Abstract
The human hnRNP C is a ubiquitous cellular protein involved in mRNA maturation. Recently, we have shown that this protein specifically recognizes uridine (U) pentamers through its single RNA recognition motif (RRM). However, a large fraction of natural RNA targets of hnRNP C consists of much longer contiguous uridine stretches. To understand how these extended sites are recognized, we studied the binding of the RRM to U-tracts of 8–11 bases. In vivo investigation of internal translation activation of unr (upstream of N-ras) mRNA indicates that the conservation of the entire hnRNP C binding site, UC(U)8, is required for hnRNP C-dependent IRES activation. The assays further suggest a synergistic interplay between hnRNP C monomers, dependent on the protein's ability to oligomerize. In vitro spectroscopic and thermodynamic analyses show that isolated RRMs bind to (U)11 oligomers as dimers. Structural modeling of a ternary double-RRM/RNA complex indicates additionally that two RRM copies can be accommodated on the canonical sequence UC(U)8. The proposed tandem RRM binding is in very good agreement with the transcriptome-wide recognition of extended U-tracts by full-length hnRNP C, which displays a cross-linking pattern consistent with a positively cooperative RRM dimer binding model.
Keywords: hnRNP C, uridine, alternative splicing, IRES, RRM
INTRODUCTION
The heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNP C) was first characterized in the 1980s as an abundant, RNA-associated nuclear protein (Choi and Dreyfuss 1984). Today, hnRNP C is established as a major player in the mRNA maturation pathway: Its function spans nascent RNA packaging (Rech et al. 1995), sorting (McCloskey et al. 2012), and splicing regulation (Venables et al. 2008; König et al. 2010). More recently, the protein was also detected in the cytoplasm, associating with internal ribosome entry sites (IRES) of several eukaryotic mRNAs (Zaidi and Malter 1995; Kim et al. 2003; Schepens et al. 2007). Located within the 5′ untranslated regions (UTR), IRES sites serve to recruit the ribosome and associated trans-acting factors, and initiate protein synthesis in a cap-independent manner.
The human hnRNP C homolog is ∼300 residues long and is present in vivo in two variants, C1 and C2; the latter includes a 13 amino acid insertion (Merrill et al. 1989). While the endogenous hnRNP C protein acts as a (C1)3(C2) oligomer (Barnett et al. 1989), it has been shown that C1 or C2 homotetramers retain the wild-type structural and functional properties (McAfee et al. 1996b). The oligomerization is dependent on a central helical domain folding into antiparallel coiled-coil tetramers (Whitson et al. 2005). Two hnRNP C subdomains are thought to be involved in RNA binding: The amino-terminal RNA recognition motif (RRM) recognizes five contiguous uridines, with the sequence stringency increasing gradually in the 5′–3′ direction (Wan et al. 2001; Cieniková et al. 2014); a basic region preceding the oligomerization domain, rich in positively charged amino acids, provides additional, sequence-independent affinity for RNA (McAfee et al. 1996a; Soltaninassab et al. 1998). One tetramer binds ∼200 to 300 ribonucleotides in a strongly cooperative fashion (McAfee et al. 1996b). A large-scale cross-linking study of hnRNP C revealed that the protein's association on RNA transcripts undergoing splicing is coordinated by regularly spaced uridine tracts (König et al. 2010). Uridine pentamers, corresponding to the sequence specifically recognized by the RRM domain, were the most represented sites among hnRNP C cellular targets. However, uridine stretches considerably longer than 5 nucleotides (nt) were also highly enriched among the identified binding sites. Further supporting the importance of longer uridine tracts, the activation of internal translation initiation of upstream of N-ras (unr) mRNA during mitosis was shown to be regulated by hnRNP C (Sella et al. 1999) migrating into the cytoplasm and binding to a UC(U)8 tract located at position −102 to −93 upstream of the translation start site (Cornelis et al. 2005; Schepens et al. 2007). An additional, weaker poly(U) hnRNP C-binding site was identified at position −190 to −180. A thermodynamic investigation of the RRM-poly(U) RNA interaction (Cieniková et al. 2014) revealed that register sampling of the RRM on extended uridine tracts leads to a gain of binding affinity which could be the reason for the conservation of such long binding sites. Alternatively, König et al. (2010) proposed that hnRNP C tetramers bind long uridine stretches via two RRMs in tandem.
To understand the recognition of extended uridine-rich sites by hnRNP C, we investigated the binding of the RRM to a (U)11 oligomer using NMR spectroscopy and isothermal titration calorimetry (ITC). Using the insights derived from these in vitro studies and from molecular modeling driven by the experimental NMR and thermodynamic data, we extend our thermodynamic model of hnRNP C binding to account for tandem RRM binding to long U tracts in vivo and characterized the functional requirements for this binding mode using hnRNP C-dependent unr IRES activation in cell culture assays.
RESULTS
Affinity enhancement due to multiregister binding of the RRM is insufficient to account for the in vivo hnRNP C occupancy on long uridine tracts
We recently developed a structurally driven thermodynamic model to predict the binding of hnRNP C RRM to poly(U) sites (Cieniková et al. 2014). This model explained very well the in vivo ligand saturation obtained by iCLIP for uridine tracts shorter than 7 nt (König et al. 2010), indicating that one RRM binds such sites in multiregister mode (Fig. 1A, green curve). In the present study, we extended this analysis to longer uridine stretches. However, we observed that the saturation deviates sharply from this model when uridine tracts exceed 8 nt in length. Since one RRM binds four to five consecutive uridines (Cieniková et al. 2014), this change of binding behavior could indicate that two hnRNP C RRMs bind side-by-side on such long tracts.
FIGURE 1.
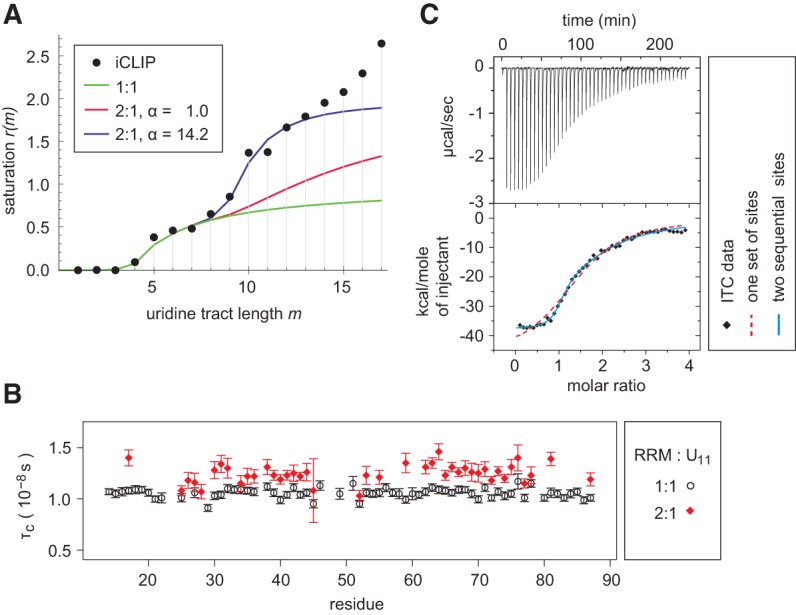
(A) Occupancy of cellular uridine tracts by hnRNP C. Experimental hnRNP C cross-link saturation (König et al. 2010) is compared with a model allowing only 1:1 RRM:RNA stoichiometry (in green), a model assuming two independent RRM ligands binding one uridine tract (in red), and a model allowing ligand cooperativity (in blue). In all three cases the scaling factor (see Materials and Methods) s = 73. (B) Rotational correlation times τC obtained from resolved amide resonances of residues 14–87 of hnRNP C RRM in presence of one or half equivalent of (U)11 RNA. (C) ITC of the RRM binding to (U)11 oligomer (320 μM of protein in the syringe, 17 μM of RNA in the cell, 6 μL/inj.). Experimental isotherm is fitted to two binding models.
