Abstract
Hepatitis B virus (HBV) infection is the predominant risk factor for chronic hepatitis B (CHB), liver cirrhosis (LC) and hepatocellular carcinoma (HCC). Recently, genome-wide association studies have identified human leukocyte antigen (HLA)-DP polymorphisms (rs3077 and rs9277535) as a new chronic HBV infection susceptibility locus. Since then, the relationship between HLA-DP polymorphisms and various outcomes of HBV infection has been reported. However, the results have been inconclusive. To derive a more precise estimation of the relationship between HLA-DP polymorphisms and various outcomes of HBV infection, a meta-analysis of 62,050 subjects from 29 case-control studies was performed. We found that rs3077 and rs9277535 in HLA-DP significantly decreased HBV infection risks and increased HBV clearance possibility in a dose-dependent manner. In the subgroup analysis by ethnicity, study design and sample size, significant associations were found for these polymorphisms in almost all comparisons. Meanwhile, haplotype analyses of the two polymorphisms revealed a significant association between the combination of these alleles and HBV infection outcomes. However, no significant results were observed in HCC development. Our results further confirm that genetic variants in the HLA-DP locus are strongly associated with reduced HBV infection and increased the likelihood of spontaneous viral clearance.
Hepatitis B virus (HBV) infection is a major global health concern, with more than 2 billion people infected, of whom 400 million are chronic carriers1,2. Although some HBV carriers spontaneously eliminate the virus, carriers are at an increased risk of developing liver cirrhosis (LC), hepatic decompensation, and hepatocellular carcinoma (HCC) development3, especially in the endemic areas of Southeast Asia, China, Japan, and Sub-Saharan Africa4. In addition to the viral and environmental factors5, results from twin studies, family clustering studies and studies of differences between ancestry groups have suggested that host factors are critical in determining the outcome of HBV infection6.
Over the past decade, many efforts have been put into the deciphering the genetic architecture for HBV persistence. From a long list of candidate genes, may variants have been inconsistently associated with HBV infection: IFNG7, TNF7, ESR18, VDR9, MBP10, CTLA411, and the human leukocyte antigen (HLA) class II12. However, because the exact mechanisms of HBV persistence are yet to be elucidated completely, the candidate-gene approach is limited in power to detect novel disease-susceptibility genes. Recently, a genome-wide association study (GWAS) identified two single-nucleotide polymorphism (SNPs) in HLA-DP (rs3077 and rs9277535) for persistent HBV infection in Japanese and Thai populations13. Replication of initial GWAS findings is considered a gold standard for reporting genotype–phenotype associations. As stated by McClellan and King, many if not most of the genetic polymorphisms that are reported to be associated with common disorders in GWA studies are factually spurious associations caused by subtle differences in ancestry between the populations being studied (known as “cryptic population stratification”)14. To date, many case–control studies have been carried out to investigate the role of the two SNPs in HLA-DP in relation to outcomes of HBV infection among various populations. Genetic association studies can be problematic to reproduce due to insufficient power, ethnic diversity, multiple hypothesis testing, population stratification, phenotypic heterogeneity and publication bias. Therefore, we carried out a comprehensive meta-analysis on all eligible studies to estimate relationship between HLA-DP polymorphisms (rs3077 and rs9277535) and HBV infection outcomes as well as to quantify the between-study heterogeneity and potential bias.
Results
Characteristics of the included studies
Based on our search strategy, the primary screening produced 369 potentially relevant articles. Study selection process was shown in Figure S1. Overall, 28 published studies and one unpublished data involving 29,790 HBV carriers, 7,804 subjects with HBV natural clearance, 4,864 HCC patients, and 24,456 healthy individuals were finally included13,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41. Of the subjects, most were Asians, only 3.1% were of other ethnic origins. The main study characteristics were summarized in Table S1. There was a wide variation in the risk allele frequency of the two polymorphisms among the controls across different ethnicities (Figure S2).
Association between HLA-DPA1 rs3077-A and HBV infection, HBV spontaneous clearance, HCC development
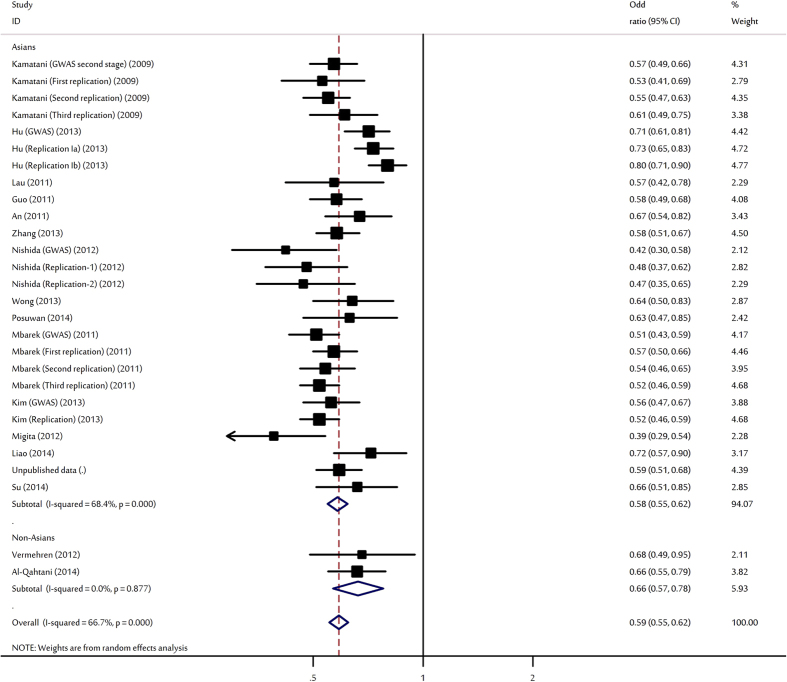
The evaluations of the association of HLA-DPA1 rs3077-A polymorphism with various outcomes of HBV infection are shown in Table 1. Overall, the HLA-DPA1 rs3077-A allele was shown to protect against HBV infection with per-allele OR of 0.59 (95% CI: 0.55–0.62, P < 10−5; Fig. 1). Significant associations were also found for AG heterozygote (OR = 0.55, 95% CI: 0.51–0.61, P < 10−5) and AA homozygote (OR = 0.39, 95% CI: 0.35–0.45, P < 10−5). In the subgroup analysis by ethnicity, study design and sample size, significantly decreased HBV infection risks were found for the polymorphism in almost all comparisons.
