Abstract
Grading cervical intraepithelial neoplasia (CIN) determines clinical management of women after abnormal cytology with potential for over-diagnosis and overtreatment. We studied a novel biomarker of HPV life-cycle completion (panHPVE4), in combination with the MCM cell-cycle marker and the p16INK4a transformation marker to improve CIN diagnosis and categorization. Scoring these biomarkers alongside CIN grading by three pathologists was performed on 114 cervical specimens with high-risk (HR-) HPV. Inter-observer agreement for histopathology was moderate (kappa (κ): 0.43 for CIN1/negative, 0.54 for CIN2/≤CIN1, and 0.36 for CIN3). Agreement was good or excellent for biomarker scoring (E4: κ=0.896; 95%CI: 0.763-0.969, p16INK4a: κ=0.798; 95%CI: 0.712-0.884, MCM: κ=0.894; 95%CI: n.c.). Biomarker expression was studied by immunofluorescence and immunohistochemistry and correlated with 104 final CIN diagnoses following histological review. All 25 histologically negative specimens were p16INK4a and panHPVE4 negative although 9 were MCM positive. There were variable extents of p16INK4a positivity in 11/11 CIN1, and extensive panHPVE4 staining in 9/11. Ten CIN2 lesions expressed panHPVE4 and p16INK4a and 13 CIN2 expressed only p16INK4a. CIN3 showed extensive p16INK4a positivity with no/minimal panHPVE4 staining. PanHPVE4, unlike MCM, distinguished CIN1 from negative. PanHPVE4 with p16INK4a separated CIN2/3 showing only expression of p16INK4a indicating transforming HR-HPV E7 expression, from CIN1/2 showing completion of HR-HPV life-cycle by E4 expression and variable p16INK4a expression. PanHPVE4 and p16INK4a staining are complementary markers that could provide simple, reliable support for diagnosing CIN. Their value in distinguishing CIN1/2 that supports HR-HPV life cycle completion (and which might ultimately regress), from purely transforming CIN2/3 needing treatment warrants further research.
Keywords: Human Papillomavirus, Cervical Intraepithelial Neoplasia, biomarkers, E4, p16INK4a, MCM, reproducibility
Introduction
Prevention of cervical cancer based on screening and treatment of cervical intraepithelial neoplasia (CIN) has proved highly effective [1-3]. An important part of clinical management of CIN is histological grading, distinguishing CIN1 lesions that generally regress from CIN2/3 that are currently treated, preferably by excision. Histological diagnosis of CIN is based on subjective interpretation of multiple cellular and architectural neoplastic changes. Effects of inflammation, repair, pregnancy and atrophy complicate the diagnosis of pre-malignant lesions, and the histological grading of CIN is subject to substantial inter- and intra-observer variability [4-6]. CIN2 is the treatment threshold but is not very reproducible [7], and is thought to include a mixture of transient human papillomavirus (HPV) infections and true cancer precursors [8]. The reproducibility of the diagnosis of CIN1 is also very poor. In the ALTS-trial, a quality control panel of pathologists reviewed 2237 colposcopically directed biopsies diagnosed at the local sites. Only 43% of biopsies initially diagnosed as CIN1 were classified as CIN1 after further review with many not considered as CIN at all [9]. Overall, although CIN3 is always considered a true cancer precursor requiring treatment, important clinical decisions based on diagnosis of CIN1 and CIN2 are made on poorly reproducible criteria, leading to extensive follow-up and overtreatment of lesions that would spontaneously regress [10, 11]. In particular it has been suggested that CIN2 in young women should not always be treated by excision [12].
Based on understanding of HPV gene expression during productive HPV infection and in neoplasia [13-18], several molecular and immunohistochemical biomarkers have been proposed for objective grading of CIN lesions. The two most studied are p16INK4a and the proliferation marker ki-67. Over-expression of p16INK4a is caused by up-regulated expression of high-risk (HR)-HPV oncogene E7, and diffuse p16INK4a expression is widely used as a biomarker for HPV induced high-grade (HG)-CIN [19-21]. A recent systematic review and meta-analysis showed that p16INK4a immunostaining correlates with the severity of cytological and histological abnormalities [22]. One limitation is that diffuse basal and parabasal expression is seen in some lesions that are histologically typical CIN1 and management of these is unclear.
Minichromosome maintenance (MCM) proteins are DNA helicases that are essential for genomic DNA replication and restrict replication to once per cell cycle [23]. Several studies have shown MCM to be a proliferation marker [24-27] similar in expression to ki-67 [28-30], which is used widely in the diagnosis of CIN, but is not a specific marker of HR-HPV [31-33]. In normal squamous epithelium MCM staining is limited to the basal and immediate parabasal cell layers. In contrast, MCM is expressed in the upper two-thirds of the epithelium in HG-CIN [34].
HPV E4 protein is expressed in infected squamous cells supporting viral genome amplification [35]. E4 is only expressed in terminally differentiated squamous cells of the intermediate or superficial cell layers of the infected epithelium [36]. With increasing precancerous grade cell differentiation is lost. As a result, transforming HPV infections fail to express the differentiation dependent E4 protein. Therefore, E4 has been suggested as a marker of the onset of the productive stage in the viral life cycle and low-grade lesions [37, 38]. Recently, a mouse monoclonal antibody against the E4 protein of 15 HR-HPV types (panHPVE4) has been developed, and we investigated the potential clinical application of this antibody using immunofluorescence and immunohistochemistry. For our initial studies, we used immunofluorescence microscopy to facilitate visualization of multiple markers in the same tissue section, followed by hematoxylin and eosin staining to confirm routine pathological diagnosis. In clinical practice, biomarker detection will be carried out by immunohistochemistry (IHC) and we show that this performs well for the visualization of E4.
The objective of this project was to study the expression patterns of panHPVE4, p16INK4a and MCM in relation to the classification of routine biopsy specimens with different histological grades of cervical lesions. The study aimed to investigate whether combining the immunohistochemical biomarker panHPVE4 with p16INK4a and/or MCM could provide a more objective and reproducible clinico-pathological classification of cervical precancerous lesions related to current concepts of the biology and natural history of HPV infection than simple histological grading of CIN or use of p16INK4a alone. Such a classification could offer a more standardized system of grading, and describe more simply the complex nature of lesions within each CIN grade. This could provide the basis for further investigations aimed at predicting the likely prognosis of different cervical lesions and identifying appropriate management.
Materials and Methods
Study population
Loop electrosurgical excision procedure (LEEP) and hysterectomy specimens from women treated for CIN at the Jagiellonian University Medical College, Krakow, Poland and cervical biopsy specimens of women obtained during colposcopy at the Gynaecological outpatient clinic of Hospital Clínic, Barcelona, Spain and Reinier de Graaf Groep, Voorburg, the Netherlands were enrolled in this study [39].