In vitro, a pair of isolated hnRNP C RRMs can bind an 11-nt uridine tract
To determine whether two RRMs of hnRNP C can simultaneously bind to a long pyrimidine tract, we characterized spectroscopically and thermodynamically the interaction of the hnRNP C RRM with an extended uridine stretch. We recorded NMR relaxation spectra of the RRM in the presence of a synthetic (U)11 oligomer at 1:1 and 2:1 stoichiometric ratios. The chemical shifts of the corresponding protein amides in the 1:1 and 2:1 RRM:(U)11 complexes are expected to mostly overlap due to the degenerate nature of the RNA sequence and therefore the relaxation behavior of these signals is a superposition of the 1:1 and 2:1 complexes. The average rotational correlation time τC of the RRM, determined from 15N relaxation data, is 10.6 ± 0.4 nsec (mean ± SD) in the 1:1 sample and it increases to 12.6 ± 1.0 nsec in the 2:1 sample (Fig. 1B). This difference is statistically significant (P-value <10−13 using Student's t-test), suggesting that in the latter sample a significant fraction of the RRM is involved in the more slowly tumbling 2:1 protein:RNA complex. However, NMR signals belonging to the free RRM form remain observable in the 1H-15N HSQC spectrum obtained at the 2:1 ratio, consistent with the interpretation that the 2:1 species is only partially saturated (Supplemental Fig. S1A).
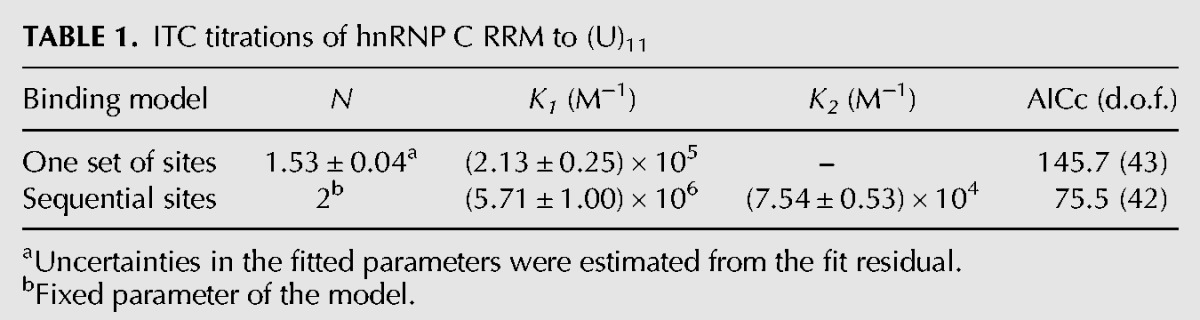
Additional insights are provided by a calorimetric titration of the RRM to the (U)11 RNA (Fig. 1C). An initial fit of the ITC data to the simplest binding model (one set of N identical sites) suggests a binding stoichiometry N ≈ 1.5. A visual inspection of the binding isotherm reveals biphasic behavior, indicating two association events of different macroscopic affinity. A model presuming two identical, sequentially bound ligands increases 106-fold the fit quality, as indicated by the value of the corrected Akaike information criterion (Table 1). The fitted macroscopic affinity constant of the first binding step displays an ∼11-fold enhancement compared with the expected microscopic single site affinity (Ks ≈ 5 × 105 M−1 [Cieniková et al. 2014]), confirming a strong multiregister binding of the RRM ligand. The second binding step manifests a weak negative cooperativity (interaction factor α ≈ 0.4), when accounting for the existence of ∼4 binding registers on the (U)11 RNA at 2:1 stoichiometry. This 2.5-fold reduction in affinity is likely to stem from adjustments required to avoid steric clashes between the adjacent RRM molecules.
TABLE 1.
ITC titrations of hnRNP C RRM to (U)11
Structural modeling suggests that a 10-nt site containing eight consecutive uridines can accommodate a dimer of hnRNP C RRMs
To obtain insight into how two tandem RRMs could be accommodated on a longer U-tract, we performed structural modeling guided by constraints derived from the solution NMR structure of a single RRM binding the five uridines of the AUUUUUC RNA (Cieniková et al. 2014). Since the cross-linking data show that hnRNP C binding is enhanced for uridine tracts of eight or more bases, and the NMR structures previously obtained indicate that one hnRNP C RRM contains 5-nt binding pockets, we calculated using torsion angle dynamics a model of two RRMs binding in tandem on a sequence of 10 nt containing a (U)8 tract. The sequence UC(U)8 was chosen because it corresponds to the known hnRNP C regulatory site in unr 5′ UTR. The proposed ternary complex model (Fig. 2A) satisfies the minimal specificity requirements of the protein (three strongly uridine-selective pockets at the 3′ end of each RRM). A cytidine occupies the second position of the upstream RRM in the model, instead of a uridine as in the solved structure. To confirm that this substitution can be accommodated by the RRM in vitro, we compared the AUCUUUC- and AUUUUUC-RRM complexes by NMR (Fig. 2B,C; Supplemental Fig. S1B). The similarity of the protein and RNA chemical shifts indicates that the AUCUUUC-RRM complex adopts a similar overall conformation and binds in the same register as the solved AUUUUUC-RRM complex, with the first cytidine positioned in the aforementioned pocket 2. The torsion angle dynamics simulation of the double-RRM-UC(U)8 complex structure (Fig. 3) shows additionally that the proposed arrangement of two RRMs on the UC(U)8 oligomer is indeed possible without major steric clashes, since we observe <40% increase in the average CYANA target function measuring the severity of violations of the NMR-derived input constraints compared with the sum of equivalent simulations with two isolated RRMs (Table 2). Our analysis therefore indicates that a sequence of 10 nt containing a tract of eight uridines constitutes the minimal specific binding site for a hnRNP C RRM dimer.
FIGURE 2.
