Abstract
RNA interference is a conserved homology-dependent post-transcriptional/transcriptional gene silencing mechanism in eukaryotes. The filamentous fungus Neurospora crassa is one of the first organisms used for RNAi studies. Quelling and meiotic silencing by unpaired DNA are two RNAi-related phenomena discovered in Neurospora, and their characterizations have contributed significantly to our understanding of RNAi mechanisms in eukaryotes. A type of DNA damage-induced small RNA, microRNA-like small RNAs and Dicer-independent small silencing RNAs were recently discovered in Neurospora. In addition, there are at least six different pathways responsible for the production of these small RNAs, establishing this fungus as an important model system to study small RNA function and biogenesis. The studies in Cryphonectria, Mucor, Aspergillus and other species indicate that RNAi is widely conserved in filamentous fungi and plays important roles in genome defense. This review summarizes our current understanding of RNAi pathways in filamentous fungi.
Keywords: RNAi, Quelling, Meiotic silencing, MicroRNA, Dicer-independent small RNAs, siRNA, qiRNA
Introduction
RNA interference (RNAi) is a conserved eukaryotic gene silencing mechanism at both post-transcriptional and transcriptional levels. It is mediated by small noncoding RNAs (sRNAs) of about 20–30 nucleotides [1–4]. In RNAi pathways, the RNase III domain-containing Dicers generate the small RNA duplexes from double-stranded RNA (dsRNA) precursors, then the small RNA duplexes are loaded onto the RNA-induced silencing complex (RISC) in which an Argonaute family protein functions as the core catalytic component. Following the removal of the passenger strand of the small RNA duplex, the RISC is activated and uses the remaining single-stranded small RNA as a guide to silence the target RNAs [5–11].
In addition to the Dicer-dependent microRNAs (miRNAs) and small interfering RNAs (siRNAs), Dicer-independent small RNAs such as PIWI-interacting RNAs (piRNAs) were also discovered (see reviews in [3, 4, 7, 12–14]). miRNAs are processed from single-stranded RNA precursor transcripts containing hairpin structures, and can function by mediating mRNA degradation, translational repression or transcriptional gene silencing. siRNAs are generated from dsRNA precursors, and can act through post-transcriptional gene silencing pathways and transcriptional gene silencing pathways. The animal piRNAs, which are derived mainly from repetitive elements, transposons and large piRNA clusters, may function to protect germline integrity, although the functions of many piRNAs are still not known.
The filamentous fungus N. crassa is one of the first eukaryotic model system for RNAi studies, and RNAi pathways in filamentous fungi have been most extensively studied in N. crassa. As an excellent experimental model system for more than 70 years, N. crassa has contributed significantly to the understanding of RNAi. RNAi is widely present in filamentous fungi, and studies and applications of RNAi in filamentous fungi have resulted in better understanding of gene regulation and RNAi functions in these organisms. This review will mainly focus on the mechanisms of the RNAi in Neurospora and discuss the studies and the applications of RNAi in other filamentous fungi.
The quelling pathway and dsRNA triggered gene silencing
Discovery of quelling
Quelling, the first transgene-induced gene silencing phenomenon described in fungi, was discovered in N. crassa by Macino and colleagues [15, 16], soon after the discovery of co-suppression in plants. It is the second post-transcriptional gene silencing mechanism reported in eukaryotes. Quelling was originally found by transforming exogenous albino-1 (al-1) or albino-3 (al-3) sequences, two of the structural genes required for biosynthesis of carotenoids, into an orange wild-type strain, which resulted in silencing of the endogenous al-1 or al-3 genes indicated by the albino (white)/pale yellow phenotype in some of the transformants and reduced al mRNA levels [15, 17, 18]. This silencing phenomenon is spontaneously reversible, as some of the albino transformants could revert back to wild-type or intermediate phenotypes, likely due to the reduction of the copy number of the exogenous sequences [15]. Although quelled strains can contain only a few ectopic copies of transgenes, high copy numbers of tandem repeats of the transgenes seem to correlate with the efficiency and stability of quelling [15, 17–20].
Quelling silences both the transgene and homologous endogenous gene in vegetative tissues with the minimum length requirement of the transgene of ~130 nt. However, the promoter region is not required for and cannot induce quelling, suggesting that silencing is mediated post-transcriptionally [15, 17, 18]. Mutations of al genes are generally recessive, but most of the al quelled transformants were heterokaryons and were dominant over wild-type strains [17]. These results indicate that silencing by quelling is not nucleus-limited and can act in-trans by diffusible molecules [17]. Consistent with this notion, a transgene-derived sense RNA derived from promoter-less al-1 transgenes was specifically found in quelled strains but absent in the reverted strains, suggesting that the transcription of transgene is required for quelling [17]. The observation of the transgene-specific sense RNA led to the hypothesis that production of aberrant RNA (aRNA) in the presence of transgenes causes post-transcriptional gene silencing. This was one of the earliest studies that suggested that the production of aberrant RNA (aRNA) is involved in gene silencing mechanisms.
Cloning of the quelling deficient genes
Using a stably al-1 quelled strain, Cogoni and Macino [18] isolated 15 quelling deficient (qde) mutants, belonging to three distinct genetic loci: qde-1, qde-2 and qde-3. The corresponding genes were subsequently cloned and were found to encode three key components in the quelling pathway and demonstrated that quelling is an RNAi-related phenomenon [18, 21–23]. QDE-1 (quelling deficient-1) is the first RNAi gene ever identified, which encodes a cellular RNA-dependent RNA polymerase (RdRP) [21]. The cloning of QDE-1 soon after the landmark study by Fire et al. [24] provided the first experimental evidence that an RdRP is involved in PTGS and suggested a model that aRNAs produced from transgenes are used as templates by an RdRP to produce dsRNA [20, 21]. The wide presence of QDE-1 homologues in plants, fungi and C. elegans indicate that a conserved PTGS mechanism involving RdRP may exist in all these organisms. Indeed, it was later shown that RdRPs in Arabidopsis and C. elegans are required for RNAi [25–27].
The cloning of qde-2 revealed that it encodes for an Argonaute protein that is homologous to the rde-1 gene in C. elegans, which is required for the dsRNA-induced silencing [23]. This result provided the first experimental evidence that RNAi and transgene-induced PTGS have a common genetic component and that they evolved from the same ancestral mechanism. Together with the requirement of an RdRP in PTGS, these results demonstrate that quelling Neurospora and RNAi in plants and animals are mechanistically linked gene silencing phenomena [28].
The qde-3 gene encodes for a RecQ DNA helicase homologous to the human Werner/Bloom’s syndrome proteins [22], suggesting that regulation of DNA structure is a critical step in quelling. Although the exact role of QDE-3 in quelling is still largely unknown, it was thought to act upstream of the dsRNA formation and is important for the generation of aRNA/dsRNA from the transgenic loci [22, 29]. QDE-3 and another RecQ DNA helicase RecQ-2 also play roles in DNA repair, although the relationship between DNA repair and quelling is not clear [22, 30, 31]. OsRecQ1, a QDE-3 homologue in rice, was later found to be required for RNA silencing induced by the introduction of inverted-repeat DNA, but not for dsRNA induced RNA silencing, which is similar to QDE-3 [32]. rRecQ-1, a homologue of QDE-3 in rats was reported to be associated with piRNA-binding complex [32–34].
Catalanotto et al. [35] later showed that small RNAs of about 25 nt were found to be specifically involved in quelling and were associated with QDE-2. The production of these small RNAs required qde-1 and qde-3, but not qde-2. Genes for two partially redundant Dicer proteins DCL-1 (Dicer-like-1) and DCL-2 (Dicer-like-2) were further identified and characterized by reverse genetics as a result of the release of the whole genome sequence of N. crassa [36, 37]. In 2007, our lab identified QIP, a QDE-2-interacting exonuclease, as another key component required for RNAi in Neurospora [8].
Mechanism of quelling and production of dsRNA
Though RNAi pathways triggered by dsRNA has been extensively studied in several systems, how repetitive sequences/transgenes are distinguished from endogenous genes and how endogenous dsRNA are made is not clear. It is believed that aRNA synthesis and its specific recognition by RdRPs result in the production of dsRNA. In N. crassa, QDE-1 and QDE-3 were proposed to be required for aRNA and dsRNA production.
The RdRP activity of QDE-1 was confirmed in vitro and the crystal structure of its catalytic core was solved [33, 38–40]. The QDE-1 RdRP activity further confirmed that QDE-1 uses aRNAs as templates to make dsRNA. One surprising feature revealed by the structural study is that the catalytic core of QDE-1 is structurally similar to eukaryotic DNA-dependent RNA polymerases but not to viral RdRPs [39]. It was originally postulated that the transcription of a transgene by RNA polymerase II produces aRNA, which is used as the substrate by QDE-1 to generate dsRNA [33]. Recent evidence suggests that QDE-1 is both an RdRP and a DNA-dependent RNA polymerase and is involved in making aRNA together with QDE-3 ([41], see below).