Table 1. Results of meta-analysis for rs3077 polymorphism with HBV infection outcomes.
| Overall and subgroups analyses | No. of data sets | No. of cases/controls | A vs. G allele |
AG vs. GG |
AA vs. GG |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P(Z) | P(Q) | I2 (%) | OR (95% CI) | P(Z) | P(Q) | I2 (%) | OR (95% CI) | P(Z) | P(Q) | I2 (%) | |||
| HBV Infection (All) | 28 | 18898/22419 | 0.59 (0.55–0.62) | <10−5 | <10−5 | 67.7 | 0.55 (0.51–0.61) | <10−5 | <10−5 | 62.3 | 0.39 (0.35–0.45) | <10−5 | 0.002 | 50.1 |
| Asian | 26 | 17918/21610 | 0.58 (0.55–0.62) | <10−5 | <10−5 | 68.4 | 0.56 (0.51–0.61) | <10−5 | <10−5 | 62.7 | 0.39 (0.35–0.44) | <10−5 | 0.005 | 48.5 |
| Non-Asians | 2 | 980/809 | 0.66 (0.57–0.78) | <10−5 | 0.88 | 0 | 0.39 (0.11–1.32) | 0.13 | 0.03 | 79.1 | 0.39 (0.13–1.13) | 0.08 | 0.05 | 75.0 |
| GWAS | 5 | 2592/5439 | 0.56 (0.48–0.65) | <10−5 | 0.007 | 71.9 | 0.51 (0.40–0.64) | <10−5 | 0.01 | 73.5 | 0.37 (0.26–0.53) | <10−5 | 0.03 | 67.4 |
| Replication study | 23 | 16306/16980 | 0.59 (0.56–0.65) | <10−5 | <10−5 | 66.7 | 0.56 (0.51–0.62) | <10−5 | <10−5 | 60.7 | 0.40 (0.35–0.46) | <10−5 | 0.008 | 48.2 |
| Large studies | 14 | 14356/15273 | 0.62 (0.57–0.67) | <10−5 | <10−5 | 75.6 | 0.61 (0.55–0.67) | <10−5 | 0.003 | 61.3 | 0.44 (0.37–0.52) | <10−5 | 0.001 | 65.3 |
| Small studies | 14 | 4542/7146 | 0.55 (0.51–0.59) | <10−5 | 0.20 | 23.4 | 0.49 (0.43–0.56) | <10−5 | 0.05 | 43.0 | 0.34 (0.29–0.39) | <10−5 | 0.73 | 0 |
| HBV Clearance (All) | 22 | 6627/14041 | 1.51 (1.35–1.68) | <10−5 | <10−5 | 76.0 | 1.66 (1.47–1.87) | <10−5 | 0.003 | 52.7 | 2.19 (1.80–2.67) | <10−5 | <10−5 | 59.9 |
| Asian | 21 | 6327/13262 | 1.53 (1.37–1.71) | <10−5 | <10−5 | 75.3 | 1.69 (1.50–1.90) | <10−5 | <10−5 | 51.7 | 2.28 (1.87–2.77) | <10−5 | <10−5 | 57.1 |
| Non-Asians | 1 | 300/779 | 1.12 (0.90–1.39) | 0.32 | NA | NA | 1.03 (0.61–1.73) | 0.91 | NA | NA | 1.17 (0.71–1.93) | 0.53 | NA | NA |
| GWAS | 1 | 185/181 | 2.25 (1.62–3.13) | <10−5 | NA | NA | 2.54 (1.61–4.01) | <10−5 | NA | NA | 3.53 (1.72–7.22) | 0.001 | NA | NA |
| Replication study | 21 | 6442/13860 | 1.48 (1.33–1.65) | <10−5 | <10−5 | 75.3 | 1.63 (1.45–1.83) | <10−5 | 0.007 | 50.3 | 2.15 (1.76–2.62) | <10−5 | <10−5 | 60.2 |
| Large studies | 8 | 4253/10412 | 1.51 (1.31–1.73) | <10−5 | <10−5 | 79.9 | 1.66 (1.37–2.00) | <10−5 | 0.001 | 74.4. | 2.09 (1.57–2.79) | <10−5 | <10−5 | 75.1 |
| Small studies | 14 | 2374/3629 | 1.52 (1.27–1.82) | <10−5 | <10−5 | 75.2 | 1.66 (1.42–1.94) | <10−5 | 0.17 | 26.7 | 2.31 (1.73–3.08) | <10−5 | <10−5 | 48.3 |
| AsC | 4 | 1142/1388 | 1.04 (0.83–1.29) | 0.75 | 0.08 | 56.0 | 1.23 (0.97–1.56) | 0.09 | 0.30 | 17.5 | 1.25 (0.93–1.68) | 0.15 | 0.55 | 0 |
| HCC development | 7 | 4475/6299 | 0.99 (0.89–1.11) | 0.90 | 0.02 | 61.3 | 0.97 (0.83–1.14) | 0.71 | 0.05 | 55.6 | 1.05 (0.78–1.42) | 0.74 | 0.02 | 62.6 |
NA: not available; AsC: asymptomatic HBsAg carrier; HCC: hepatocellular carcinoma.
Figure 1. Forest plot for the meta-analysis of the association between rs3077 polymorphism and HBV infection stratified by ethnicity.