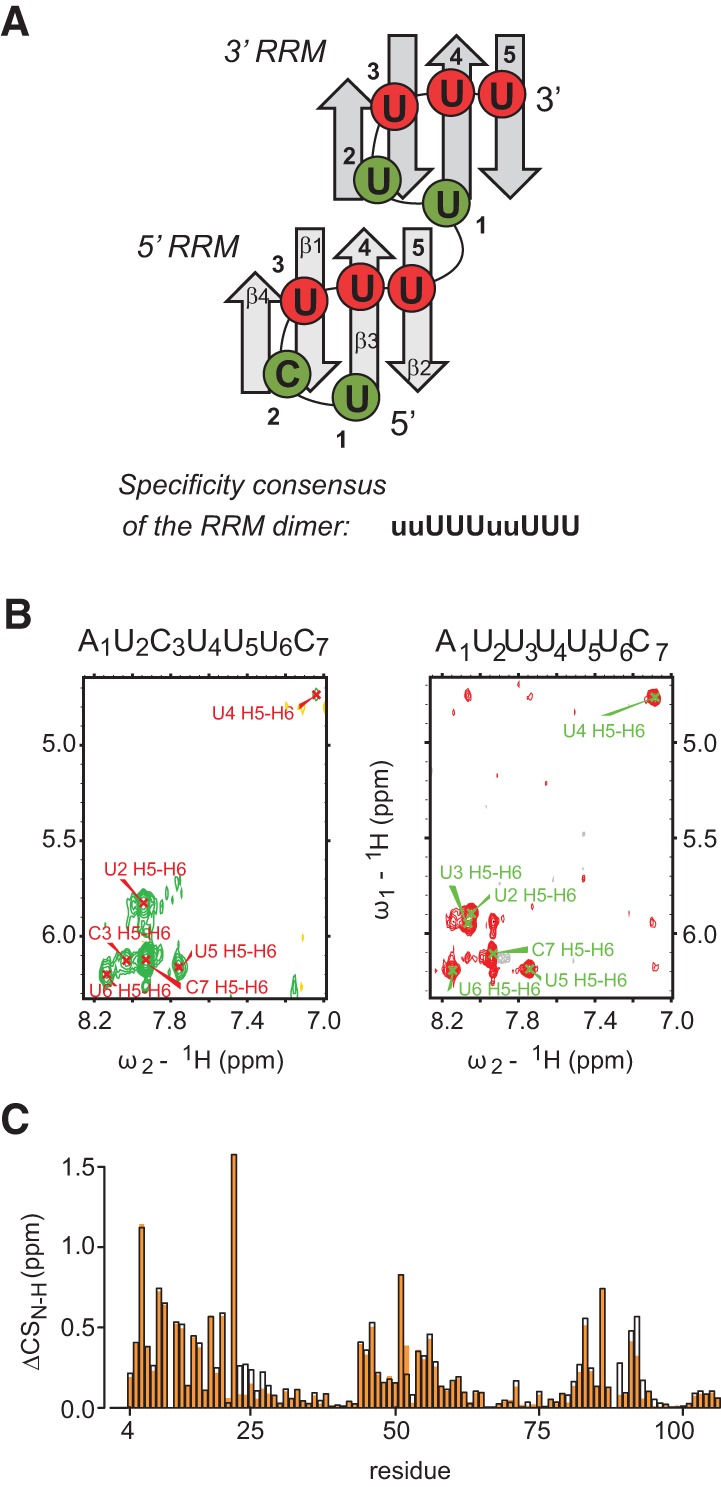
Model for recognition of a 10-nt RNA by two hnRNP C RRMs. (A) Scheme of two tandem RRMs binding the UC(U)8 oligomer. Strictly uridine-specific binding pockets 3–5 (equivalent to positions 2–4 in Cieniková et al. 2014) are colored in red in the graphical model and marked as “U” in the specificity consensus, weakly specific pockets 1 and 2 are in green and labeled with “u.” (B) 2D 1H-1H TOCSY spectra (H5-H6 region) of the RRM-bound AUCUUUC and AUUUUUC RNAs. (C) 1H-15N chemical shift differences (ΔCS) between the free and the RNA-bound RRM. Complex with AUUUUUC in black outline and AUCUUUC in orange.
FIGURE 3.
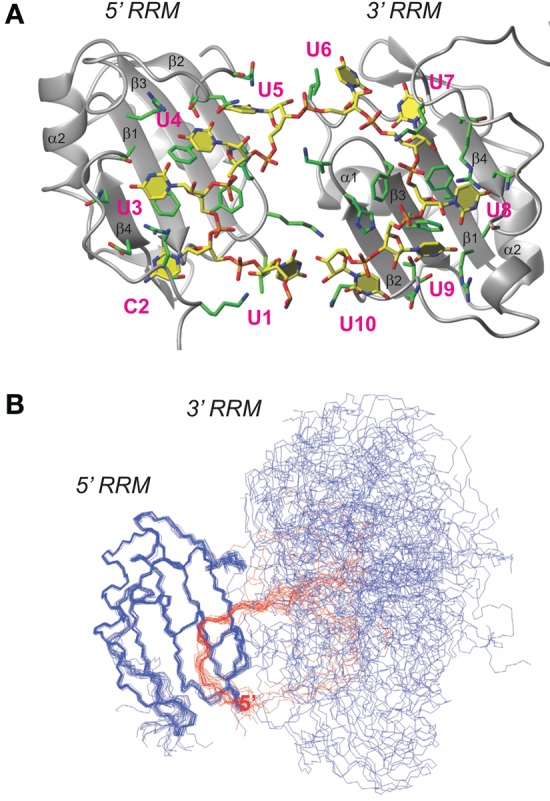
Structural model of the RRM dimer. (A) Representative structure of the double-RRM/UC(U)8 model obtained by torsion angle dynamics calculation. Protein is shown in ribbon and RNA in stick representation. Nucleotides are numbered in magenta. (B) Bundle of 20 structures with the lowest CYANA target function. Protein backbone shown in blue and RNA in red. The ensemble is aligned to the backbone coordinates of the 5′ end-binding RRM in order to show the inter-RRM flexibility. Backbone pairwise RMSD of the 5′ end UC(U)3 RNA–RRM complex is 0.56 Å, that of the 3′ end (U)5 RNA–RRM complex is 0.52 Å, and the RMSD of the 2:1 RRM: UC(U)8 complex is 8.16 Å.
TABLE 2.
CYANA calculation statistics for hnRNP C RRM structural models
In vivo, tandem hnRNP C RRM binding is positively cooperative
The in vitro experiments support the hypothesis of double-RRM binding on sites carrying at least eight contiguous uridines. To address whether this binding behavior reflects how hnRNP C interacts with cellular transcripts, we implemented a structure-based thermodynamic model of uridine lattice saturation by tandem RRMs and then compared the transcriptome-wide distribution of hnRNP C cross-links on uridine tracts up to 12-nt long found by iCLIP (König et al. 2010) with the predictions of this model. The saturation model is an extension of the uridine lattice binding model that we used previously to describe the single RRM's interaction with short tracts (Cieniková et al. 2014). It was parameterized with the affinity weight values obtained for equimolar binding but extended to account for 2:1 RRM:RNA stoichiometry. The introduction of the 2:1 stoichiometry species manifests itself by an inflexion point on the saturation curve for tracts long enough to allow binding of two RRM ligands (m ≈ 8 or 9) (Fig. 1A, red curve). The gain in saturation due to the bipartite binding improves the overall fit to the in vivo data but does not fully account for the observed saturation. A good fit to the in vivo cross-link data for uridine lattice lengths between 1 and 12 nt is obtained only after the introduction of a positive interaction α between the two RRMs (Fig. 1A, blue curve). The experimental data are therefore consistent only with a model assuming that the cross-links found in tracts of 8–12 uridines originate from cooperative binding of two RRMs, each occupying five contiguous nucleotides. The value of the affinity enhancement at 2:1 RRM:RNA stoichiometry obtained from the fit to the iCLIP data set is α ≈ 14. The binding at uridine tract lengths above 13 nt cannot be quantitatively investigated, since the experimental data for very long uridine tracts are sparse and therefore the cross-link frequencies unreliable. A more recent iCLIP of hnRNP C, performed with increased sequencing depth, shows however that the uridine site occupancy saturates at tract lengths of 11 or more uridines (Supplemental Fig. S1B of Zarnack et al. 2013), suggesting that there is no positive cooperativity driving binding of additional RRMs beyond the first dimer. However, fitting to this data set is hindered by the fact that it also includes cross-links with only one detected cDNA, which appear to predominantly correspond to nonspecific interactions.
Further insights into the arrangement of the two side-by-side binding RRMs can be gained by analyzing the position-specific distribution of hnRNP C cross-links within the uridine tracts (Fig. 4; Supplemental Fig. S2). Our thermodynamic model for the binding of two RRMs in tandem predicts correctly the existence of two binding maxima and their absolute and relative position. However, while the modeled maxima are of similar height, the experimental iCLIP data show a decrease or broadening of cross-link deposition at the 5′ end of long uridine tracts, indicating that a feature in the binding of the upstream RRM is not captured by the model. Introducing an additional dynamic component into this upstream binding which permits Gaussian sampling of registers centered on the predicted position broadens the maximum and improves considerably the fit to the observed cross-link distribution.
FIGURE 4.