Histological diagnosis
Serial paraffin sections were obtained from formalin-fixed and paraffin-embedded (FFPE) histological specimens. Three pathologists independently classified all haematoxylin and eosin (H&E) sections as negative, CIN1, CIN2 or CIN3. The CIN classification was used in order to permit exploration of the biomarker pattern seen in CIN2 and the possibility of further study in relation to controversies over the outcome of CIN1 and CIN2. It is acknowledged that the LAST guidelines indicate a two-tier classification in which CIN2 and 3 are combined as high-grade precancer for safe clinical practice [40]. Only specimens that were adequate for grading according to the pathologists were included. The pathologists were blinded to the HPV status and biomarker results. Specimens with total (3/3 pathologists) and partial (2/3 pathologists) agreement on histological diagnosis were combined into a consensus diagnosis group. Specimens showing discrepant results when the p16INK4a and histological diagnosis were compared, were subjected to additional histological review. As advocated by the LAST guidelines [40] a final diagnosis was made based on further pathological review with knowledge of the consensus diagnosis and the p16INK4a immunohistochemistry results.
HPV detection
DNA from all specimens was isolated by a proteinase K procedure as described previously [41] and HPV DNA detection and genotyping was performed with the SPF10-PCR-DEIA-LiPA25 system (SPF10 HPV LiPA25 version 1; Labo Bio-Medical Products, Rijswijk, The Netherlands) as described elsewhere [42, 43]. DEIA is an ELISA-based hybridization assay detecting at least 54 HPV types using a cocktail of 9 different probes. DEIA positive amplimers were analysed by LiPA25. The LiPA25 can identify 25 HPV genotypes (6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70, and 74) by reverse hybridization on a line probe assay.
DEIA-negative samples were spiked with HPV16 DNA and analysed with type specific PCR to exclude inhibition [44] and if necessary the PCR and DEIA were repeated on a 1/10 diluted sample. With this an additional 5 samples became HPV positive. Each run contained negative and positive controls. Contamination or failure of analyses was not encountered. No additional type-specific testing was done.
PanHPVE4 antibody development and validation
Purified maltose binding protein (MBP)-HPV18 E1^E4 fusion proteins were generated according to the manufacturer’s instructions (New England Biolabs, Beverly, USA). HPV18 E1^E4-MBP fusion proteins were used to immunize female BALB/c mice. Standard enzyme-linked immunosorbent assay (ELISA) was used to select the specific monoclonal antibody (mAb). The ability of this mAb, FH1.1, to detect HPV E1^E4 protein of different HPV types was tested with MBP-E4 fusion proteins prepared from a panel of 10 HPV types (HPV16, 18, 31, 33, 35, 39, 45, 52, 58 and 59) by ELISA and western blotting, and by rafts from NIKS cell lines containing HPV16, 18, 31, 45 and 58 [38]. The FH1.1 mAb was reactive to all these E4 proteins tested by ELISA, western blot analyses and in the HPV-containing rafts. Furthermore, the FH1.1 mAb was assessed on biopsies from cervical lesions containing HPV33, 35, 39, 51, 52, 53, 56, 66, 67 or 70. The FH1.1 mAb stained positively to E4 proteins of all these HPV types with comparable signal strength. The newly generated panHPVE4 mAb FH1.1 is thus capable of detecting at least 16 HR-HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67 and 70 (Zhonglin Wu, National Institute for Medical Research, London, UK - manuscript in preparation). It is anticipated that DDL Diagnostic Laboratory will distribute the validated panHPVE4 mAb (FH1.1). In the first instance, enquiries should be made either to John Doorbar (jd121@cam.ac.uk) or Wim Quint (wim.quint@ddl.nl).
Immunofluorescence
Four-micrometer (μm) thick paraffin sections were cut, slides were dried overnight at 37°C, deparaffinised in xylene and rehydrated in a descending alcohol series. For epitope retrieval, slides were autoclaved in solution D pH6.0 (Dako, Glostrup, Denmark) for 2 min at 121°C. The primary antibodies against E4 (panHPVE4: FH1.1) and MCM (Abcam, Cambridge, UK) were applied 1:100, and incubated overnight at 4°C. Visualization was performed with 150-fold diluted Alexa-488 (green) conjugated anti-mouse secondary antibody against E4 and Alexa-594 (red) conjugated anti-rabbit secondary antibody against MCM (both Invitrogen, Paisley, UK). Nuclear counterstaining was performed with DAPI (blue) (Sigma, St-Louis, MO, USA).
Immunohistochemistry
P16INK4a staining was performed on one 4 μm FFPE-section using heat-induced epitope retrieval with citrate buffer (Dako) and a primary mouse monoclonal antibody anti-p16INK4a clone JC8 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). Reactivity was visualized using the EnVision™ Detection System (Dako) for the biopsies and 3-amino-9-ethylcarbazol (AEC, Sigma, St-Louis, MO, USA) for the LEEP and hysterectomy specimens. A subset of 48 slides was also stained with the panHPVE4 mAb FH1.1 antibody using immunohistochemistry according to the above protocol to allow comparison of immunofluorescence with immunohistochemistry.
Quantification of immunohistochemical results
PanHPVE4 immunoreactivity was scored as (1) negative, (2) superficial - restricted to the upper quarter of the epithelium, (3) extensive - upper half of the epithelium or more.
P16IKN4a immunostaining was classified as (1) no or focal p16IKN4a positivity, (2) diffuse p16IKN4a staining restricted to the lower third of the epithelium, (3) diffuse p16IKN4a positivity more than a third of the epithelium including full thickness staining.
MCM score was classified as (1) basal and parabasal staining only, (2) diffuse staining restricted to the lower third of the epithelium, (3) diffuse MCM positivity more than a third of the epithelium including diffuse full thickness staining.
Scoring was based on the highest category present in a specimen.
A subset of lesional areas (n=102) was scored by two of the researchers (HG and RvB). Discordant scorings were reviewed with an expert pathologist (DJ) and final scoring was determined in consultation. The remaining lesions were scored individually by HG and RvB.
Data and statistical analyses
We excluded cases with HPV types 6, 43 or 68/73, because the panHPVE4 antibody is not validated for these types. Quadratic weighted kappa statistics were used to assess agreement on the four possible histological diagnoses between the pathologists and for the p16INK4a and MCM scoring. Unweighted kappa-values were calculated for dichotomized categories and for the panHPVE4 scoring. Strength of agreement was judged according to Landis and Koch [45]: kappa (κ)<0 as no agreement, 0-0.20 as slight, 0.21-0.40 as fair, 0.41-0.60 as moderate, 0.61-0.80 as substantial, and 0.81-1 as almost perfect agreement. A pathologist annotated regions of interest on the H&E slides. The three pathologists also independently graded all regions. These diagnoses were used to correlate the extent of the E4 expression to the CIN grade. The Chi-square test was used to analyze the association between panHPVE4 positivity and lesion grade and to calculate the relation between the extension of the panHPVE4 expression and the CIN-grade. P-values below 0.05 were considered significant. Immunostaining results of p16INK4a, panHPVE4 and MCM were related to the consensus and final diagnoses.