The efficiency of transgene-induced quelling is usually around 20–30% of the total transformants [15, 33]. However, overexpression of QDE-1 could dramatically elevate the quelling efficiency. Furthermore, a few copies of transgenes were sufficient to induce silencing when QDE-1 was overexpressed. These results suggest that dsRNA production is the limiting factors for quelling. On the other hand, the activation and maintenance of transgene-induced silencing may depend on both the cellular level of QDE-1 and the copy number of the integrated transgenes, which can influence the amount of dsRNA produced [29, 33, 42, 43].
Nolan et al. [44] showed that QDE-1 interacts with RPA-1, the Neurospora homologue of the largest subunit of Replication Protein A, a single-stranded DNA-binding protein important for DNA replication, repair and recombination. In addition, QDE-1 was found to be enriched at the transgenic al-1 locus, demonstrating that QDE-1 is recruited to the transgenic locus [44]. The accumulation of siRNAs appears to be DNA synthesis dependent, as hydroxyurea treatment of mycelia, which inhibits DNA replication, abolished siRNAs accumulation [44]. Thus, it was proposed that, during replication, repetitive transgenes were distinguished by QDE-1 and RPA-1 from endogenous genes and then were targeted for silencing [44]. However, the physiological importance of RPA in gene silencing is not clear because rpa-1 is an essential gene.
We have recently shown that QDE-1 is both an RdRP and a DNA-dependent RNA polymerases (DdRP) and is required for aRNA production as QDE-3 (see below and [41]). This raises the possibility that QDE-3, the DNA helicase, and RPA may facilitate QDE-1 to bind to ssDNA at the transgenic region (QDE-3 may resolve the complex DNA structures at the transgenic locus and RPA could recruit QDE-1 to ssDNA). Afterwards, QDE-1 could act as a DdRP to generate aRNA which will be further converted into dsRNA by the RdRP activity of QDE-1 [33, 41, 44]. Future genetic and biochemical studies will be needed to test this hypothesis.
Despite the importance of QDE-1 and QDE-3 in quelling, they are not required for dsRNA-induced gene silencing [29, 41, 45]. Expressing an inverted repeat-containing transgene, which can result in the production of dsRNA, can totally bypass QDE-1 and QDE-3 to induce gene silencing with high efficiency [29, 41]. In addition, unlike some RdRPs in some other organisms, QDE-1 is not involved in the amplification and production of secondary small RNAs [41]. These results further support the notion that dsRNA is a necessary intermediate for quelling and that QDE-1 and QDE-3 both function upstream of the dsRNA production pathway. On the other hand, QDE-2 and DCLs are essential for gene silencing induced by dsRNA [29].
Generation of siRNA and the activation of RISC
Neurospora has two partially redundant Dicer proteins: Dicer-like-1 (DCL-1) and DCL-2. Both DCLs can process dsRNA into about 25-nt small RNAs in an ATP-dependent manner [36]. The double mutant of dcl-1 and dcl-2 completely abolished quelling and disrupted the processing of dsRNA into siRNA in vivo and in vitro, but the single dcl mutants had comparable quelling frequencies to the wild-type. These results suggest that the two Dicers are functionally redundant, explaining why they escaped the earlier screening for quelling defective mutation [18, 36]. However, the accumulation of siRNA was significantly reduced in the dcl-2 mutant compared to that of the wild-type, indicating that the DCL-2 is the major dsRNA processing enzyme [36].
The Argonaute protein QDE-2 is the core component of the RISC complex and is associated with siRNA [23, 35]. The siRNA associated with QDE-2 in siRNA duplex form and the RISC is inactive [8]. To activate the RISC, the passenger strand of the siRNA duplex needs to be removed. We showed that QDE-2 and its slicer activity are required for gene silencing and the generation of single-stranded siRNA from siRNA duplexes in vivo [8]. While the wild-type QDE-2 was associated with single-stranded siRNA, mutation of the qde-2 gene or the catalytic residue of QDE-2 abolished gene silencing and single-stranded siRNA production. These results provide the first in vivo experimental evidence that the cleavage of the passenger strand by Argonautes is required for single-stranded siRNA generation and RISC activation.
However, QDE-2 cleavage of the passenger strand alone is not sufficient for single-stranded siRNA production and RISC activation. Purification of QDE-2 leads to the identification of QIP, a QDE-2-interacting protein with an exonuclease domain [8]. Disruption of the qip gene resulted in the accumulation of siRNA duplexes and impairment of gene silencing. Further analyses showed that QIP functions as an exonuclease by removing the nicked passenger strand from the siRNA duplex in a QDE-2-dependent manner [8]. Thus, QIP is a critical player in dsRNA-induced gene silencing and is also the first identified eukaryotic exonuclease required for efficient RNAi. These results established that both the cleavage and removal of the passenger strand from the siRNA duplex are important steps in RNAi pathways. Recently, a Drosophila ribonuclease C3PO was demonstrated biochemically to activate RISC by removing the cleaved siRNA passenger strand in a manner that is very similar to QIP [46].
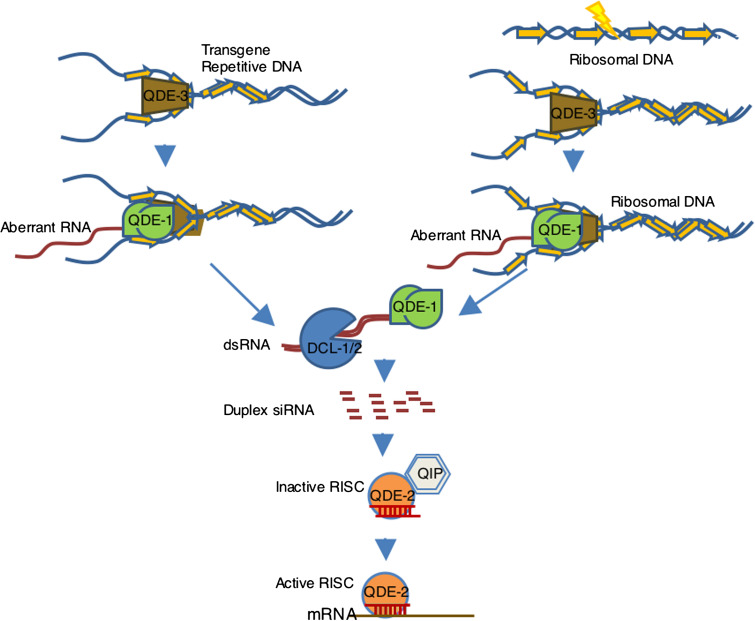
Based on these studies, a model for the Neurospora RNAi pathway was proposed [8]: dsRNA is processed into siRNA duplexes by Dicer protein(s), which are loaded onto the RISC; the Argonaute protein QDE-2 cleaves the passenger strands of the siRNA duplexes, and the exonuclease QIP removes the nicked passenger strands with the guide strands remaining in the RISC complexes; and the activated RISCs guided by the single-stranded siRNA cleaves homologous mRNAs, resulting in gene silencing (Fig. 1).
Fig. 1.
A model for the quelling and qiRNA pathway in vegetative cells in N. crassa repetitive transgenes (quelling) or the rDNA locus after DNA-damage induce the synthesis of aberrant RNAs by the DdRP activity of QDE-1 facilitated by QDE-3. The aberrant RNA is converted into dsRNAs by the RdRP activity of QDE-1. The Dicer proteins DCL-1 and DCL-2 cleave the dsRNAs into siRNAs or qiRNAs, which are then loaded onto the RNA-induced silencing complex (RISC) containing QDE-2 and QIP. QDE-2 and QIP convert the siRNA duplex into the mature siRNA, resulting in RISC activation and gene silencing of homologous RNAs
Meiotic silencing by unpaired DNA
Discovery of MSUD
Another RNAi-related mechanism in N. crassa is the meiotic silencing by unpaired DNA (MSUD) discovered by Metzenberg and colleagues. MSUD is similar to quelling but only occurs during meiosis [47–49]. N. crassa is generally haploid, although a transient diploid ascus cell is present when the two nuclei of opposite mating types fuse (karyogamy) [50]. The diploid cell quickly goes through two rounds of meiosis and then one round of mitosis, resulting in eight ascospores each containing one nucleus [47, 49, 50]. MSUD functions in the first meiotic prophase by silencing all copies of the unpaired gene during the pairing of homologous chromosomes, though the silencing effects seem to be contained within the ascus (or asci) where the unpaired DNA is present [48, 49]. MSUD was originally named meiotic transvection (or meiotic trans-sensing), referring to the phenomenon that proper function of the ascospore maturation 1 gene asm-1 and the proper maturation of ascospores requires asm-1+ being in proximity or paired to its allelic counterpart in the transient diploid zygote [47, 48, 51–53]. Further experiments demonstrated that the absence of unpaired copies of asm-1+ in the genome is required for ascospore maturation, thus the renaming of the phenomenon as MSUD [48, 53]. It was recently proposed that meiotic trans-sensing and meiotic silencing are two different steps in MSUD: the trans-sensing mechanism scans the presence or absence of paired homologous genes and the presence of unpaired gene results in the meiotic silencing of all homologous copies present in the genome [54]. This proposal is supported by the observation that DNA methylation affects meiotic trans-sensing without influencing meiotic silencing [55].