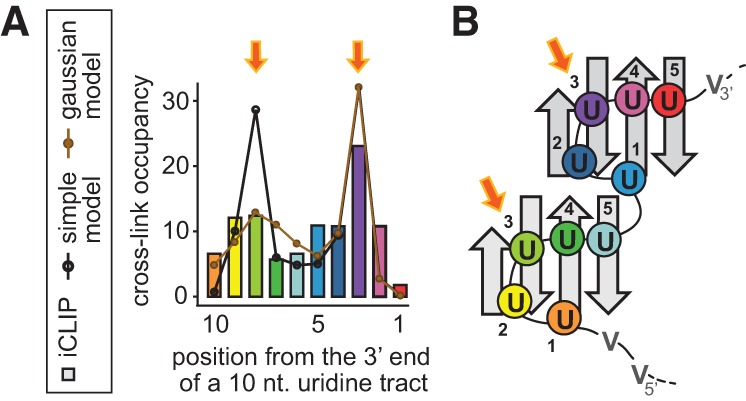
Comparison between the cross-link frequencies observed within a uridine tract of 10 nt and the predicted register occupancy of such a tract by the RRM dimer. (A) The two thermodynamic models are superimposed on the iCLIP data (figure adapted with permission from Macmillan Publishers Ltd. © 2010 [König et al. 2010]). The iCLIP positions are color-coded to match the uridines with nucleotide pockets in the structural model (B) of the RRM dimer binding the (U)10 tract in the favored register. The two cross-linking maxima, highlighted by orange arrows, match the RRM pockets where the cross-link formation was predicted (pocket 3, Cieniková et al. 2014).
Disruptions within the extended poly(U) site in unr IRES site abolish the translation activation by hnRNP C
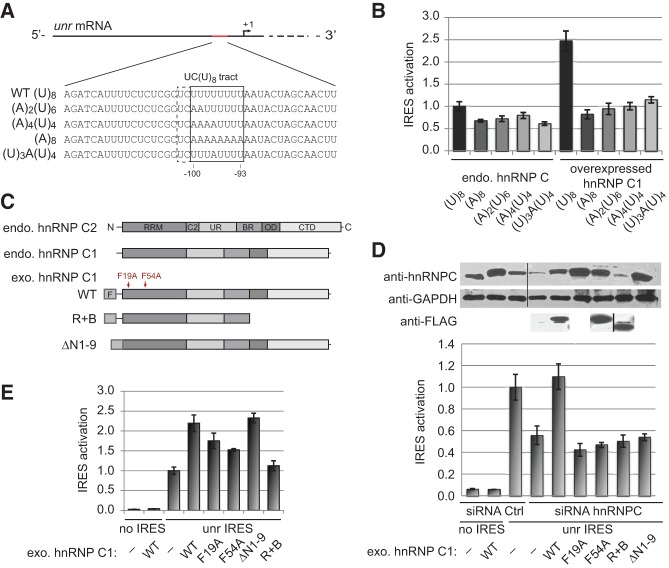
We have shown that two hnRNP C RRM domains are able to bind in tandem on a uridine stretch of ∼8 nt. We observed a synergistic binding behavior in the context of the full-length, oligomerizing protein interacting with cellular transcripts in vivo, in contrast to isolated RRM domains binding to (U)11 in vitro which did not manifest positive cooperativity at 2:1 stoichiometry. To understand how the requirement for side-by-side binding of hnRNP C RRMs affects IRES activity in vivo, we studied the nucleotide requirements of hnRNP C-dependent internal translation activation of unr mRNA in HEK293T cells (Schepens et al. 2007). We designed mRNAs with mutations within the unr IRES UC(U)8 site (Fig. 5A). The wild-type variant (WT) carried the central (U)8 tract; in the mutants (A)2(U)6 and (A)4(U)4, the first two, respectively four uridines of the (U)8 site were replaced by adenosines; in (U)3A(U)4, the fourth uridine was replaced with an adenosine; finally, the (A)8 mRNA, with adenosines instead of the eight uridines, was used as our negative control. We show that shortening or interrupting the (U)8 tract leads to a 20%–30% decrease in translation activation (Fig. 5B). Moreover, these altered IRES variants are unresponsive to overexpressed ectopic hnRNP C1, in contrast to the WT unr IRES where the translation initiation is strongly promoted. All these mRNA variants (except the negative control) were designed to contain between four and six contiguous uridines, which is sufficient for binding one hnRNP C RRM, as indicated by the results of electrophoretic mobility shift assays (Supplemental Fig. S3). Nevertheless, the introduced modifications interfere with the activator role of hnRNP C, indicating that an interaction spanning the full uridine-rich binding site is required to promote UNR translation initiation.
FIGURE 5.
Mutations within the hnRNP C or the UC(U)8 site on the unr IRES impair the internal translation initiation of the unr mRNA. (A) Unr IRES RNAs. (B) Translation activation of WT and mutated unr IRES sequences by hnRNP C. The activity of the IRES represented in the bar chart was calculated as the normalized averaged ratio of the Firefly over Renilla luminescence; standard deviations (SD) were estimated from a minimum of four independent replicates. (C) Schematic representation of hnRNP C variants. (RRM) RNA recognition motif; (C2) 13 amino acid insertion in the C2 variant; (UR) unstructured region; (BR) basic region, (OD) oligomerization domain; (CTD) carboxy-terminal domain; (F) Flag tag. Red arrows indicate the position of mutated residues. (D) Unr IRES activation with wild-type and mutated ectopic hnRNP C1 in the absence of endogenous hnRNP C protein. The quantity of soluble hnRNP C in the cellular extracts was assessed by Western blot against the carboxy-terminal epitope 4F4 and/or against the Flag tag. (E) Effect of hnRNP C1 mutants on the IRES activity in the presence of endogenous hnRNP C.
Intact hnRNP C RRM and oligomerization domains are required for the translation activation
To understand the role of the hnRNP C subdomains in IRES-dependent UNR translation initiation, we overexpressed hnRNP C1 deletion variants or full-length constructs containing point mutations within the RRM domain (Fig. 5C,D). When endogenous hnRNP C is silenced, the IRES activity decreases to levels comparable to those observed with an abolished unr uridine site in the absence of silencing (Fig. 5B). Wild-type ectopic hnRNP C1 rescues translation initiation, while this activation is not observed for the ectopically expressed amino-terminal hnRNP C1 deletion variant ΔN1-9 as well as for RRM domain mutants F19A and F54A lacking aromatic side-chains at the binding interface. This demonstrates that the RRM and the N-terminal residues are essential for internal translation initiation of UNR. Previously we showed (Cieniková et al. 2014) that the mutated or deleted residues are involved in uridine binding; the loss of function seems therefore to be connected to the inability of the mutants to recognize their uridine target sites in the unr IRES. Additionally, we observe that a shortened hnRNP C1 variant retaining only the RRM domain and the basic region (“R + B”, residues 1 to 179) is unable to promote translation initiation, despite the fact that it comprises all subdomains thought to drive the binding to RNA. Since this variant lacks the central helical domain and is therefore unable to multimerize, these data suggest that oligomerization is required for IRES activation.
To investigate the role of cooperative interactions between hnRNP C monomers in IRES activation, we designed an experiment where the full-length exogenous WT hnRNP C1 or one of the following mutants, F19A, F54A, or ΔN1-9, were coexpressed alongside the endogenous hnRNP C. As expected, overexpression of the WT variant promotes IRES activation (Fig. 5E). On the other hand, because the mutants do not activate translation at all when the endogenous hnRNP C is depleted (Fig. 5D), we anticipated little or no response when expressing them in the presence of the endogenous hnRNP C. Contrary to expectations however, the expression of these hnRNP C1 mutants leads to a partial or full activation of the unr IRES relative to the activation level observed upon overexpression of WT hnRNP C1. Clearly, the effects of these overexpressed mutants and of the intact endogenous protein are nonadditive, meaning that the wild-type and mutant full-length hnRNP C monomers act synergistically. It is very likely that this effect is achieved through their interaction within the tetramer.