Results
HPV detection
Initially, 114 specimens were included in the study from patients with a median age of 39 years (range: 19-80 years). HPV was detected in 100 (88%) specimens, with 14 (12%) of the specimens being HPV negative. HPV types 16, 18, 31, 33, 35, 39, 51, 52, 53, 56, 58, 59 and 66 were detected amongst the samples analyzed. The HPV type distribution by diagnosis is shown in Supplemental Digital Content 1. HPV16 was the most prevalent HPV type (n=44; 39%), followed by HPV33 (n=16; 14%), HPV52 (n=11; 10%), and HPV31 (n=9; 8%). Multiple HPV infections were apparent in 11/100 (11%) of the HPV positive biopsies. HR-HPV was detected in 59/61 (94.3%) of women diagnosed with CIN2 or 3. Twelve of the HPV negative specimens were histologically completely negative and two HPV negative specimens were CIN3 at final consensus diagnosis.
Interobserver agreement on histological diagnoses
A total of 104 specimens with a consensus diagnosis were included in our analyses. In 53 specimens, the pathologist panel achieved total agreement with regard to the diagnosis of the worst CIN grade, whereas in 51 specimens, the diagnosis was based on agreement of 2/3 pathologists. Ten specimens were excluded because there was total disagreement amongst the pathologist panel. Total pathologist agreement was achieved for only two CIN2 specimens.
CIN grading showed substantial inter-observer variation. The mean inter-observer agreement ranged from fair (κ=0.357 for diagnosis of CIN3) to moderate (κ=0.536 for CIN2 versus CIN1 or less) with an overall weighted κ-value of 0.568 (Table 1).
Table 1.
Agreement in histological diagnoses between pairs of pathologists
| Histological categories | Mean kappa value (weighted) |
|---|---|
| All histological diagnoses | 0.568 |
| Mean kappa value (unweighted) | |
| Negative versus ≥CIN1 | 0.427 |
| ≤CIN1 versus CIN2/3 | 0.536 |
| ≤CIN2 versus CIN3 | 0.357 |
Agreement in biomarker scoring
There was excellent agreement in panHPVE4 immunofluorescence scoring between the two researchers (κ=0.896; 95%CI: 0.763-0.969) with 93.8% concordance. The concordance in p16INK4a scoring was 72.9%, with good agreement above chance (κ=0.798; 95%CI: 0.712-0.884) and a concordance of 81.0% in scoring of MCM with κ=0.894 (95%CI: n.c.) (Table 2).
Table 2.
Agreement in biomarker scoring
| Biomarker scoring | Kappa values | 95% CI |
|---|---|---|
| E4 | 0.896 | 0.763 - 0.969 |
| p16 | 0.798 | 0.712 - 0.884 |
| MCM | 0.894 | n.c. |
As shown in Figure 1, the pattern of E4 expression observed in the different samples was broadly similar, irrespective of whether immunofluorescence of immunohostochemical detection was used for visualization, which is of particular importance given the possible utility of E4 antibodies for routine diagnostic purposes. A 100% agreement was observed between the two researchers on the subset of specimens stained using panHPVE4 immunohistochemistry, with a κ-value of 0.898 (95%CI: 0.807-0.989) on the extent of the staining.
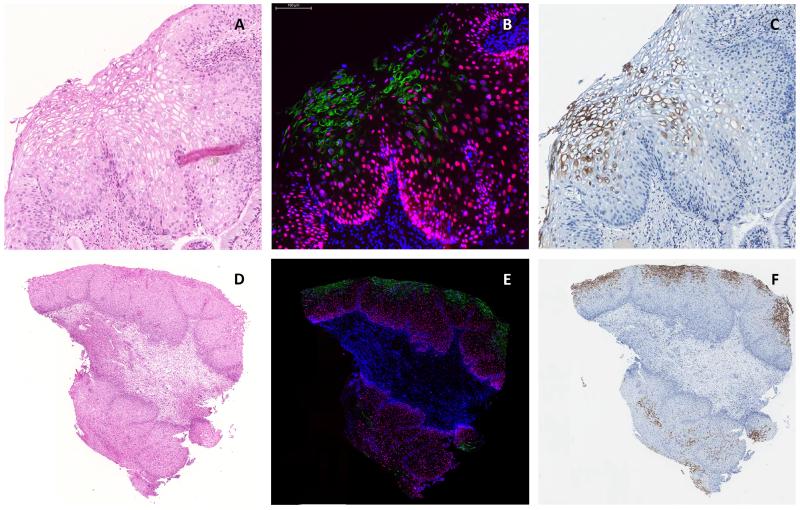
Figure 1.
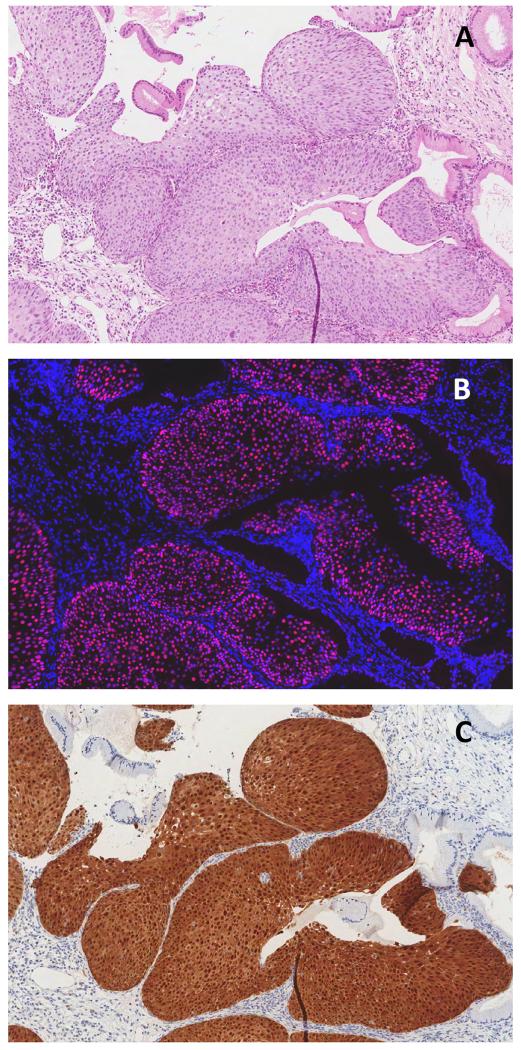
Comparison of detection of expression of panHPVE4 by immunofluorescence (IF; in green; nuclei counterstained using DAPI, in blue) (B, E) and immunohistochemistry (in brown) (C, F) in a productive CIN1 lesion. Images A, B, and C were captured at higher magnification than those shown in D, E, and Fto illustrate the wide distribution of HPVE4 that can sometimes be seen in low-grade CIN1 lesions.
Biomarker expression patterns and HPV status in specimens with consensus diagnosis
Table 3 shows the biomarker expression patterns and HPV positivity rates in relation to the consensus diagnosis. All CIN lesions were HPV positive, with the exception of two of the CIN3 lesions, and 12/26 histologically negative specimens, which were also HPV negative.
Table 3.