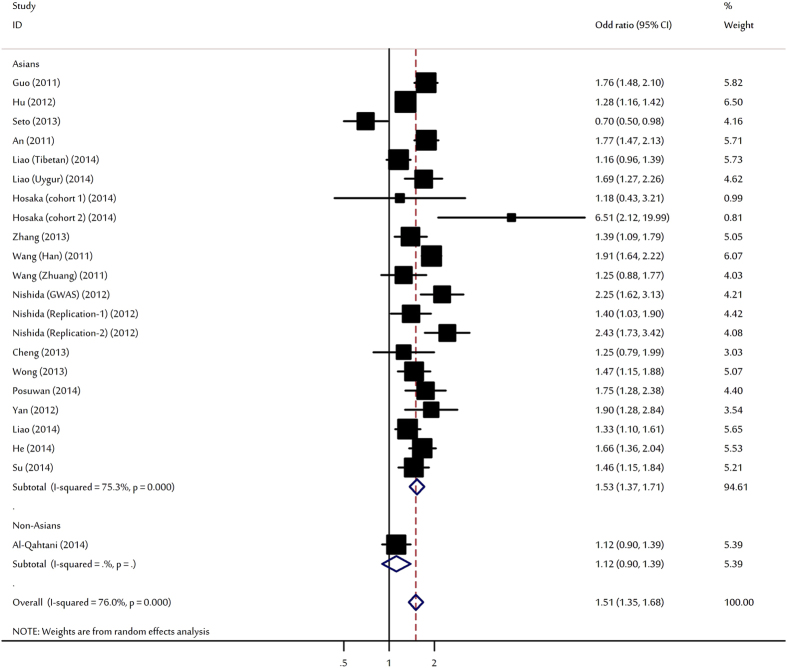
For HBV clearance, our meta-analysis shown that individuals carrying the HLA-DPA1 rs3077-A allele had a significantly higher chance of spontaneous clearance upon HBV infection (OR = 1.51, 95% CI: 1.35–1.68, P < 10−5; Fig. 2). Significant increased HBV natural clearances were also observed for heterozygote and homozygote. When stratifying for ethnicity, significant associations were only found among Asians. Subsidiary analyses according to study design and sample size, significant associations were also maintained for all comparisons (Table 1).
Figure 2. Forest plot for the meta-analysis of the association between rs3077 polymorphism and HBV clearance stratified by ethnicity.
Compared with active symptomatic HBV-carriers (e.g., LC, CHB), there is no significant difference in rs3077 genotype distribution in asymptomatic HBV-carriers (Table 1).
The data on genotypes of rs3077 among HBV-induced HCC patients and HBV carriers were available in 7 studies. No evidence of any gene-disease association was obtained (Table 1).
Association between HLA-DPB1 rs9277535-A and HBV infection, HBV spontaneous clearance, HCC development
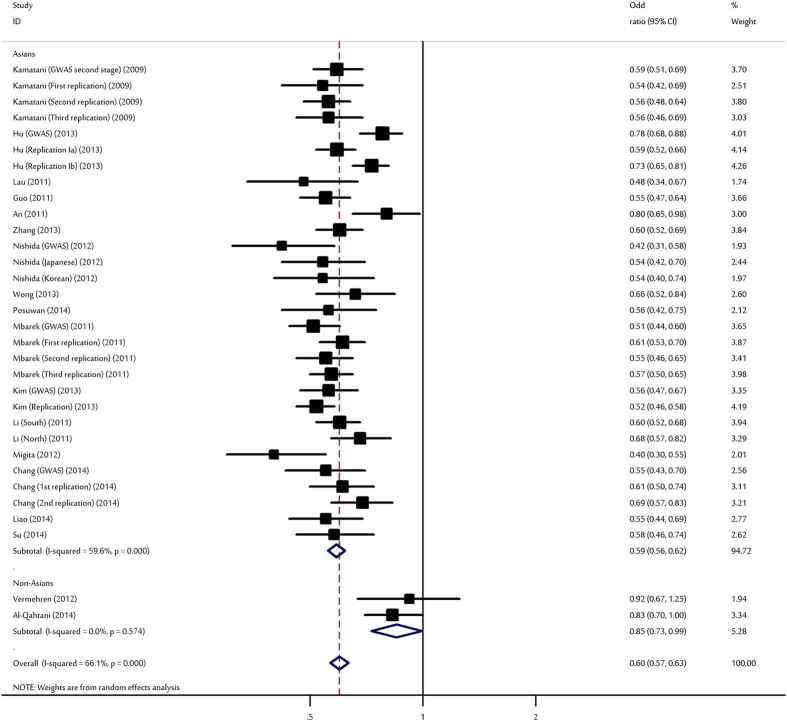
Overall, there was evidence of an association between the decreased risk of HBV infection and the rs9277535-A variant in different genetic models when all the eligible studies were pooled into the meta-analysis (Table 2). Using random-effects model, the summary per-allele OR of the rs9277535-A variant for HBV infection was 0.60 (95% CI: 0.57–0.63, P < 10−5; Fig. 3). Significantly decreased HBV infection risks were also found for those heterozygote (OR = 0.56, 95% CI: 0.52–0.60, P < 10−5) and homozygous for the minor A allele (OR = 0.39, 95% CI: 0.35–0.43, P < 10−5) when compared with the wild type genotype. In the stratified analysis by ethnicity, study design and sample size, significant associations were detected in all genetic models for the polymorphism (Table 2).
Table 2. Results of meta-analysis for rs9277535 polymorphism with HBV infection outcomes.