In conclusion, our results show that to promote the internal translation initiation of UNR, hnRNP C requires an intact U8 stretch in the UC(U)8 site on human unr mRNA. Combining structural modeling, calorimetry and NMR dynamics measurements, we deduce that the wild-type, 10-nt-long uridine-rich site can accommodate two hnRNP C RRMs binding side-by-side. The RRM domains in isolation are however unable to drive their own multimerization on the poly(U) RNA. Therefore, the positive cooperativity observed in vivo between two hnRNP C monomers must arise from the tetramerization of the full-length protein.
DISCUSSION
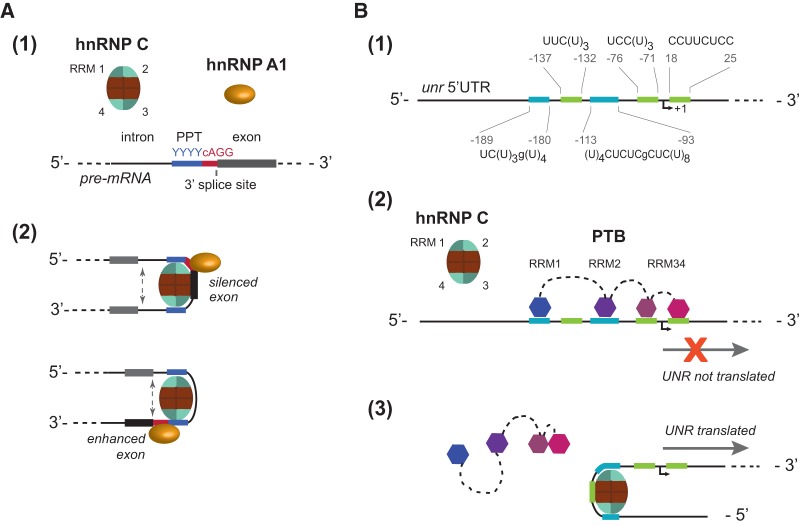
To date, about a dozen structures of RRMs bound to poly(U) sequences have been solved (Table 3). It is noteworthy that, with the exception of Daz-l, Nab3, and hnRNP C, all characterized proteins contain tandem RRM domains that act in concert and bind side-by-side on uridine-rich tracts of ∼7 nt. Moreover, although the Daz-l and Nab3 proteins carry a single copy of the RNA-binding domain, they are reported to form dimers with a second RRM; Daz-l homodimerizes (Ruggiu and Cooke 2000) and Nab3 heterodimerizes with the single RRM domain of Nrd1 (Carroll et al. 2007). Both the Daz-l homodimer and the Nab3-Nrd1 heterodimer bind 9 nt uridine-rich sequences (Jenkins et al. 2011; Hobor et al. 2011). Similar to Daz-l and Nab3, hnRNP C contains a single RRM but forms oligomers in vivo. While the isolated RNA-binding domain of hnRNP C has been shown to recognize uridine pentamers (Cieniková et al. 2014), studies of the hnRNP C binding targets in vitro (Görlach et al. 1994) and in vivo (König et al. 2010) revealed an enrichment of tracts with eight or more contiguous uridines. This observation led to the hypothesis that the RRM domains assembled within the tetramer bind their RNA target sites in pairs. Our structural model of the ternary complex formed by two RRMs bound to UC(U)8 RNA shows that the minimal binding site containing eight contiguous uridines is able to accommodate two tandem RRM domains. However, a preference for uridine over cytidine in the second pocket of the 5′ end-binding RRM (supported by the 10-fold weaker affinity of the RRM for AUCUUUC RNA compared to AUUUUUC [Cieniková et al. 2014]) and possibly steric hindrance between the domains indicated by the moderate increase in NMR constraint violations in our calculation hint that efficient tandem binding likely requires slightly longer uridine tracts. This is experimentally supported by the results of the transcriptome-wide UV cross-linking study (König et al. 2010): Within the pool of enriched RNA sequences, only targets carrying 10 or more contiguous uridines manifest two clear cross-linking maxima. However, the constant 5–6 nt spacing between these maxima supports the compact arrangement of the RRM dimer proposed in our structural model. Further insights into tandem RRM binding on extended uridine tracts are provided by the thermodynamic interaction model. The model reproduces very well in vivo cross-link saturation (Fig. 1A) and position probabilities (Fig. 4; Supplemental Fig. S2). Additionally, broadening in the cross-link position distribution suggests that the upstream-binding RRM is more dynamic than the downstream RRM, occupying a larger number of registers. Similar behavior was observed previously in the case of Sxl and U2AF65 tandem RRMs binding to degenerate polypyrimidine tracts, where the 5′-end RRM2 was shown to sample a larger nucleotide range than the 3′-end RRM1 (Banerjee et al. 2003). While the origin of this difference in interaction behavior is unclear, it is possible that the downstream RRM is tethered to the 3′ end of the tract by protein or RNA contacts unaccounted for in our model. In the case of U2AF65 binding to the polypyrimidine tract preceding the 3′ splice site, U2AF35 could act as such an anchoring protein (Jenkins et al. 2013). For hnRNP C, hnRNP A1 could have an analogous function. Both proteins are constitutive components of the 40S hnRNP core particle (Lothstein et al. 1985), and there is evidence that they act in similar molecular contexts, in particular in 3′ splice site definition (Fig. 6A; Domsic et al. 2003; Tavanez et al. 2012; Zarnack et al. 2013).
TABLE 3.
Poly(U)-binding RRMs with known atomic structures
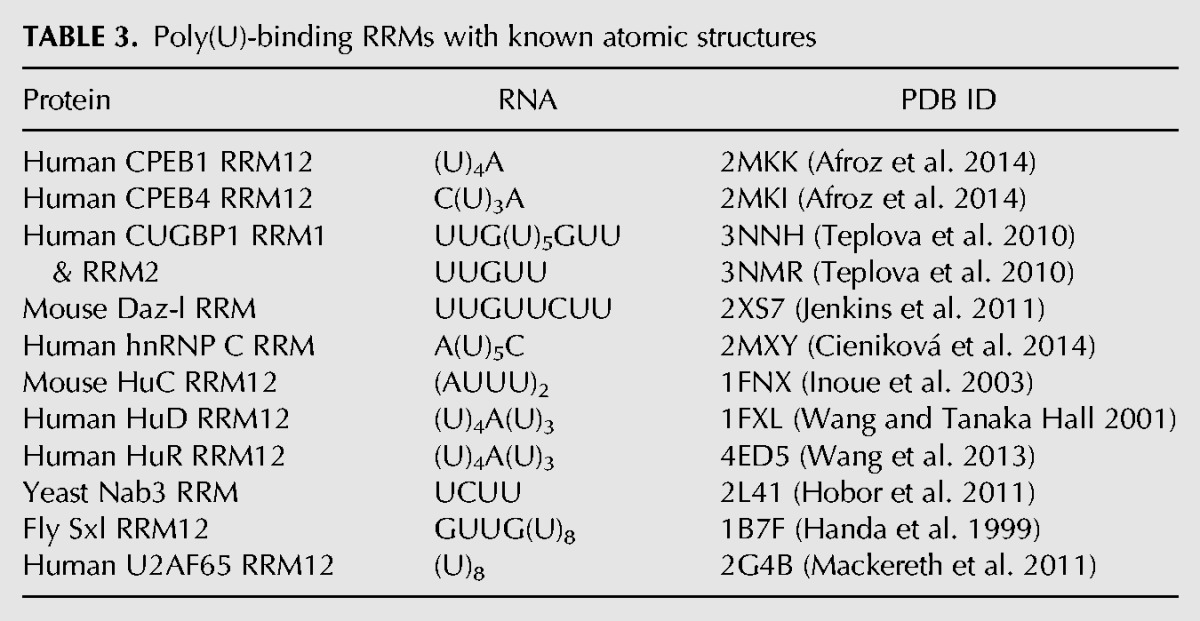
FIGURE 6.
