SUMMARY
The proper organization of the microtubule-based mitotic spindle is proposed to depend on nanometer-sized motor proteins generating forces that scale with a micron-sized geometric feature, such as microtubule overlap length. However, it is unclear if such regulation can be achieved by any mitotic motor protein. Here, we employ an optical trap- and TIRF-based assay to show that ensembles of kinesin-5, a conserved mitotic motor protein, can push apart overlapping antiparallel microtubules to generate force whose magnitude scales with filament overlap length. We also find that kinesin-5 can produce overlap length-dependent ‘brake-like’ resistance against relative microtubule sliding in both parallel and antiparallel geometries, an activity that has been suggested by cell biological studies, but had not been directly measured. Together, these findings, along with numerical simulations, reveal how a motor protein can function as an analog converter, ‘reading’ simple geometric and dynamic features in cytoskeletal networks to produce regulated force outputs.
Graphical Abstract
INTRODUCTION
Successful cell division in eukaryotes requires the assembly of a bipolar spindle. The overall size and shape of this microtubule-based structure depends on the activities of molecular motors that generate forces to push or pull microtubules (Compton, 2000; Sharp et al., 2000; Wittmann et al., 2001). Long-standing models suggest that the magnitudes of these forces are proportional to a geometric feature, such as the length of overlap between adjacent filaments, within the dynamic microtubule network (McIntosh et al., 1969; Mogilner et al., 2006). However, it is currently unclear if the forces generated by any molecular motor required for bipolar spindle assembly are regulated in this manner.
Kinesin-5 is a conserved homo-tetrameric microtubule-based motor protein required for cell division in eukaryotes (Blangy et al., 1995; Heck et al., 1993; Sawin et al., 1992). Kinesin-5 localizes along spindle microtubules and is enriched at spindle poles. Metaphase spindle microtubules overlap in parallel orientations near spindle poles and in antiparallel orientations at the spindle equator (Mastronarde et al., 1993; McDonald et al., 1977; Sharp et al., 1999). In vitro studies reveal that kinesin-5 can crosslink parallel and antiparallel microtubules (Hentrich and Surrey, 2010; Kapitein et al., 2005; Kashina et al., 1996; van den Wildenberg et al., 2008). In the case of antiparallel microtubules, kinesin-5 has been shown to slide apart filaments by stepping towards the plus-ends of each of the two microtubules it crosslinks (Hentrich and Surrey, 2010; Kapitein et al., 2005; van den Wildenberg et al., 2008). This activity is proposed to push centrosomes apart to establish a bipolar spindle. On the other hand, kinesin-5 localizes at spindle poles and can crosslink parallel microtubules (Gaglio et al., 1996; Sawin et al., 1992; Sharp et al., 1999), but its exact contributions to parallel microtubule sliding and organization are poorly understood.
The force-generating properties of kinesin-5 constructs attached to optically trapped plastic beads have been studied using single molecule methods (Korneev et al., 2007; Valentine and Block, 2009; Valentine et al., 2006). However, unlike other well-studied kinesins, kinesin-5 uses microtubules as both its track and cargo (Kapitein et al., 2008). In order to understand how kinesin-5 generates forces needed for spindle assembly, it is necessary to measure the force output across the sliding microtubule pairs as opposed to linking the motor protein directly to a bead. In addition, the size of parallel and antiparallel microtubule overlaps that are found in spindles is sufficient to recruit multiple kinesin-5 molecules (Sharp et al., 1999). Therefore, it is important that force generation by kinesin-5 is not only analyzed for microtubules in the parallel or antiparallel configuration, but also for different overlap lengths that will recruit different numbers of active motor protein molecules. The micromechanics of multi-motor cargo transport, wherein two or more kinesin molecules simultaneously carry a bead along a single microtubule, have been examined (Furuta et al., 2013; Gross et al., 2007; Jamison et al., 2012; Vershinin et al., 2007). These studies suggest that increasing the number of motor proteins may not necessarily lead to a larger force output. However, given the appropriate cargo geometry and motor protein mechanics, it may be possible for the magnitude of sustained force output to scale with increasing motor protein number. These different outcomes will depend on how force is transmitted across the cargo and how frequently kinesins detach from the microtubule (Gross et al., 2007). It is currently unclear how forces produced by ensembles of kinesin-5 will depend on filament overlap length or motor protein number.
In addition to generating force to slide microtubules apart, recent studies have suggested that kinesin-5 may also act to oppose relative microtubule motion. For example, loss of kinesin-5 activity in dividing cells results in a faster rate of spindle elongation in anaphase (Rozelle et al., 2011; Saunders et al., 2007), suggesting that kinesin-5 could act as a molecular brake within overlapping antiparallel filaments. Microtubule gliding assays that employed a mixture of surface-bound kinesins suggest kinesin-5 can reduce the velocity of microtubule sliding driven by kinesin-1 (Crevel et al., 2004). However, the magnitude of the braking force generated by kinesin-5 across two microtubules has not been directly measured and is unclear how these forces scale with filament overlap, orientation or relative velocity.
To examine whether and how kinesin-5 can achieve different force-generating functions within overlapping microtubules, we have developed an optical trap- and fluorescence-based in vitro reconstitution assay, in which the relative sliding velocity and the polymer polarity can be controlled and the force across the microtubule pair measured. Along with the force measurement, the motion and localization of microtubules and kinesin-5 molecules can be directly analyzed. Using this assay we show that the magnitude and direction of force developed within microtubule pairs crosslinked by kinesin-5 depends on the extent of filament overlap, the relative sliding velocity and the relative polarity of these filaments. Together with numerical model simulations, our data reveal how distinct mechanical outputs of this motor protein ensembles can be regulated by simple geometric features of the microtubule network, such as filament overlap length.
RESULTS
Counting the number of kinesin-5 molecules crosslinking microtubules at overlap regions
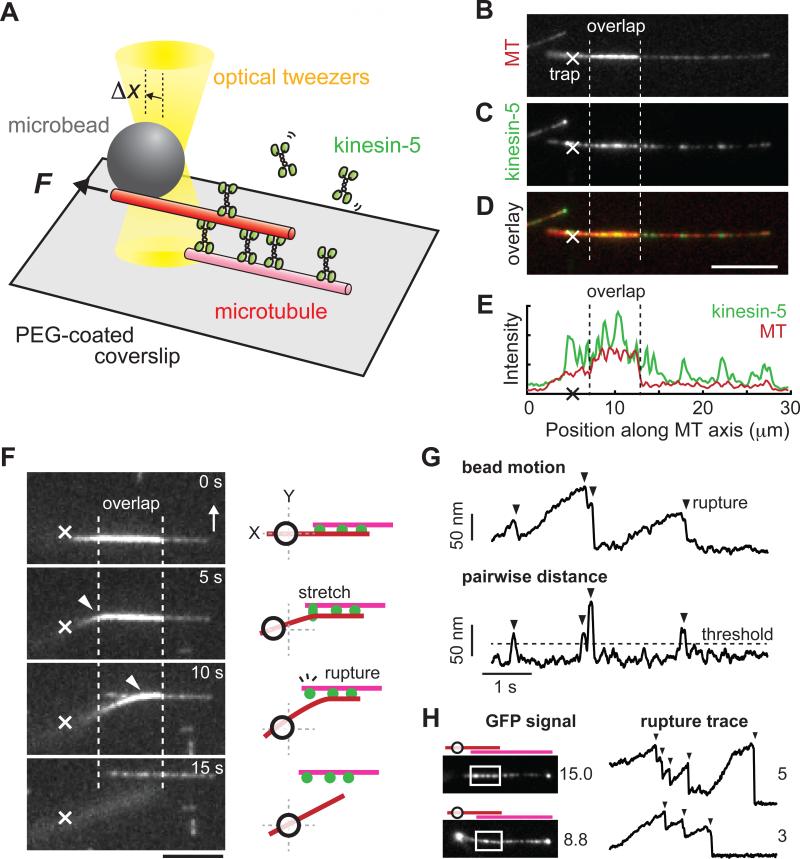
To analyze force generation by ensembles of kinesin-5 molecules we need to measure the forces generated across a pair of microtubules and to determine how many motor protein molecules are pushing the filaments apart. For these measurements we devised an assay that combines high-resolution fluorescence imaging and force-calibrated optical tweezers (Figure 1A). Briefly, microtubules doubly-labeled with X-rhodamine fluorescent dye and biotin were immobilized on a glass coverslip surface by means of biotin-neutravidin linkages. The coverslip surface was pre-coated by polyethylene glycol (PEG) to prevent non-specific adsorption of proteins. Addition of non-biotinylated, X-rhodamine-labeled microtubules (~2× more brightly labeled than the biotinylated filaments), recombinant GFP-tagged full-length Xenopus kinesin-5 (hereafter, kinesin-5-GFP; 1 nM) that we have previously characterized (Kwok et al., 2006; Weinger et al., 2011) and MgATP (2 mM) resulted in the formation of microtubule ‘sandwiches’. The free-end of the non-biotinylated microtubule in the ‘sandwich’ was then attached to an optically-trapped bead. Under this condition, kinesin-5 crosslinked two adjacent microtubules and displaced the trapped bead away from its equilibrium position (depicted as Δx, Figure 1A).