Consensus diagnosis in relation to the staining patterns and HPV status
| Consensus diagnosis | p16 | E4 | MCM | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| n | score | n | score | n | score | n | HPV status | Review | |
| Negative | 26 | Negative | 23 | Negative | 23 | Negative | 15 | 8/15 positive | |
|
| |||||||||
| Lower third | 5 | 2/5 positive | |||||||
|
| |||||||||
| Extensive | 3 | 1/3 positive | |||||||
|
| |||||||||
| Extensive | 3 | Negative | 3 | Extensive | 3 | 3/3 positive | CIN3 | ||
|
| |||||||||
| CIN1 | 17 | Negative | 2 | Negative | 2 | Negative | 1 | 17/17 positive | Negative |
|
|
|
||||||||
| Lower third | 1 | Negative | |||||||
|
|
|
||||||||
| Lower third | 3 | Negative | 1 | Lower third | 1 | ||||
|
|
|
||||||||
| Extensive | 2 | Extensive | 2 | ||||||
|
|
|
||||||||
| Extensive | 12 | Negative | 4 | Extensive | 12 | 3 CIN2 | |||
|
|
|
||||||||
| Superficial | 1 | CIN2 | |||||||
|
|
|
||||||||
| Extensive | 7 | ||||||||
|
| |||||||||
| CIN2 | 19 | Extensive | 19 | Negative | 10 | Extensive | 19 | 19/19 positive | |
|
|
|
||||||||
| Superficial | 3 | ||||||||
|
|
|
||||||||
| Extensive | 6 | ||||||||
|
| |||||||||
| CIN3 | 42 | Extensive | 42 | Negative | 33 | Extensive | 42 | 31/33 positive | |
|
|
|
||||||||
| Superficial | 8 | 8/8 positive | |||||||
|
|
|
||||||||
| Extensive | 1 | 1/1 positive | |||||||
Of 26 specimens judged to be histologically negative for dysplasia, 23 (88%) were negative for p16INK4a and panHPVE4 (Figure 2). Three histologically negative specimens showed regions of extensive p16INK4a and MCM staining but were negative for panHPVE4. On re-examination, these were recognized as small CIN3 lesions that had been missed during the initial assessment. In addition, 8 specimens were MCM positive but negative for both p16INK4a and panHPVE4 and were not reclassified as CIN. Eight out of 15 MCM negative specimens were HPV positive by PCR.
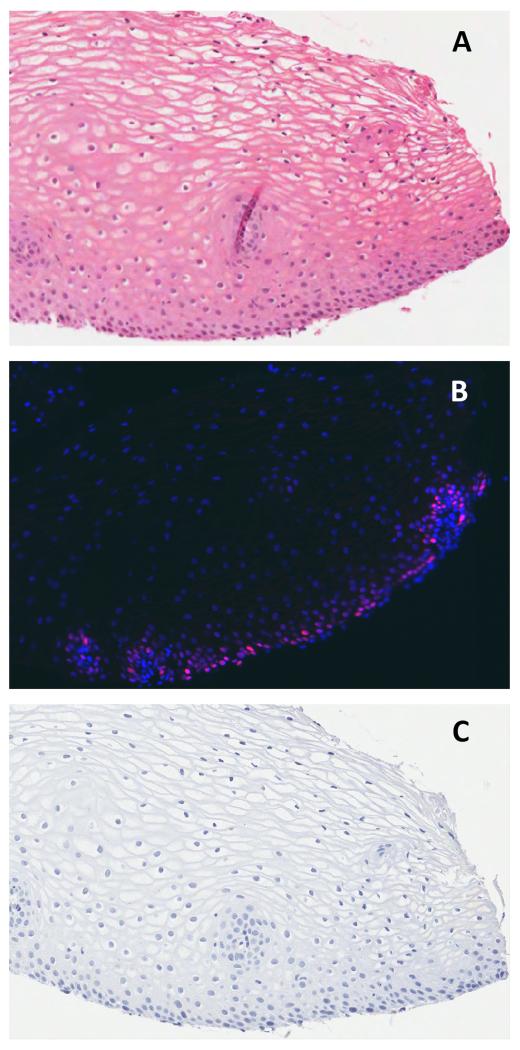
Figure 2.
Cervical squamous epithelium, negative for HPV DNA and by consensus histology (A): there is no panHPVE4 detected by IF and only parabasal MCM staining (in red; nuclei counterstained using DAPI, in blue) (B), and absent p16INK4a by IHC (C).
Fifteen of the 17 CIN1 specimens were p16INK4a positive, of which three had diffuse lower third p16INK4a staining. Twelve showed diffuse p16INK4a staining, which extended through more than one-third of the epithelium. Four of these 12 were subsequently reclassified as CIN2 after histological review. Two of the 17 CIN1 specimens were p16INK4a and panHPVE4 negative and were downgraded to negative for CIN on review. One of these two specimens was MCM positive and most likely represents normal metaplasia.
All CIN2 and CIN3 specimens showed extensive p16INK4a and a corresponding pattern of MCM staining which extended through more than one-third of the epithelium. Nine CIN2 were panHPVE4 positive (extensive or superficial) and 10 were negative, compared with 8 CIN3 which showed only focal panHPVE4 positivity in a few superficial cells, and 33 which were panHPVE4 negative. One CIN3 specimen showed more extensive panHPVE4 positivity of the upper half (extensive).
Biomarker expression patterns and HPV status in relation to final diagnosis
Table 4 shows the expression patterns and HPV status of p16INK4a and panHPVE4 staining in relation to the final diagnoses after additional review. Four patterns of staining were identified.
Table 4.
p16 and panE4 staining in relation to the final histological grade
| Final histological grade (p16+consensus+review) | p16 | E4 | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| n | score | n | score | n | HPV status | |
| Negative | 25 | Negative | 25 | Negative | 25 | 13/25 positive |
|
| ||||||
| CIN1 | 11 | Lower third | 3 | Negative | 1 | 11/11 positive |
|
| ||||||
| Extensive | 2 | |||||
|
| ||||||
| Extensive | 8 | Negative | 1 | |||
|
| ||||||
| Extensive | 7 | |||||
|
| ||||||
| CIN2 | 23 | Extensive | 23 | Negative | 13 | 23/23 positive |
|
| ||||||
| Superficial | 4 | |||||
|
| ||||||
| Extensive | 6 | |||||
|
| ||||||
| CIN3 | 45 | Extensive | 45 | Negative | 36 | 43/45 positive |
|
| ||||||
| Superficial | 8 | |||||
|
| ||||||
| Extensive | 1 | |||||
NEGATIVE
All 25 specimens that were histologically negative on final diagnosis were negative for p16INK4a and panHPVE4 (see Figure 2), although 9 were MCM positive.
