Modified Eagle's Medium (GIBCO/Life Technologies) supplemented with 10% FBS (Atlanta Biologicals), 1% penicillin/streptomycin and 2.5 μg/mL amphotericin B (Sigma-Aldrich) and maintained at 37°C and 5% CO2.
Transfection and luciferase reporter assays
Equimolar amounts (totaling 500 ng) of three plasmids (different HP GLuc reporters, Csy4 and a control plasmid containing the tdTomato reporter driven by the CBA promoter) were transfected into HEK293 cells using PEI Max seeded at a density of 5 × 104 per well in a 24-well plate. Media (50 μL) were collected from each well at different time intervals and diluted (1:100) before assessing luciferase reporter activity. For measuring GLuc activity, native coelenterazine (Nanolight) was dissolved in methanol to a concentration of 1 mg/mL and diluted (1:200) in 600 mM NaCl–Tris–EDTA buffer, following which, 50 μL of the substrate solution was added to 50 μL of collected media. Luminometric analysis was carried out using a PerkinElmer Victor 3 plate reader.
Fluorescence microscopy
HEK293 cells were transfected with different HP GFP reporter cassettes as described earlier and the cells imaged at different time intervals post-transfection using an EVOS FL epifluorescence cell imaging system (AMC/Life Technologies) using the GFP light cube (excitation 470 nm, emission 510 nm).
mRNA analysis
Processing of mRNA was analyzed by monitoring the levels of DNA obtained through reverse transcriptase (RT) PCR. Briefly, HEK293 cells seeded overnight in 6-well plates at a density of 3 × 105 per well were transfected with a total of 2.5 μg of different plasmids (HP GFP reporters, Csy4wt or tdTomato control) as outlined earlier. Total RNA from each well was isolated at 48 h post-transfection using the Total RNA Purification Kit (#17200, Norgen Biotek). The purified RNA was then treated with DNase using the Turbo DNA-free kit (Ambion/Life Technologies). Equal nanogram amounts of the purified total RNA product were utilized as the template for reverse transcriptase PCR using the High Capacity RNA-to-cDNA kit (Applied Biosystems/Life Technologies). Products of this reaction were used as the template for further PCR amplification with gene-specific primers and visualized on an agarose gel. Forward primer for amplifying ATG-HP-GFP cDNA: 5′-GCCACCATGAGTTCACTGCCG-3′; forward and reverse primers for amplifying all other GFP reporter cDNAs: 5′-GAAATGTGAGCAAGGGCGAGGAGC-3′; 5′-GCGGACTTGAAGAAGTCGTGCTGC-3′; forward and reverse primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA: 5′-CCACTCCTCCACCTTTGAC-3′; 5′-ACCCTGTTGCTGTAGC-3′.
Detection of poly-adenylated mRNA
HEK293 cells seeded overnight in 6-well plates at a density of 3 × 105 cells per plate were transfected with a total of 3 μg DNA as indicated. Prior to transfection, DNA was prepared by digestion with ClaI, BglII, and CIP (New England BioLabs) to remove the GLuc expression cassette from its backbone and to prevent interference from potential downstream cryptic poly(A) signals. At 24 h post-transfection, RNA was harvested using the Total RNA Purification Kit (#17200, Norgen Biotek) and DNase treated using the Turbo DNA-free kit (Ambion/Life Technologies). A total of 1.3-μg DNase-treated RNA was converted to cDNA using SMARTScribe reverse transcriptase (Clontech) according to the manufacturer's protocol. An oligo-dT20 primer was used to prime the reverse transcription reactions. Products of this reaction were utilized as templates for further PCR amplification with gene-specific primers and visualized on an agarose gel. Forward and reverse primers for reporter cDNAs (GLuc) 5′-CAACTTCGCGACCACGGATCTCG-3′; 5′-CGGCAGCCACTTCTTGAGCAGG-3′. Forward and reverse primers for GAPDH are listed above.
Northern blot
HEK293 cells seeded overnight in 10-cm plates at a density of 2.2 × 106 cells per plate were transfected with a total of 6 μg DNA as indicated. RNA was purified using TRIzol reagent (Invitrogen/Life Technologies) and treated with DNase using the Turbo DNA-free kit (Ambion/Life Technologies). One microgram of RNA was separated on a 4% polyacrylamide/8 M urea gel and transferred to Hybond N+ membrane (GE Healthcare). To generate radiolabeled probe, cGFP was digested out of the cGFP-MALAT1-3′ backbone and used as template for the incorporation of radiolabeled CTP via a Random Primed DNA Labeling Kit (Roche Diagnostics). Hybridization of radiolabeled probe to the membrane was carried out using Rapid-Hyb buffer (GE Healthcare).
Western blot
HEK293 cells seeded overnight in 6-well plates at a density of 3 × 105 cells per plate were transfected with a total of 2.5 μg DNA as indicated. Lysates were recovered 48 h post-transfection using Passive Lysis Buffer (Promega). Lysates were diluted 1:50 and separated on a 10% Bis–Tris gel. Blots were probed with mouse monoclonal anti-GFP antibody (1:000 dilution, Santa Cruz) or mouse monoclonal anti-Actin (1:2000 dilution, Abcam) as the primary antibody. Stabilized peroxidase-conjugated goat anti-mouse antibody (1:5000 dilution, ThermoScientific/Life Technologies) was used as secondary antibody. Blots were developed using SuperSignal West Femto substrate (ThermoScientific/Life Technologies).
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants awarded to A.A. (R01HL089221, P01HL112761). In addition, this study was supported in part by a grant from the National Institute of General Medical Sciences under award 5T32 GM007092. We thank Chengwen Li for help generating radiolabeled probe for use in Northern blotting. We thank Dr. Deirdre Tatomer for her insight pertaining to the MALAT1 and mascRNA constructs.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.051227.115.
REFERENCES
- Babendure JR, Babendure JL, Ding JH, Tsien RY. 2006. Control of mammalian translation by mRNA structure near caps. RNA 12: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. 2012. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci 109: 19202–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, O'Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193: 3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, Wolf JJ, Smolke CD. 2012. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol 23: 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. 2012. Engineering RNA endonucleases with customized sequence specificities. Nat Commun 3: 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C, Straathof K, Liu E, Durett AG, Grilley B, et al. 2011. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med 365: 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329: 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA. 2015. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci 40: 58–66. [DOI] [PubMed] [Google Scholar]
- Ketzer P, Kaufmann JK, Engelhardt S, Bossow S, von Kalle C, Hartig JS, Ungerechts G, Nettelbeck DM. 2014. Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc Natl Acad Sci 111: E554–E562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci 83: 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 9: 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Haurwitz RE, Apffel A, Zhou K, Smart B, Wenger CD, Laderman S, Bruhn L, Doudna JA. 2013. RNA–protein analysis using a conditional CRISPR nuclease. Proc Natl Acad Sci 110: 5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. 2012. Novel 3′ ends that support translation. Genes Dev 26: 2457–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330: 1244–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174–182. [DOI] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. 2012. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 20: 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151: 653–663. [DOI] [PubMed] [Google Scholar]
- Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. 2012. RNA processing enables predictable programming of gene expression. Nat Biotechnol 30: 1002–1006. [DOI] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. 2013. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32: 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Haurwitz RE, Doudna JA. 2012. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Hüttenhofer A. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 55: 469–481. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z, Tanaka Hall TM. 2013. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J 280: 3755–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108: 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]