Figure 1. Measuring forces generated and counting kinesin-5 molecules within overlapping microtubules.
(A) Schematic of the in vitro assay. Shown are a biotinylated microtubule (pink, 1X X-rhodamine-labeled) immobilized on a PEG-coated coverslip surface and a nonbiotinylated microtubule (red, 2X X-rhodamine-labeled) held by force-calibrated optical tweezers (yellow) via a bead (grey). Kinesin-5-GFP (green) in solution can crosslink these microtubules to generate a ‘sandwich’. The force (F) generated results in bead displacement (Δx).
(B-E) Fluorescence images of the ‘sandwich’. Microtubules (MT) were imaged using epi-fluorescence (B); kinesin-5-GFP was imaged using TIRF (C); Overlay (D). Trap center position is marked (X). Scale bar, 5 μm. Line-scans along the microtubules (red) reveal kinesin-5 (green) localized both on single microtubules and within the overlap region (between the two dotted lines) (E).
(F-H) Rupture-based analysis to count the number of crosslinking motor molecules. (F) Sample time-lapse images and the corresponding schematics, with the representation similar to Figure 1A. A bead-attached microtubule was pulled nearly orthogonal (white arrow) to the surface microtubule by moving the sample stage at constant velocity. The bead-held microtubule bent (white arrowheads) and then detached from the surface microtubule. Scale bar, 5 μm. Time between images, 5 s. The bead position recording was processed and used to count the number of such rupture events (vertical arrowheads) (G). (H) Comparison of the number of kinesin-5 molecules determined by fluorescence and rupture-based methods. Data from two different microtubule pairs are shown. Region of microtubule overlap (white rectangle) and motor number (adjacent to image) are indicated.
Fluorescence images of X-rhodamine and GFP revealed regions of microtubule overlap and localization of kinesin-5 (Figure 1B-E). Using the average GFP signal of single kinesin-5 homotetramers (0.7 ± 0.3 ×104 a.u., n = 410) (Figure S1A), we estimated that the number of motor protein molecules within overlaps (typically 1-6 μm) ranged from 3 to 26. Consistent with previous studies (Hentrich and Surrey, 2010; Kapitein et al., 2005), we observed kinesin-5 molecules also binding single microtubules (Figure 1B-E). It is therefore likely that some fraction of kinesin-5 localized within the overlap may also bind only to single microtubules. To distinguish such ‘passenger’ molecules from those that actively crosslink two adjacent filaments, we used the optical trap to rapidly disrupt motor-filament interactions and count ‘rupture’ events (Figure 1F-H), a modification of the approach used to analyze surface-bound myosin molecules interacting with a single actin filament (Nishizaka et al., 1995). Rapidly pulling (within seconds, for details see Methods) one end of the filament orthogonal to the other in the ‘sandwich’ resulted in their separation (Figure 1F and S1B). The position of the optically trapped bead revealed a series of sawtooth-like responses that are consistent with multiple ‘rupture’ events (Figure 1G). Independent experiments revealed that our bead-microtubule linkage could sustain large (~30 pN) forces for many tens of seconds, and would remain intact throughout these measurements without detachment (Figure S1C). We therefore conclude that the ‘rupture’ events likely corresponded to the sequential loss of individual kinesin-5 mediated microtubule crosslinks. Our rupture-based analyses yielded measurements of motor protein number that were less than estimates made from GFP fluorescence data, consistent with there being both crosslinking and ‘passenger’ kinesin-5 molecules within overlap regions (Figure 1H; for full data set, see Figure S1D). Together, our assay allows us to monitor force, measure microtubule overlap length, and count kinesin-5 molecules, distinguishing between those molecules crosslinking filaments to push them apart and those bound only to a single filament.
Kinesin-5-dependent forces pushing two antiparallel microtubules apart scale with microtubule overlap
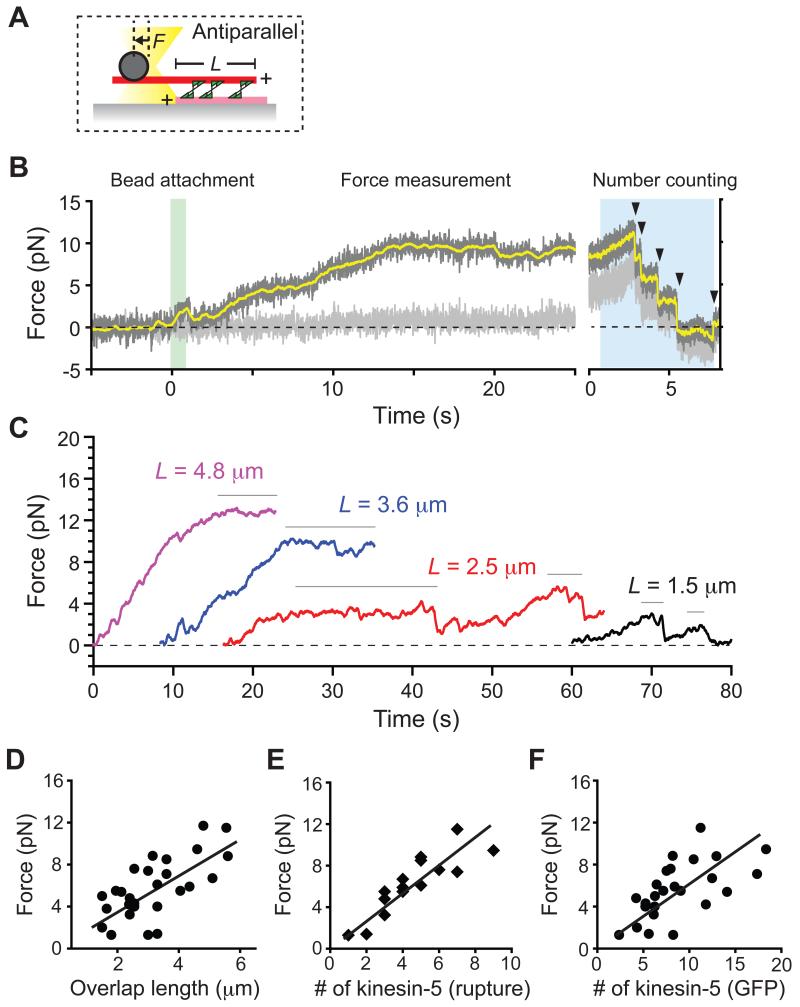
Using this assay, we first measured how much force is generated by kinesin-5 molecules crosslinking antiparallel microtubules (Figure 2A). We recorded the position of an optically-trapped bead that was attached to the free microtubule in the ‘sandwich’ and used this to calculate the force produced across the antiparallel microtubule pair. The polarity of microtubules was determined from the X-rhodamine fluorescence signal prior to the bead attachment, as antiparallel filaments slid apart at >40 nm/s velocity while parallel filaments appeared relatively static, as expected (Kapitein et al., 2005). We observed that, soon after the trapped bead was attached to the leading end (i.e., the minus-end) of a sliding microtubule in a ‘sandwich’, the force built up gradually along the microtubule axis until it reached a plateau (Figure 2B). After the force plateau was apparent the number of crosslinking kinesin-5 molecules was estimated using the rupture-based counting method (blue area, Figure 2B). The fluctuations in the force signal, most frequently observed at lower forces and for microtubule pairs with short overlap lengths, are likely due to stochastic dissociation and re-association of crosslinking kinesin-5 molecules from the microtubule (Figure S2A-C). The maximum force developed within the antiparallel microtubule pair was determined by averaging the force at plateaus that lasted for >1 s before dissociation. At the force plateau, the length of microtubule overlap was also determined based on the X-rhodamine fluorescence signal (Figure 2C). We analyzed a total of 27 antiparallel microtubule pairs and found that the maximum developed force increased with the amount of filament overlap (Figure 2D). Therefore, ensembles of kinesin-5 can push apart antiparallel microtubules against opposing load, and the pushing force scales with filament overlap length.