Mechanism of MSUD
MSUD appears to be a pathway that is parallel to quelling and only functions during meiosis. To understand the mechanism of MSUD, UV mutagenesis was carried out and the resulting mutants were crossed with a strain with unpaired asm-1+ to identify mutants with impaired MSUD [48, 49]. Sad-1 (suppressor of ascus dominance-1) was the first mutant identified. The cloning of sad-1 revealed that it encodes a paralog of qde-1. The mutations of sad-1 by UV mutagenesis, repeat-induced point mutation or homologous deletion suppress MSUD [48, 49]. SAD-1 shares high identities with cellular RdRPs involved in gene silencing. In addition, an unpaired DNA can trigger silencing of paired copies as well as the unpaired copy. These results indicate that MSUD is an RNAi-related phenomenon and dsRNA synthesis is required for MSUD [48].
sms-2 (suppressor of meiotic silencing-2) was identified from the Neurospora genome based on its homology to the Argonaute proteins and was demonstrated to be required for MSUD by analyzing the sms-2 loss-of-function mutants [37, 56].
sad-2, another dominant suppressor of meiotic silencing, is required for the proper localization of SAD-1 and the mutation of sad-2 suppresses MSUD [57, 58]. SAD-1 and SAD-2 co-localize in the perinuclear region, and most likely interact with each other physically in vivo based on the bimolecular fluorescence complementation (BiFC) analysis [57, 58]. Because the localization of SAD-2 in the perinuclear region is independent of SAD-1, SAD-2 may function by recruiting SAD-1 to the perinuclear region [57, 58].
DCL-1 (also called SMS-3) is one of the two Dicer proteins in N. crassa. Although it is not a dominant meiotic silencing suppressor, it is found to be required for MSUD [37, 54, 59]. A homozygous cross of dcl-1 deletion mutants is barren (the dcl-2 deletion mutants is normal), which is also true for the homozygous cross of sad-1 and sad-2 mutants, respectively [48, 57, 59]. However, although asci were observed for the homozygous cross of sad-1 and sad-2 mutants, no perithecium was observed for the dcl-1 mutants, indicating that sexual development is defective at an earlier stage for the dcl-1 mutant compared to sad-1 and sad-2 mutants [59]. Single mutants of sad-1 and sad-2 function as dominant suppressors of meiotic silencing, but none of the dcl-1 deletion mutant, the dcl-2 deletion mutant, or the dcl-1 dcl-2 double mutant could function as dominant suppressors of meiotic silencing [48, 57, 59]. By expressing dcl-1 at an early stage in the sexual cycle but not at later stages, Alexander et al. [59] demonstrated that the dcl-1 deletion mutation, but not the dcl-2 deletion mutation, suppressed the silencing of unpaired hH1-gfp, thus establishing an important role of DCL-1 in MSUD. Interestingly, for quelling, although DCL-2 is responsible for the production of most of the siRNA, DCL-1 and DCL-2 play a redundant role. In contrast, MSUD only requires DCL-1 but not DCL-2, suggesting that DCL-1 but not DCL-2 is specifically expressed during meiosis [59]. Thus, different sets of RNA-related proteins are required for MSUD and quelling, respectively, supporting the notion that two separate RNAi pathways are present in N. crassa [37, 60].
DCL-1, SMS-2, SAD-1 and SAD-2 were demonstrated to co-localize in the perinuclear region where siRNAs were shown to be accumulated in other organisms, suggesting that the perinuclear region is an active center for MSUD and small RNAs might be involved in MSUD [57, 59]. On the other hand, the requirement of the RdRP SAD-1, the Argonaute protein SMS-2 and the Dicer DCL-1 indicates that dsRNA and small RNAs are involved in the MSUD [54, 59].
Lee et al. [61] demonstrated that only the unpaired regions with homology to the reporter transcript could trigger the meiotic silencing of the reporter gene, and efficiency of silencing increases as the size and homology of the unpaired region increase. In addition, there was an unpairing-dependent loss of a reporter transcript correlated with the induction of meiotic silencing, further supporting that MSUD is post-transcriptional. Although a lot more work is needed to understand the MSUD pathway, a simple model can be proposed for MSUD [54, 59, 60] (Fig. 2): an unpaired DNA is a signal to initiate the transcription of aRNAs from the unpaired DNA region during meiosis, and the RdRP SAD-1 converts aRNA into dsRNA, which is processed by DCL-1 into small RNAs. Small RNAs are then loaded onto a SMS-2-based RISC complex, which then results in the post-transcriptional silencing of homologous genes. SAD-2 may function in the MSUD pathway by recruiting SAD-1 to the proper location to perform its activity. The future identification of small RNAs associated with SMS-2 and the understanding of the meiotic sensing mechanism will be critical to the understanding of MSUD.
Fig. 2.
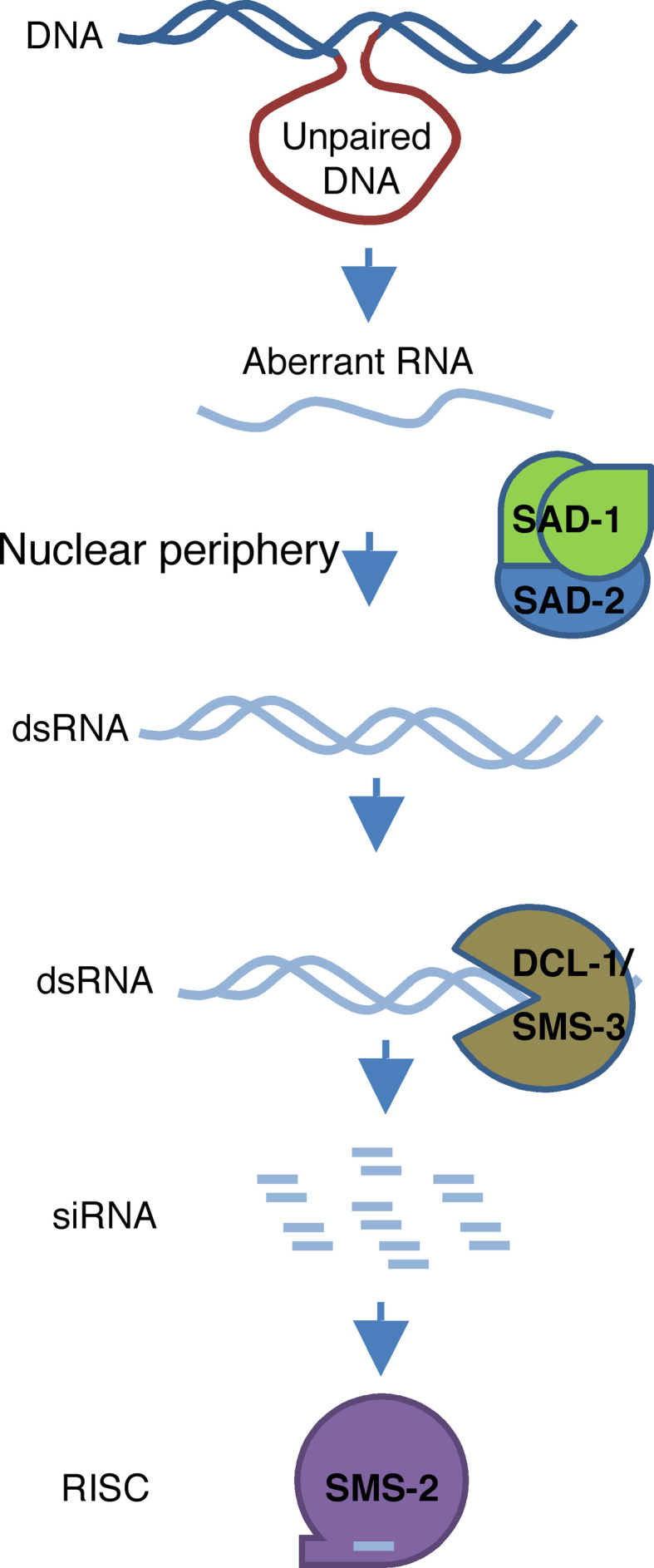
A model for meiotic silencing (MSUD) in N. crassa. During the first meiotic prophase, an unpaired DNA triggers the transcription of aberrant RNAs from an unpaired DNA region, and the RdRP SAD-1converts aberrant RNA into dsRNAs, which are processed by DCL-1 into small RNAs, and small RNAs are then loaded onto a RISC complex with the Argonaute SMS-2 as the core component. The activation of the SMS-2 complex results in the silencing of homologous RNA. SAD-2 may function in the pathway by recruiting SAD-1 to its proper location to perform its activity
Functions of RNAi in Neurospora
RNAi play roles in genome defense against viruses and transposons, development regulation and chromosomal segregation in animals and plants [2, 3, 62, 63]. Quelling in Neurospora can function in silencing the transgenes by detecting and targeting the transgenic DNA, and thus is a potent mechanism that represses the expression and expansion of transposons [64, 65]. siRNAs against transposons were detected in N. crassa, and the transcript levels and copy number of the LINE1-like transposon Tad were significantly elevated in qde-2 mutants. In addition, the Tad transcripts were also found to be up-regulated in the dcl-1 dcl-2 double mutants. Similarly, MSUD may also function as a mechanism that silences transposon expansion during meiosis because the replication of transposon will generate unpaired DNA.
