Summary
A subset of individuals infected with human immunodeficiency virus 1 (HIV-1) develops broadly neutralizing antibodies (bNAbs) that can prevent infection, but it has not yet been possible to elicit these antibodies by immunization. To systematically explore how immunization might be tailored to produce them, we generated mice expressing a diverse repertoire of light chains and predicted germline or mature heavy chains of a potent bNAb to the CD4 binding site (CD4bs) on the HIV-1 envelope glycoprotein (Env). Immunogens specifically designed to activate B cells bearing germline antibodies are required to initiate immune responses, but they do not elicit bNAbs. In contrast, native-like Env trimers fail to activate B cells expressing germline antibodies but elicit bNAbs by selecting for a restricted group of light chains bearing specific somatic mutations that enhance neutralizing activity. The data suggest that vaccination to elicit anti-HIV-1 antibodies will require immunization with a succession of related immunogens.
Keywords: HIV-1 vaccine, bNAbs, 3BNC60, Knock-in, HIV-1 envelope glycoprotein, HIV-1 neutralization
Graphical abstract
Introduction
A fraction of HIV-1 infected individuals develop potent bNAbs that target several independent sites on gp160, the viral envelope glycoprotein (Env) (West et al., 2014). When passively transferred into non-human primates or into genetically engineered or humanized mice, these antibodies can protect against challenge with chimeric simian/human immunodeficiency virus (SHIV) or HIV-1 viruses, respectively (Burton et al., 2012; Klein et al., 2013; Mascola and Haynes, 2013; West et al., 2014). Antibodies were also the only correlate of protection in a recent phase 3 human HIV-1 vaccine trial that showed limited efficacy (Karasavvas et al., 2012; Rerks-Ngarm et al., 2009). Thus, one of the goals of the HIV-1 vaccine effort has been to elicit bNAbs by immunization. However, this goal has not been achieved despite over 25 years of concerted vaccination efforts.
Why it is so difficult to elicit these antibodies was only fully appreciated after the advent of single cell antibody cloning techniques (Klein et al., 2013; West et al., 2014). Antibody cloning revealed that anti-HIV-1 antibodies are unusual in that they carry large numbers of somatic hypermutations that are required for binding to most recombinant HIV-1 Env antigens and for broad neutralization (Mouquet et al., 2010; Scheid et al., 2011; Wu et al., 2010). These mutations are likely to arise as a result of multiple rounds of hypermutation and selection in the germinal center in response to rapidly evolving escape mutations in the HIV-1 Env (Mouquet et al., 2010; Scheid et al., 2009). This idea is supported by the observation that bNAbs co-evolve with HIV-1 in the host through multiple rounds of HIV-1 escape from antibody pressure (Doria-Rose et al., 2014; Klein et al., 2013; Liao et al., 2013; Wu et al., 2015). Considered together, these findings have led to the hypothesis that eliciting such antibodies may require using a series of engineered or naturally arising antigens to direct the antibody response (Dimitrov, 2010; Doria-Rose et al., 2014; Jardine et al., 2013; Klein et al., 2013; Liao et al., 2013; Wu et al., 2011). According to this idea, an antigen that activates B cells carrying a germline antibody would initially be used to expand a reactive B cell clone and produce a group of somatic variants by hypermutation. To shepherd the antibody response towards broad neutralization, the initial immunization would be followed by one or a series of related antigens.
To test this hypothesis, we produced Ig heavy chain knock-in mice expressing the predicted germline (GLVH) or mature mutated (MuVH) version of 3BNC60, a bNAb that targets the CD4 binding site (CD4bs) of HIV-1 (Scheid et al., 2011). 3BNC60 is one of a closely related group of potent antibodies referred to as VRC01-class antibodies (West et al., 2012), arising in several different individuals, all of which are derived from IgHV1-2*02 (West et al., 2014). In addition to the shared origin of their heavy chains, this group of antibodies all carry light chains that have short (5 amino acid) third complementarity determining regions (CDR3s) (West et al., 2012; Zhou et al., 2013).
Mice that carry heavy chain knock-in genes have a restricted B cell repertoire because the heavy chain is fixed. Nevertheless, the repertoire remains relatively diverse because the antibody light chain is produced by random VJ recombination in developing B cells. Thus, only a small fraction of the B cells carry heavy and light chains that combine to produce antibodies able to bind to the HIV-1 Env (see below). Immunization of GLVH mice affords the opportunity to evaluate antigens for their ability to select B cells expressing light chains that show features that could support bNAb evolution. In contrast, MuVH mice represent a synthetic intermediate since the human heavy chain carries all of the required mutations, but the mouse light chain is germline.
To track the evolution of the HIV-1 antibody response in GLVH and MuVH mice, we immunized them with antigens designed to bind to the predicted unmutated precursor of 3BNC60, or with BG505 SOSIP trimers that resemble the native HIV-1 Env.
Results
3BNC60 knock-in mice
GLVH and MuVH mice were produced by gene targeting using C57Bl-6 embryonic stem cells (Pelanda et al., 1997; Shih et al., 2002) (Figure S1A). GLVH reverts all of the heavy chain residues outside of CDRH3 (Hoot et al., 2013). CDRH3 cannot be reverted with certainty because the sequence represents the combined effects of random V(D)J recombination, N addition, deletion and somatic mutation. Both strains showed near normal frequencies of immature and recirculating B cells in the BM (Figure S1B) and of mature B cell populations in spleen (Figure S1C).