Figure 2. Kinesin-5-dependent forces that push apart two antiparallel microtubules scale with the length of filament overlap and the number of motor protein molecules.
(A) Schematic of the assay, with the representation similar to Figure 1A. L, length of overlap; F, developed force. Microtubule plus-ends are marked (+).
(B) A representative time record of force developed within an antiparallel microtubule pair crosslinked by kinesin-5. The force along the microtubule's long axis (dark grey, sampled at 0.1 kHz; yellow, filtered using 200-ms moving average) and its short axis (light grey: sampled at 0.1 kHz) are shown. At t = 0, an optically-trapped bead was brought into contact with the sliding microtubule in a ‘sandwich’ (green highlight). After reaching a steady force plateau (t > 15 s), the rupture-based counting method was employed to determine motor protein number (blue highlight). Arrowheads indicate identified rupture events.
(C) Time records of force (filtered using 200-ms moving average) developed within antiparallel microtubule pairs of various overlap lengths. The force developed predominantly parallel to the microtubule axis and reached a steady state at indicated overlap length, L. For clarity, each trace is offset along the time-axis.
(D-F) The maximum developed force versus the microtubule overlap length or the number of kinesin-5 molecules. The maximum force, determined by averaging the forces at plateaus (denoted with horizontal bars in (C)), is plotted against the length of filament overlap (D). Pearson's r = 0.68 (n = 27). For several microtubule pairs in (D), the number of kinesin-5 at the overlap was estimated based on the rupture counting method (n = 15) (E) and from GFP signal intensity (n = 25) (F). Linear regressions yield slopes of 1.6 (R2 = 0.45), 1.34 (R2 = 0.76) and 0.49 (R2 = 0.45) for data shown in panels (D-F), respectively.
For 15 of the 27 antiparallel microtubule pairs examined we could estimate the number of kinesin-5 molecules crosslinking two microtubules using the rupture-based method. We found that the force generated by kinesin-5 to push apart antiparallel microtubules is additive; i.e., the magnitude of maximal force increases linearly with the number of crosslinking motor protein molecules (R2 = 0.76) (Figure 2E). Consistent with these rupture-based analyses, the maximum force also scaled with the fluorescence intensity of kinesin-5-GFP within the overlap (Figure 2F). These results suggest that individual kinesin-5 molecules can transmit forces across microtubules and the developed force output can be linearly summed up over several microns across the filament pair.
From the slope of this linear regression analysis, we can estimate that the average maximum force generated by individual tetrameric kinesin-5 motors within the ensemble is ~1.5 pN per molecule (1.3 ± 0.3 pN, mean ± S.D., n = 15). This value is 4-5 times smaller than that reported for a truncated human kinesin-5 dimeric construct (Valentine et al., 2006), or vesicle-transporting kinesin-1 (Svoboda and Block, 1994). In order to confirm that individual kinesin-5 molecules were producing this amount of force, we performed two independent experiments to directly measure the maximum force that a single kinesin-5 tetramer could generate. First, we measured the force across a crosslinked microtubule ‘sandwich’ that was held at a small angle to minimize the length of filament overlap. In this geometry, only one or a few motors could crosslink the filaments (Figure 3A). We observed rare events of sustained interaction between the two microtubules, detecting a transient force signal that developed gradually and then suddenly dropped (Figure 3B). We characterized such events from several microtubule pairs and found that the maximum developed force, measured at each peak before the sudden force drop, was 1.7 ± 0.6 pN (mean ± S.D., n = 11) (Figure 3C). Second, we attached our kinesin-5 construct directly to an optically-trapped bead by means of anti-GFP antibody (Figure 3D), following an experimental methodology similar to that previously reported for a tetrameric kinesin-5 construct that was linked to beads via a poly-histidine tag (Korneev et al., 2007). The bead-motor conjugate was brought into contact with a single surface-immobilized microtubule and the developed forces were measured (Figure 3E). We observed frequent events during which time the force increased to a locally maximum value and persisted for several seconds (Figure 3E). At the highest density of motors bound to the bead, we measured a range of maximum developed force values (Figure 3F). As the density of motor protein on the bead was decreased, a peak in the histograms of maximum force values was observed at 1-2 pN, consistent with this force having been generated by a single kinesin-5 molecule (Figure 3F). We next calculated the force-velocity relationship from individual force generation events taken in the lowest motor protein density condition. We found this relationship to be well-fit (R2 = 0.93) by a linear function, with a zero-velocity force value of 1.7 pN (Figure 3G). Together, these results indicate that single kinesin-5 molecules can exert ~1.5 pN of force before stalling and the total force output across an antiparallel ‘sandwich’ is the linear sum of the forces produced by individual crosslinking kinesin-5 molecules within the region of overlap.
Figure 3. Single tetrameric kinesin-5 molecules produce approximately 1.5 pN of force.
(A-C) Measurement of kinesin-5 force within microtubule pairs at crossed geometry. (A) Fluorescence image of a microtubule pair and the corresponding assay schematic, with the representation similar to Figure 1A. (B) Time record of force along the long axis of the bead-attached microtubule (grey: sampled at 0.1 kHz; black: filtered using 200-ms moving average) developed by kinesin-5 within a microtubule pair in a crossed geometry. The maximum developed force was determined at each force peak just prior to the large force drop (black horizontal dashes). (C) Histogram showing the distribution of maximum developed forces measured from different microtubule pairs (1.7 ± 0.6 pN (mean ± S.D.); n = 11 events from 7 microtubule pairs).
(D-G) Single molecule measurements of bead-attached kinesin-5 molecules. (D) Schematic of the assay. Full-length GFP-tagged kinesin-5 (green) was attached to a bead via antibody to GFP. The bead was optically trapped and held near a surface-immobilized microtubule, and forces produced by kinesin-5 were measured. (E) Time records of force (grey: sampled at 0.1 kHz; black: filtered using 200-ms moving average) developed at different kinesin-5 concentrations. (F) Histograms showing the distribution of the average maximum force measured for different kinesin-5 concentrations used during bead-coating preparation (n = 60-86 events per condition). (G) Force-velocity relationship calculated for individual force generation events acquired with the 0.25 nM kinesin-5 bead-coating preparation (black points: mean ± S.E.M.; red line: linear fit, R2 = 0.93; n = 60 events).