| Overall and subgroups analyses | No. of data sets | No. of cases/controls | A vs. G allele |
AG vs. GG |
AA vs. GG |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P(Z) | P(Q) | I2 (%) | OR (95% CI) | P(Z) | P(Q) | I2 (%) | OR (95% CI) | P(Z) | P(Q) | I2 (%) | |||
| HBV Infection (All) | 32 | 22065/23500 | 0.60 (0.57–0.63) | <10−5 | <10−5 | 66.1 | 0.56 (0.52–0.60) | <10−5 | 0.01 | 40.7 | 0.39 (0.35–0.43) | <10−5 | 0.01 | 40.6 |
| Asian | 30 | 21099/22694 | 0.59 (0.56–0.62) | <10−5 | <10−5 | 59.6 | 0.56 (0.52–0.60) | <10−5 | 0.009 | 43.6 | 0.38 (0.35–0.42) | <10−5 | 0.02 | 39.0 |
| Non-Asians | 2 | 966/806 | 0.85 (0.73–0.99) | 0.04 | 0.57 | 0 | 0.62 (0.41–0.94) | 0.02 | 0.32 | 0 | 0.59 (0.39–0.89) | 0.01 | 0.35 | 0 |
| GWAS | 6 | 2917/5947 | 0.57 (0.48–0.67) | <10−5 | <10−5 | 80.9 | 0.49 (0.38–0.63) | <10−5 | 0.001 | 77.6 | 0.38 (0.28–0.53) | <10−5 | 0.01 | 69.9 |
| Replication study | 26 | 19148/17553 | 0.60 (0.57–0.64) | <10−5 | <10−5 | 61.7 | 0.57 (0.54–0.61) | <10−5 | 0.22 | 17.4 | 0.39 (0.35–0.43) | <10−5 | 0.06 | 32.7 |
| Large studies | 17 | 17211/15611 | 0.64 (0.59–0.68) | <10−5 | <10−5 | 70.1 | 0.60 (0.57–0.64) | <10−5 | 0.29 | 15.0 | 0.43 (0.38–0.48) | <10−5 | 0.03 | 45.5 |
| Small studies | 15 | 4854/7889 | 0.54 (0.51–0.58) | <10−5 | 0.16 | 27.1 | 0.49 (0.44–0.54) | <10−5 | 0.24 | 19.3 | 0.32 (0.28–0.37) | <10−5 | 0.77 | 0 |
| HBV Clearance (All) | 25 | 7753/17089 | 1.54 (1.43–1.66) | <10−5 | <10−4 | 62.8 | 1.64 (1.50–1.79) | <10−5 | 0.08 | 30.7 | 2.31 (1.99–2.67) | <10−5 | 0.003 | 50.4 |
| Asian | 24 | 7451/16324 | 1.57 (1.46–1.68) | <10−5 | 0.001 | 55.6 | 1.65 (1.51–1.80) | <10−5 | 0.10 | 29.0 | 2.37 (2.06–2.73) | <10−5 | 0.02 | 42.3 |
| Non-Asians | 1 | 302/765 | 1.02 (0.82–1.26) | 0.85 | NA | NA | 1.08 (0.64–1.81) | 0.78 | NA | NA | 1.07 (0.65–1.77) | 0.78 | NA | NA |
| GWAS | 1 | 185/181 | 1.95 (1.42–2.69) | <10−5 | NA | NA | 2.65 (1.68–4.17) | <10−5 | NA | NA | 2.48 (1.28–4.82) | 0.007 | NA | NA |
| Replication study | 24 | 7568/16908 | 1.53 (1.41–1.65) | <10−5 | <10−5 | 62.7 | 1.60 (1.48–1.74) | <10−5 | 0.18 | 21.6 | 2.30 (1.98–2.69) | <10−5 | 0.002 | 52.5 |
| Large studies | 10 | 4443/12359 | 1.47 (1.36–1.60) | <10−5 | 0.04 | 50.1 | 1.52 (1.38–1.68) | <10−5 | 0.22 | 25.3 | 2.12 (1.78–2.54) | <10−5 | 0.04 | 51.4 |
| Small studies | 15 | 3310/4730 | 1.61 (1.40–1.85) | <10−5 | <10−5 | 67.1 | 1.82 (1.59–2.08) | <10−5 | 0.36 | 8.7 | 2.52 (1.99–3.17) | <10−5 | 0.04 | 44.7 |
| AsC | 6 | 1751/2706 | 0.98 (0.87–1.09) | 0.66 | 0.33 | 13.9 | 0.97 (0.84–1.12) | 0.65 | 0.93 | 0 | 1.04 (0.71–1.52) | 0.85 | 0.06 | 53.1 |
| HCC development | 8 | 4398/7358 | 1.03 (0.97–1.09) | 0.35 | 0.59 | 0 | 0.92 (0.84–1.02) | 0.10 | 0.93 | 0 | 1.11 (0.95–1.29) | 0.19 | 0.66 | 0 |
NA: not available; AsC: asymptomatic HBsAg carrier; HCC: hepatocellular carcinoma.
Figure 3. Forest plot for the meta-analysis of the association between rs9277535 polymorphism and HBV infection stratified by ethnicity.
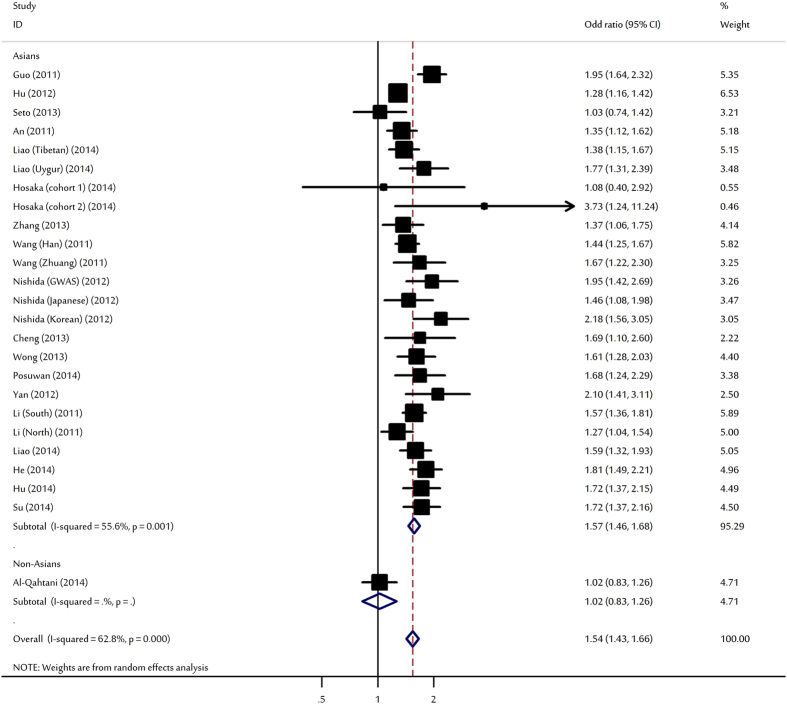
Meta-analyses showed that rs9277535-A was significantly associated with increased HBV clearance with per-allele OR of 1.54 (95% CI: 1.43–1.66, P < 10−5; Fig. 4). Significant associations were found for heterozygote and homozygote. Subgroup analysis for rs9277535 and HBV clearance was also performed to explore the sources of heterogeneity. In the subgroup analyses by ethnicity, the SNP significantly increased natural HBV clearances only among Asians. Subsidiary analyses by study design yielded OR of 1.95 (95% CI: 1.42–2.69) and 1.53 (95% CI: 1.41–1.65) for GWAS and replication studies, respectively. After stratification for sample size, significant results still maintained irrespective to sample size (Table 2).