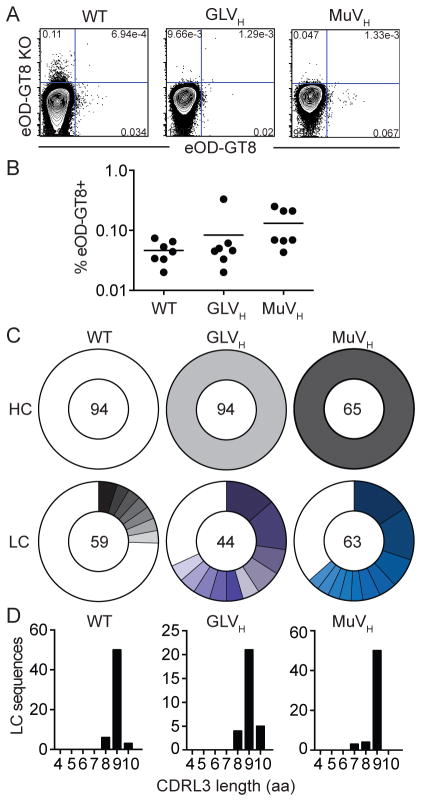
To determine the frequency of naive B cells that bind to Env-based antigens in the knock-in mice, we stained splenocytes with an improved version (eOD-GT8) of the previously described immunogen eOD-GT6, an engineered outer domain of gp120 designed to bind to both germline and mature versions of 3BNC60 and other VRC01-class antibodies (see below and (Jardine et al., 2013)). The average number of naive B cells that bind to eOD-GT8 in GLVH and MuVH mice (0.08% and 0.13%, respectively) was similar to that found in wild type C57Bl6 mice (0.05%) indicating that these cells are rare and found at frequencies similar to those in wild type C57Bl6 mice (Figure 1A–B). To examine the antibody repertoire in naive B cells in these mice we isolated single cells by flow cytometry and sequenced their Ig genes. As expected, the heavy chain was always the product of the respective knock-in allele and the mouse light chain sequences were less diverse than in wild type C57Bl6 B cells (51/59, 24/44 and 33/63 unique sequences for wild type C57Bl6, GLVH and MuVH respectively) (Figure 1C). Consistent with the rare occurrence of antigen-binding cells in the naive knock-in mice (Figure 1A and B), none of the mouse light chains showed the 5-residue CDRL3 signature typical of authentic 3BNC60 or of other VRC01-class antibodies (Figure 1D).
Figure 1. Characterization of B cells in GLVH and MuVH mice.
(A) Representative FACS plots show binding of eOD-GT8 and eOD-GT8 CD4bs knock-out (KO) proteins by mature naive B cells in naive wild type C57Bl6 (WT), GLVH and MuVH mice. (B) Graph shows frequency of eOD-GT8-binding B cells in naive WT, GLVH and MuVH mice. Each dot represents one mouse. (C) Pie charts show heavy (HC) and light chain (LC) sequences cloned from purified single cell sorted naive B cells from naive mice. The number in the center of the pie chart is the number of sequences analyzed, each colored slice represents one clone (identical V gene and CDRL3) and its size is proportional to the size of the clone. White indicates unique sequences. (D) CDRL3 amino acid (aa) lengths from LCs in C. See also Figure S1.
Immune responses in 3BNC60 GLVH knock-in mice
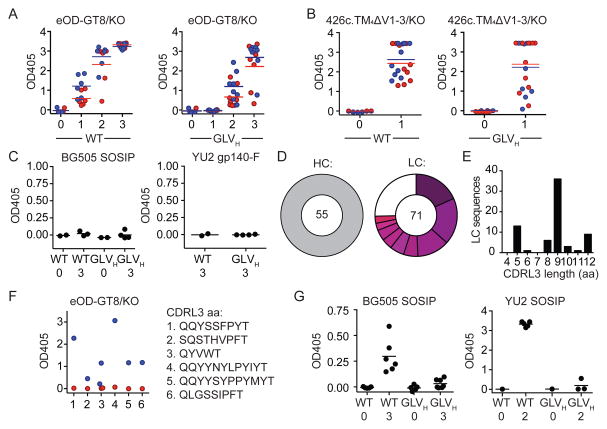
Like most other potent bNAbs, reversion of the somatic mutations in the 3BNC60 antibody to the predicted germline antibody results in complete loss of affinity and neutralizing activity for all recombinant HIV-1 Env proteins and viruses tested (Hoot et al., 2013; Jardine et al., 2013; McGuire et al., 2013; Mouquet et al., 2010; Scheid et al., 2011; Xiao et al., 2009) (Figure S2A and S2B). To determine whether naive GLVH B cells can be stimulated with antigens that are specifically engineered to activate cells expressing germline VRC01-class antibodies (Figure S2C), we immunized these mice and wild type C57Bl6 controls with eOD-GT8 60mers or with multimerized 426c.TM4ΔV1-3 ((McGuire et al., 2014) and see methods). Immunization produced a robust antibody response in wild type C57Bl6 and GLVH mice after one or two immunizations (Figure 2A and B and Figure S3A and S3B). Although the response was not restricted to the CD4bs, as determined by ELISA, sera from the eOD-GT8 60mer-immunized mice showed some preference for the wild type vs. a control eOD-GT8 CD4bs knock-out protein (Figure 2A and Figure S3A). Finally, none of the sera from the eOD-GT8 60mer-immunized mice showed cross-reactivity to a native-like BG505 SOSIP (Julien et al., 2013; Sanders et al., 2013) or a YU2 gp140 foldon trimer (YU2 gp140-F) (Yang et al., 2002) (Figure 2C and Figure S3C).
Figure 2. Antibody responses to immunization by GLVH mice.
(A) Graphs show ELISA results of serum (1/900 dilution) for individual mice against eOD-GT8 (blue) and eOD-GT8 CD4bs knock-out (KO, red) from wild type C57Bl6 (WT) and GLVH mice. Naive serum (0) and serum after one, two or three (1, 2 or 3) immunizations with eOD-GT8 60mers. (B) Graphs show ELISA results of serum (1/900 dilution) for individual mice against 426c.TM4ΔV1-3 (blue) or 426c.TM4ΔV1-3 CD4bs knock-out (KO, red) from wild type C57Bl6 (WT) and GLVH mice. Naive serum (0) and serum after one (1) immunization with multimerized 426c.TM4ΔV1-3. (C) Graphs show ELISA results of serum (1/300 dilution) for individual mice against BG505 SOSIP and YU2gp140-F in naive mice (0) or after three immunizations (3) with eOD-GT8 60mers. (D) Heavy (HC) and light chain (LC) sequences from single antigen binding B cells isolated from four eOD-GT8 60mer-immunized GLVH mice. Pie charts as in Figure 1A. A clone is defined by identical V gene and similar CDRL3. (E) CDRL3 amino acid (aa) lengths from LCs in (D). (F) Graph shows ELISA results for monoclonal antibodies (5μg/ml) cloned from eOD-GT8 60mer-immunized GLVH mice against eOD-GT8 (blue) and eOD-GT8 CD4bs knock-out (KO, red). (G) Graphs show ELISA results of serum (1/300 dilution) for individual mice against BG505 SOSIP or YU2 SOSIP in wild type C57Bl6 (WT) and GLVH mice after immunization with BG505 SOSIP or YU2 SOSIP respectively. Naive serum (0) and serum after two-(2) or three (3) immunizations. See also Figure S2, Figure S3 and Table S1.