The force developed by ensembles of kinesin-5 is overlap length-dependent for sliding antiparallel microtubule pairs
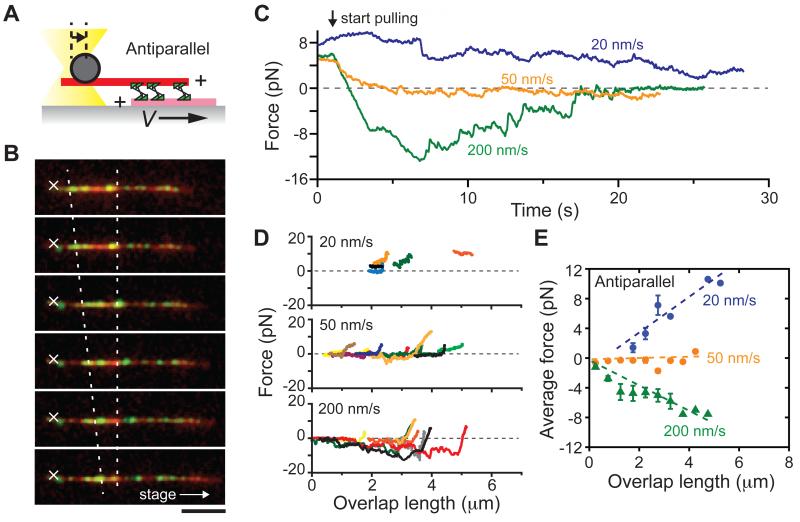
At the equator of the metaphase spindle, antiparallel microtubules continuously slide apart at ~2-3 μm/min (~30-50 nm/s), with minus ends pointing towards opposite spindle poles (Sawin and Mitchison, 1991). To test whether kinesin-5 can generate overlap-length dependent forces between microtubules that are moving relative to each other, we moved the sample stage at constant velocity to pull apart pairs of antiparallel microtubules crosslinked by kinesin-5 (Figure 4A, B and S3). When microtubule minus-ends in the ‘sandwich’ were moved apart at a velocity of 20 nm/s, we found that kinesin-5 generated substantial pushing force that assisted filament sliding (blue, Figure 4C). Further, the magnitude of this pushing force decreased as the two microtubules moved apart and filament overlap shortened. In contrast, when microtubules were moved at faster relative velocities (200 nm/s), resisting forces were developed that oppose filament motion (green, Figure 4C). The magnitudes of these resisting forces also decreased as the two microtubules moved apart and filament overlap shortened. A similar response was observed for several microtubule pairs with different initial overlap lengths (n = 5-14, Figure 4D). The average force, calculated using 0.5 μm bins of filament overlap length, increased with antiparallel microtubule overlap for both assisting and resisting forces generated by kinesin-5 (Figure 4E). Interestingly, when the sliding velocity was set to 50 nm/s, which is close to the unloaded rate of kinesin-5 driven relative microtubule sliding (Kapitein et al., 2005), we observed a force value near zero for a duration of many seconds (orange, Figure 4C-E). Together, these data indicate that within dynamically moving antiparallel microtubule pairs, ensembles of kinesin-5 can generate either pushing or resisting forces depending on the filament sliding velocity, and the magnitudes of these forces also scale with filament overlap length.
Figure 4. Kinesin-5 can generate overlap-length dependent force within sliding antiparallel microtubule pairs.
(A) Schematic of the assay, with representation similar to Figure 1A. The stage was moved at constant velocity (V) in the direction that moved microtubule minus-ends apart.
(B) Representative fluorescence images obtained at different time points during the assay. Merged images of microtubules (red) and kinesin-5 (green) are shown. The trap center is shown (white ‘X’). The stage was moved along the indicated direction (white arrow) at 200 nm/s, resulting in a decrease in the length of filament overlap (two dotted lines). Scale bar, 5 μm. Time between images, 5 s.
(C) Sample time records of force (filtered using 200-ms moving average) developed within kinesin-5-crosslinked antiparallel microtubule pairs that were moved apart at the indicated velocities. The force value at t = 0 corresponds to the maximum plateau force developed for non-moving antiparallel microtubule, as in Figure 2.
(D) Relationships between the developed force and overlap length at different velocities. At each time point in (C), the length of microtubule overlap was calculated based on the extent of initial filament overlap and the travel distance of the stage, and plotted against the measured force value (colors indicate data from different microtubule pairs, n = 5-14 per condition).
(E) Average developed forces for different overlap lengths and sliding velocities. For each sliding velocity examined, the forces measured for microtubule pairs in (D) were pooled for 0.5 μm bins of overlap length and averaged (mean ± S.D. are shown). Results of linear regression analyses are shown (dotted lines); slopes are 2.5, 0.4 and - 1.3 (R2 values: 0.88, 0.26 and 0.90) at 20, 50 and 200 nm/s, respectively.
Parallel microtubules experience small bidirectional fluctuations whose amplitude is nearly independent of the amount of filament overlap
Within metaphase spindles, kinesin-5 also accumulates at spindle poles (Compton, 2000; Gaglio et al., 1996; Sawin et al., 1992) where microtubules are arranged in a predominantly parallel geometry. We therefore examined how forces develop within parallel microtubules crosslinked by kinesin-5 (Figure 5A). Consistent with earlier studies (Hentrich and Surrey, 2010; Kapitein et al., 2005), fluorescence imaging revealed no clear directional motion of parallel filament sliding (data not shown). Remarkably, however, the optically-trapped bead attached to a microtubule in the parallel ‘sandwich’ revealed a dynamic back-and-forth motion around the trap center (Figure 5B). This motion requires the activity of kinesin-5, as the fluctuations were significantly larger than thermal noise and were not seen in the presence of a slow-hydrolyzing ATP analog, AMPPNP (2 mM) (bottom trace, Figure 5B). In this geometry, the microtubule motion and the associated forces are in marked contrast to those within an antiparallel configuration (Figure 2C).
Figure 5. Kinesin-5 force generation within static and sliding parallel microtubule pairs.
(A-G) Measurement of force developed within non-moving parallel microtubule pairs crosslinked by kinesin-5. (A) Schematic of the assay, with the representation similar to Figure 1A. The optically-trapped bead captured either the plus- or the minus-end of the microtubule. (B) Time records of force (filtered using 200-ms moving average) developed within parallel microtubule pairs crosslinked by kinesin-5. Example traces from microtubule pairs of different overlap lengths are shown (top three traces: 2mM MgATP; bottom trace: 2 mM MgAMPPNP). (C) Microtubule ‘flipping’ assay. Fluorescence images show an antiparallel microtubule pair (upper image) to which a bead was attached and the force was measured (upper trace). The microtubule was then rotated by 180° and re-attached to the same su rface microtubule (lower image). The force record obtained from the ‘flipped’ filament pair is shown (lower trace). (D-F) The maximum developed force versus microtubule overlap length or the number of kinesin-5 molecules. The maximum developed force (Figure S4A), was plotted against the overlap length (D). Pearson's r = 0.01 (n = 21). Following a force measurement of >30 seconds, the number of kinesin-5 molecules at the overlap was estimated for several microtubule pairs using the rupture counting method (n = 13) (E) and from the GFP signal intensity (n = 18) (F). Linear regressions yield slopes of 0.006, 0.06 and 0.004 for (D-F), respectively. (G) Fourier spectral density obtained from force development records of parallel microtubule pairs. Shown are the analysis results for data in (B) (2 mM MgATP).