RNAi has been shown to be involved in heterochromation formation and/or DNA methylation in fission yeast, plants and animals [13, 66–69]. However, the known RNAi components in Neurospora, including three RdRPs (QDE-1, SAD-1 and RRP-3), two Argonaute proteins (QDE-2 and SMS-2), two dicer-like proteins (DCL-1 or SMS-3, DCL-2) and two RecQ helicases (QDE-3 and RecQ-2), are not required for the initiation or maintenance of heterochromatin formation and DNA methylation [70]. Chicas et al. [64] also demonstrated that the transgenic siRNA production/quelling is also not required for histone H3 Lys9 methylation. Thus, the RNAi pathway does not appear to function in transcriptional gene silencing in Neurospora. However, the mutation of the histone Lys9H3 methyltransferase gene dim-5 caused a low quelling efficiency and frequent reversion of the quelled transformants due to rapid loss of the integrated transgenic copies [64]. Thus, DIM-5 and histone methylation play an important role in stabilizing tandom copies of the transgene.
More recently, it was reported that the rDNA gene copy numbers in the quelling mutants qde-1, qde-2 and qde-3 are all reduced comparing to the wild-type strain, suggesting that quelling may play a role in maintaining the rDNA locus integrity and stability [71].
A dsRNA-induced transcriptional program important for RNAi
In vertebrates, dsRNA is known to trigger the transcription-based antiviral interferon response, an important part of innate immunity. We have shown that a similar response exists in Neurospora and that the expression of dsRNA in Neurospora can significantly induce transcriptional activation of key RNAi components including qde-2 and dcl-2 [72]. Such a transcriptional response was regulated by dsRNA instead of siRNA, as the transcriptional activation of dsRNA-activated genes (DRAGs) was maintained in the dcl double mutant, in which siRNA production was completely abolished. In addition, QDE-1 and QDE-2 are not required for the induction of DRAGs by dsRNA. However, dsRNA regulates QDE-2 both transcriptionally and post-transcriptionally. In the dcl double mutant, despite the induction of qde-2 mRNA by dsRNA, QDE-2 protein level stayed at a low level, suggesting that the production of siRNA or the association of siRNA with QDE-2 is important for QDE-2 stabilization [72]. In mutants where qde-2 expression cannot be induced by dsRNA, gene silencing efficiency is reduced, indicating that this response is important for the efficiency of dsRNA-triggerred gene silencing [72].
Genome-wide analyses by microarray and quantitative PCR showed that dsRNA could activate the expression of ~60 genes, including additional RNAi components and homologs of antiviral and interferon-stimulated genes. The induction of the latter genes by dsRNA suggests that the dsRNA-induced activation of RNAi components is part of a conserved ancient host defense response to counter against viral infection and transposons [72]. The signaling pathway in Neurospora responsible for the dsRNA response is not known. Since the key RNAi genes are not required for the dsRNA-induced transcriptional activation of DRAGs and Neurospora does not encode homologs of the known mammalian dsRNA sensors, there should be a novel dsRNA-sensing and transcriptional activation pathway in Neurospora [72]. Interestingly, dcl-2 and agl2 expression levels were also found to be significantly increased in response to viral infection and expression of hairpin RNA in the chestnut blight fungus Cryphonectria paracitica [73, 74], suggesting that the dsRNA-induced transcriptional response is conserved in filamentous fungi.
The DNA damage-induced qiRNA and its relationship with quelling
In addition to the dsRNA-induced qde-2 expression, we found that the treatment of Neurospora culture with various DNA damage agents resulted in significantly elevated levels of qde-2 mRNA and QDE-2 protein [41]. The induction of QDE-2 expression by DNA damage regents requires functional QDE-1 and QDE-3. In addition, QDE-2 levels were found to be elevated to high levels in many DNA repair mutants in the absence of DNA damage agents. Since dsRNA can induce the expression of QDE-2, and QDE-1 and QDE-3 are required for dsRNA generation from transgenes in quelling, DNA damage should induce the production of endogenous dsRNA and sRNAs which result in the induction of QDE-2 expression [41, 72].
Analysis of QDE-2-associated small RNAs uncovered a novel class of ~21-nt-long small RNAs that is significantly induced by DNA damage [41]. The levels of these small RNAs were low under normal growth conditions and were induced robustly after the treatment with DNA-damaging agents. These small RNAs are shorter than the regular 25-nt siRNAs and were named qiRNAs for their interaction with QDE-2. qiRNAs originate mainly from the highly repetitive rDNA locus, have a strong 5′ uridine preference and a 3′ preference for adenine, and depend on QDE-1, QDE-3 and the Dicers for their production, indicating that qiRNAs are not nonspecific degradation products of rRNAs[41].
qiRNAs correspond to both sense and antisense rDNA strands and their long precursor RNAs accumulated to high levels in the dcl-1 dcl-2 double mutant, indicating that qiRNAs are processed from long dsRNAs. Although DNA damage induces the expression of QDE-2, qiRNA production is not dependent on QDE-2. In addition, qiRNAs not only match to the transcribed rRNA regions they also match to normally untranscripted intergenic spacer regions, suggesting that qiRNAs originate from aRNAs. Indeed, aRNA transcripts with sizes from a few hundred nucleotides to about 2 kb from the intergenic spacer regions were found to be robustly induced by DNA damage. Surprisingly, a potent inhibitor (thiolutin) of RNA polymerases I, II and III did not block the induction of aRNA by DNA damage, suggesting that aRNAs are produced by another polymerase. In contrast, aRNA was completely abolished in both qde-3 and qde-1 mutants, indicating that the RecQ DNA helicase QDE-3 and the RdRP QDE-1 are required for the synthesis of the DNA damage-induced aRNA [41]. Using partially purified QDE-1 from Neurospora showed that QDE-1 can produce RNA products using either ssRNA or ssDNA as a template, indicating that QDE-1 is both an RdRP and DdRP [41]. These results suggest that, in addition to its role in converting aRNA into dsRNA, QDE-1 is also an RNA polymerase that generates aRNA.
QDE-3 was previously shown to play a role in DNA damage response [22, 30, 31]. In addition, other RNAi mutants in which qiRNA production is abolished were also shown to have increased sensitivity to DNA damage [41]. These results suggest that qiRNAs may contribute to the DNA damage response by inhibiting protein translation.
Both quelling and the qiRNA production pathway share the same key components, such as QDE-1, QDE-2, QDE-3, and Dicers, and both require aRNA production and dsRNA production, suggesting that these two pathways are mechanistically linked. Although quelling is triggered by repetitive transgene and qiRNA is induced by DNA damage, both the transgene specific siRNA and qiRNA originate from highly repetitive DNA loci. In a normal wild-type Neurospora strain, the rDNA locus is the only highly repetitive DNA locus and the transgene-mediated quelling generates a second such locus. Therefore, the repetitive nature of the rDNA and transgene loci is very likely to be the common trigger for aRNA and small RNA production for both processes. Since DNA damage induces qiRNA from the rDNA region and repetitive transgenic locus is known to have frequent recombination and rearrangement [41, 65], the generation of aRNA and siRNA from the quelled loci is also likely the result of DNA replication stress caused by the repetitive transgene,.
miRNA-like RNA and diverse sRNA biogenesis pathways in Neurospora
miRNA-like RNAs
miRNAs are small non-coding RNAs originated from single-stranded RNA precursor containing hairpin structures [63, 75]. miRNAs have been found in animals, plants, and algae [76–82], and have been widely considered to be absent in the fungal kingdom. However, our recent studies of Neurospora QDE-2-associated sRNAs have led to the discovery of miRNA-like RNAs (milRNAs) in this filamentous fungus [83], indicating that the miRNA-like gene regulatory mechanism evolved early in the eukaryotic lineage.