eOD-GT8-binding IgG+ B cells are rare in GLVH naive mice but increase to 5.8% on average after immunization (Figure S3D). A fraction of these cells appeared to express CD4bs-specific antibodies as measured by flow cytometry (Figure S3E). To further characterize the B cell responses to eOD-GT8 60mers in GLVH mice, we sorted single cells that bound to eOD-GT8, but not eOD-GT8 CD4bs knock-out, and cloned their antibody genes (Figure S3E). 71 light and 55 heavy chain sequences were obtained from 4 mice. As expected, all of the heavy chains were GLVH. The 71 antibodies include 8 clones ranging in size from 2–13 clonal members, one of which appeared in more than one mouse. In addition there were 13 cells that carried unique sequences (Figure 2D, Figure S3F and Table S1A). The CDRL3 lengths of the light chains showed a near normal distribution when compared to naive GLVH or control wild type C57Bl6 mice, with only one of the expanded clones showing the 5 residue CDRL3 signature typical of 3BNC60 and related antibodies (Figure 2E, Figure S3G and Table S1A).
Out of 20 antibodies cloned from B cells expressing antibodies that bound to eOD-GT8 but not eOD-GT8 CD4bs knock-out by FACS, 7 were CD4bs specific as measured by ELISA (Figure 2F). However, none of the 17 monoclonal antibodies tested, neutralized any of the tier 1 and 2 viruses tested (Table S1B). We conclude that the eOD-GT8 60mer, an antigen designed to activate B cells expressing germline 3BNC60 antibody, and select only for early somatic mutations, elicits B cell responses in mice that carry germline knock-in antibody heavy chains including B cells expressing antibody light chains with 5-residue CDRL3s, but these antibodies do not neutralize HIV-1.
To determine whether GLVH B cells can respond to a more native appearing HIV-1 antigen in vivo, we immunized these mice and wild type C57Bl6 mice with BG505 SOSIP (Julien et al., 2013; Sanders et al., 2013) or YU2 SOSIP. Wild type C57Bl6 mice developed detectable antibody responses as measured by ELISA. In contrast, GLVH mice failed to respond to these antigens (Figure 2G and Figure S3H). We conclude that naive B cells expressing GLVH paired with a heterogenous group of mouse light chains fail to mount significant immune responses to BG505 SOSIP or YU2 SOSIP.
Immune responses in 3BNC60 MuVH knock-in mice
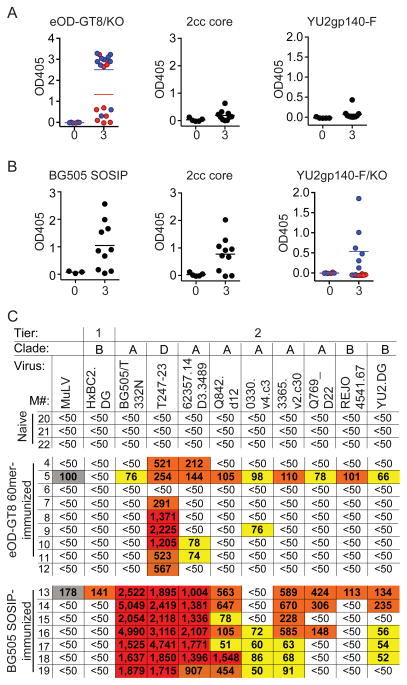
Serum from naive MuVH mice has no detectable HIV-1 Env-binding activity in ELISA or neutralizing activity in TZM-bl assays (Figure 3A–C and Figure S4A–B). To determine whether anti-HIV-1 antibodies can be elicited in these mice they were immunized three times, with eODGT8 60mers or BG505 SOSIP. The eOD-GT8 60mer immunization produced strong serologic responses in MuVH mice but the sera were not cross-reactive against other HIV-1 proteins such as 2cc-core (an engineered gp120 (Dey et al., 2009)) or YU2 gp140-F, and a fraction of the mice showed CD4bs specificity as measured by ELISA on eOD-GT8 and eOD-GT8 CD4bs knock-out (Figure 3A and Figure S4A). MuVH mice also developed strong serologic responses to BG505 SOSIP, but in contrast to eOD-GT8 immunization, there was serologic cross-reactivity to 2cc core, as well as YU2 gp140-F. Moreover, a significant fraction of the response was CD4bs specific as determined by ELISA against YU2 gp140-F and a YU2 gp140-F CD4bs knock-out mutant protein (Figure 3B and Figure S4B).
Figure 3. Antibody responses to immunization by MuVH mice.
(A) Graphs show ELISA results of serum (1/300 dilution) for individual mice against eOD-GT8 (blue) and eOD-GT8 CD4bs knock-out (KO, red), 2cc core and YU2 gp140-F. Naive serum (0) and serum after three (3) immunizations with eOD-GT8 60mer. (B) As in (A) but after BG505 SOSIP-immunization. ELISA against BG505 SOSIP, 2cc core and YU2 gp140-F (blue) and YU2 gp140-F CD4bs knock-out (KO, red). (C) Neutralizing activity in TZM-bl assays of serum from individual naive, eOD-GT8 60mer- or BG505 SOSIP -immunized mice (M) against a panel of HIV-1 viruses. Numbers indicate the reciprocal dilution of serum at the median inhibitory concentration (IC50): red, >1000; orange, 100–1000; yellow, 50–100 and white, was not neutralized at any dilution tested. Grey indicates background activities against the control MuLV virus. See also Figure S4.
To determine whether MuVH mice develop HIV-1 neutralizing responses we assayed mouse serum on a panel of 9 HIV-1 viruses (8 tier 2, and 1 tier 1) in TZMbl-assays (Li et al., 2005). We found that the eOD-GT8 60mer-immunized MuVH mice only showed modest levels of activity mainly against tier 2 Clade D (T247-23) and A (62357.14.D3.3489) viruses that lack glycosylation at position 276 (Genebank entry ACD63071 and ABY50658.1 respectively) (Figure 3C). In contrast, all seven out of ten BG505 SOSIP-immunized MuVH mice that showed a robust response to BG505 SOSIP in ELISA, developed strong neutralizing responses against the autologous Clade A virus. In addition, high levels of neutralization were also evident against T247-23, and 62357.14.D3.3489, and more modest levels of neutralization were seen against three other clade A tier 2 viruses. Finally, four of the seven also showed a low level of neutralizing activity against the clade B tier 2 strain YU2.DG (Figure 3C). Thus, BG505 SOSIP trimers elicit potent responses with some cross-clade breadth in MuVH mice but eOD-GT8 60mer does not.
eOD-GT8-binding IgG+ B cells cannot be detected in naive MuVH mice but increase to 15% and 30% after immunization with eOD-GT8 60mers or BG505 SOSIP, respectively (Figure S4C). Thus, both antigens were able to induce clonal expansion and antigen-specific memory B cells. To characterize the antibodies that develop in the MuVH mice, we isolated and cloned the antibodies from antigen-specific memory B cells (Figure S4D and S4E).