(H-L) Measurement of force developed within kinesin-5-crosslinked parallel microtubules that are moved apart. (H) Schematic of the assay, with the representation similar to Figure 1A. The stage was moved at constant velocity (V). (I) Representative fluorescence images obtained at different time points (interval: 5 s) during the assay. Merged images of microtubules (red) and kinesin-5 (green) are shown. The stage was moved along the indicated direction (white arrow) at 200 nm/s. (J) Time records of force (filtered using 200-ms moving average) developed within crosslinked parallel microtubule pairs that slide apart. (K) Relationships between the developed force and overlap length shown for different sliding velocities (colors indicate data from different microtubule pairs, n = 12-20 per condition). (L) Average developed force at various overlap lengths and sliding velocities. For each sliding velocity examined, the forces measured for microtubule pairs in (K) were pooled for 0.5 μm bins of overlap length and averaged (mean ± S.D. are shown). Results from linear regression analyses are shown. Slopes are −0.20, −0.22 and −1.52 (R2 values: 0.26, 0.47 and 0.89) at 20, 50 and 200 nm/s, respectively. In all images, trap center (white ‘X’) and filament overlap regions (dashed lines) are indicated. Scale bar, 5 μm.
To ensure that the observed force fluctuations were associated with crosslinked parallel microtubules, we used a micromanipulation strategy to capture and flip a microtubule in a ‘sandwich’ for which we had previously observed relative sliding and unidirectional force generation (Figure 5C), consistent with an antiparallel geometry. Therefore, after mechanically flipping the microtubule, the resulting orientation would be parallel. We found that 7 of the 9 flipped pairs analyzed exhibited clear bidirectional force fluctuations; in the remaining two cases, no motion was observed. Importantly, when we took a microtubule from a ‘sandwich’ that was fluctuating, and flipped by 180°, we observed unidirectional force generation, consistent with kinesin-5 sliding apart antiparallel microtubules (n = 5) (Figure S4A). Together, these data indicate that the bidirectional force fluctuation is linked to the activity of kinesin-5 crosslinking parallel microtubules.
We performed force measurements for several parallel microtubule pairs (n = 20) of different overlap lengths and found that the maximally developed force, determined from the extreme values of calculated force histograms (Figure S4B), was largely independent of the length of parallel microtubule overlap and reached values of 1-3 pN, which was comparable in magnitude to the maximum force generated by single kinesin-5 molecules (Figure 5D). Importantly, the average filament overlap length, crosslinking motor number and GFP signal distributions (Figure 5D-F) were comparable to those we observed for antiparallel microtubule pairs (Figure 2D-F). Further, unlike myosin ensembles interacting with actin filaments (Placais et al., 2009), there was no characteristic single frequency of the observed fluctuations (Figure 5G). We conclude that kinesin-5 molecules are actively stepping and can generate forces while crosslinking two parallel microtubules, but the developed forces were small relative to the antiparallel case, not unidirectional, and largely independent of the length of filament overlap.
Kinesin-5 can generate substantial resistive force within parallel microtubule pairs
We next examined the force production by kinesin-5 molecules that crosslinked parallel microtubules moving relative to one another (Figure 5H, I). Remarkably, when parallel filaments were pulled, kinesin-5 generated substantial resisting forces that scaled with the filament overlap length across a wide range of velocities (Figure 5J). Time courses of the measured force exhibited prominent sawtooth-like peaks consistent with crosslinking kinesin-5 molecules being subjected to load and then unbinding, in contrast to our observations of kinesin-5 behavior in antiparallel microtubule pairs (Figure 4C). From these data we obtained two parameters: a peak force, which was the maximum value within each sawtooth-like event, and an average force, which was calculated for 0.5 μm bins of overlap length and averaged across all microtubule pairs examined. The peak and the average force both increased with overlap length (Figure 5K and L, Figure S4C). The average force also increased with the sliding velocity, and importantly, resisting forces were generated even when the filaments were moved at velocities less than kinesin-5's unloaded velocity (Figure 5L). These data indicate that multiple kinesin-5 molecules can crosslink sliding parallel microtubules and transmit a resistive force across the adjacent filaments in an overlap length- and velocity-dependent manner.
Numerical simulations suggest molecular mechanics of the kinesin-5 tetramer
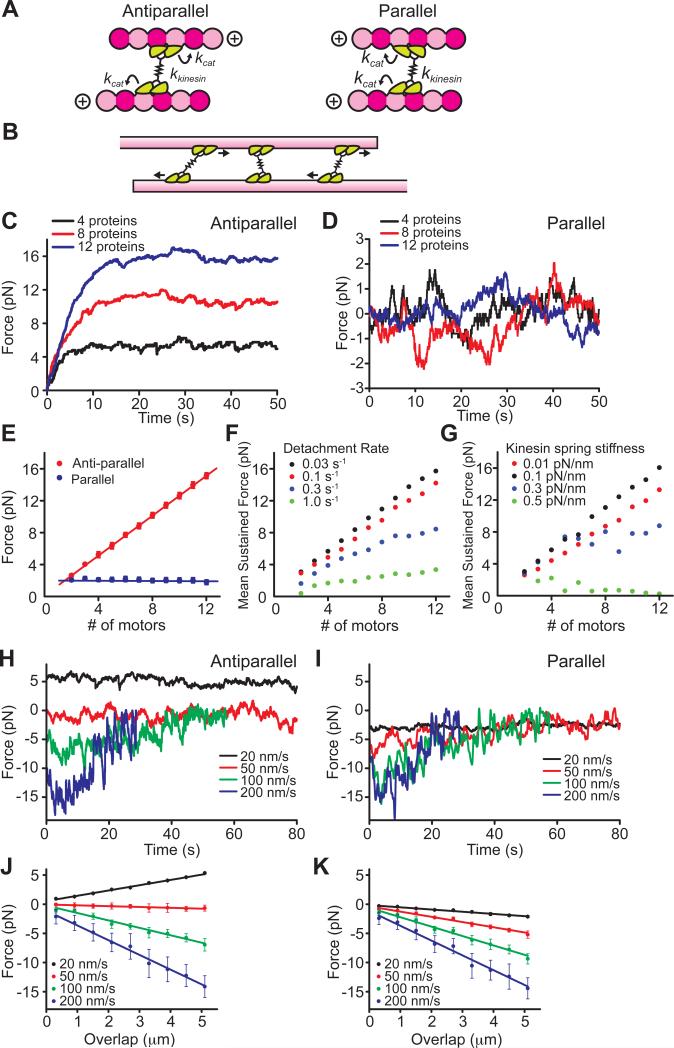
To understand how these distinct mechanical outputs could arise from ensembles of kinesin-5 motors crosslinking two microtubules, we performed numerical model simulations similar to previous reports (Grill et al., 2005; Kunwar and Mogilner, 2010; Kunwar et al., 2008) (Figure 6). In a microtubule overlap region, kinesin-5 molecules can be thought of as a series of force-generating elements that must each step towards the plus-end of a microtubule. Full-length kinesin-5 has been shown to have a ‘dumbbell’ shape, where pairs of motor domains are linked by a tetramerization domain that extends >60 nm (Figure 6A) (Kashina et al., 1996). However, in the framework of a crosslinked microtubule pair, the stochastic stepping behavior of any one homo-tetramer will be influenced by the forces generated by other molecules within the overlap region (Figure 6B). These forces would be transmitted along each microtubule, and across the kinesin-5 tetramerization domain. Microtubules are known to be extremely stiff on the micron length-scale and would readily transmit forces without dissipating mechanical energy. In contrast, the mechanism of force transmission across the tetramerization domain is unclear, but is approximated here as a linear mechanical spring. Monte Carlo simulations were performed using parameters measured in this work and reported values for the force-dependent modulation of dimeric kinesin-5 motor domain activity (Valentine et al., 2006). We also incorporate force-dependent diffusion and unbinding terms that account for the generation of frictional resistance along and detachment from the microtubule surface (see Supplemental Information for details and model parameters). The simulation was performed over a given range of motor numbers, possible ‘spring’ stiffness values and detachment frequencies (Figure S5).
Figure 6. Numerical model simulations for force generation of kinesin-5 ensembles.
(A, B) Schematics of the model simulation. A kinesin-5 tetramer (green) is represented as a pair of dimeric motor domains connected by a linear mechanical spring (kkinesin) (A). Between parallel and antiparallel pairs, the direction of one of the two pairs of motor domains of a kinesin-5 is rotated by 180°. The individual dimers step towards the plus-ends of the microtubule tracks in a load-dependent manner (kcat). Multiple tetrameric motor molecules can crosslink two adjacent microtubules (B).