Figure 4. Forest plot for the meta-analysis of the association between rs9277535 polymorphism and HBV clearance stratified by ethnicity.
When HBV infection outcomes as asymptomatic HBsAg carrier (AsC) and HCC development were considered, no significant associations were detected (Table 2).
Haplotype analysis
Haplotype analyses between rs3077 and rs9277535 polymorphisms were performed in the 11 articles, involving 4,044 HBV carriers, 5,357 HBV natural clearances and 2,730 healthy controls. When compared with the most frequent G-G haplotype, all other haplotypes containing variant alleles of the two SNPs were associated with decreased HBV infection (OR range from 0.57 to 0.82, Table S2). In addition, these haplotypes were also associated with higher chance of HBV natural clearance (OR range from 1.32 to 1.65, Table S3). Results from Haplotype analyses were consistent with the single SNP analysis.
Credibility of genetic association
To assess the credibility of genetic associations, we considered the BFDP (Table S4–S5) and the Venice criteria (Table S6). Applying these filters indicate that the two variants were graded strong for cumulative evidence of association with HBV infection and HBV clearance. In addition, associations of HBV infection and HBV clearance were maintain statistically significant after Bonferroni correction for multiple genetic models for the 2 SNPs.
Heterogeneity analyses
In view of significant heterogeneity and to seek for its potential sources, we performed a panel of meta-regression analysis. In meta-regression analysis, sample size, study quality, mean age of cases and controls, sex distribution among cases and controls, ethnicity, study design, did not significantly correlated with the magnitude of the genetic effect for rs3077 and rs9277535 (P > 0.05 for all). Furthermore, Galbraith plot analyses of all included studies were used to assess the potential sources of heterogeneity (Figure S3–S6).
Association of rs3077-A variant with HLA-DPA1 mRNA expression
To further explored potential function, mRNA expression level of HLA-DPA1 by rs3077 genotypes from peripheral blood mononuclear cells (PBMC) and brain tissues of European descent was obtained from SNPExpress. When pooled all available data together, significantly increased of transcript expression levels by A allele carriers was found for HLA-DPA1 in PBMC (P = 0.017; Figure S7a) and in brain tissues (P = 0.003; Figure S7b).
Sensitivity analyses and publication bias
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual dataset to the pooled ORs, and the corresponding pooled ORs were not qualitatively altered for rs3077 (Figure S8–S9) and rs9277535 (Figure S10–S11), suggesting that the results of this meta-analysis are stable. Funnel plot and Egger’s test were performed to access the publication bias of the literatures. The shapes of the funnel plot for the per-allele comparison of the A allele and the G allele seemed symmetrical (Figure S12–S15). The statistical results still did not show publication bias in these studies (Egger’s test: P > 0.05, for all).
Discussion
The nature history of HBV infection is complicated and identifying biomarkers could facilitate prediction and prevention of vulnerable populations with higher risk to develop CHB and even worse outcomes, such as LC and HCC. Accumulating evidence indicated that host genetic factors play a major role in the persistence of HBV infection42. Recent GWAS studies have suggested that certain variations in the HLA-DP regions are associated with protection against chronic hepatitis B as well as viral clearance13,20,24. After that, many replications studies have been conducted to explore the relationship between HLA-DP polymorphisms (rs3077 and rs9277535) and various outcomes of HBV infection. As significant differences in allele frequencies and the prevalence of HBV infection among various populations exist, it is, therefore, important to quantitatively assess the effects of the GWAS-identified markers in different ethnic populations and explore potential heterogeneity of published data. This is the most comprehensive meta-analysis examining the association of rs3077 and rs9277535 polymorphisms on HLA-DP regions and its relationship to outcomes HBV infection. Its strength was based on the accumulation of data giving greater information to detect significant differences. In total, the meta-analysis involved 29 studies including 62,050 subjects.
In this large-scale meta-analysis, the combined evidence confirmed that two SNPs (rs3077 and rs9277535) at HLA-DP locus were significantly associated with decreased HBV infection risk as well as increased spontaneous viral clearance. A panel of subgroup analysis based on ethnicity, sample size and study design were performed and significant associations maintained in almost all comparisons for the two SNPs. However, the heterogeneity of OR is high in our data, especially in the studies for Asian populations. Indeed, the Asian population reports in the subgroup analysis include a mixture of populations from very distant areas. The presence of heterogeneity can result from differences in environmental factors, lifestyle and host-related physical factors43. Furthermore, HBV genotype, viral activity, duration of infection may also contribute such heterogeneity28,44. Applying Venice criteria and the BFDP indicate that associations with the 2 SNPs represent the most credible findings.
When stratified by ethnicity, inconsistent association results for the two SNPs were observed in Asians and non-Asian populations. In fact, differences in genetic backgrounds may attribute to these results. For example, the risk allele distribution of rs3077 varies between Asians, and non-Asians, with a prevalence of 47, and 76%, respectively. Such a result could also be due to the limited number of studies among non-Asians, which had insufficient statistical power to detect a slight effect. On the other hand, different populations usually have different linkage disequilibrium (LD) patterns. A polymorphism may be in close linkage with another nearby causal variant in one ethnic population, but not in another. Furthermore, it is possible that variation at this locus has modest effects on outcomes of HBV infection, but environmental factors may predominate in the progress of HBV infection, and mask the effects of this variation. Specific environmental factors like aflatoxin B1 exposure2 and prevalence of HBV3 have been already well studied in recent decades.