PRODUCTIVE
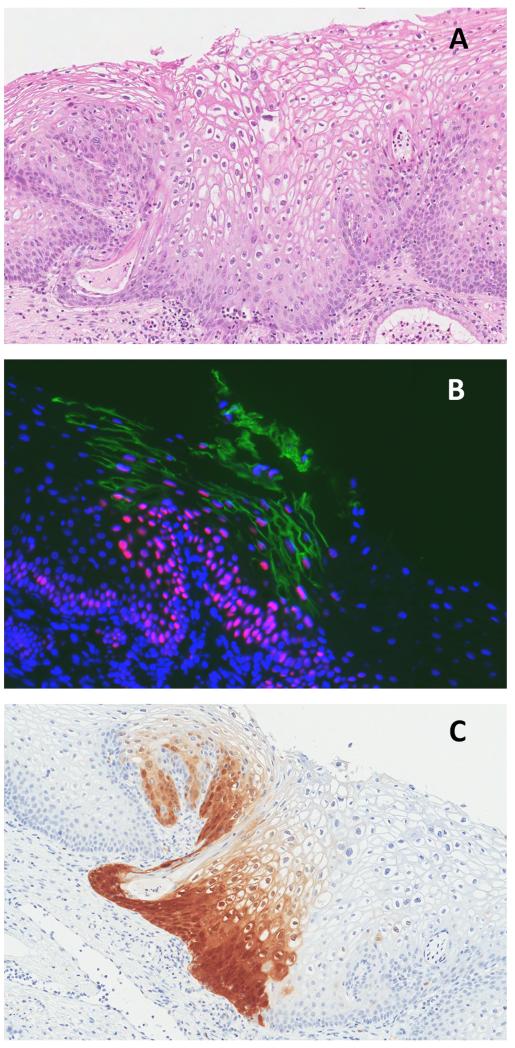
This pattern showed strongly positive panHPVE4 staining and diffuse p16INK4a staining that was restricted to the lower third of the epithelium. Such staining was seen in 2/11 CIN1. One CIN1 showed only lower-third p16INK4a staining with no panHPVE4 staining. Figure 3 shows the biomarker expression pattern typical of productive CIN1 lesion.
Figure 3.
CIN1 lesion by consensus histology (A): strongly positive for panHPVE4 by IF (in green), widespread MCM staining (in red; nuclei counterstained using DAPI, in blue) (B), and p16INK4a staining of the lower third of the epithelium by IHC (C).
INTERMEDIATE
Seven of the 11 CIN1 showed diffuse p16INK4a staining of two-thirds or more of the epithelium with extensive panHPVE4 positivity. A similar pattern was seen in 6/23 CIN2 cases that were panHPVE4 positive. One out of 45 CIN3 cases showed this pattern (see Figure 4).
Figure 4.
CIN2 lesion by consensus histology (A): panHPVE4 staining of upper quarter of the epithelium by IF (green; MCM red; DAPI blue) (B) and extensive p16INK4a staining by IHC (brown) (C).
TRANSFORMING
This pattern showed limited or absent E4 expression, with p16INK4a expression in two-thirds of the epithelium or full thickness. This was seen in 44/45 CIN3 lesions, of which only 8 showed any panHPVE4. In such lesions, staining was limited to one or two cell layers or even to just a few cells. Absence of E4 expression was seen in 13/23 CIN2 lesions. This pattern was seen in only one of 11 CIN1 lesions. An example of a typical biomarker expression pattern of CIN3 specimens is shown in Figure 5, with the broad patterns seen in different lesions being shown in Figure 6.
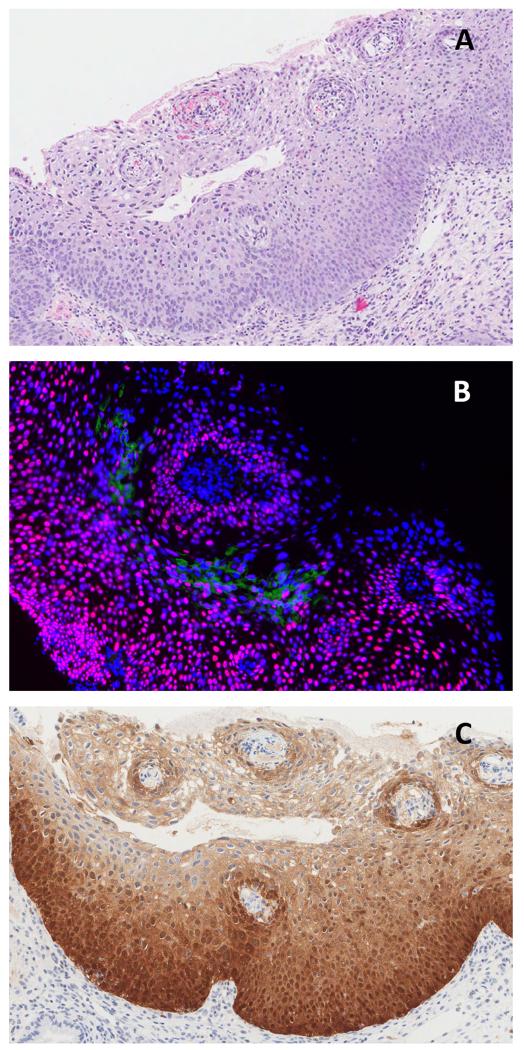
Figure 5.
CIN3 lesion by consensus histology (A): nopanHPVE4 expression by IF (green) with full thickness MCM (red; DAPI blue) (B) and p16INK4a expression by IHC (brown) (C).
Figure 6.
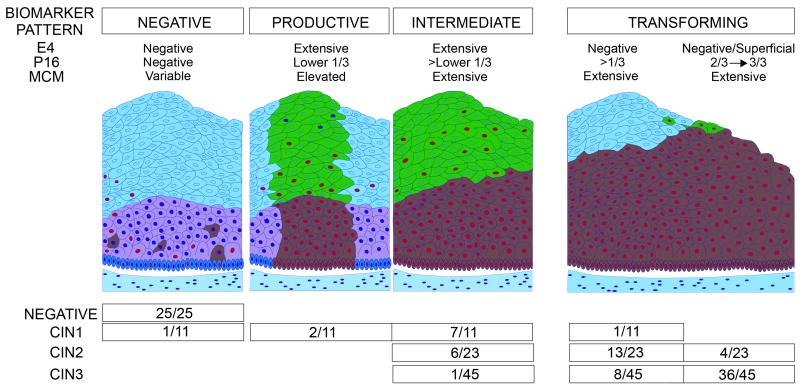
Schematic diagram summarizing the panHPVE4, p16INK4a and MCM biomarker expression patterns in relation to histological diagnoses by CIN classification.
Cells expressing E4 are shown in green, while those expressing p16 are shown in brown. Cells expressing MCM are indicated by the presence of red nuclei. Normal squamous epithelium does not express p16INK4a or E4 and has only (para)basal MCM staining. The productive pattern is extensively positive for E4 with widespread MCM staining, and p16INK4a staining typically restricted to the lower third of the epithelium. The intermediate pattern shows E4 staining of upper quarter or less of the epithelium and p16INK4a in lower two-thirds of epithelium. The transforming pattern shows limited or no E4 expression, with p16INK4a expression in two-thirds or more of the epithelium or full thickness.