(C-K) The force-generating characteristics of parallel and antiparallel microtubule pairs crosslinked by ensembles of kinesin-5. (C, D) Example time courses of force development from simulations performed using N = 4, 8, and 12 motors. The simulation was run in both antiparallel (C) and parallel (D) geometries, and the average maximum developed force was plotted against the motor number (E). (F, G) Simulations of antiparallel microtubule force generation performed while varying two model parameters. Force versus motor protein number relationships are shown for simulations run using different motor detachment rates (F) and stiffness constants of the kinesin-5 linear spring (G). (H, I) Example time courses of force development simulated at constant relative sliding velocities. For both antiparallel (H) and parallel (I) geometries, simulations were run at indicated sliding velocities. The average force developed was calculated at each 0.5 μm bins of overlap and plotted against the filament overlap length (J, K). Plots are mean ± S.D. (n = 100 simulation runs per condition).
An essential outcome of these simulations is that identical model assumptions and parameter values are sufficient to recapitulate the qualitative and quantitative characteristics of the force-generating properties of kinesin-5 crosslinking two microtubules in both antiparallel and parallel geometries. Without assuming any torque or angular orientation-dependence of the linker spring, the model reproduced the experimentally observed persistent, unidirectional force development with fluctuations in antiparallel microtubule pairs (Figure 6C), and also the bidirectional force fluctuations about a mean force of zero in parallel pairs (Figure 6D). Remarkably, the maximally developed force exhibited the same dependencies on overlap length both for parallel and antiparallel microtubule pairs, without requiring changes in the stepping or unbinding parameters (Figure 6E). Our simulations suggest that the choice of several of the parameters was particularly critical in order to recapitulate the experimental observations. First, the force-dependent rate of motor unbinding (kdetach) must be small enough to ensure prolonged (e.g. ~30 s) attachment with the microtubule. If the value chosen was too large, the motor domains detached rapidly and sustained force generation could not be achieved (Figure 6F and S5C, D). Second, the constant describing the spring-like tetramerization domain needed to be large enough to ensure force transmission, but not so large as to generate interference and promote stalling or detachment of the other kinesin-5 molecules when individual motor proteins took steps (Figure 6G). The brake-like resistive forces developed across microtubule pairs also exhibited similar dependencies on relative sliding velocity (Figure 6H-K) as observed experimentally. Together, our simulations suggest that individual kinesin-5 proteins do not significantly alter their mechano-chemical stepping properties when crosslinking different geometries, maintain long-lived contacts with microtubules during crosslinking, and are sufficiently compliant to transmit forces across microtubule pairs.
DISCUSSION
Successful cell division requires the proper organization of the bipolar mitotic spindle, which is accomplished in part by the microtubule sliding activity of kinesin-5. We find that the force developed by ensembles of kinesin-5 can be regulated by the geometry and relative motion of the microtubule filaments that they crosslink. Kinesin-5 can push apart overlapping antiparallel microtubules with forces that scale linearly with both overlap length and motor protein number. Kinesin-5 can also generate brake-like forces that scale with overlap length to resist filament sliding motions in both parallel and antiparallel geometries. These properties differ significantly from other classes of motor protein ensembles, such as those transporting vesicles or those pulling actin filaments, and may play important roles in proper assembly and function of the mitotic spindle.
Force generation by ensembles of kinesin-5 molecules crosslinking two microtubules
Studies of force generation by ensembles of other kinesins indicate that the magnitude of sustained force production can be largely unaffected by changes in motor protein number (Furuta et al., 2013; Shubeita et al., 2008; Vershinin et al., 2007). Current models suggest that this could be due to interference between motors, wherein the force generation by one protein can promote the detachment of other motor proteins from the microtubule track (Gross et al., 2007; Holzbaur and Goldman, 2010).
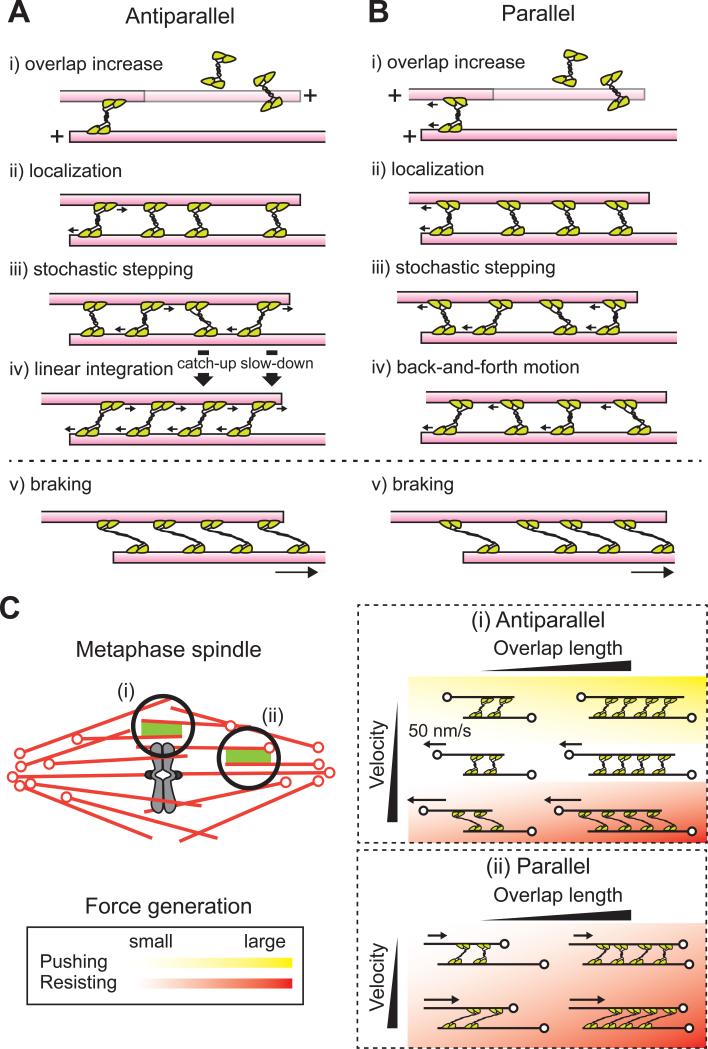
Based on our findings we propose a simple mechanism for how overlap length-dependent pushing and braking forces are produced by kinesin-5. Kinesin-5 molecules can bind single microtubules and may crosslink a second microtubule when encountering a region of filament overlap. More crosslinking kinesin-5 proteins will be recruited to regions with longer overlap lengths. Crosslinking motor proteins step stochastically towards plus-ends according to load-dependent kinetics, with stepping probability reduced by opposing force. When one kinesin-5 takes a force-producing step, this force is transmitted along the rigid microtubule and is distributed along the other kinesin-5 proteins in the ensemble. Differences in force across the kinesin-5 molecules will allow ‘leading’ or faster motor molecules in the ensemble to slow down and ‘hindering’ or slower motor molecules to step more frequently to catch-up with other molecules. Within antiparallel overlap regions, each kinesin-5 steps towards plus-ends until the amount of force across each molecule reaches the stall force and a linear integration of force output is achieved. In parallel overlap regions, kinesin-5 molecules step towards the plus-ends of microtubules they crosslink and stochastic changes in forces across the molecules give rise to a fluctuating total force output. When microtubules are sliding in either antiparallel or parallel geometries, kinesin-5 can maintain its association with both of the filaments for a sustained period of time and generate mechanical resistance. Fewer crosslinking proteins are available at shorter filament overlap, leading to an overall decrease in the braking force. This range of scenarios that we have examined is summarized in Figure 7A and B.