(A) Model of tandem hnRNP C RRM interaction with hnRNP A1 at the 3′ splice site (ss). (1) Regulatory elements surrounding the intron/exon boundary. The 3′ ss is delimited by a conserved cAG/G site preceded by a polypyrimidine tract (PPT) of 10–15 nt (Senapathy et al. 1990). (2) Two interacting RRMs of hnRNP C recognize the PPT. At the exon boundary, the hnRNP A1 RRMs recognizing the YAGG motif would interact with the neighboring hnRNP C RRM, anchoring it to the 3′ end of the PPT. Depending on the relative position of the spliced exon with respect to the hnRNP C tetramer, the exon would be either enhanced or silenced. (B) Model of unr IRES activation by hnRNP C through binding site competition and displacement of PTB. (1) Localization of polypyrimidine tracts (PPT) within human unr 5′ UTR. (2) PTB was shown to bind PPTs within the unr IRES, repressing translation initiation. Binding to the PPT sites in cyan was confirmed experimentally (Cornelis et al. 2005); green PPTs are putative binding sites. Since these PPTs occur on both sides of the start codon, the binding of PTB would prevent the assembly of translation machinery. (3) HnRNP C tetramer displaces PTB at the beginning of mitosis. Its two confirmed poly(U) binding sites (overlapping with the PTB sites in cyan) are localized upstream of the start site (Schepens et al. 2007). The binding of hnRNP C and removal of PTB would therefore expose the start site for translation.
Our translation assays on unr IRES as well as the modeling of the iCLIP data suggest a synergistic interaction between two hnRNP C RRMs binding side-by-side in vivo. Such positive cooperativity between tandem binding RRMs is frequently observed (Kanaar et al. 1995; Park et al. 2000; Sickmier et al. 2006; Kuo et al. 2009; Teplova et al. 2010; Kuwasako et al. 2014). Indeed, structural data indicate that two RRMs binding in tandem to RNA make more extensive contacts to their RNA targets than single RRMs (57.7% ± 3.6% of RNA surface area contacting the protein interface in the former case, compared to 44.6% ± 9.1% in the latter, P-value < 10−4 based on 13 tandem RRM and 26 single RRM poly(U) RNA complexes deposited in PDB [see Materials and Methods]). In most of the examples studied structurally to date, the interacting RRMs are in cis within the same protein molecule. However, a recent study of the ASD-1/SUP-12/RNA ternary complex (Kuwasako et al. 2014) showed that a synergistic RNA recognition can be achieved via RRMs provided by two separate protein monomers, although the measured affinity enhancement is rather moderate (one order of magnitude). In this respect, it is similar to the value of the intermolecular cooperativity factor (α ≈ 14) estimated here for hnRNP C RRMs on the basis of the thermodynamic modeling of the iCLIP data. Investigation of the unr IRES activation indicates that the ability to oligomerize is likely to be responsible for the cooperative behavior of tandem hnRNP C RRMs. We therefore propose that the oligomerization of hnRNP C serves a similar purpose as the RRM multiplicity in other proteins. It facilitates the cooperative recognition of extended low-complexity binding sites. It remains to be determined whether the arrangement within the tetramer serves simply to favorably orient the RRMs so as to prime them for the dual site recognition, or whether other domains of hnRNP C, such as the basic region, also directly contribute to sequence-specific RNA binding.
The analysis of the saturation pattern of the endogenous hnRNP C in the human transcriptome using the RRM binding model reveals that binding of two RRMs is accompanied by a strong enhancement of overall affinity, whereas no further enhancement is detected from the binding of additional RRM domains. This observation is consistent with the proposed orientation of the RRM subdomains within the hnRNP C tetramer, where the four RRMs are arranged in two interacting pairs (König et al. 2010). In agreement with this, the unr 5′ UTR carries two extended uridine-rich sites (Fig. 6B): In addition to the described UC(U)8 site, the UC(U)3G(U)4 motif located 87 nt upstream of it also fits the double-RRM sequence consensus and was shown to bind hnRNP C and contribute to translation activation (Schepens et al. 2007). While the spacing between these sites is only half of the 165 nt suggested previously as the preferred span between the two RRM pairs in the tetramer model, it is still sufficient considering the known dimensions of the tetramer (hydrated diameter of 13.6 nm [Huang et al. 1994]) and assuming an average distance between consecutive phosphorus atoms in a single-stranded nucleic acid of 6.3 Å (Murphy et al. 2004). The proposed positioning of the hnRNP C entirely within the 5′ UTR would leave the start codon free for ribosome binding, and is therefore in agreement with the translation activator role of the hnRNP C protein.
MATERIALS AND METHODS
Cloning and hnRNP C RRM purification for in vitro studies
The vector containing the sequence coding for the human hnRNP C RRM subdomain (AA 2-106), and the protocol of protein purification were described previously (Cieniková et al. 2014).
RNA production and purification
RNA oligomers of ∼26 nt were in vitro transcribed with T7 polymerase from purchased DNA templates annealed to 18 nt-long T7 primers. The target was purified by anion-exchange chromatography under denaturing conditions, then desalted by two rounds of butanol precipitation, lyophilized, and resuspended in the appropriate buffer.
The short single-stranded RNAs (U)11, AUUUUUC and AUCU-UUC purchased from Dharmacon were deprotected according to manufacturer's instructions and then purified as described above.
NMR spectroscopy and dynamics
NMR samples consisted of 1 mM 15N-labeled protein and unlabeled RNA present in 1:1 or 2:1 protein:RNA molar ratio, and were prepared in 250 μL of NMR buffer (20 mM Na2HPO4, 10 mM NaCl, pH 6.5, 10% D2O). Spectra were measured at 293 K on Bruker AVIII-600 MHz, AVIII-700 MHz, and AVIII-750 MHz spectrometers. All instruments except the AVIII-750 were equipped with cryoprobes. NMR data were processed with Topspin 2.1 (Bruker); the analysis of the spectra was performed in Sparky (Goddard and Kneller 1997). Chemical shift assignments were derived from 2D 1H-1H TOCSY and 1H-15N HSQC spectra recorded on the AUCUUUC and (U)11 RNA–RRM complexes and were based on the published assignments of the AUUUUUC–hnRNP C RRM complex (Cieniková et al. 2014). Longitudinal amide relaxation rates R1 = 1/T1 were extracted from a series of 2D 1H-15N HSQC-based Nz relaxation experiments (Kay et al. 1992) recorded on the AVIII-700 MHz spectrometer with nine mixing times ranging from 20 to 1500 msec. The rates for each residue were obtained by fitting the amide signal intensities to an exponential decay using the least-squares minimization method; the corresponding standard deviations (SD) were estimated with the jackknife approach (Efron and Stein 1981). Transverse relaxation rates R2 = 1/T2 were obtained on the AVIII-700 MHz spectrometer from measurements of the 15N CSA/1H-15N dipolar cross-correlated relaxation rates ηxy using the expression R2 = κηxy, where κ = 1.33 is a spectrometer field-dependent constant relating ηxy to the transverse relaxation rate R2 devoid of the contribution from chemical exchange (Tjandra et al. 1996; Wang and Palmer 2003). We recorded a pair of 2D spectra detecting the NxHz magnetization due to cross-relaxation (ICC) and magnetization remaining in the initial state Nx (IREF), respectively, with mixing times τhe of 16, 32, and 50 msec, and fitted these data with the relation ICC/IREF = tanh(ηxy/τhe). Duplicate spectra for one of the mixing times were measured to estimate the standard deviation of the fitted cross-relaxation rates. The mean rotational correlation time τC of a molecule was then calculated according to Fushman et al. (1994), using the data from resolved backbone amides belonging to the RRM globular domain (residues 14–87) having R1/R2 ratio falling within one standard deviation of the mean. To test whether the determined τC obtained for the RRM at 1:1 and 2:1 protein:RNA ratios were the same, we used a paired one-sided t-test with the data from amide signals resolved in both conditions used as matched observations.