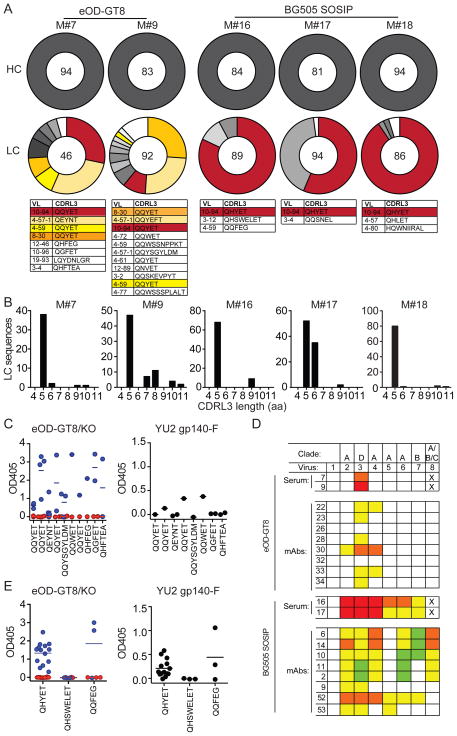
As expected, all of the heavy chains obtained carried the MuVH knock-in sequence. A total of 138 and 269 light chains were cloned from mice immunized with eOD-GT8 60mers and BG505 SOSIP respectively (Figure 4A, Table S2 and S4). The antibodies obtained from eOD-GT8 60mer-immunized mice represent 15 independent clones ranging in size from 2–24 members, 4 of the clones appeared independently in both eOD-GT8 60mer-immunized mice. In addition there were 13 cells that carried unique sequences (Figure 4A and Table S2). In contrast to the antibodies obtained from eOD-GT8 60mer-immunized GLVH mice, the light chains from MuVH immunized mice were highly biased to express 5 aa-residue CDRL3s. Moreover, the fourth residue in the CDRL3 was highly selected for an E, which is an important residue in the 3BNC60 antibody and other VRC01 class antibodies. It forms a contact with the backbone of the V5 loop of HIV-1 Env (Scheid et al., 2011; Wu et al., 2010; Zhou et al., 2013) (Figure 4A and B and Table S2). When assayed by ELISA, the majority of these antibodies were CD4bs specific to eOD-GT8, and a fraction was cross-reactive to YU2 gp140-F (Figure 4C). However, despite CD4bs specificity, only two viruses (T247-2 and 62357.14.D3.3489, which lack glycosylation at amino acid position 276) were neutralized by the monoclonal antibodies tested (Fig 4D and Table S3). Thus, eOD-GT8 60mer-immunization of MuVH mice is highly selective for a subset of mouse light chains with short CDRL3s that allow binding to the CD4bs of eOD-GT8, but this is not sufficient to produce broad neutralizing activity against tier 2 viruses.
Figure 4. Monoclonal antibodies from immunized MuVH mice.
(A) Pie charts show heavy (HC) and light (LC) sequences cloned from two eODGT8 60mer-immunized (M#7 and 9) and three BG505 SOSIP-immunized (M#16, 17 and 18) MuVH mice. VL gene usage and CDRL3 sequences are shown for each clone, colors correspond to colors in pie charts. The same color indicate clones shared by different mice. A clone is defined by identical V gene and similar CDRL3. (B) CDRL3 amino acid (aa) lengths of the LCs in (A). (C) Graphs show ELISA results for individual monoclonal antibodies with the indicated CDRL3s cloned from eOD-GT8 60mer-immunized MuVH mice against eOD-GT8 (blue) and eOD-GT8 CD4bs knock-out (KO, red) (at 0.55 μg/ml) or against YU2 gp140-F (at 5 μg/ml). (D) Neutralizing activity of a set of representative eOD-GT8 60mer- or BG505 SOSIP- elicited monoclonal antibodies (mAbs) against 7 HIV-1 tier-2 viruses (1=MuLV control, 2=BG505/T 332N, 3=T247-23, 4=62357.14.D3.3489, 5=Q842.d12, 6=3365.V2.c30, 7=YU2.DG, 8=0815.v3.c3) compared to the serum of the mouse they were cloned from. Colors for mAbs indicate concentration of mAb at the median inhibitory concentration (IC50): orange, 0.1 to 1 μg/ml; yellow, 1 to 10 μg/ml; green, 10–25 or 50 μg/ml and white, not neutralized at any concentration tested. Lowest dilution tested for serum was 1:50, highest concentration tested for mAbs were between 10–50 μg/ml (see Table S3 and S5). X indicates sample not tested. The numbering of the antibodies corresponds to the numbering of the antibodies in Table S3 and S5. Colors and numbering for serum as in Figure 3C. (E) Graphs show ELISA results for individual mAbs with the indicated CDRL3s cloned from BG505 SOSIP-immunized MuVH mice against eOD-GT8 (blue) and eOD-GT8 CD4bs knock-out (KO, red) (at 0.55 μg/ml) or against YU2 gp140-F (at 5 μg/ml). See also Figure S4 and Table S2, S3, S4 and S5.
Antibodies cloned from BG505 SOSIP-immunized MuVH mice were far more clonally restricted than those obtained after eOD-GT8 60mer immunization. Light chains were cloned from 269 cells from 3 separate mice, and again, in all cases analyzed the heavy chain was from the knock-in. In contrast to eOD-GT8 60mer, BG505 SOSIP-immunization was dominated by only six highly expanded clones, one of which arose independently in all three mice (Figure 4A and Table S4). Similar to eOD-GT8 60mer-immunization, the B cell clones expanded by BG505 SOSIP-immunization were highly biased to express light chains with 5 aa-residue CDRL3s that carry the E at the fourth position of CDRL3 (Figure 4A and B and Table S4) When tested by ELISA, all but 4 of the 20 antibodies elicited by BG505 SOSIP were CD4bs specific on eOD-GT8 and a fraction of these were cross-reactive to YU2 gp140-F (Figure 4E). Finally, some of these monoclonal antibodies showed neutralizing activity against several tier 2 viruses (Figure 4D and Table S5). We conclude that in contrast to eOD-GT8−, BG505 SOSIP-immunization elicits neutralizing antibodies against a diverse group of tier 2 viruses in MuVH mice.