The extent of E4 expression declines with lesion grade
Interestingly, there was a significant difference in E4 positivity between CIN grades. Nine of the 11 final CIN1 cases were E4 positive (82%), 10/23 CIN2 (43%) and 9/45 CIN3 (20%) (p<0.0001). For panHPVE4 positive cases, E4 expression was limited to the upper quarter of the epithelium in 8/9 (89%) CIN3 cases, in 4/10 (40%) CIN2 cases, and in 0/9 (0%) CIN1 cases. Of the panHPVE4 positive CIN1 cases, 6 (67%) showed E4 occupying half the depth of the epithelium and in 3 (33%) E4 was even more extensive. Overall, the extent of E4 expression declined with increasing lesion grade (p=0.001).
Discussion
Our findings show that the scoring of biomarkers panHPVE4, MCM and p16INK4a can be reproducibly achieved, and that consistent expression patterns of panHPVE4 and p16INK4a can be defined and related to grade of CIN in lesions associated with HR-HPV. Specimens that were finally agreed to be normal showed no expression of either panHPVE4 or p16INK4a, although MCM was sometimes positive. These data show that panE4 expression can be detected by immunofluorescence and IHC, and suggest that a combination of panHPVE4 and p16INK4a, detected using standard IHC techniques in routine clinical practice, may be particularly useful in distinguishing between normal events (such as metaplasia or inflammation), and true HPV-associated CIN. In CIN caused by HPV, the extent of panHPVE4 and p16INK4a expression generally showed an inverse correlation. When CIN1 was agreed, there was almost always extensive panHPVE4-positivity in the upper epithelial layers, and either lower-third or more extensive p16INK4a staining. CIN2 typically showed extensive p16INK4a staining and was divided into two categories, which could be defined by the presence or absence of panHPVE4 expression. For hematoxylin and eosin diagnosis there was greatest disagreement (kappa= 0.36) over CIN3, especially versus CIN2. Most (80%) of CIN3 cases were completely panHPVE4 negative and when E4 was detected it was mostly confined to a few superficial cells. All CIN3 cases showed diffusely full thickness p16INK4a staining. P16INK4a biomarker patterns also identified some difficult and small high-grade lesions that were missed even by multiple experienced pathologists, but confirmed on pathological review. The combination of panHPVE4 and p16INK4a proved to be most useful in separating normal and CIN, as increased MCM staining above the basal layer was sometimes seen in cervical lesions not considered as CIN on consensus pathological diagnosis. Cell cycle markers such as MCM and ki-67 are not specific for dysplasia; they identify also cells proliferating because of inflammation, epithelial repair or metaplasia. We do not provide evidence to support the suggestion that the distribution of MCM differs importantly in cervical neoplasia from that described for ki-67, or suggest it is specific for neoplasia. In other tissues, MCM and ki-67 can give non-identical staining patterns, with MCM being proposed as a more sensitive marker of high-grade disease [28, 30].
The proportion of panHPVE4 positive cases and the extent of panHPVE4 expression decreased significantly with increasing lesion-grade, consistent with our previous analysis [38]. This correlates with observations in animal models and HPV16-positive biopsies showing loss of E4 with loss of surface epithelial differentiation [17, 35]. Many of these previous studies used immunofluorescence to visualize protein distributions, but in this study we also applied standard immunohistochemistry methodologies to 48 of the cases. We found 100% agreement between immunofluoresence and immunohistochemical staining for panHPVE4 and p16INK4a. Establishing this is of some importance, as it suggests that panHPVE4 antibody is suitable for routine immunohistochemistry, in addition to research-based studies aimed at understanding disease biology (see Figure 1).
The combination of panHPVE4 biomarker used here with p16INK4a offers a reliable, objective approach to grading CIN lesions associated with HR-HPV, identifying productive, intermediate and transforming lesions, thus adding to the information provided by p16INK4a alone. The productive pattern corresponds closely to the concept of “classical” CIN1 in which the full life-cycle of the virus is supported (see Fig. 6), but was seen in only a minority of CIN1 associated with HR-HPV in this study. This may be partly because only HR-HPV positive cases with high-grade, low-grade or repeated ASC-US cytology or being treated for CIN2+ were included. This pattern, however, blends into the intermediate pattern of expression, with both more extensive p16INK4a positivity and widespread panHPVE4 expression seen in most (64%) CIN1 and 26% (6/23) of CIN2. The transforming category, with no or minimal evidence of a HPV productive infection and panHPVE4 expression, but with extensive p16INK4a expression, was seen in 44/45 CIN3, in 17/23 CIN2 and one CIN1. These patterns therefore confirm that CIN1 and CIN2 are not homogeneous categories, and show that CIN2, particularly, is a mixture of lesions.
Strong expression of panHPVE4 in some CIN2 with the intermediate pattern shows that the HR-HPV productive cycle has been initiated, with the extensive expression of p16INK4a suggesting that this is combined with an elevated activity of the transforming HR-HPV gene E7. In this intermediate pattern there is substantial overlap between CIN2 and morphological CIN1, with seven CIN1 lesions showing extensive p16INK4a expression whilst also showing strong panHPVE4 staining.
The loss or minimal extent of HR-HPV life cycle completion associated with strong p16INK4a staining indicating HR-HPV E7 gene activity, was seen in all CIN3 and also 17/23 (74%) CIN2 and one CIN1. This clearly indicates that CIN2 is not biologically homogeneous, with some aligning with CIN1 (intermediate pattern) and some with CIN3 (transforming pattern).
The diagnosis of CIN2 and its distinction from CIN1 and from CIN3 is a well-recognised problem. Our finding that the CIN2 category is a mixture of intermediate (productive life-cycle initiating) and transforming infections is in line with previous findings that CIN2 is the least reproducible grade of CIN [5-7], and with the paper from Castle et al. which suggests CIN2 to be a mixture of transient HPV infections and true cancer precursors [8]. This, together with a relatively high rate of regression of CIN2 [11, 12] suggests that the biomarkers panHR-HPV E4 and p16INK4a in combination could provide a reliable basis for separating CIN2 into subcategories and investigating the frequency and natural history of these subcategories in relation to age, incident and persistent HPV infection, or other molecular markers of neoplastic progression, regression and treatment response which might be used to improve patient management. The different biomarker patterns seen in CIN2 contrast with the transforming pattern that was always seen in CIN3. CIN1 also was not homogeneous, although most of these lesions in “older” women (median age 39 years) were in the intermediate category.
PanHPVE4 could also contribute to avoiding over-diagnosis of normal epithelial areas as CIN. None of the final histologically negative specimens were panHPVE4 positive and both consensus CIN1 lesions that were downgraded to normal (Table 3) were also panHPVE4 negative. The markers used in this study have a strong rationale for their use, and are based on well-characterized patterns of HPV gene expression that have been validated at both the protein and mRNA level [13-18]. The distribution of E6/E7 mRNA in the suprabasal epithelial layers, and the elevation of transcripts that span the E4 region during productive infection was first observed during the late 1980s and early 1990s [13, 15, 18]. Although the relatively low levels of the viral early gene products compromises routine detection [46], the use of surrogate biomarkers of their presence (such as p16INK4a, MCM and ki-67) has gained acceptance in recent years. In addition, we now know that the E4 protein has the ability to accumulate in the form of amyloid fibres at high levels [47, 48], which allows the protein to be easily detected in the upper epithelial layers during productive infection [37, 38].