If genetic susceptibility to HBV infection is, in part, mediated through gene polymorphisms, it is possible that the combinations of certain genotypes may be more discriminating as risk factors for HBV infection than a single locus genotype. Haplotypes analyses of the rs3077-A and rs9277535-A alleles reveal the association between the combination of these alleles in protection against HBV infection as well as beneficial effect of spontaneous viral clearance.
The rs3077 and rs9277535 SNPs are located within 3′-UTR of HLA-DPA1 and HLA-DPB1 gene, respectively. It is possible that they act as the binding site of microRNA and thus affect both the translation and stability of mRNA. SNPs located at miRNA-binding site are likely to disrupt miRNA-target interaction, and result in the deregulation of target gene expression45. We therefore compared the mRNA expression levels of HLA-DPA1 by the rs3077 genotypes and found that rs3077-A allele is associated with higher HLA-DPA1 expression. More recently, the A alleles of HLA-DP rs3077 and rs9277535 were reported to be strongly associated with increased levels of mRNA expression of HLA-DPA1 and HLA-DPB1, respectively, in normal liver tissues46. Higher levels of HLA-DPA1 on target cell surfaces might be more effective in presenting viral antigen to CD4 + T helper cells, leading to an impaired immune response to viral invasion or to the resolution of HBV infection47. Furthermore, rs3077 showed protective effects for response to hepatitis B vaccination48. On the other hand, these two SNPs may be in close linkage with another nearby causal variant. Therefore, re-sequencing and fine mapping of this region to identify putative causal variants, combined with functional evaluation, are required.
Chronic HBV infection seems to be the most important risk factor for HCC4,49. However, no significant associations were observed for asymptomatic HBsAg carrier or HBV-related HCC for rs3077 and rs9277535. One of the possible reasons could be the high complexity of multivariate interactions between the genomic information and the phenotype that is manifesting. HCC development is a multiple process which links to causative factors such as environmental toxins (e.g., Aflatoxin B1), alcohol drinking and smoking habits (two of the main recognized HCC risk factors), lifestyle (e.g., vegetables, fruit consumption), and HBV genotype variations50,51.
Compared with the previous meta-analysis52, the present study is much larger, with more than three times as many subjects as the earlier study. In addition, we assessed not only the effect on HBV infection and viral clearance but also the effects on HBV activity and HCC development. Furthermore, we also investigated whether the haplotypes were associated with HBV infection or clearance. Moreover, we explored potential sources of heterogeneity across studies and the possibility of publication bias.
In interpreting the results, some limitations of this meta-analysis should be addressed. Firstly, the vast majority of subjects in the study are of East Asian descent, and statistical power for analyses in other ethnicities is limited. Because the sample size was relatively smaller for Caucasian studies, the main conclusions from this manuscript are based on analyses among East Asian populations. Further studies including a wider spectrum of ethnic populations are necessary. Secondly, our results were based on unadjusted estimates, while a more precise analysis should be conducted if all individual raw data were available, which would allow for the adjustment by other co-variants including alcohol abuse, aflatoxin B1 exposure, cigarette smoking and other lifestyle. Finally, lacking the original data limited our further evaluation of potential interactions clinical outcomes (e.g., ALT level, AST level, Albumin level) and viral backgrounds (e.g., HBV genotype, viral load).
Despite these limitations, findings of the present study showed that SNPs rs3077 and rs9277535 at HLA-DP locus protected against HBV infection and increased chance of HBV clearance; while the importance of these polymorphisms as a predictor of HCC may be limited.
Methods
Identification and screening of relevant studies
The present meta-analysis was performed according to the guideline of PRISMA statement. Genetic association studies published before the end of March 2015 on various outcomes of HBV infection and the two SNPs (rs3077 and rs9277535) at HLA-DP were identified through a search of PubMed, ISI Web of Knowledge, EMBASE, SCOPUS, and Cochrane databases without language restriction. Search term combinations were keywords relating to HBV (e.g., “chronic HBV infection”, “chronic hepatitis B”, “hepatitis B Virus”, “HBV clearance”, “liver cirrhosis”, “hepatocellular carcinoma”) in combination with words related to HLA-DP (e.g., “rs3077”, “rs9277535”, “HLA”, “human leukocyte antigen-DP”). The titles and abstracts of potential articles were screened to determine their relevance, and any clearly irrelevant studies were excluded. The full texts of the remaining articles were read to determine whether they contained information on the topic of interest. Reference lists of included studies and relevant reviews were hand searched for additional eligible studies.
Criteria for inclusion
The included studies have to meet the following criteria: (1) case–control or cohort studies to evaluate the association between polymorphisms at HLA-DP and various outcomes of HBV infection; (2) original papers containing independent data; (3) Identification of HBV infected cases was confirmed pathologically; (4) available genotype distribution information or odds ratios (ORs) with its 95% confidence intervals (CIs) and P value; (5) genotype distribution of control group must be consistent with Hardy–Weinberg equilibrium (HWE). The major reasons for exclusion of studies were (1) overlapping data, (2) case-only studies, and (3) review articles.
Quality assessment and data extraction
For association studies with inconsistent results on the same polymorphisms, the methodological quality should be assessed by appropriate criteria to limit the risk of introducing bias into meta-analyses. A procedure known as ‘Newcastle – Ottawa Scale (NOS)’ has been used to assess the quality of association studies. Detailed procedure of the quality assessment was previously described53.