Somatic mutations
Only the antibodies arising from BG505 SOSIP-immunization neutralized tier 2 viruses with an intact glycosylation site at position 276 (Figure 4D, Table S3 and S5). To gain further insights into this phenomenon we analysed the monoclonal antibodies and somatic mutations that arose during the immunization.
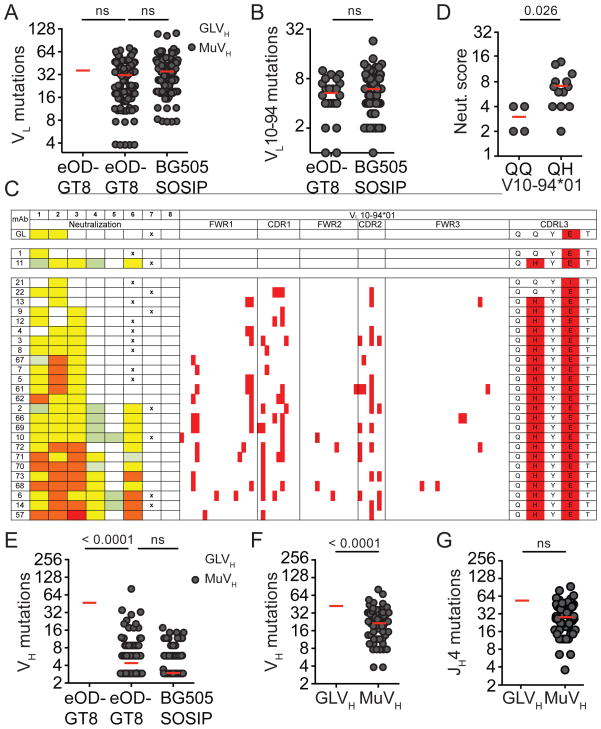
The total number of light chain mutations arising as a result of immunization by the eOD-GT8 60mer or BG505 SOSIP was similar in both strains of mice, irrespective of whether the knock-in heavy chain was germline or mature (Figure 5A). In addition, both immunogens selected for B cells bearing the same VL germline gene (VL10-94*01) carrying similar number of somatic mutations and nearly identical CDRL3s in MuVH mice (Figure 4A, 5B, Table S2 and S4). Thus, the nature of the immunogen did not influence the rate of somatic mutations in the light chains.
Figure 5. Somatic mutations in immunized mice.
(A) Graph shows number of light chain (LC) somatic hypermutations in individual sequences obtained after eOD-GT8 60mer- or BG505 SOSIP immunization in GLVH or MuVH mice. (B) LC somatic hypermutations in VL10-94*01 in individual sequences obtained after eOD-GT8 60mer- or BG505 SOSIP immunization in MuVH mice. (C) VL10-94*01 germline (GL) and 17 monoclonal antibodies (mAbs) from immunized MuVH mice sorted by neutralization strength. Neutralization data for each antibody tested against virus 1=T247-23, 2=62357.14.D3.3489, 3=BG505/T332N, 4=3365.v2.c30, 5=YU2.DG, 6=0815.v3.c3, 7=HxBC2.DG and 8=MuLV. Colors for mAbs indicate concentration of mAb at the median inhibitory concentration (IC50): red, <0.1 μg/ml; orange, 0.1 to 1 μg/ml; yellow, 1 to 10 μg/ml; green, 10–25 or 50 μg/ml and white, not neutralized at any concentration tested (see also Table S3 and 5). Position of amino acid mutations in the LCs compared to germline are highlighted in red. X indicates sample not tested. (D) Graph shows neutralization score for VL10-94*01 cloned antibodies with Q90Q or Q90H CDRL3 from immunized MuVH mice. Neutralization score: sum of viruses tested IC50 0.1–1 μg/ml: 3, 1–10 μg/ml: 2, 10–50 μg/ml: 1 and >50 μg/ml: 0. (E) Somatic hypermutations in individual heavy chain sequences obtained after eOD-GT8 60mer- or BG505 SOSIP-immunization in GLVH or MuVH mice. Data in (A), (B) and (E) are pooled from 9 mice (M#1, 2, 3, 4, 7, 9, 16, 17 and 18). (F) Somatic hypermutations in individual germinal center B cell heavy chain (HC) sequences obtained from naive GLVH or MuVH mice. (G) Somatic hypermutations in individual germinal center B cell JH4 intron sequences obtained from naive GLVH or MuVH mice. Data in (F) and (G) are pooled from 2 independent experiments with 2–3 mice each. 80 clones were sequenced for GLVH or MuVH germinal center B cells HC and JH4. See also Figure S5, Table S3 and 5.
However, some mutations in VL10-94*01-expressing B cells appeared to be specifically selected by immunization with BG505 SOSIP because they were over-represented (Figure S5A). For example, Q90H was found in the 2nd position of CDRL3 of nearly all members of the VL10-94 clone obtained independently from three MuVH mice immunized with BG505 SOSIP but rarely in the VL10-94 clone that arose after immunization with eOD-GT8 60mers (Figure 4A, Table S2 and S4, Figure S5A). While others such as the E mutation found in the 4th residue of CDRL3 was selected by both immunogens (Figure 4A and Table S2 and S4). Thus immunization with BG505 SOSIP and eOD-GT8 60mers led to differential selection of somatically mutated light chains.
To determine whether light chain mutations might be responsible for increasing neutralizing activity after BG505 SOSIP immunization, we produced an antibody that carries the MuVH and the predicted germline light chain of VL10-94*01 (GL in Figure 5C, Table S3 and S5). When this antibody was compared to BG505 SOSIP-derived monoclonal antibodies (mAbs), the reverted antibody showed only modest activity. The reverted antibody was similar to the eOD-GT8 60mer-derived VL10-94*01 mAbs indicating that only BG505 SOSIP-immunization selects for light chain somatic mutations that increase neutralizing antibody activity in MuVH mice (Figure 5C). Moreover, the Q90H found in the CDRL3 of nearly all VL10-94-expressing clones derived from BG505 SOSIP-immunized MuVH mice was sufficient to increase neutralizing activity (Figure 5C and D, Table S3 and S5). Additional mutations that further enhanced or interfered with neutralizing activity appeared randomly (Figure 5C). Finally, there was no evidence for selection of specific mutations in the heavy chain in MuVH B cells as seen by the absence of significant clonal expansion of any particular mutation (Figure S5B).