Follow-up studies are required to identify the risk related to the different biomarker patterns in CIN1 and CIN2, and to decide on the appropriate treatment or follow-up of lesions that show evidence of life-cycle completion (productive/intermediate), compared to those expressing only p16INK4a as evidence that there is only HR-HPV transforming gene activity. Recent guidelines from the USA suggest treatment of all p16INK4a positive CIN lesions with any suspicion of being high-grade [40]. However, a substantial number of CIN1 lesions show p16INK4a over-expression [49]. There are several studies that show an increased progression risk of p16INK4a positive CIN1 [50-54]. Still the majority of women did not progress. These published studies however, did not distinguish extent of p16INK4a expression or take account of the differences between high-risk and low-risk HPV in relation to progression, which makes further study necessary.
The combination of panHPVE4 and p16INK4a provides a simple approach to distinguishing CIN associated with HR-HPV from normal and also clearly demonstrates the complexity of CIN1 and CIN2. The robust staining obtained with both markers by IHC, further suggests that E4 could be used alongside p16INK4a during routine analysis and IHC double staining is being developed. CIN1 showed both productive, intermediate (mainly) and occasionally transforming patterns. In this study all CIN3 and half the CIN2 were transforming lesions by panHPVE4 and p16INK4a, with the other half of the CIN2 being intermediate lesions. Development of IHC double staining is in progress and further studies are required to understand the biology of these lesion categories, and to investigate whether the combination of panHPVE4 and p16INK4a can predict progression or regression of lesions and provide a basis for refining management decisions about follow-up and treatment of CIN1 and CIN2.
Supplementary Material
Acknowledgements
We would like to thank Jan Lindeman (in memoriam), DDL Diagnostic Laboratory, for his critical review of some of the pathology slides. We wish to thank Hans Berkhof for his assistance with the statistical analysis, Annemiek Kasius for her help with the analysis of the panHPVE4 immunohistochemical staining, and Frank van der Panne for his support in the graphical artwork. Thanks also to Albert Singer (UCLH, London) and Margaret Stanley (University of Cambridge) for their ongoing advice and support during the course of the project.
Source of Funding: This research was partly funded by the Stichting Pathologie Ontwikkeling en Onderzoek (SPOO) Foundation, The Netherlands. Funding was also provided from the UK Medical Research Council to HG, YS, ZW and JD.
List of abbreviations
- CIN
Cervical intraepithelial neoplasia
- DEIA
DNA enzyme immuno assay
- FFPE
Formalin fixed paraffin embedded
- H&E
Hematoxylin and Eosin
- HPV
Human papillomavirus
- HR-HPV
High-risk Human papillomavirus
- HG-CIN
High-grade cervical intraepithelial neoplasia
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- LEEP
Loop electrosurgical excision procedure
- LiPA
Line probe assay
- MCM
Minichromosome maintenance protein
- PCR
Polymerase chain reaction
- SPF
Short PCR fragment
Footnotes
Conflicts of Interest: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
Reference List
- 1.Richart RM. Causes and management of cervical intraepithelial neoplasia. Cancer. 1987;60:1951–1959. doi: 10.1002/1097-0142(19901015)60:8+<1951::aid-cncr2820601505>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.van der Aa MA, Pukkala E, Coebergh JW, et al. Mass screening programmes and trends in cervical cancer in Finland and the Netherlands. Int J Cancer. 2008;122:1854–1858. doi: 10.1002/ijc.23276. [DOI] [PubMed] [Google Scholar]
- 4.de Vet HC, Knipschild PG, Schouten HJ, et al. Interobserver variation in histopathological grading of cervical dysplasia. J Clin Epidemiol. 1990;43:1395–1398. doi: 10.1016/0895-4356(90)90107-z. [DOI] [PubMed] [Google Scholar]
- 5.Ismail SM, Colclough AB, Dinnen JS, et al. Observer variation in histopathological diagnosis and grading of cervical intraepithelial neoplasia. BMJ. 1989;298:707–710. doi: 10.1136/bmj.298.6675.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson AJ, Anderson JM, Beck JS, et al. Observer variability in histopathological reporting of cervical biopsy specimens. J Clin Pathol. 1989;42:231–238. doi: 10.1136/jcp.42.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carreon JD, Sherman ME, Guillen D, et al. CIN2 is a much less reproducible and less valid diagnosis than CIN3: results from a histological review of population-based cervical samples. Int J Gynecol Pathol. 2007;26:441–446. doi: 10.1097/pgp.0b013e31805152ab. [DOI] [PubMed] [Google Scholar]
- 8.Castle PE, Stoler MH, Solomon D, et al. The relationship of community biopsy-diagnosed cervical intraepithelial neoplasia grade 2 to the quality control pathology-reviewed diagnoses: an ALTS report. Am J Clin Pathol. 2007;127:805–815. doi: 10.1309/PT3PNC1QL2F4D2VL. [DOI] [PubMed] [Google Scholar]
- 9.Stoler MH, Schiffman M. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA. 2001;285:1500–1505. doi: 10.1001/jama.285.11.1500. [DOI] [PubMed] [Google Scholar]
- 10.Ostor AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993;12:186–192. [PubMed] [Google Scholar]
- 11.Castle PE, Schiffman M, Wheeler CM, et al. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol. 2009;113:18–25. doi: 10.1097/AOG.0b013e31818f5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moscicki AB, Ma Y, Wibbelsman C, et al. Rate of and risks for regression of cervical intraepithelial neoplasia 2 in adolescents and young women. Obstet Gynecol. 2010;116:1373–1380. doi: 10.1097/AOG.0b013e3181fe777f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crum CP, Nuovo G, Friedman D, et al. Accumulation of RNA homologous to human papillomavirus type 16 open reading frames in genital precancers. J Virol. 1988;62:84–90. doi: 10.1128/jvi.62.1.84-90.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorbar J, Foo C, Coleman N, et al. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology. 1997;238:40–52. doi: 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- 15.Durst M, Glitz D, Schneider A, et al. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology. 1992;189:132–140. doi: 10.1016/0042-6822(92)90688-l. [DOI] [PubMed] [Google Scholar]
- 16.Freeman A, Morris LS, Mills AD, et al. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–2132. [PubMed] [Google Scholar]
- 17.Middleton K, Peh W, Southern S, et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoler MH, Rhodes CR, Whitbeck A, et al. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992;23:117–128. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- 19.von Knebel Doeberitz M, Reuschenbach M, Schmidt D, et al. Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev Proteomics. 2012;9:149–163. doi: 10.1586/epr.12.13. [DOI] [PubMed] [Google Scholar]