By analyzing the QDE-2-associated sRNAs, at least 25 potential milRNA-producing loci were discovered [83]. These milRNAs share many similarities with conventional miRNAs from animals and plants: they come from highly specific stem-loop RNA precursors; most of the milRNAs require Dicer for the biogenesis; and milRNAs may silence endogenous targets with imperfect complementarity as the animal miRNAs. For each milRNA locus, the vast majority of small RNA sequences matched one arm of the hairpin (called the milRNA arm) and much lower numbers of sRNA matched to the complementary arm (named as milRNA*). Although nearly all milRNAs from each locus share the same 5′ U position, there is heterogeneity at 3′ termini of milRNAs as miRNAs in other eukaryotes [84].
Diverse milRNA production pathways
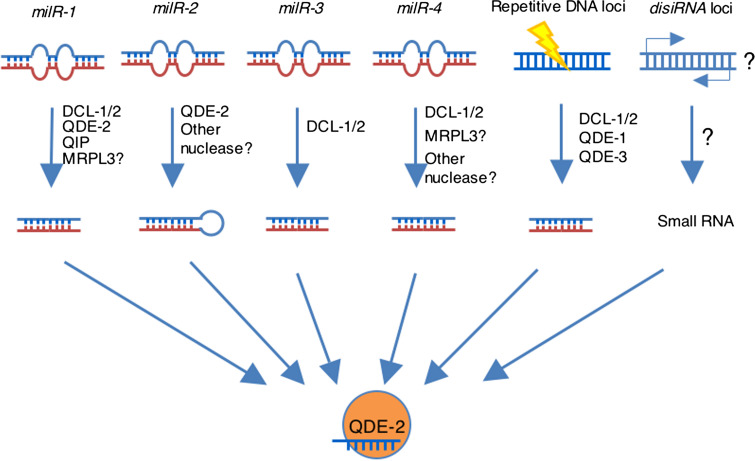
The most surprising finding of this study is the discovery of diverse pathways for milRNA biogenesis. The animal/plant miRNAs are known to be produced by a well-defined Dicer-dependent pathway [63, 75]. However, our close examination of four milRs uncovered four different pathways for milRNAs production and revealed an important role of Argonaute proteins in sRNA production [83] (Fig. 3). Unlike qiRNAs, production of milRNAs is independent of QDE-1 and QDE-3. Among these four milRs, only the biogenesis of milR-3 milRNAs resembles that in plants, which require only Dicers for pre-milRNA and milRNA processing [85]. On the other hand, milR-4 milRNAs biogenesis is only partially dependent on Dicer, indicating the existence of a novel nuclease.
Fig. 3.
A diagram depicting six endogenous small RNA production pathways in vegetative tissues of Neurospora crassa. There are at least six different endogenous sRNA biogenesis pathways in N. crassa: a Dicer, QDE-2 and QIP-dependent pathway for milRNA-1; b Dicer-independent, Argonaute QDE-2-dependent pathway for milR-2; c Dicer-dependent pathway for milR-3; d partially Dicer-dependent pathway for milR-4; e DNA damage-induced Dicer-dependent pathway for qiRNA; f Dicer and QDE-2-independent pathway for disiRNA
The milR-1 milRNAs production pathway is currently the best understood [83]. Although it is completely dependent on Dicers for the production of pre-milRNA and mature milRNAs production, the biogenesis of mature milRNAs requires QDE-2 (but not its catalytic activity) and the exonuclease activity of QIP. These results and the association of pre-milRNA with QDE-2 led to the following model for milR-1 biogenesis: the pri-milRNA is first processed by Dicer to generate double-stranded pre-milRNAs, and afterwards QDE-2 binds to pre-milRNA and recruits the exonuclease QIP to process the pre-milRNAs into mature milRNAs. This study established a novel sRNA biogenesis pathway in which the Argonaute protein functions as an adaptor by binding to pre-milRNA and recruiting other factor(s) to mediate milRNA maturation.
Although the production of milR-1, milR-3 and milR-4 milRNAs is completely or partially dependent on Dicer, surprisingly, the biogenesis of milR-2 milRNAs is completely independent of Dicer. Instead, its biogenesis requires QDE-2 and its catalytic activity. The predicted milR-2 pri-milRNA forms a hairpin structure with both milRNA strand and milRNA* on the stem that is close to the loop. In addition to the mature milRNAs, its pre-miRNAs are associated with QDE-2. These results suggest that, for milR-2, QDE-2 binds to a long pre-milRNA and cleaves the milRNA* strand of the pre-milRNAs. Afterwards, another unknown nuclease is involved in the further cleavage and maturation of milRNAs. The milR-2 pathway is the first known example of Dicer-independent but Argonaute-dependent mechanism for small RNA biogenesis, and it extends the boundary of miRNA since all miRNAs are previously regarded as Dicer-dependent.
The mechanistic diversity observed for milRNA biogenesis in Neurospora stands in stark contrast with the well-defined miRNA production pathways in animals and plants. Our results suggest that eukaryotic small RNA generation mechanisms are more diverse than previously thought. Soon after the publication of our study, the mouse miR-451 was shown to be produced by a Dicer-independent but Argonaute-dependent mechanism that is very similar to that of milR-2 [86, 87], suggesting that the other novel sRNA biogenesis pathways in Neurospora may also exist in other eukaryotes. Our results also raise the possibility that miRNA-like RNAs may also be found in eubacteria and archea, which lack homologs of Dicer but encode Argonaute-like proteins [88].
An RNAse III domain-containing protein MRPL3 in milRNA processing
The presence of Dicer-independent mechanisms for milRNA generation indicates the involvement of novel nucleases in milRNA biogenesis. We demonstrated that the Neurospora homolog of the yeast mitochondrial ribosomal protein MRPL3 is an important factor in milRNA production [83]. MRPL3 contains a putative RNAse III domain and a dsRNA recognition motif, although its RNAse III domains has little sequence similarity to those of Dicers and Drosha. In both a heterokaryotic knock-out strain and a mrpl3 knock-down strain in which mrpl3 is silenced by dsRNA, the production of milR-1 and milR-4 milRNAs were significantly decreased. In addition, the in vitro miRNA cleavage activity was also reduced in these mutants. These results indicate that the putative nuclease MRPL3 is an important component for the production of some milRNAs, although its mode of action is still unclear.
milRNAs mediate gene silencing in Neurospora
Like the animal/plant miRNAs, milRNAs regulate gene expression in Neurospora [83]. By introducing a milRNA reporter construct into a wild-type and qde-2 strains, we showed that milR-1 expression resulted in robust gene silencing of its complementary target. However, despite the dramatic up-regulation of the reporter expression at the protein level, only a modest effect was observed at the mRNA level, suggesting that milRNA may mostly mediate gene silencing by repression of translation. In addition, the mRNA levels of several predicted targets of the milRNAs are up-regulated in the dcl and qde-2 mutants. Furthermore, QDE-2 was found to specifically associate with the mRNA of these target genes, suggesting that milRNAs directly regulate their expression in vivo. However, milRNAs do not appear to play a prominent role in cell growth/development in Neurospora as miRNAs in animals and plants since the disruption of key RNAi components does not result in severe phenotypes. Future studies are still needed to establish the physiological importance of milRNAs in Neurospora.
Dicer-independent small interfering RNAs (disiRNAs)
In addition to milRNAs, disiRNAs was shown to be another novel type of sRNA in Neurospora [83]. disiRNAs were symmetrically matched to both strands of DNA, and are averaged about 22 nt long with a strong 5′ U preference. They are derived from ~50 non-repetitive DNA loci which contain genes and intergenic regions with no apparent shared sequence motifs. Interestingly, based on available EST data, nearly 80% of the disiRNA loci have overlapping sense and antisense transcripts, which suggests that these disiRNAs are likely processed from dsRNA made from naturally occurring complementary sense and antisense transcripts.
disiRNAs do not depend on QDE-1, QDE-2, or QDE-3 for the biogenesis, and surprisingly, their levels were not significantly changed in the dcl-1/dcl-2 double mutant, thus they are a class of Dicer-independent sRNAs. Moreover, their levels were not affected by mutations of Argonaute genes or other known RNAi genes. These results indicate that disiRNAs are distinct from the animal Dicer-independent piRNAs and are produced by a novel sRNA biogenesis pathway. Although the functions of disiRNAs are not known, these small RNAs interact with QDE-2, the core component of the RISC, suggesting that they may function via the RNAi pathway.
RNAi studies in Cryphonectria, Aspergillus and Mucor
In addition to N. crassa, Cryphonectria paracitica, Aspergillus nidulans and Mucor circinelloides are also important model organisms for studying RNAi pathways and their functions in filamentous fungi.
