Abstract
Bone-targeting N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer-PGE1 conjugates, containing cathepsin K sensitive spacers, were incubated with induced osteoclasts and osteoblasts, their precursors, and control non-skeletal cells. The release of PGE1 was monitored by an HPLC assay. In both murine and human cell lines, osteoclasts appeared to be the most active cells in the cleavage (PGE1 release). Incubation with osteoblasts also resulted in fast PGE1 release, whereas precursor and control cells released PGE1 with a substantially slower rate than bone cells (apparently through ester bond cleavage). Experiments in the presence of inhibitors revealed that other enzymes, in addition to cathepsin K, were participating in the cleavage of the conjugate. Confocal fluorescence studies exposed internalization of the conjugate by endocytosis with ultimate localization in the lysosomal/endosomal compartment.
Keywords: bone cell, drug delivery systems, enzymes, HPMA copolymer conjugate, prostaglandin
Introduction
Osteoporosis is a common metabolic bone disease characterized by reduction of bone mineral density, disruption of bone microarchitecture, and an increased risk of fracture. Bone resorption is mainly mediated by osteoclasts, whereas osteoblasts are cells that synthesize the bonematrix. In healthy individuals the balance between bone resorption and bone building is well coordinated. However, disturbances of this equilibrium occur in numerous bone diseases, including osteoporosis.[1]
Osteoclasts are large multinucleated cells, polarized on the bone surface. At bone resorption sites, the osteoclasts form a ruffled border facing the bone and create a resorption lacuna surrounded by a sealing zone.[2] The resorption lacuna acidifies through a proton pump; the resulting pH of about 4.5 mediates the dissolution of the inorganic component of the bone (mainly hydroxyapatite) and provides an optimal microenvironment for organic matrix degradation[3] by cathepsin K, highly expressed in osteoclasts.[4,5] Cathepsin K is a cysteine proteinase of the papain superfamily;[6] it is highly homologous in amino acid sequence to cathepsins L and S.[4]
Many therapeutic agents are currently being used for the treatment of osteoporosis such as hormones[7] and selective estrogen-receptor modulators (SERMs).[8] However, they are not very effective in rebuilding lost skeletal mass, or have significant side-effects.[9,10] Antiresorptive therapies, such as with bisphosphonates, have been very effective in osteoporosis management,[11] but limiting side-effects include diarrhea, nausea, constipation, mild intestinal upset, and severely suppressed bone turnover.[12] The anabolic agents, prostaglandin E1 (PGE1) and prostaglandin E2 (PGE2), have been shown to increase the formation of new bone after systemic and local administration.[13,14] PGEs act on a variety of cells via cell-surface receptors, however, their potential clinical use has been limited by deleterious side-effects after systemic administration. The using of a bone targeted polymeric carrier is a way to localize PGE1, thereby increasing the drug efficiency with concomitant decrease in systemic steroid toxicity.
Recently, we have designed and synthesized novel HPMA copolymer-PGE1 conjugates.[15] Their activity is based on the bone-specific delivery of an anabolic agent, PGE1, mediated by a bone-seeking moiety, D-aspartic acid octapeptide or alendronate.[16] PGE1 was linked to the N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer via an ester bond connected to 1,6-elimination 4-aminobenzyl alcohol group, and a cathepsin K sensitive tetrapeptide (Gly-Gly-Pro-Nle) spacer. The conjugate was designed to release PGE1 by the action of cathepsin K in the osteoclasts.