In contrast to the light chains, mutations in the knock-in VDJH sequence were significantly higher in GLVH than in MuVH B cells (Figure 5E and Figure S5B) even when comparing Peyer’s Patch germinal center B cells in unimmunized mice that are not responding to injected antigens (Figure 5F). However, the mutation rate in the germline intron downstream of JH4, which is contiguous to the VDJH, was similar in the two mouse strains (Figure 5G). We conclude that the somatic mutation machinery is equally active in the two knock-in B cell types, but the highly mutated MuVH is less susceptible to mutation than its unmutated GLVH counterpart.
Discussion
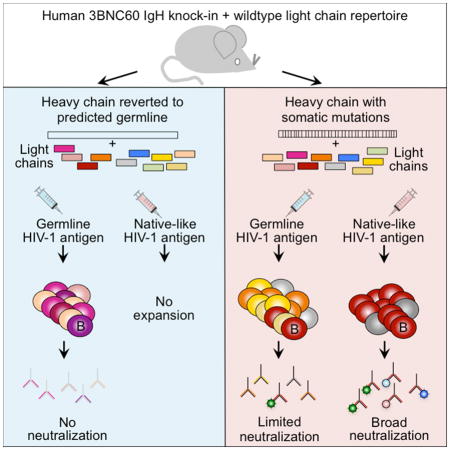
Immunoglobulin knock-in mice such as GLVH and MuVH provide a unique opportunity to examine the effects of specific immunogens and trace the evolution of an anti-HIV-1 antibody starting from predicted germline or from a synthetic intermediate composed of a mutated human heavy V region chain and a mouse germline light chain. The B cells in these mice show reduced repertoire diversity because the heavy chain is fixed, nevertheless the light chains are free to recombine randomly and the resulting B cell repertoire contains only very rare cells that bind to the HIV-1 immunogens tested.
Immunization revealed that B cells bearing predicted germline antibodies only responded to antigens specifically designed to bind to these antibodies, and not to more native appearing HIV-1 Env trimers. This result is consistent with the observation that the germline version of 3BNC60 and related antibodies fail to bind to nearly all tested recombinant HIV-1 Env antigens and supports the idea that specifically designed antigens will be required to elicit B cells bearing these antibodies (Hoot et al., 2013; Jardine et al., 2013; McGuire et al., 2013). Nevertheless, it remains possible that a more diverse repertoire of light chains would include a rare pairing that might be triggered by the native like BG505 SOSIP trimer.
One of the objectives of immunization with a germline-targeting antigen is to expand and further diversify rare clones of B cells that can serve as precursors of bNAbs. For 3BNC60 and related CD4bs antibodies this would require selection of B cells expressing antibodies with short CDRL3s and somatic hypermutation in germinal centers. Although rare in the pre-immune repertoire, GLVH B cells expressing antibodies with short CDRL3s were enriched after immunization with the eOD-GT8 60mer, providing proof of concept for selection of germline precursors of this class of antibodies by germline targeting antigens. In contrast both the engineered antigens and the native-like BG505 SOSIP trimers elicited antigen-specific B cell responses in mice bearing MuVH. However, only BG505 SOSIP produced a significant neutralizing response.
Both types of antigens selected rare MuVH B cells expressing light chains with short CDRL3s and both induced similar levels of somatic hypermutation. It would be interesting to determine whether these cells would also be selected when competing with a complete repertoire of B cells in a wild type mouse. Nevertheless, the responses to the immunogens differed in two important ways each of which likely contributed to the eventual selection of tier 2 neutralizing antibodies to HIV-1 by the BG505 SOSIP trimer. First, the repertoire of light chains that was selected to undergo clonal expansion by the trimer was far more restricted than for the eOD-GT8 60mer. This restriction is likely to be related to the glycan at position 276 that enforces the relatively narrow geometry of the recessed CD4 binding pocket in the trimer (Julien et al., 2013; Kwong et al., 1998; Lyumkis et al., 2013). Under physiologic conditions this cavity accommodates the single terminal Ig-like domain in CD4. Significant alterations including somatic mutations, deletions, or insertions must be made for the bulkier paired heavy and light chain Ig domains in 3BNC60 and related antibodies to access the CD4bs (West et al., 2014). Many of these alterations are already present in the heavy chain of MuVH B cells, facilitating extensive contacts with gp120 (West et al., 2014). Nevertheless, the steric restrictions imposed by the glycan at 276 in BG505 SOSIP limit the number of different light chains that can be recruited into the immune response against the trimer. In contrast, eOD-GT8 was designed to be a more open structure that is missing the glycan at position 276 and can accommodate germline antibodies. It therefore recruits a more diverse but overlapping set of antibodies in mice bearing the MuVH, none of which has significant breadth against tier 2 viruses in TZM-bl assays.
The second difference between the two immunogens was in their ability to select for IgVL mutations that enhance breadth and potency. In particular the Q90H mutation in the CDRL3 of the VL10-94 light chains that are selected in the BG505 SOSIP immunized mice may aid in rigidifying the light chain contacts to the CD4bs. Structural modeling of the VL10-94 light chain indicates that the histidine side chain, unlike the glutamine side chain, can adopt a commonly observed, low energy conformation that also makes stabilizing interactions within the light chain (Figure S5C). The planar packing with Y91, the close proximity to the N276 glycan, a hydrogen bond with CDRL1 and a potential for interacting with a gp120-bridging water molecule may help preconfigure the antibody for engagement of the CD4bs. The modeling and neutralization data indicate that the mutation could cause structural changes that influence binding to gp120 and may be necessary for the activity of the BG505 SOSIP elicited antibodies (Figure S5C).
Although only a small number of the mutations are likely to be crucial for breadth and potency, achieving the right combination of amino acid substitutions by random mutagenesis is a low probability event. Finding that the mature, fully mutated 3BNC60 VH suffers fewer mutations after immunization than the predicted germline 3BNC60 despite equal rates of somatic mutation in the light chains and in the JH4 intron suggests that the sequence of the VH is an important determinant of its susceptibility to hypermutation. Consistent with this idea, the fully mutated 3BNC60 VH has many fewer AID target sites than the predicted germline VH(Betz et al., 1993; Longerich et al., 2006; Neuberger et al., 1998). RGYW/WRCY are preferred targets for somatic hypermutation: 9 out of 12 RGYW and 7 out of 8 WRCY motifs found in GLVH sequence are missing in MuVH (Figure S5D).