RNAi is an antiviral defense mechanism in Cryphonectria paracitica
Although RNAi is known as a key innate immunity mechanism in antiviral defense for plant and animal viruses, the antiviral role of RNAi in fungi was only shown quite recently [2, 89–92]. RNAi pathways in filamentous fungi are best understood in Neurospora, but there is no experimental viral system that can infect this organism. The filamentous Ascomycete fungus C. paracitica, the chestnut blight fungus, has a well-established experimental system for the study of hypoviridae family of mycoviruses that reduces its pathogenicity and can support the viral replication of five different RNA virus families. These make C. paracitica an ideal model organism to study the role of RNAi silencing as an antiviral defense mechanism in fungi [93, 94].
p29 is a papain-like protease encoded by the mycovirus Cryphonectria hypovirus 1 (CHV1), which is similar to the plant potyvirus-encoded suppressor of RNA silencing HC-Pro [95–98]. Using the CHV1-EP713/C. parasitica system, Segers et al. demonstrated that p29 suppressed the hairpin RNA-induced silencing in C. parasitica. In addition, p29 also suppressed both the virus-induced and agroinfiltration-induced RNA silencing and systemic spread of silencing in GFP-expressing transgenic Nicotiana benthamiana [96]. These results suggest that the antiviral defense mechanism of RNA silencing is conserved in both fungi and plants [96].
Segers et al. [93] further cloned the dicer-like genes dcl-1 and dcl-2 in C. parasitica, which are homologous to the Neurospora dcl-1 and dcl-2, respectively. Although the single mutants of dcl-1 and dcl-2 have no obvious phenotype compared to the wild-type C. parasitica, the dcl-2 and dcl-1/dcl-2 mutant strains are highly susceptible to the infection of hypovirus CHV1-EP713 or reovirus MyRV1-Cp9B21. On the other hand, infection of the dcl-2 mutant by a hypovirus CHV1-EP713 mutant lacking the suppressor of RNA silencing p29 and the wild-type reovirus MyRV1-Cp9B21 exhibited elevated viral RNA levels compared to the wild-type. These results demonstrate that a fungal Dicer can function to regulate virus infection and RNAi plays an important role in antiviral defense in fungi [93, 96]. This conclusion is further supported by the findings that dcl-2 is required for defective viral RNA production and recombinant virus vector RNA instability for hypovirus [73]. In addition, the dcl-2 expression levels are significantly increased after viral infection and are further increased when the suppressor p29 is mutated in the virus. Moreover, the DCL-2 processes hypovirus RNAs into virus-derived small interfering RNAs (vsRNAs) as part of an inducible RNA silencing antiviral response [73, 74].
There are four Argonaute-like proteins (AGL1, AGL2, AGL3 and AGL4) in C. parasitica, but only AGL2 is required for the antiviral defense response [94]. Similar to dcl-2, the agl2 transcriptional level is also induced in response to viral infection [94]. In addition, agl2 and dcl2 transcripts were accumulated to much higher levels in response to infection by a mutant CHV1-EP713 hypovirus without p29, further supporting that a virus-encoded RNA silencing suppressor suppresses the RNA silencing components. Similar to the dsRNA-induced response in Neurospora, expression of a hairpin RNA also significantly induces the expression of agl2 and dcl2 to high levels [94], consistent with the notion that the dsRNA-induced response is part of the host defense mechanism [72]. Interestingly, the viral infection induced dcl-2 expression is blocked in the agl2 mutant [94], suggesting that AGL2 plays a regulatory role in the activation of the RNA silencing pathway.
In addition to the role of RNAi components in suppressing viral RNA replication, they also promote viral RNA recombination that generates internally deleted mutant RNAs, called defective interfering (DI) RNAs [73, 74, 94], DI RNAs are derived from the parental viral genomic RNA by recombination and can function to suppress viral RNA accumulation, leading to the instability and deletion of foreign viral RNA vectors. While DI RNAs were observed in the wild-type C. parasitica, in both dcl-2 and agl2 mutants, the hypovirus defective-inferencing (DI) RNAs were abolished and the hypovirus vector expressing EGFP became stable. These results demonstrate that the host RNAi pathway can also control a single-strand positive sense RNA virus by promoting viral recombination and DI RNA formation [73, 74, 94]. Although how RNAi pathway functions to promote viral recombination is not known, future studies will shed light on the mechanism of new virus emergence and the mechanisms of virus–host interaction [73, 74, 94].
RNAi as a viral defense mechanism in A. nidulans
Expression of inverted repeat-containing transgene also results in robust RNA silencing in A. nidulans, demonstrating that dsRNA-triggered RNAi is present in this model filamentous fungus [99, 100]. Interestingly, this fungus has only one intact Dicer and Argonaute, respectively, which are both required for RNAi. A. nidulans contains two genes encoding for RdRPs but they are not required for the inverted repeat triggered RNAi, suggesting that, as in Neurospora, RdRPs are not involved in siRNA amplification [99, 101]. By stable infection of A. nidulans with three mycoviruses, Hammond and Keller demonstrated that the Aspergillus virus 1816 could suppress the inverted repeat transgene-induced RNA silencing. Although the mechanism for the suppression is not clear, this result suggests the existence of an RNA silencing suppressor encoded by this virus. On the other hand, the virus 341-derived siRNA was detected at a high level in an Argonaute mutant, indicating that this virus is targeted and processed into siRNA by the RNA silencing machinery. Together, these results suggest that there is an antagonistic relationship between mycoviruses and RNA silencing mechanism in A. nidulans and RNAi functions as a viral defense mechanism.
RNAi and small RNA studies in M. circinelloides
Compared to Cryphonectria and Aspergillus, M. circinelloides, a basal fungus of the clade zygomycete, is distantly related to Neurospora and has emerged as an important fungal model system to study RNA silencing mechanism. Both transformation of M. circinelloides with self-replicative plasmids and expression of inverted-repeat transgenes result in post-transcriptional gene silencing [102, 103]. Two Dicer genes (dcl1and dcl2) and two RdRP genes (rdrp1 and rdrp2) were identified and characterized in M. circinelloides [102–105]. RNA silencing results in the production of both sense and antisense small RNAs, with two different size classes, 21 and 25 nt, respectively. Unlike in Neurospora, secondary sense and antisense RNAs were detected in M. circinelloides, suggesting a sRNA amplification step is present for RNA silencing in this fungus. DCL-2 is the major player in the transgene-induced silencing and the production of the two classes of antisense siRNAs [103]. On the other hand, RdRP1, but not RdRP2, is important for the transgene-induced silencing [104].
Most recently, by small RNA sequencing, four classes of endogenous small RNAs have been identified in this fungus. Most of these sRNAs match to exons and regulate mRNA levels of many protein coding genes [104]. The biogenesis of the largest class of these exonic-siRNAs (ex-siRNA) requires both RdRP1 and DCL-2, indicating RdRP1 converts the corresponding mRNA into dsRNA which is then further processed by DCL2. They target the protein-coding mRNAs where they are produced. A second group of exonic-siRNAs requires DCL-2 and RdRP2 but not RdRP1 for the biogenesis. The third group requires both RdRP1 and RdRP2 for the biogenesis, and the two Dicers seem to play redundant roles. For the fourth group, the small RNA production is mainly DCL-1- but not DCL-2-dependent, although both RdRPs are required for their biogenesis. In addition to ex-siRNAs, some Dicer-dependent endogeneous small RNAs were also mapped to transposons or repetitive sequences, but no miRNA-like RNAs were found [104]. The lack of miRNA-like RNAs suggests that either they are not present in this fungus or are not expressed abundantly under the experimental condition used. The discovery of these endogenous sRNAs from M. circinelloides further indicates the diversity of small RNA bigenesis pathways in the filamentous fungus.
RNAi studies and applications in other filamentous fungi
RNAi has been shown to function in most filamentous fungi examined and has been frequently used as a tool to study gene function [60, 106, 107]. A co-suppression phenomenon like that in plants and Neurospora was observed in Cladosporium fulvum in 1998 [60, 108]. In addition, a homology-dependent silencing phenomenon was found in Schizophyllum commune [109]. On the other hand, gene silencing by utilizing a dsRNA-expressing system containing inverted repeats and/or two opposing promoters has been successfully applied in many pathogenic and non-pathogenic fungi including Ascomycota, Basidiomycota, and Zygomycota, such as Magnaporthe oryzae [110, 111], Sclerotinia sclerotiorum [112], Aspergillus fumigatus [113–116], Aspergillus oryzae [110, 117], Aspergillus flavus [118], Aspergillus parasiticus [118], Bipolaris oryzae [119], Colletotrichum lagenarium [120], Colletotrichum gloeosporioides [121], Coprinus cinereus [122], Fusarium graminearum [118], Fusarium solani [123], Fusarium verticillioides [124], Moniliophthora perniciosa [125], Coniothyrium minitans [126], Stagonospora nodorum [127], Ophiostoma floccosum and Ophiostoma piceae [128], Botrytis cinerea [129], Penicillium chrysogenum [130, 131], Venturia inaequalis [132], Coprinopsis cinerea [133], Cryptococcus neoformans [134] and S. commune [60, 106, 107, 109, 135].