The evaluation of biological effects of novel bone targeted therapies has been hindered by lack of ideal model systems of osteoclasts and osteoblasts. Isolation and preparation of primary cells has been a low yield process. However, several osteoclast and osteoblast precursor cell lines have been established and widely used. The mature osteoclasts and osteoblasts, derived from precursors through induction, have been demonstrated to be well representative. Murine macrophage RAW 264.7 cells are able to differentiate into multinucleated osteoclasts (RAW-OCs), under stimulation of RANKL.[17,18] Brandi and coworkers established osteoclasts-like cells from human leukemic blasts (FLG 29.1), induced by 12-O-tetradecanoylphorbol 13-acetate (TPA).[19] MC3T3-E1 cells, generated from newborn mouse calvaria, have demonstrated to have the capacity of osteoblastogenesis.[20] The cells display a fibroblastic morphology in the active growing phase. They exhibit osteoblast characteristics, both morphologically and physiologically, when reaching confluence. High differentiation subclones require ascorbic acid and inorganic phosphate, whereas low differentiation subclones do not.[21] Saos-2 is an established human osteosarcoma (osteoblastic sarcoma) cell line that has osteoblastic properties.[22] In culture, the osteoblastic properties could be detected with prolonged time of incubation and elevated cell density. For example, the alkaline phosphatase reaches high expression levels at confluence.[22] These cell lines have been widely used in mechanistic studies of osteogenesis and in the evaluation of therapeutic agents.[18,23–25]
Here, we evaluated the cleavability of the HPMA copolymer-PGE1 conjugate when incubated with induced osteoclasts and osteoblasts. Selected inhibitors were used to identify the enzymes responsible for cleavage of the conjugate. The internalization and subcellular trafficking of the conjugate was evaluated by confocal fluorescence microscopy.
Experimental Part
Materials
HPMA,[26] 5-[3-(methacryloylaminopropyl)thioureidyl]fluorescein (MA-FITC)[27] and HPMA copolymer-PGE1 conjugates (Figure 1)[15] were synthesized as described previously. PGE1 was obtained from Hawkins (Minneapolis, MN). Prostaglandin A1 (PGA1‘), prostaglandin B1 (PGB1), 1,5-bis(4-allyldimethylammoniumphenyl)pentan-3-one dibromide (BW 284c51), tetraisopropyl pyrophosphoramide (iso-OMPA), bis(4-nitrophenyl) phosphate (BNPP), E-64, orlistat, ascorbic acid, glycerol 2-phosphate, phorbol 12-myristate 13-acetate (PMA), Hoechst 33342, and fetal bovine serum (FBS) were purchased from Sigma (Saint Louis, MO). 2-Chlorotrityl chloride resin (100–200 mesh, 1.1 mmol · g−1), N-Fmoc protected amino acids, benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP), and N-hydroxybenzotriazole (HOBt) were purchased from EMD Biosciences (San Diego, CA). cPLA2 inhibitor (cPLA2in), sPLA2-IIA inhibitor I (sPLA2in), and cathepsin K inhibitor III (catK III) were purchased from EMD Biosciences (La, Jolla, CA). All other reagents and solvents were purchased from VWR International (West Chester, PA).
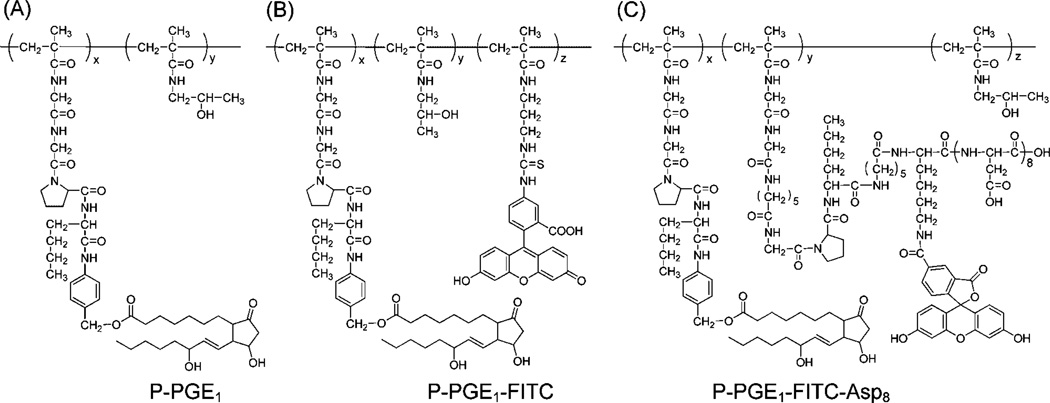
Figure 1.