Structural modeling
Distance and angle restraints used in the molecular dynamics calculations were identical to those published for the AUUUUUC–hnRNP C RRM complex (Cieniková et al. 2014). Torsion angle dynamics was performed with CYANA 3.0 (Herrmann et al. 2002) using calculations initialized with 200 random structures of the UC(U)3–RRM, the (U)5–RRM, or the UC(U)8–double-RRM complex, referred to as A, B, and C, respectively. Constraints used for the five uridine nucleotides of A(U)5C in the structure calculation described previously were assigned to the 5 nt of UC(U)3, (U)5, and duplicated for the nucleotides 1–5 and 6–10 of UC(U)8. Intra-molecular constraints were the same as in the previous work. Statistical evaluations were performed on the 50 best structures ranked by the lowest target function. The ratio of the average target function (TF) of the double-RRM complex to the average target functions of the two single RRM complexes was calculated as TFC/(TFA + TFB).
Estimation of RNA interface area
The relative buried surface area of RNA oligomers was calculated using the PDBePisa service at http://www.ebi.ac.uk/pdbe/prot_int/pistart.html, version 1.51 (Krissinel and Henrick 2007). Only the nucleotides contacting the protein were taken into account in the calculation. The mean and SD reported for single RRM–RNA complexes was determined from 26 structures (PDB IDs 1AUD, 1URN, 2AD9, 2ADB, 2ERR, 2I2Y, 2KFY, 2KG0, 2KG1, 2KH9, 2L41, 2LEB, 2LEC, 2M8D, 2MB0, 2RQC, 2RRA, 2RS2, 2RU3, 2XS2, 2XS5, 2XS7, 3IWN, 3MOJ, 4CIO, 4I67); for tandem RRM–RNA complexes the coordinates from 13 structures were considered (1B7F, 1CVJ, 1FNX, 1FXL, 1FXL, 1G2E, 2CJK, 2MGZ, 2MKI, 2MKK, 4BS2, 4ED5, 4F02).
ITC measurement and data analysis
Isothermal titration calorimetry measurements were conducted in ITC buffer (50 mM Na2HPO4, 100 mM NaCl, pH 6.5) at 303 K on a VP-ITC calorimeter (MicroCal). We followed the published protocol concerning the sample handling (Cieniková et al. 2014). Data analysis was performed with the MicroCal module of Origin 7.0 (OriginLab) using the interaction models assuming either one set of independent binding sites, or two sequential sites. Binding model selection was based on minimizing the corrected Akaike information criterion (AICc) (Burnham and Anderson 2002). In the case of the sequential, 2:1 binding model, the macroscopic constants Ki are expressed as
| (1) |
where i = 1 and 2 refer to ligand:RNA stoichiometries 1:1 and 2:1, respectively, α is the cooperativity factor at 2:1 stoichiometry, and Bi the expected number of distinct molecular species binding a uridine tract of 11 nt at stoichiometry i, assuming that a single RRM ligand recognizes five consecutive uridines with a microscopic affinity constant KS. In the context of multiregister binding, B1 and B2 are assumed to be ∼8 (Cieniková et al. 2014) and 4 (see following section), respectively.
Model of multistoichiometric RRM binding to a finite uniform tract
The analysis of the multivalent binding of hnRNP C RRM on uridines lattices is an extension of the previously published model (Cieniková et al. 2014). The RRM binding consensus was defined as 5′-uUUUU-3′, with U and u representing pockets with strong and weak preferences for uridines and w and v corresponding to the affinity penalties for U and u pocket mismatch, respectively. Analogously to the ITC analysis, the binding of two interacting protein ligands was allowed; any putative higher stoichiometries were ignored since we focused on tracts shorter than 13 uridines. Under these conditions, the saturation r at tract length m can be expressed as
| (2) |
where KS and α are the affinity and the cooperativity factors defined in the previous paragraph, [F] the concentration of the free protein ligand and Bi the affinity-weighted number of distinct molecular species at stoichiometry i = [1, 2]. The expression for B1 was described previously (Cieniková et al. 2014); the expression for B2 is
The scaled saturation function s × r(m) was fitted to the experimental hnRNP C cross-link saturation rexp(m) for m = [1, 11]. rexp(m) was calculated as the ratio of cross-link distribution on uridine m-mers, over the transcriptome-wide uridine m-mer frequency (Fig. 3B in König et al. 2010). The values of the w, v and KS[F] parameters were previously determined and set to 19.6, 5.1, and 0.33, respectively (Cieniková et al. 2014); only the cooperativity α and the scaling factor s were fitted.
The probability of binding in a given register within a uridine tract was calculated as
| (3) |
with i ≤ 2 the ligand/RNA stoichiometry. For the 1:1 species,
| (4) |
where Wq = f(w,v) is the affinity penalty of the register q. At i = 2, for species defined by registers {q,u}, the probability is
| (5) |
Dynamic binding behavior of the 5′ ligand was implemented as a Gaussian distribution of binding probability p(q2), centered on the position q2, with SD σ minimizing the residual sum of squares between the model and the data.
In vivo assays
The following plasmids were ordered from BCCM/LMBP (http://bccm.belspo.be/bccmt/lmbp.htm): pCAGGS-Flag-hnRNPC1 expression vector (acc. no. 5271), pSV-Sport-di-RF (5253) negative control vector carrying the Firefly and Renilla luciferases, and pSV-Sport-di-RF-UNR (5247) carrying the unr IRES inserted between the luciferases (Cornelis et al. 2005; Schepens et al. 2007). The Flag-hnRNPC1 was recloned into pcDNA3.1 + vector using HindII/XhoI restriction sites. The silencing duplex RNAs were purchased from Dharmacon: The siRNA targeting hnRNP C corresponds to the “C-3” sequence (Venables et al. 2008), the negative control to the “NS” sequence (Wagner and Garcia-Blanco 2002).
IRES activity assays were conducted in the HEK 293T cell strain as previously reported (Cornelis et al. 2005; Schepens et al. 2007). Briefly, the cells were seeded on day 0 by splitting a confluent plate into 2 mL wells (1/5 dilution) containing Gibco D-MEM medium supplemented with Glutamax, antibiotics, and fetal bovine serum. They were transfected on day 1 by the calcium phosphate precipitation method with 0.5 μg of luciferase reporter plasmid and 1 μg of hnRNP C expression plasmid, and harvested on day 2. The cell lysis was performed according to the Promega Dual Luciferase Reporter assay protocol; the Firefly and Renilla luminescence levels were measured on a Berthold MicrolumatPlus luminometer.
For the RNA interference assays, the above protocol was modified as follows: The cells were seeded on day 0 (split 1/10 into 500 μL wells), transfected on day 1 with 50 μL of Gibco Opti-MEM-based mixture containing 1 μL of Lipofectamine 2000 (Invitrogen), 20 pmol of siRNA, and 250 ng of reporter and 500 ng of expression plasmid, and harvested on day 5. The efficiency of the protein silencing and expression was evaluated by Western blotting with antibodies against hnRNP C (clone 4F4, Millipore) or the Flag-tag (Sigma). The GAPDH Western blot signal (antibody from Sigma) was used as expression and loading efficiency control.