As more fungal genomes being sequenced, homologues of RdRP, Argonaute and Dicer proteins in various filamentous fungi from Ascomycota, Basidiomycota, and Zygomycota were identified although they seem to be absent in the basidiomycete Ustilago maydis [60, 106]. Genes involved in the RNA silencing in the ascomycete fungi usually have three RdRPs, two Argonautes and two Dicer-like proteins. Basidiomycete fungi have similar numbers of the protein classes involved in RNAi to the ascomycetes, although they have a wider diversity and more extensive gene expansion. In the zygomycete Rhizopus oryzae, five RdRP genes were identified, indicative of more extensive gene expansion and diverse roles of RdRP in silencing mechanisms.
Conclusions
Neurospora crassa is one of the first eukaryotic model organisms for RNAi studies. The discoveries of quelling and meiotic silencing by unpaired DNA demonstrated the importance and diversity of RNAi phenomenon. The identification and characterization of components in these two pathways have contributed significantly to our understanding of the RNAi mechanism in general and will continue to do so in the future. The recent discovery of qiRNAs, milRNAs and disiRNAs and the demonstration of diverse small RNA biogenesis pathways in Neurospora established this organism as an important eukaryotic model system for studying sRNA production. Future studies on these sRNAs will shed lights on the mechanism and evolutionary origins of eukaryotic sRNA production. On the other hand, the demonstration and understanding of RNAi functions in viral or transposon defense in Neurospora, Cryphonectria and Aspergillus or M. circinelloides indicate that filamentous fungi are also important eukaryotic systems to uncover RNAi functions.
References
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 7.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 8.Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 10.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 11.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 12.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 13.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 16.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 18.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa . Proc Natl Acad Sci USA. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cogoni C, Romano N, Macino G. Suppression of gene expression by homologous transgenes. Antonie Van Leeuwenhoek. 1994;65:205–209. doi: 10.1007/BF00871948. [DOI] [PubMed] [Google Scholar]
- 20.Cogoni C, Macino G. Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 1997;2:438–443. doi: 10.1016/S1360-1385(97)90028-5. [DOI] [Google Scholar]
- 21.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 22.Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 23.Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 24.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 25.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/S0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 26.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/S0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 27.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans . Curr Biol. 2000;10:169–178. doi: 10.1016/S0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 28.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldoni M, Azzalin G, Macino G, Cogoni C. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa . Fungal Genet Biol. 2004;41:1016–1024. doi: 10.1016/j.fgb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Pickford A, Braccini L, Macino G, Cogoni C. The QDE-3 homologue RecQ-2 co-operates with QDE-3 in DNA repair in Neurospora crassa . Curr Genet. 2003;42:220–227. doi: 10.1007/s00294-002-0351-6. [DOI] [PubMed] [Google Scholar]
- 31.Kato A, Akamatsu Y, Sakuraba Y, Inoue H. The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr Genet. 2004;45:37–44. doi: 10.1007/s00294-003-0459-3. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Samadder PP, Tanaka Y, Ohira T, Okuizumi H, Yamaoka N, Miyao A, Hirochika H, Tsuchimoto S, Ohtsubo H, Nishiguchi M. OsRecQ1, a QDE-3 homologue in rice, is required for RNA silencing induced by particle bombardment for inverted repeat DNA, but not for double-stranded RNA. Plant J. 2008;56:274–286. doi: 10.1111/j.1365-313X.2008.03587.x. [DOI] [PubMed] [Google Scholar]
- 33.Fulci V, Macino G. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa . Curr Opin Microbiol. 2007;10:199–203. doi: 10.1016/j.mib.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 35.Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa . Mol Cell Biol. 2004;24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa . Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 38.Makeyev EV, Bamford DH. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell. 2002;10:1417–1427. doi: 10.1016/S1097-2765(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 39.Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurila MR, Salgado PS, Makeyev EV, Nettelship J, Stuart DI, Grimes JM, Bamford DH. Gene silencing pathway RNA-dependent RNA polymerase of Neurospora crassa: yeast expression and crystallization of selenomethionated QDE-1 protein. J Struct Biol. 2005;149:111–115. doi: 10.1016/j.jsb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forrest EC, Cogoni C, Macino G. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa . Nucleic Acids Res. 2004;32:2123–2128. doi: 10.1093/nar/gkh530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziv C, Yarden O. Gene silencing for functional analysis: assessing RNAi as a tool for manipulation of gene expression. Methods Mol Biol. 2010;638:77–100. doi: 10.1007/978-1-60761-611-5_6. [DOI] [PubMed] [Google Scholar]
- 44.Nolan T, Cecere G, Mancone C, Alonzi T, Tripodi M, Catalanotto C, Cogoni C. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008;36:532–538. doi: 10.1093/nar/gkm1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng P, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996;86:103–113. doi: 10.1016/S0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 48.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 49.Shiu PK, Metzenberg RL. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics. 2002;161:1483–1495. doi: 10.1093/genetics/161.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Springer ML. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa . Bioessays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- 51.Aramayo R, Peleg Y, Addison R, Metzenberg R. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cogoni C. Unifying homology effects. Nat Genet. 2002;30:245–246. doi: 10.1038/ng0302-245. [DOI] [PubMed] [Google Scholar]
- 53.Kasbekar DP. Sex and the single gene: meiotic silencing by unpaired DNA. J Biosci. 2002;27:633–635. doi: 10.1007/BF02708370. [DOI] [PubMed] [Google Scholar]
- 54.Kelly WG, Aramayo R. Meiotic silencing and the epigenetics of sex. Chromosome Res. 2007;15:633–651. doi: 10.1007/s10577-007-1143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pratt RJ, Lee DW, Aramayo R. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics. 2004;168:1925–1935. doi: 10.1534/genetics.104.031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee DW, Pratt RJ, McLaughlin M, Aramayo R. An argonaute-like protein is required for meiotic silencing. Genetics. 2003;164:821–828. doi: 10.1093/genetics/164.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiu PK, Zickler D, Raju NB, Ruprich-Robert G, Metzenberg RL. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc Natl Acad Sci USA. 2006;103:2243–2248. doi: 10.1073/pnas.0508896103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bardiya N, Alexander WG, Perdue TD, Barry EG, Metzenberg RL, Pukkila PJ, Shiu PK. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics. 2008;178:593–596. doi: 10.1534/genetics.107.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander WG, Raju NB, Xiao H, Hammond TM, Perdue TD, Metzenberg RL, Pukkila PJ, Shiu PK. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol. 2008;45:719–727. doi: 10.1016/j.fgb.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 60.Nakayashiki H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Lee DW, Seong KY, Pratt RJ, Baker K, Aramayo R. Properties of unpaired DNA required for efficient silencing in Neurospora crassa . Genetics. 2004;167:131–150. doi: 10.1534/genetics.167.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 63.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 64.Chicas A, Forrest EC, Sepich S, Cogoni C, Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa . Mol Cell Biol. 2005;25:3793–3801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nolan T, Braccini L, Azzalin G, De Toni A, Macino G, Cogoni C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa . Nucleic Acids Res. 2005;33:1564–1573. doi: 10.1093/nar/gki300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matzke M, Matzke AJM. RNAi extends its reach. Science. 2003;301:1060–1061. doi: 10.1126/science.1089047. [DOI] [PubMed] [Google Scholar]