- 20.Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315–330. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergeron C, Ronco G, Reuschenbach M, et al. The clinical impact of using p16 immunochemistry in cervical histopathology and cytology: An update of recent developments. Int J Cancer. 2014 doi: 10.1002/ijc.28900. [DOI] [PubMed] [Google Scholar]
- 22.Tsoumpou I, Arbyn M, Kyrgiou M, et al. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev. 2009;35:210–220. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 24.Davidson EJ, Morris LS, Scott IS, et al. Minichromosome maintenance (Mcm) proteins, cyclin B1 and D1, phosphohistone H3 and in situ DNA replication for functional analysis of vulval intraepithelial neoplasia. Br J Cancer. 2003;88:257–262. doi: 10.1038/sj.bjc.6600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Facoetti A, Ranza E, Grecchi I, et al. Immunohistochemical evaluation of minichromosome maintenance protein 7 in astrocytoma grading. Anticancer Res. 2006;26:3513–3516. [PubMed] [Google Scholar]
- 26.Lee YS, Ha SA, Kim HJ, et al. Minichromosome maintenance protein 3 is a candidate proliferation marker in papillary thyroid carcinoma. Exp Mol Pathol. 2010;88:138–142. doi: 10.1016/j.yexmp.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Wang L, Qiu M, et al. The protein levels of MCM7 and p63 in evaluating lesion severity of cervical disease. Int J Gynecol Cancer. 2013;23:318–324. doi: 10.1097/IGC.0b013e31827f6f06. [DOI] [PubMed] [Google Scholar]
- 28.Giaginis C, Vgenopoulou S, Vielh P, et al. MCM proteins as diagnostic and prognostic tumor markers in the clinical setting. Histol Histopathol. 2010;25:351–370. doi: 10.14670/HH-25.351. [DOI] [PubMed] [Google Scholar]
- 29.Lampert IA, Horncastle D, Dilworth S, et al. The expression of minichromosome maintenance protein-2 in normal and abnormal megakaryocytes and comparison with the proliferative marker Ki-67. Br J Haematol. 2005;131:490–494. doi: 10.1111/j.1365-2141.2005.05801.x. [DOI] [PubMed] [Google Scholar]
- 30.Szelachowska J, Dziegiel P, Jelen-Krzeszewska J, et al. Mcm-2 protein expression predicts prognosis better than Ki-67 antigen in oral cavity squamocellular carcinoma. Anticancer Res. 2006;26:2473–2478. [PubMed] [Google Scholar]
- 31.Bulten J, van der Laak JA, Gemmink JH, et al. MIB1, a promising marker for the classification of cervical intraepithelial neoplasia. J Pathol. 1996;178:268–273. doi: 10.1002/(SICI)1096-9896(199603)178:3<268::AID-PATH482>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Nam EJ, Kim JW, Hong JW, et al. Expression of the p16 and Ki-67 in relation to the grade of cervical intraepithelial neoplasia and high-risk human papillomavirus infection. J Gynecol Oncol. 2008;19:162–168. doi: 10.3802/jgo.2008.19.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirog EC, Baergen RN, Soslow RA, et al. Diagnostic Accuracy of Cervical Low-Grade Squamous Intraepithelial Lesions Is Improved With MIB-1 Immunostaining. Am J Surg Pathol. 2002;26:70–75. doi: 10.1097/00000478-200201000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Henderson D, Hall L, Prpic N, et al. The selection and characterization of antibodies to minichromosome maintenance proteins that highlight cervical dysplasia. J Immunol Methods. 2011;370:1–13. doi: 10.1016/j.jim.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Peh WL, Middleton K, Christensen N, et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol. 2002;76:10401–10416. doi: 10.1128/JVI.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 37.Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98. doi: 10.1016/j.virol.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Griffin H, Wu Z, Marnane R, et al. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974. doi: 10.1371/journal.pone.0049974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Marel J, van Baars R, Quint WG, et al. The impact of human papillomavirus genotype on colposcopic appearance: a cross-sectional analysis. BJOG. 2014;121:1117–1126. doi: 10.1111/1471-0528.12668. [DOI] [PubMed] [Google Scholar]
- 40.Darragh TM, Colgan TJ, Cox JT, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-Associated Lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205–242. doi: 10.1097/LGT.0b013e31825c31dd. [DOI] [PubMed] [Google Scholar]
- 41.Quint WG, Scholte G, van Doorn LJ, et al. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF(10) PCR and HPV genotyping. J Pathol. 2001;194:51–58. doi: 10.1002/path.855. [DOI] [PubMed] [Google Scholar]
- 42.Kleter B, van Doorn LJ, ter Schegget J, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleter B, van Doorn LJ, Schrauwen L, et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol. 1999;37:2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Doorn LJ, Molijn A, Kleter B, et al. Highly effective detection of human papillomavirus 16 and 18 DNA by a testing algorithm combining broad-spectrum and type-specific PCR. J Clin Microbiol. 2006;44:3292–3298. doi: 10.1128/JCM.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 46.Doorbar J, Cubie H. Molecular basis for advances in cervical screening. Mol Diagn. 2005;9:129–142. doi: 10.1007/BF03260081. [DOI] [PubMed] [Google Scholar]
- 47.Khan J, Davy CE, McIntosh PB, et al. Role of calpain in the formation of human papillomavirus type 16 E1^E4 amyloid fibers and reorganization of the keratin network. J Virol. 2011;85:9984–9997. doi: 10.1128/JVI.02158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McIntosh PB, Martin SR, Jackson DJ, et al. Structural analysis reveals an amyloid form of the human papillomavirus type 16 E1--E4 protein and provides a molecular basis for its accumulation. J Virol. 2008;82:8196–8203. doi: 10.1128/JVI.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001;92:276–284. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- 50.Cortecchia S, Galanti G, Sgadari C, et al. Follow-up Study of Patients With Cervical Intraepithelial Neoplasia Grade 1 Overexpressing p16Ink4a. Int J Gynecol Cancer. 2013;23:1663–1669. doi: 10.1097/IGC.0b013e3182a80b14. [DOI] [PubMed] [Google Scholar]
- 51.del Pino M, Garcia S, Fuste V, et al. Value of p16(INK4a) as a marker of progression/regression in cervical intraepithelial neoplasia grade 1. Am J Obstet Gynecol. 2009;201:488, e481–487. doi: 10.1016/j.ajog.2009.05.046. [DOI] [PubMed] [Google Scholar]
- 52.Liao GD, Sellors JW, Sun HK, et al. p16 immunohistochemical staining and predictive value for progression of cervical intraepithelial neoplasia grade 1: A prospective study in China. Int J Cancer. 2013 doi: 10.1002/ijc.28485. [DOI] [PubMed] [Google Scholar]
- 53.Nishio S, Fujii T, Nishio H, et al. p16(INK4a) immunohistochemistry is a promising biomarker to predict the outcome of low grade cervical intraepithelial neoplasia: comparison study with HPV genotyping. J Gynecol Oncol. 2013;24:215–221. doi: 10.3802/jgo.2013.24.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pacchiarotti A, Ferrari F, Bellardini P, et al. Prognostic value of p16-INK4A protein in women with negative or CIN1 histology result: A follow-up study. Int J Cancer. 2014;134:897–904. doi: 10.1002/ijc.28407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.