HIV-1 and SHIV infection can be prevented by passive infusion of potent bNAbs into mice or macaques, but despite significant efforts, including large clinical trials, all attempts to elicit such antibodies in humans by immunization have failed to date (Klein et al., 2013; Mascola and Haynes, 2013; West et al., 2014) Antibody cloning experiments revealed that these bNAbs differ from most other antibodies in that they are significantly more mutated, leading to the suggestion that eliciting them would require immunization with a sequential series of different antigens selected or designed to shepherd the antibody response through several successive rounds of mutation and selection in germinal centers (Klein et al., 2013; Mouquet et al., 2010; Scheid et al., 2009). Consistent with this notion, elegant studies of human HIV-1 infection show a correlation between HIV-1 viral evolution and the antibody response (Doria-Rose et al., 2014; Liao et al., 2013; Wu et al., 2015). Our experiments demonstrate that specific immunogens are required to target B cells at different stages of evolution of the anti-HIV-1 neutralizing response and lend support to the idea that vaccination for HIV-1 will require a strategy that involves sequential immunization with different antigens.
Experimental procedures
Mice and immunizations
Knock-in mice were produced by gene targeting using the human VDJH sequences of the mature and germline 3BNC60 heavy chain (Hoot et al., 2013). The targeting vector was designed with homologous regions flanking the endogenous J segments, which results in the deletion of the J segments and thereby minimizes rearrangement of the wild type locus (Pelanda et al., 1997; Shih et al., 2002). Mice were immunized once every two weeks with 10 μg of protein in Alum Imject (Thermo Scientific). Serum was collected two weeks after each immunization. All experiments were performed according to the protocols approved by the IACUC at Rockefeller University.
Protein production
eOD-GT8 is an improved version of eOD-GT6 (Jardine et al., 2013). The eOD-GT8 CD4bs knock-out carries mutations to reintroduce the N276 glycan and also includes D368R and N279A. eOD-GT8 60-mers for immunization and monomers for ELISA detection were produced as previously described (Jardine et al., 2013). The 426c.TM4ΔV1-3 immunogen (+/− AviTag) is a derivative of the 426c.NLGS.TMΔV1-3 (McGuire et al., 2014) with the following additional modifications: D276N, S278R, G471S (HXB2 numbering). The protein was produced as previously described (McGuire et al., 2014). Multimerized 426c.TM4ΔV1-3 was generated as follows; Avi-tagged and biotinylated 426c.TM4ΔV1-3 was mixed with streptavidin and biotinylated dextran (Life Technologies) at a 3:1 ratio of Env to biotin, with the assumption that the biotin had 77 biotin molecules/dextran. Streptavidin (New England Biolabs) was added to achieve a 3:1:1 Env to streptavidin to biotin ratio. To generate YU2 SOSIP.664, a gene encoding YU2 SOSIP.664 was constructed to include the ‘SOS’ substitutions (A501Cgp120, T605Cgp41), the ‘IP’ substitution (I559Pgp41); changing the gp120-gp41 cleavage site to 6R (REKR to RRRRRR), and introducing a stop codon after residue 664gp41 (HXB2 numbering) analogous to mutations introduced to generate BG505 SOSIP.664 as described (Sanders et al., 2013). YU2 SOSIP.664, BG505 SOSIP.664 and B41 SOSIP.664 (referred to as YU2 SOSIP, BG505 SOSIP and B41 SOSIP) were produced and purified by the 2G12/SEC method as previously described (Chung et al., 2014; Pugach et al., 2015; Sanders et al., 2013). YU2 gp140-F CD4bs knock-out is a YU2 gp140-foldon (F) HIV-1 Env with A281T and D368K mutations and proteins were produced as previously described (Forsell et al., 2008).
ELISA
ELISA for eOD-GT8, eOD-GT8 CD4bs knock-out, YU2 gp140-F, YU2 gp140-F knock-out, YU2 SOSIP and B41 SOSIP was performed by coating high binding 96-well plates (Corning Incorporated) with 200 ng/well of protein. After incubation overnight (ON) at 4 °C the plates were washed in wash buffer (PBS with 0.05% TWEEN 20 (Sigma)) and incubated in blocking buffer (PBS with 2% milk). Serum samples or monoclonal antibodies were added in dilutions. Plates were washed and secondary antibody, HRP conjugated anti-mouse or anti-human IgG (Jackson Immuno Research), was added. Plates were developed by the addition of HRP substrate (Life Technologies) and the absorbance was measured at 405 nm. ELISA specific for BG505 SOSIP was performed using the D7324-capture ELISA as described in (Sanders et al., 2013).
TZM-bl neutralization assay
Serum samples and monoclonal antibodies were tested for neutralization against a panel of selected HIV-1 pseudoviruses using a TZM-bl neutralization assay as previously described (Li et al., 2005).
B cell enrichment
B cells were enriched from single cell suspensions of total splenocytes prior to flow cytometry staining by magnetic bead separation using anti-CD19 or anti-CD43 MicroBeads (Miltenyi Biotec). The separation was performed on MACS separation LS columns according to the manufacturers instructions.
Flow Cytometry
Single cell suspensions of total splenocytes, enriched B cells or BM were stained with different combinations of the following antibodies: Anti-CD4 APC-eFluor 780, anti-CD8 APC-eFluor 780, anti-Gr1 APC-eFluor 780, anti-F4/80 APC-eFluor 780, anti-B220 APC, anti-B220 APC-eFlour 780, anti-B220 FITC, anti CD19 PeCy7, anti-CD38 Alexa Fluor 700, anti-CD93 APC, anti-IgM PerCP-eFluor 710, anti CD21/CD35 eFluor 450 (eBiosciences), anti-CD23 PE (BioLegend), anti-CD4 PE-CF594, anti-CD8 PE-CF594, anti-Ly-6G and Ly-6C PE-CF594 and anti-IgG1 BV421 (BD biosciences). Live dead aqua stain was added to separate dead cells (Life Technologies) and eOD-GT8-specific cells were visualized by the addition of FITC-conjugated eOD-GT8 and PE-conjugated eOD-GT8 CD4bs knock-out. BG505 SOSIP- (Sok et al., 2014) and 2cc-specific memory B cells were visualized by the addition of biotinylated protein with the addition of streptavidin conjugated PE and APC respectively (BD Biosicences).