Radioactive gel mobility shift assays
Transcribed RNAs were γ-dephosphorylated at the 5′ end using Antarctic phosphatase (NEB), then re-phosphorylated with T4 PNK (NEB) using γ32P ATP. The RNAs (20 fmol/condition) were then incubated for 30 min at room temperature in the NMR buffer with increasing concentrations of hnRNP C RRM in the presence of RNase inhibitor (Ambion SUPERase-In 1%), glycerol (5%) and tRNA (0.1 μg/μL). The samples were loaded onto a nondenaturant 6% polyacrylamide gel and run for 45 min at 100 V in a precooled EMSA buffer (TBE 0.5×, pH 8.3, 0.5% glycerol).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Research of FH-TA is supported by the Swiss National Science Foundation (SNF), grant numbers 3100A0-118118, 31003ab-133134, and 31003A-149921. We thank K. Zarnack and J. Ule for providing and discussing the iCLIP data.
Author contributions: C.M., Z.C., and F.H.-T.A. designed the project; Z.C. prepared protein and RNA samples for NMR and in vitro experiments; Z.C. and F.F.D. recorded and analyzed the NMR data; Z.C. performed the molecular dynamics calculations; Z.C. conducted the binding assays and analysis; Z.C. and S.J. performed the in vivo assays; Z.C., C.M., F.F.D., and F.H.-T.A. wrote the manuscript; all authors discussed the results and approved the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.052373.115.
REFERENCES
- Afroz T, Skrisovska L, Belloc E, Guillén-Boixet J, Méndez R, Allain FH-T. 2014. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev 28: 1498–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee H, Rahn A, Davis W, Singh R. 2003. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA 9: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SF, Friedman DL, LeStourgeon WM. 1989. The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol Cell Biol 9: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. Springer, New York. [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL. 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. 1984. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci 81: 7471–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieniková Z, Damberger FF, Hall J, Allain FH-T, Maris C. 2014. Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif. J Am Chem Soc 136: 14536–14544. [DOI] [PubMed] [Google Scholar]
- Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. 2005. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res 33: 3095–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsic J, Wang Y, Mayeda A, Krainer A, Stoltzfus M. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol 23: 8762–8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Stein C. 1981. The jackknife estimate of variance. Ann Stat 9: 586–596. [Google Scholar]
- Fushman D, Weisemann R, Thuring H, Rüterjans H. 1994. Backbone dynamics of ribonuclease T1 and its complex with 2′GMP studied by two-dimensional heteronuclear NMR spectroscopy. J Biomol NMR 4: 61–78. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. 1997. SPARKY 3. UCSF, San Francisco. [Google Scholar]
- Görlach M, Burd CG, Dreyfuss G. 1994. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem 269: 23074–23078. [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. 1999. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature 398: 579–585. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319: 209–227. [DOI] [PubMed] [Google Scholar]
- Hobor F, Pergoli R, Kubicek K, Hrossova D, Bacikova V, Zimmermann M, Pasulka J, Hofr C, Vanacova S, Stefl R. 2011. Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J Biol Chem 286: 3645–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, Krainer AR, LeStourgeon WM. 1994. The C-protein tetramer binds 230 to 240 nt of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol 14: 518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Hirao M, Kasashima K, Kim IS, Kawai G, Kigawa T, Sakamoto H, Muto Y, Yokoyama S. 2003. Solution structure of mouse HuC RNA-binding domains complexed with an AU-rich element reveals determinants of neuronal differentiation. Protein Data Bank ID: 1FNX. 10.2210/pdb1fnx/pdb. [DOI]
- Jenkins H, Malkova B, Edwards T. 2011. Kinked β-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc Natl Acad Sci 108: 18266–18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J, Agrawal A, Gupta A, Green M, Kielkopf C. 2013. U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res 41: 3859–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Lee AL, Rudner DZ, Wemmer DE, Rio DC. 1995. Interaction of the sex-lethal RNA binding domains with RNA. EMBO J 14: 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA. 1992. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J Magn Reson 97: 359–375. [Google Scholar]
- Kim JH, Paek KY, Choi K, Kim TD, Hahm B, Kim KT, Jang SK. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol 23: 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. 2009. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K, Takahashi M, Unzai S, Tsuda K, Yoshikawa S, He F, Kobayashi N, Güntert P, Shirouzu M, Ito T, et al. 2014. RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing. Nat Struct Mol Biol 21: 778–786. [DOI] [PubMed] [Google Scholar]
- Lothstein L, Arenstorf HP, Chung SY, Walker BW, Wooley JC, LeStourgeon WM. 1985. General organization of protein in HeLa 40S nuclear ribonucleoprotein particles. J Cell Biol 100: 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcárcel J, Sattler M. 2011. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475: 408–411. [DOI] [PubMed] [Google Scholar]
- McAfee JG, Shahied-Milam L, Soltaninassab SR, LeStourgeon WM. 1996a. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA 2: 1139–1152. [PMC free article] [PubMed] [Google Scholar]
- McAfee JG, Soltaninassab SR, Lindsay ME, LeStourgeon WM. 1996b. Proteins C1 and C2 of heterogeneous nuclear ribonucleoprotein complexes bind RNA in a highly cooperative fashion: support for their contiguous deposition on pre-mRNA during transcription. Biochemistry 35: 1212–1222. [DOI] [PubMed] [Google Scholar]
- McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. 2012. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science 335: 1643–1646. [DOI] [PubMed] [Google Scholar]
- Merrill BM, Barnett SF, LeStourgeon WM, Williams KR. 1989. Primary structure differences between proteins C1 and C2 of HeLa 40S nuclear ribonucleoprotein particles. Nucleic Acids Res 17: 8441–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. 2004. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J 86: 2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA. 2000. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol Cell Biol 20: 4765–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech JE, Huang MH, LeStourgeon WM, Flicker PF. 1995. An ultrastructural characterization of in vitro-assembled hnRNP C protein-RNA complexes. J Struct Biol 114: 84–92. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Cooke HJ. 2000. In vivo and in vitro analysis of homodimerisation activity of the mouse Dazl1 protein. Gene 252: 119–126. [DOI] [PubMed] [Google Scholar]
- Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, Cornelis S. 2007. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J 26: 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella O, Gerlitz G, Le S-Y, Elroy-Stein O. 1999. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol 19: 5429–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P, Shapiro MB, Harris NL. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol 183: 252–278. [DOI] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltaninassab SR, McAfee JG, Shahied-Milam L, LeStourgeon WM. 1998. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res 26: 3410–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanez J, Madl T, Kooshapur H, Sattler M, Valcárcel J. 2012. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol Cell 45: 314–329. [DOI] [PubMed] [Google Scholar]
- Teplova M, Song J, Gaw H, Teplov A, Patel D. 2010. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure 18: 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N, Szabo A, Bax A. 1996. Protein backbone dynamics and 15N chemical shift anisotropy from quantitative measurement of relaxation interference effects. J Am Chem Soc 118: 6986–6991. [Google Scholar]
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. 2008. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol 28: 6033–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol Cell 10: 943–949. [DOI] [PubMed] [Google Scholar]
- Wan L, Kim JK, Pollard VW, Dreyfuss G. 2001. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J Biol Chem 276: 7681–7688. [DOI] [PubMed] [Google Scholar]
- Wang C, Palmer AG III. 2003. Solution NMR methods for quantitative identification of chemical exchange in 15N-labeled proteins. Magn Reson Chem 41: 866–876. [Google Scholar]
- Wang X, Tanaka Hall TM. 2001. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol 8: 141–145. [DOI] [PubMed] [Google Scholar]
- Wang H, Zeng F, Liu Q, Liu H, Liu Z, Niu L, Teng M, Li X. 2013. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr D Biol Crystallogr 69: 373–380. [DOI] [PubMed] [Google Scholar]
- Whitson SR, LeStourgeon WM, Krezel AM. 2005. Solution structure of the symmetric coiled coil tetramer formed by the oligomerization domain of hnRNP C: implications for biological function. J Mol Biol 350: 319–337. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Malter J. 1995. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem 270: 17292–17298. [DOI] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.