Figure 7. Model for force generation by ensembles of kinesin-5 crosslinking microtubules.
(A, B) A model for how ensembles of kinesin-5 generate and transmit force within overlapping microtubules. Antiparallel (A) and Parallel (B) cases are shown. An increase in the extent of filament overlap leads to the increased localization of kinesin-5 molecules (i-ii). These motors can step stochastically towards the plus-end of each microtubule while experiencing strains from the stepping of other motors. Because individual motors follow the same load-dependent kinetics, ‘leading’ motors slow down while ‘hindering’ motors speed up, and all motors maintain attachment to microtubules for many seconds (iii). In the antiparallel case, all the motors can reach their maximum force-generating state and the unitary motor force can be linearly summed up across the micron-sized overlap (iv). In the parallel case, the filament experiences back-and-forth motion (iv). The crosslinking motors can maintain their association with the sliding filament pairs and generate resisting force in both parallel and antiparallel geometries (v).
(C) Model for how kinesin-5's force-generating function is regulated by microtubule motion and geometry in the metaphase spindle. (i) Antiparallel case. For velocities less than kinesin-5's unloaded rate (~50 nm/s), pushing forces are generated. For faster velocities, resisting forces are generated and the magnitudes increase with velocity. Both pushing and resisting forces increase with filament overlap length. (ii) Parallel case. Kinesin-5 generates resisting forces whose magnitudes increase with overlap length and filament sliding velocity. Heat map shows the magnitude and direction of the developed force. Green: pushing force; red: resisting force; darker colors: higher magnitudes; clear area: essentially no force. Arrows indicate filament motion. Microtubule minus-ends are marked (o).
The best-characterized case of motor protein-dependent force generation that scales with a simple geometric feature, similar to what we observe for kinesin-5 sliding two antiparallel microtubules, is for myosin in the muscle. To generate force outputs that scale with microtubule overlap depends on the linear integration of forces generated by multiple motor protein molecules that must perform work without experiencing interference by other proteins in the ensemble. In the case of the muscle, myosin's ‘weak’ affinity for actin is likely to play a key role. Within an ensemble, a single myosin motor immediately detaches from the actin filament lattice upon completion of its force-generating power stroke, and will therefore not influence the activities of other motors that generate force in the subsequent time period. This is not likely to be the case for kinesin-5 ensembles. While previous force measurements of a truncated dimeric construct of human kinesin-5 attached to an optically-trapped bead have shown that the motor domains detach from the microtubule lattice after an average of only 8 steps (Valentine et al., 2006), we have shown that full-length kinesin-5 binds microtubules via its motor domain and a second, non-motor domain that together greatly enhance its microtubule-association lifetime (Weinger et al., 2011). Therefore, multiple kinesin-5 molecules are likely to be simultaneously associated with the microtubule such that the forces generated by one active motor will be transmitted to the others along the rigid microtubules. Studies of Drosophila kinesin-5 have reported the detailed architecture of its tetramerization domain (Scholey et al., 2014). Our analyses suggest that this domain is likely rigid enough to transmit forces across the molecule's pairs of motor domains, but is compliant enough to avoid detachment due to the force-producing activities of other motor proteins in the ensemble. In addition, this tetramerization domain may have considerable torsional flexibility, allowing pairs of motor domains at either end of the molecule to achieve stereospecific interactions with microtubules in both antiparallel and parallel geometries. Together, the structural and kinetic properties of kinesin-5 appear to provide an ideal framework for efficient transmission of forces from one molecule to another across their rigid microtubule substrate.
Implications for kinesin-5's spindle assembly functions
Within mitotic spindles, antiparallel microtubules overlap at the equator of the bipolar structure, and long-standing models for the spindle assembly have posited that the force pushing apart those overlapping filaments is somehow regulated in a length-dependent mechanism (McIntosh et al., 1969; Scholey et al., 2003). Here we have demonstrated that an ensemble of kinesin-5 can achieve this function. With this property, the length of the bipolar structure can be set at a single stable point where pushing and pulling forces are balanced. Notably, this force was found to be overlap length-dependent even within dynamically sliding microtubule pairs, not simply for filaments that do not move once steady-state size is achieved (Figure 7C, inset (i)). It has been known that antiparallel microtubules overlapping at the spindle equator exhibit a persistent bidirectional flow towards each spindle pole (Sawin and Mitchison, 1991). The microtubule flux within the bipolar spindle moves at varying velocities over time and space, with the average flux velocity of 2-3 microns per minutes (Yang et al., 2007). We predict that stochastic reduction or increase in filament sliding relative to kinesin-5's unloaded velocity would lead to either assisting or braking forces due to kinesin-5, regulating filament sliding velocity depending on the extent of filament overlap.
In vertebrate meiotic spindles microtubule minus-ends have been shown to distribute along the length of the bipolar structure (Burbank et al., 2006), suggesting that both parallel and antiparallel overlap geometries are found throughout the entire structure. Nearer the poles, microtubules are predominantly parallel and, in the steady-state structure, continuously translocate polewards at matching velocities (Yang et al., 2007). How such coordination is achieved across the >10 micron width of the spindle has not been clear. Our findings suggest that kinesin-5 could couple the motion of these polewards fluxing parallel microtubules (Figure 7C, inset (ii)). If stochastic force fluctuations arise between two parallel filaments, kinesin-5 would generate a braking force that would be greater at higher relative velocity. As the braking force also scales with parallel filament overlap, the motions of longer microtubules will be more strongly coupled. Conversely, shorter microtubules may be more readily transported at velocities that are higher than that of polewards flux, consistent with analysis of short microtubule ‘seed’ dynamics in the metaphase spindle (Heald et al., 1996). Additionally, consistent with our proposal, it has been shown that shorter microtubules are transported to spindle poles with higher probability than longer microtubules, and this ‘sorting’ mechanism is linked to the activity of kinesin-5 (Brugues et al., 2012). Together, our findings suggest that kinesin-5 may contribute to spindle size regulation, and also govern the velocity at which microtubules slide in the steady-state metaphase spindle. It is also possible that the kinesin-5-dependent crosslinking of kinetochore and nonkinetochore microtubules, which have parallel orientations near the poles, may contribute to the length-dependent forces pulling chromosomes polewards (Hays et al., 1982). Recent cell biological studies suggest that the targeting of kinesin-5 to spindle microtubules involves TPX2, a non-motor MAP (Eckerdt et al., 2008; Gable et al., 2012; Helmke and Heald, 2014). It is likely that our ‘minimal spindle’ assay will help dissect how this regulatory mechanism contributes to relative microtubule sliding.
In summary, our findings reveal how kinesin-5 molecules working in concert can ‘read’ micron-scale cues to generate a mechanical signal, essentially converting a geometric feature within a dynamic filament network into an analog force output. Micron-scale patterns of post-translational modification of protein substrates (Fuller et al., 2008) or filament-length dependent protein localization patterns of nanometer-sized proteins (Leduc et al., 2012; Subramanian et al., 2013) have been characterized and can play important roles in regulating cytoskeletal self-organization. We propose that micron-scale mechanical regulation supplements such chemical regulation in the metaphase spindle. It is also likely that ensembles of other enzymes, required for the assembly and function of biological polymers ranging from cytoskeletal filaments to DNA, also function in distinct modes that depend on the relative geometry and dynamics of their substrates.
EXPERIMENTAL PROCEDURES
Proteins
Full-length Xenopus laevis kinesin-5 tagged at the C-terminus with EGFP was expressed and purified as previously described (Kwok et al., 2006). Microtubules were polymerized from a mixture of X-rhodamine tubulin, biotinylated tubulin and unmodified tubulin with 1 mM GMPCPP and stabilized by Taxol. A rigor kinesin mutant, used for bead-microtubule attachment, was expressed and purified as previously described (Rice et al., 1999).