- 67.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SCR. Heterochromatic silencing and HP1 localization in drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 68.Chan SW-L, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 69.Chinnusamy V, Zhu JK. RNA-directed DNA methylation and demethylation in plants. Sci Chin C Life Sci. 2009;52:331–343. doi: 10.1007/s11427-009-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freitag M, Lee DW, Kothe GO, Pratt RJ, Aramayo R, Selker EU. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- 71.Cecere G, Cogoni C. Quelling targets the rDNA locus and functions in rDNA copy number control. BMC Microbiol. 2009;9:44. doi: 10.1186/1471-2180-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choudhary S, Lee HC, Maiti M, He Q, Cheng P, Liu Q, Liu Y. A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol. 2007;27:3995–4005. doi: 10.1128/MCB.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Nuss DL. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc Natl Acad Sci USA. 2008;105:16749–16754. doi: 10.1073/pnas.0807225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol. 2008;82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 77.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 78.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 79.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii . Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 81.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii . Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, Liu Y. Diverse pathways generate MicroRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 85.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 86.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson N, Wolfe S, Giraldez AJ. A Novel miRNA processing pathway independent of dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 89.Li HW, Ding SW. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li F, Ding SW. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun Q, Choi GH, Nuss DL. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci USA. 2009;106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suzuki N, Maruyama K, Moriyama M, Nuss DL. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J Virol. 2003;77:11697–11707. doi: 10.1128/JVI.77.21.11697-11707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot Cell. 2006;5:896–904. doi: 10.1128/EC.00373-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzuki N, Chen B, Nuss DL. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J Virol. 1999;73:9478–9484. doi: 10.1128/jvi.73.11.9478-9484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi GH, Pawlyk DM, Nuss DL. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology. 1991;183:747–752. doi: 10.1016/0042-6822(91)91004-Z. [DOI] [PubMed] [Google Scholar]
- 99.Hammond TM, Keller NP. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics. 2005;169:607–617. doi: 10.1534/genetics.104.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barton LM, Prade RA. Inducible RNA interference of brlAbeta in Aspergillus nidulans . Eukaryot Cell. 2008;7:2004–2007. doi: 10.1128/EC.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hammond TM, Bok JW, Andrewski MD, Reyes-Dominguez Y, Scazzocchio C, Keller NP. RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans . Eukaryot Cell. 2008;7:339–349. doi: 10.1128/EC.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Haro JP, Calo S, Cervantes M, Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides . Eukaryot Cell. 2009;8:1486–1497. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, Torres-Martinez S, Moulton V, Ruiz-Vazquez RM, Dalmay T (2010) Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. doi:10.1093/nar/gkq301 [DOI] [PMC free article] [PubMed]
- 105.Nicolas FE, de Haro JP, Torres-Martinez S, Ruiz-Vazquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 106.Nakayashiki H, Kadotani N, Mayama S. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- 107.Nakayashiki H, Nguyen QB. RNA interference: roles in fungal biology. Curr Opin Microbiol. 2008;11:494–502. doi: 10.1016/j.mib.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 108.Hamada W, Spanu PD. Co-suppression of the hydrophobin gene HCf-1 is correlated with antisense RNA biosynthesis in Cladosporium fulvum . Mol Gen Genet. 1998;259:630–638. doi: 10.1007/s004380050857. [DOI] [PubMed] [Google Scholar]
- 109.Schuurs TA, Schaeffer EA, Wessels JG. Homology-dependent silencing of the SC3 gene in Schizophyllum commune . Genetics. 1997;147:589–596. doi: 10.1093/genetics/147.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kadotani N, Nakayashiki H, Tosa Y, Mayama S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae . Mol Plant Microbe Interact. 2003;16:769–776. doi: 10.1094/MPMI.2003.16.9.769. [DOI] [PubMed] [Google Scholar]
- 111.Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, Nakayashiki H. Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol. 2008;68:1348–1365. doi: 10.1111/j.1365-2958.2008.06242.x. [DOI] [PubMed] [Google Scholar]
- 112.Erental A, Harel A, Yarden O. Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum . Mol Plant Microbe Interact. 2007;20:944–954. doi: 10.1094/MPMI-20-8-0944. [DOI] [PubMed] [Google Scholar]
- 113.Mouyna I, Henry C, Doering TL, Latge JP. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus . FEMS Microbiol Lett. 2004;237:317–324. doi: 10.1016/j.femsle.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 114.Khalaj V, Eslami H, Azizi M, Rovira-Graells N, Bromley M. Efficient downregulation of alb1 gene using an AMA1-based episomal expression of RNAi construct in Aspergillus fumigatus . FEMS Microbiol Lett. 2007;270:250–254. doi: 10.1111/j.1574-6968.2007.00680.x. [DOI] [PubMed] [Google Scholar]
- 115.Henry C, Mouyna I, Latge JP. Testing the efficacy of RNA interference constructs in Aspergillus fumigatus . Curr Genet. 2007;51:277–284. doi: 10.1007/s00294-007-0119-0. [DOI] [PubMed] [Google Scholar]
- 116.Bromley M, Gordon C, Rovira-Graells N, Oliver J. The Aspergillus fumigatus cellobiohydrolase B (cbhB) promoter is tightly regulated and can be exploited for controlled protein expression and RNAi. FEMS Microbiol Lett. 2006;264:246–254. doi: 10.1111/j.1574-6968.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 117.Yamada O, Ikeda R, Ohkita Y, Hayashi R, Sakamoto K, Akita O. Gene silencing by RNA interference in the koji mold Aspergillus oryzae . Biosci Biotechnol Biochem. 2007;71:138–144. doi: 10.1271/bbb.60405. [DOI] [PubMed] [Google Scholar]
- 118.McDonald T, Brown D, Keller NP, Hammond TM. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact. 2005;18:539–545. doi: 10.1094/MPMI-18-0539. [DOI] [PubMed] [Google Scholar]
- 119.Moriwaki A, Ueno M, Arase S, Kihara J. RNA-mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae . FEMS Microbiol Lett. 2007;269:85–89. doi: 10.1111/j.1574-6968.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- 120.Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, Mayama S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol. 2005;42:275–283. doi: 10.1016/j.fgb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 121.Chague V, Maor R, Sharon A. CgOpt1, a putative oligopeptide transporter from Colletotrichum gloeosporioides that is involved in responses to auxin and pathogenicity. BMC Microbiol. 2009;9:173. doi: 10.1186/1471-2180-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Namekawa SH, Iwabata K, Sugawara H, Hamada FN, Koshiyama A, Chiku H, Kamada T, Sakaguchi K. Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology. 2005;151:3669–3678. doi: 10.1099/mic.0.28209-0. [DOI] [PubMed] [Google Scholar]
- 123.Ha YS, Covert SF, Momany M. FsFKS1, the 1,3-beta-glucan synthase from the caspofungin-resistant fungus Fusarium solani . Eukaryot Cell. 2006;5:1036–1042. doi: 10.1128/EC.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tinoco ML, Dias BB, Dall’astta RC, Pamphile JA, Aragao FJ. In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 2010;8:27. doi: 10.1186/1741-7007-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Caribe dos Santos AC, Sena JA, Santos SC, Dias CV, Pirovani CP, Pungartnik C, Valle RR, Cascardo JC, Vincentz M. dsRNA-induced gene silencing in Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao. Fungal Genet Biol. 2009;46:825–836. doi: 10.1016/j.fgb.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 126.Gong X, Fu Y, Jiang D, Li G, Yi X, Peng Y. l-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet Biol. 2007;44:1368–1379. doi: 10.1016/j.fgb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 127.Solomon PS, Jorgens CI, Oliver RP. Delta-aminolaevulinic acid synthesis is required for virulence of the wheat pathogen Stagonospora nodorum . Microbiology. 2006;152:1533–1538. doi: 10.1099/mic.0.28556-0. [DOI] [PubMed] [Google Scholar]
- 128.Tanguay P, Bozza S, Breuil C. Assessing RNAi frequency and efficiency in Ophiostoma floccosum and O. piceae . Fungal Genet Biol. 2006;43:804–812. doi: 10.1016/j.fgb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 129.Patel RM, van Kan JA, Bailey AM, Foster GD. RNA-mediated gene silencing of superoxide dismutase (bcsod1) in Botrytis cinerea . Phytopathology. 2008;98:1334–1339. doi: 10.1094/PHYTO-98-12-1334. [DOI] [PubMed] [Google Scholar]
- 130.Janus D, Hoff B, Kuck U. Evidence for Dicer-dependent RNA interference in the industrial penicillin producer Penicillium chrysogenum . Microbiology. 2009;155:3946–3956. doi: 10.1099/mic.0.032763-0. [DOI] [PubMed] [Google Scholar]
- 131.Ullan RV, Godio RP, Teijeira F, Vaca I, Garcia-Estrada C, Feltrer R, Kosalkova K, Martin JF. RNA-silencing in Penicillium chrysogenum and Acremonium chrysogenum: validation studies using beta-lactam genes expression. J Microbiol Methods. 2008;75:209–218. doi: 10.1016/j.mimet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 132.Fitzgerald A, Van Kan JA, Plummer KM. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet Biol. 2004;41:963–971. doi: 10.1016/j.fgb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 133.Costa AM, Mills PR, Bailey AM, Foster GD, Challen MP. Oligonucleotide sequences forming short self-complimentary hairpins can expedite the down-regulation of Coprinopsis cinerea genes. J Microbiol Methods. 2008;75:205–208. doi: 10.1016/j.mimet.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 134.Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL. RNA interference in the pathogenic fungus Cryptococcus neoformans . Genetics. 2002;160:463–470. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Jong JF, Deelstra HJ, Wosten HA, Lugones LG. RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune . Appl Environ Microbiol. 2006;72:1267–1269. doi: 10.1128/AEM.72.2.1267-1269.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]