Not all researchers use the same HLA-DP SNPs, and most articles reported results for multiple SNPs (uniquely identified by their rs number). We report herein 2 common SNPs (rs3077, rs9277535) that were included in all but 3 articles27,31,34. The remaining 3 articles used 1 additional SNP (rs9277378), as this SNP had a high level of linkage disequilibrium with rs9277535 (D’ = 1.00, R2 = 0.954) in the HapMap Han Chinese in Beijing (CHB) and Japanese in Tokyo (JPT) Populations54. Data extraction was performed independently by two reviewers. For each study, the following variables were collected according to a fixed protocol: the first author, published year, ethnicity, identification of cases, HBV genotype, viral activity, duration of infection, ALT (alanine aminotransferase) level, AST (aspartate aminotransferase) level, Albumin level, bilirubin level, definitions of control groups, age, sex, study design, source of controls, Hardy–Weinberg equilibrium (HWE) status among controls, number of cases and controls, outcomes of HBV infection (CHB, natural clearance, AsC, LC, HCC), number of genotypes and genotyping methods. For studies including subjects of different ethnic groups, data were extracted separately and categorized. Meanwhile, different case-control groups in one study were considered as independent studies. Review reports from the two were then compared to identify any inconsistency, and differences were resolved by further discussion among all authors.
Genotype and gene expression correlation analysis
The data on rs3077 genotype and HLA-DPA1 expression levels were available by SNPExpress tool55. The transcript (mRNA) expression data were detected by using genome-wide expression arrays from peripheral blood monocytes of 80 healthy individuals and brain tissues of 93 healthy individuals. Genome-wide genotyping was performed using genechips.
Statistical analysis
The data from each SNP was divided into three groups: chronic HBV infection vs. healthy controls; spontaneous clearance individuals vs. chronic HBV infection and HCC vs. HBV carriers. The strength of the association between various outcomes of HBV infection and the two polymorphisms (rs3077, rs9277535) was estimated using ORs, with the corresponding 95% CIs. The per-allele OR of the risk allele of these polymorphisms was compared between cases and controls. Then, we estimated the risks of the heterozygote and homozygous genotypes compared with the wild-type homozygote56. Cochran’s chi-square-based Q statistic test and I2 statistics was performed to evaluate possible heterogeneity between the individual studies. Random effects and fixed effect summary measures were calculated as inverse-variance-weighted average of the log odds ratio57,58. The results of random effects summary were reported in the text because it takes into account the variation between studies. Sources of heterogeneity were investigated by stratified meta-analyses based on ethnicity (Asians or Non-Asians), study design (GWAS or replication study), and sample size (≥500 cases or, <500 cases). Furthermore, ethnic group, study design, sample size, mean age of cases and controls and sex distribution in cases and controls were analysed as covariates in meta-regression. Sensitivity analysis was performed by removing each individual study in turn from the total and re-analysing the remainder. Publication bias was assessed with the funnel plot and Egger test. All P values are two-sided at the P = 0.05 level. Statistical analyses were carried out using the STATA software version 10.0 (Stata Corporation, College Station, TX, USA) and SAS (version 9.1; SAS Institute, Cary, NC, USA).
Credibility of genetic association
For statistically significant associations identified by meta-analyses, Venice criteria and BFDP were applied to assess the credibility of the evidence59. Venice criteria details are published elsewhere60. The BFDP threshold for noteworthiness was set up to be equal to 0.20, based on the assumption that a false discovery would be four times more costly than a false non-discovery. We chose to calculate BFDP values for two levels of prior probabilities: at a medium or low prior level (0.05 to 10−3) that would be close to what would be expected for a candidate gene; and at a very low prior level (10−4 to 10−6) that would be close to what would be expected for a random SNP.
Additional Information
How to cite this article: Yu, L. et al. Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci. Rep. 5, 14933; doi: 10.1038/srep14933 (2015).
Supplementary Material
Footnotes
Author Contributions Conceived and designed the study: L.Y., Y.J.C., M.L.C. and G.Z.Y.; Performed the experiments: L.Y., Y.J.C., Y.M.Y., Q.Z., X.K.Z., H.J.L., Y.X.H., M.M., B.W., L.L.Z. and S.Z.; Contributed material/analysis tools: L.Y., Y.J.C., Y.M.Y., Q.Z., X.K.Z., H.J.L., Y.X.H., M.M., B.W., L.L.Z. and S.Z.; Statistical analyses and paper writing, revising: L.Y., Y.J.C., M.L.C. and G.Z.Y.
References
- Lai C. L. & Yuen M. F. Chronic hepatitis B–new goals, new treatment. N Engl J Med 359, 2488–2491 (2008). [DOI] [PubMed] [Google Scholar]
- Kao J. H. & Chen D. S. Global control of hepatitis B virus infection. Lancet Infect Dis 2, 395–403 (2002). [DOI] [PubMed] [Google Scholar]
- Ganem D. & Prince A. Hepatitis B virus infection–natural history and clinical consequences. N Engl J Med 350, 1118–1129 (2004). [DOI] [PubMed] [Google Scholar]
- Parkin D. M., Bray F., Ferlay J. & Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 55, 74–108 (2005). [DOI] [PubMed] [Google Scholar]
- Yu M. W. et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst 97, 265–272 (2005). [DOI] [PubMed] [Google Scholar]
- Shen F. M., Lee M. K., Gong H. M., Cai X. Q. & King M. C. Complex segregation analysis of primary hepatocellular carcinoma in Chinese families: interaction of inherited susceptibility and hepatitis B viral infection. Am J Hum Genet 49, 88–93 (1991). [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Z. et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol 98, 144–150 (2003). [DOI] [PubMed] [Google Scholar]
- Deng G. et al. Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology 40, 318–326 (2004). [DOI] [PubMed] [Google Scholar]
- Bellamy R. et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis 179, 721–724 (1999). [DOI] [PubMed] [Google Scholar]
- Thomas H. C. et al. Mutation of gene of mannose-binding protein associated with chronic hepatitis B viral infection. Lancet 348, 1417–1419 (1996). [DOI] [PubMed] [Google Scholar]
- Thio C. L. et al. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J Virol 78, 11258–11262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz M. R. et al. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med 332, 1065–1069 (1995). [DOI] [PubMed] [Google Scholar]
- Kamatani Y. et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41, 591–595 (2009). [DOI] [PubMed] [Google Scholar]
- McClellan J. & King M. C. Genetic heterogeneity in human disease. Cell 141, 210–217 (2010). [DOI] [PubMed] [Google Scholar]