Assays
Measurement of kinesin-5 force development was performed on an inverted microscope equipped with force-calibrated optical tweezers and dual-mode TIRF and Epifluorescence imaging optics (Forth et al., 2014). To generate microtubule ‘sandwiches’ crosslinked by kinesin-5, a flow chamber assembled with a PEG-coated coverslip (Subramanian et al., 2013) was incubated with reagents in the following order: 0.5 mg/ml α-casein for 3 min, 0.2 mg/ml neutravidin for 5 min, biotinylated microtubules for 10 min, and motility sample (non-biotinylated microtubules, 1 nM kinesin-5-GFP, 2 mM ATP, 3 mM MgCl2, 1 mM EGTA, 60 mM PIPES (pH 6.8), 4.5 mg/ml glucose, 350U/ml glucose oxidase, 34U/ml catalase, 0.5 mg/ml α-casein, 1 mM DTT, 20 μM Taxol and ~0.1 pM beads). After sealing the chamber with VaLaP, a microtubule pair was selected and an optically-trapped bead was attached to the non-biotinylated microtubule within 2-5 microns of the region of microtubule overlap to prevent filament buckling. The bead position was recorded using a quadrant photodiode (0.1-0.5 kHz sampling rate). Microtubules and kinesin-5-GFP were imaged (interval: 1-5 s; exposure time: 200-300 ms), in parallel with the force measurement. For measurements within sliding microtubule pairs, the piezo stage was moved along the microtubule axis at constant velocity once the kinesin-5 dependent force reached a plateau. The trapped bead was held ~1 μm above the coverslip surface, where the trap stiffness was determined to be 0.065 pN/nm. For all experiments, beads were held within ±250 nm of the trap center to maintain a linear trap stiffness.
The rupture-based counting method was performed by moving the sample stage orthogonal to the surface microtubule at a speed of 0.3 μm/s while holding the trap center at a fixed position. The motion of the trapped bead was recorded at 0.5 kHz and processed as described to estimate the rupture number.
For microtubule flipping experiments, the bead-held microtubule was detached from the surface microtubule and then flipped by 180° using fluid flow induced by motion of the sample stage. The flipped filament was then brought into contact with the same surface microtubule.
Data analysis
The maximum force developed within antiparallel microtubule pairs was determined at the plateau of the force development record, defined as the region where the force signal persisted for more than 1 s without significant fluctuation or sudden drop (<10% change in force). For parallel microtubule pairs, the force record was used to generate a histogram (0.25 pN bin width) and the values of its two end columns were averaged.
The microtubule overlap length was determined from images of microtubules taken during the force plateau. The region where the X-rhodamine signal along the microtubule axes was more than twice that of single microtubules was determined to be the region of overlap. Within moving microtubule pairs, the filament overlap length at time t was calculated according to L = L0 - V•t + Δx, where L0, V, Δx are the initial overlap length, stage velocity and displacement of the bead from the trap center, respectively. To estimate the number of kinesin-5 molecules within a microtubule overlap region, the total intensity of GFP signal within the overlap was divided by the average intensity of single kinesin-5-GFP tetramers.
Model simulations
A detailed description of the model and simulation parameters can be found in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Sarah Rice (Northwestern University) for the rigor kinesin plasmid, JST PRESTO (Y.S.), NIH NRSA fellowship GM099380 (S.F.) and NIH/NIGMS GM65933 (T.M.K.) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.S. and T.M.K. designed experiments. Y.S. and S.F. built the microscope system. Y.S. and S.F. performed experiments and analyzed data. S.F. performed computer simulations. Y.S., S.F. and T.M.K. wrote the manuscript.
The authors declare no competing financial interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and extended experimental procedures, and can be found with this article online.
REFERENCES
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Brugues J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Burbank KS, Groen AC, Perlman ZE, Fisher DS, Mitchison TJ. A new method reveals microtubule minus ends throughout the meiotic spindle. J Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA. Spindle assembly in animal cells. Annu Rev Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Crevel IM, Alonso MC, Cross RA. Monastrol stabilises an attached low-friction mode of Eg5. Curr Biol. 2004;14:R411–412. doi: 10.1016/j.cub.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr Biol. 2008;18:519–525. doi: 10.1016/j.cub.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S, Hsia KC, Shimamoto Y, Kapoor TM. Asymmetric Friction of Nonmotor MAPs Can Lead to Their Directional Motion in Active Microtubule Networks. Cell. 2014;157:420–432. doi: 10.1016/j.cell.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K, Furuta A, Toyoshima YY, Amino M, Oiwa K, Kojima H. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci U S A. 2013;110:501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable A, Qiu M, Titus J, Balchand S, Ferenz NP, Ma N, Collins ES, Fagerstrom C, Ross JL, Yang G, et al. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol Biol Cell. 2012;23:1254–1266. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Kruse K, Julicher F. Theory of mitotic spindle oscillations. Phys Rev Lett. 2005;94:108104. doi: 10.1103/PhysRevLett.94.108104. [DOI] [PubMed] [Google Scholar]
- Gross SP, Vershinin M, Shubeita GT. Cargo transport: two motors are sometimes better than one. Curr Biol. 2007;17:R478–486. doi: 10.1016/j.cub.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Hays TS, Wise D, Salmon ED. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J Cell Biol. 1982;93:374–389. doi: 10.1083/jcb.93.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J Cell Biol. 2014;206:385–393. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. Journal of Cell Biology. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur EL, Goldman YE. Coordination of molecular motors: from in vitro assays to intracellular dynamics. Curr Opin Cell Biol. 2010;22:4–13. doi: 10.1016/j.ceb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison DK, Driver JW, Diehl MR. Cooperative responses of multiple kinesins to variable and constant loads. J Biol Chem. 2012;287:3357–3365. doi: 10.1074/jbc.M111.296582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev MJ, Lakamper S, Schmidt CF. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur Biophys J. 2007;36:675–681. doi: 10.1007/s00249-007-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar A, Mogilner A. Robust transport by multiple motors with nonlinear force-velocity relations and stochastic load sharing. Phys Biol. 2010;7:16012. doi: 10.1088/1478-3975/7/1/016012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar A, Vershinin M, Xu J, Gross SP. Stepping, strain gating, and an unexpected force-velocity curve for multiple-motor-based transport. Curr Biol. 2008;18:1173–1183. doi: 10.1016/j.cub.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- Leduc C, Padberg-Gehle K, Varga V, Helbing D, Diez S, Howard J. Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc Natl Acad Sci U S A. 2012;109:6100–6105. doi: 10.1073/pnas.1107281109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K, Pickett-Heaps JD, McIntosh JR, Tippit DH. On the mechanism of anaphase spindle elongation in Diatoma vulgare. J Cell Biol. 1977;74:377–388. doi: 10.1083/jcb.74.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Hepler PK, Van Wie DG. Model for mitosis. Nature. 1969;224:659–663. [Google Scholar]
- Mogilner A, Wollman R, Civelekoglu-Scholey G, Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Nishizaka T, Miyata H, Yoshikawa H, Ishiwata S, Kinosita K., Jr. Unbinding force of a single motor molecule of muscle measured using optical tweezers. Nature. 1995;377:251–254. doi: 10.1038/377251a0. [DOI] [PubMed] [Google Scholar]
- Placais PY, Balland M, Guerin T, Joanny JF, Martin P. Spontaneous oscillations of a minimal actomyosin system under elastic loading. Phys Rev Lett. 2009;103:158102. doi: 10.1103/PhysRevLett.103.158102. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rozelle DK, Hansen SD, Kaplan KB. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J Cell Biol. 2011;193:285–294. doi: 10.1083/jcb.201011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife. 2014;3:e02217. doi: 10.7554/eLife.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Brust-Mascher I, Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, Welte MA, Gross SP. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154:377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Block SM. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys J. 2009;97:1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci U S A. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Yang G, Houghtaling BR, Gaetz J, Liu JZ, Danuser G, Kapoor TM. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.