Single B cell sorting
Staining and single-cell sorting of naïve and memory B cells was performed as follows, for naïve mice CD4−, CD8−, Gr-1−, F4/80−, B220+ and IgM+ B cells were sorted. For immunized mice CD4−, CD8−, Gr-1−, F4/80−, B220+, CD38−, IgM−, IgG+, eOD-GT8 CD4bs knock-out- and eOD-GT8+ or CD4−, CD8−, Gr-1−, F4/80−, B220+, CD38−, IgM−, IgG+, BG505 SOSIP+ and 2cc+ memory B cells were single cell sorted into 96-well plates using a FACSAria III sorter (Becton Dickinson). The cells were lysed with 4 μl lysis buffer containing RNASin (Promega) 40 U/μl (0,3 μl), DPBS (Dulbecco) 10 x (0.2 μl), DTT (Invitrogen) 100 mM (0.4 μl) and nuclease-free water (3.1 μl). The sorted plates were stored at −80 °C until further processing (Tiller et al., 2009). Sequencing and cloning primers are listed in Table S6. Antibodies were cloned using a modified sequence and ligation-independent cloning (SLIC) approach as described in (Li and Elledge, 2007). Antibodies were produced in HEK 293-6E cells as previously described (Klein et al., 2014).
Structural modeling
To create a structural model of one of the antibodies elicited in the MuVH mouse by BG505 SOSIP, we integrated information from HIV bNAb structures and mouse light chain structures. The antibody 3BNC117 has very high sequence similarity to 3BNC60 (10 mutations in heavy chain and 3 mutations in light chain (Scheid et al., 2011) and has a co-crystal structure available (PDB id: 4JPV). We made an initial model of 3BNC60 by running 50 ROSETTA-Fixbb (Leaver-Fay et al., 2011) simulations to model the mutations from the 3BNC117 structure. The elicited antibodies contain a mutated mouse light chain from the VL10-94*01 germline gene, here we focused on the Q90H mutation. To model the light chain, we used an unrelated crystal structure of a mouse VL10-94*01 antibody (PDB id: 1EMT). We superimposed the mouse antibody to the 3BNC60 model and created a hybrid of 3BNC60-mature heavy chain and VL10-94*01 light chain to mimic what was elicited in MuVH. The 1EMT structure did not contain a 5 amino acid CDRL3, so we aligned the light chain from 3BNC117 and grafted the 5 amino CDRL3 onto our model. The final model was obtained by adding an interfacial water molecule (as found in the structurally similar VRC-CH31 light chain, PDB id: 4LSP) and by running 100 ROSETTA-Relax simulations to allow for slight movements of the backbone.
Statistical analysis
Statistical differences were analysed by the Mann-Whitney test. GraphPad Prism software was used for analysis and data were considered significant at * for p≤0.05, ** for p≤0.01 and *** for p≤0.001.
Supplementary Material
Acknowledgments
We thank Thomas Eisenreich, David Bosque and Susan Hinklein for assistance with the mice, Klara Velinzon and Neena Thomas for single cell FACS sorting, Zoran Jankovic for laboratory support, Alex Robles for technical assistance with the HIV-1 TZMbl neutralization assays and Anthony West for advice regarding the selection of the HIV-1 virus panels. This work was supported by grants from NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) UM1 AI100663 (M. C. N., W. R. S., J. P. M. and R. W. S.), Bill and Melinda Gates Foundation grant OPP1033115 (M. C. N.) and NIH grant AI037526 (M.C.N). This work was also supported by NIH grant U19 AI109632 (M. C. N. and L. S.), HIVRAD/P01 AI094419-01 (L. S.), HIVRAD/P01 AI082362 (J. P. M. and R. W. S), HIVRAD/P01 AI100148 (P.J.B. and M.C.N.), Bill and Melinda Gates Foundation grant 1032144 (M. S. S.) and the Aids Fonds Netherlands, grant #2012041 (R. W. S.). P.D. is supported by an international postdoctoral fellowship from the Swedish Research Council. L.v.B. is supported by The Rockefeller University Center for Clinical and Translational Science grant UL1 TR000043/KL2TR000151 from the National Center for Advancing Translational Sciences (NCATS). A.D.G. is supported by MSTP grant T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. A. M. is supported by a Canadian Institutes of Health Research Fellowship. R.W.S. is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research (NWO) and a Starting Investigator Grant from the European Research Council (ERC-StG-2011–280829-SHEV). M.C.N. and P. J. B. are Howard Hughes Medical Institute Investigators.
Footnotes
Author contributions
P. D. and L. v. B. planned and performed experiments, analyzed data and wrote the manuscript. A. E. performed experiments and analyzed data. J. J. and D. W. K. generated the eOD-GT8 proteins. N. T. F., A. D. G. and J. P. generated the GLVH and MuVH mice and performed and provided technical advice regarding the characterization of the mice. A. T. M. and M. D. G generated the 426c.TM4ΔTM1-3 proteins. T. O. generated the antibody database. L.S. generated the YU2 SOSIP protein. A. C. and M. J. v. G. produced the BG505- and B41 SOSIP proteins. K.- H. Y. performed experiments, C. L. and A. G produced monoclonal antibodies. M. S. S. planned and analyzed neutralization experiments. P. J. B., R. W. S and J. P. M. critically read and contributed to the manuscript preparation. L. S., W. R. S. planned experiments, analysed data and critically read and contributed to the manuscript preparation. M. C. N. planned experiments, analyzed data and wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Betz AG, Neuberger MS, Milstein C. Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunology today. 1993;14:405–411. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NP, Matthews K, Kim HJ, Ketas TJ, Golabek M, de Los Reyes K, Korzun J, Yasmeen A, Sanders RW, Klasse PJ, et al. Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies. Retrovirology. 2014;11:33. doi: 10.1186/1742-4690-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. mAbs. 2010;2:347–356. doi: 10.4161/mabs.2.3.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell MN, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, et al. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods in enzymology. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger MS, Ehrenstein MR, Klix N, Jolly CJ, Yelamos J, Rada C, Milstein C. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Pelanda R, Schwers S, Sonoda E, Torres RM, Nemazee D, Rajewsky K. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Do Kwon Y, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.