- An P. et al. A common HLA-DPA1 variant is a major determinant of hepatitis B virus clearance in Han Chinese. J Infect Dis 203, 943–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Evaluation of genetic susceptibility loci for chronic hepatitis B in Chinese: two independent case-control studies. PLoS One 6, e17608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K. C. et al. Non-invasive screening of HLA-DPA1 and HLA-DPB1 alleles for persistent hepatitis B virus infection: susceptibility for vertical transmission and toward a personalized approach for vaccination and treatment. Clin Chim Acta 412, 952–957 (2011). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case-control study. PLoS One 6, e24221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. et al. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 53, 422–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarek H. et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 20, 3884–3892 (2011). [DOI] [PubMed] [Google Scholar]
- Migita K. et al. HLA-DP gene polymorphisms and hepatitis B infection in the Japanese population. Transl Res 160, 443–444 (2012). [DOI] [PubMed] [Google Scholar]
- Hu L. et al. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 55, 1426–1431 (2012). [DOI] [PubMed] [Google Scholar]
- Vermehren J. et al. A common HLA-DPA1 variant is associated with hepatitis B virus infection but fails to distinguish active from inactive Caucasian carriers. PLoS One 7, e32605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N. et al. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One 7, e39175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. et al. Relationship between HLA-DP gene polymorphisms and clearance of chronic hepatitis B virus infections: case-control study and meta-analysis. Infect Genet Evol 12, 1222–1228 (2012). [DOI] [PubMed] [Google Scholar]
- Jiang D. K. et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 45, 72–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto W. K. et al. HLA-DP and IL28B polymorphisms: influence of host genome on hepatitis B surface antigen seroclearance in chronic hepatitis B. Clin Infect Dis 56, 1695–1703 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol 87, 12176–12186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. R. et al. Host genetic factors affecting spontaneous HBsAg seroclearance in chronic hepatitis B patients. PLoS One 8, e53008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet 45, 1499–1503 (2013). [DOI] [PubMed] [Google Scholar]
- Wong D. K. et al. Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. PLoS One 8, e66920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J. et al. A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum Mol Genet 22, 4233–4238 (2013). [DOI] [PubMed] [Google Scholar]
- Al-Qahtani A. A. et al. Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One 9, e80445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posuwan N. et al. Genetic association of human leukocyte antigens with chronicity or resolution of hepatitis B infection in thai population. PLoS One 9, e86007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. et al. Association of HLA-DP/DQ and STAT4 Polymorphisms with HBV Infection Outcomes and a Mini Meta-Analysis. PLoS One 9, e111677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T. et al. HLA-DP genes polymorphisms associate with hepatitis B surface antigen kinetics and seroclearance during nucleot(s)ide analogue therapy. Liver Int 35, 1290–302 (2015). [DOI] [PubMed] [Google Scholar]
- Chang S. W. et al. A genome-wide association study on chronic HBV infection and its clinical progression in male Han-Taiwanese. PLoS One 9, e99724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. et al. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int 35, 886–96 (2015). [DOI] [PubMed] [Google Scholar]
- He D. et al. Interaction of TLR-IFN and HLA polymorphisms on susceptibility of chronic HBV infection in Southwest Han Chinese. Liver Int 10.1111/liv.12756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. et al. HLA-DPB1 Variant Effect on Hepatitis B Virus Clearance and Liver Cirrhosis Development Among Southwest Chinese Population. Hepat Mon 14, e19747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M. et al. Studies on the association of single nucleotide polymorphisms of HLA-DP and DQ genes with the outcome of chronic hepatitis B virus infection. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 31, 765–9 (2014). [DOI] [PubMed] [Google Scholar]
- Thursz M. Genetic susceptibility in chronic viral hepatitis. Antivir Res 52, 113–116 (2001). [DOI] [PubMed] [Google Scholar]
- Hohler T. et al. Differential genetic determination of immune responsiveness to hepatitis B surface antigen and to hepatitis A virus: a vaccination study in twins. Lancet 360, 991–995 (2002). [DOI] [PubMed] [Google Scholar]
- Lam Y. F. et al. HLA-DP and γ-interferon receptor-2 gene variants and their association with viral hepatitis activity in chronic hepatitis B infection. J Gastroenterol Hepatol 29, 533–539 (2014). [DOI] [PubMed] [Google Scholar]
- Yu Z. et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res 35, 4535–4541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T. R. et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12, 428–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbani S. et al. Acute phase HBV-specific T cell responses associated with HBV persistence after HBV/HCV coinfection. Hepatology 41, 826–831 (2005). [DOI] [PubMed] [Google Scholar]
- Pan L. et al. A genome-wide association study identifies polymorphisms in the HLA-DR region associated with non-response to hepatitis B vaccination in Chinese Han populations. Hum Mol Genet 23, 2210–2219 (2014). [DOI] [PubMed] [Google Scholar]
- Yang J. D. & Roberts L. R. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatology 7, 448–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. I. et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 100, 1134–1143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Increased Intake of Vegetables, But Not Fruit, Reduces Risk for Hepatocellular Carcinoma: A Meta-analysis. Gastroenterology 147, 1031–42 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang X. L. et al. Association of the rs3077 and rs9277535 polymorphisms in HLA-DP with hepatitis B virus infection and spontaneous clearance: a meta-analysis. Scand J Gastroenterol 48, 736–744 (2013). [DOI] [PubMed] [Google Scholar]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25, 603–605 (2010). [DOI] [PubMed] [Google Scholar]
- International HapMap C. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen E. L. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 6, e1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. X., Jiang G. N., Zheng H., Duan L. & Ding J. A. Common genetic variants on 3q28 contribute to non-small cell lung cancer susceptibility: evidence from 10 case-control studies. Mol Genet Genomics 290, 573–84 (2015). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Wakefield J. A. Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 81, 208–227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J. P. et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol 37, 120–132 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.