Structure of HPMA copolymer-PGE1 conjugates. (A) P-PGE1 (M̅w 21.0 kDa, polydispersity 1.4, PGE1 3.1 mol-%); (B) P-PGE1-FITC (M̅w 27.9 kDa, polydispersity 1.5, PGE1 3.1 mol-%, FITC 2.0 mol-%); (C) P-PGE1-FITC-Asp8 (M̅w 37.3 kDa, polydispersity 1.3, PGE1 3.7 mol-%; FITC 2.7 mol-%, d-Asp8 2.7 mol-%).
Cell Lines
The cell lines RAW 264.7, MC3T3-E1, C2C12, Saos-2, and HS-5 were purchased from ATCC, the FLG 29.1 cell line was established in house (M.L. Brandi).[19] Recombinant murine soluble RANK ligand (sRANKL) was purchased from PeproTech (Rocky Hill, NJ). Cell culture media were purchased from Invitrogen (Frederick, MD).
Methods
UV-Vis spectra were measured on a Varian Cary 400 Bio UV-visible spectrophotometer. The molecular weight and molecular weight distribution of polymers were measured on the ÄKTA FPLC system (GE Healthcare) equipped with UV and RI detectors using a Superose 6 HR10/30 column with PBS (pH 7.3) and PBS/acetonitrile (70/30) as the mobile phase. The average molecular weights were calculated using a calibration with polyHPMA fractions.
HPLC analyses of PGE1 samples were performed on the Agilent 1100 series HPLC apparatus equipped with a reverse-phase column (ZORBAX 300SB-C18 4.6 × 250 mm, 5 mm) and a diode-array detector. The mobile phase was a mixture of phosphate buffer (0.02 m, pH 5.0) and acetonitrile, 65:35 at a constant flow rate of 1 ml · min−1. PGE1 was detected at 205 nm (elution time 9.2 min), PGA1 was detected at 230nm(elution time 19.1 min), and PGB1 was detected at 280 nm (elution time 20.1 min). The PGE1 released was calculated as total amount of PGE1 and its metabolized products PGA1 and PGB1.[15] Cortisone (elution time 5.3 min) was used as the internal standard.[15] A calibration curve for PGE1 was obtained using the corresponding peak area versus concentration.
The cell images were captured by laser confocal microscope (FV 1000 Olympus IX81). The HPMA copolymer-PGE1 conjugates labeled with fluorescein (FITC) were added to culture medium of cells with or without induction and incubated for 2 h at 37 °C. Subsequently, Hoechst 33342 (50 × 10−9 m) and Lysotracker Red DND-99 (50 × 10−9 m) were added into medium and incubated for 30 min. After incubation, cells were washed with PBS twice and live cell fluorescence imaging was performed immediately.
Cell Culture and Induction Conditions
All cultures were maintained at 37 °C and 5% CO2. Murine osteoclast precursors, RAW 264.7 cells, were maintained with DMEM supplemented with 10% FBS. They were treated with sRANKL (35 ng · ml−1) upon induction.[28] Mature osteoclasts developed after one week. RAW-OCs were purified by serum density gradient fractionation.[28] Human osteoclast precursors, FLG 29.1 cells, were maintained with RPMI-1640 supplemented with 10% FBS and induced with PMA (0.1 × 10−6 m).[19] Morphological evidence initially appeared after 3 d exposure to PMA and continued to increase. The cells were harvested after one-week induction. Murine osteoblast precursors, MC3T3-E1 cells (subclone 4), were maintained in a-MEM medium supplemented with 5% FBS. l-ascorbic acid (50 mg · ml−1) and glycerol 2-phosphate disodium salt (4 × 10−3 m) were added to promote the rate of differentiation.[20] A well-mineralized extracellular matrix was formed after 10 d and cells were harvested for experiment. Human osteoblast precursors, Saos-2 cells, were grown in McCoy’s 5a medium containing 15% FBS. Before experiment, cells were subcultured in culture dishes and growth medium was changed until cells reached complete confluence.[22] Mouse myoblast C2C12 cells and human bone marrow stromal HS-5 cells were cultured in RPMI-1640 supplemented with 10% FBS as non-skeletal control cells.
Release of PGE1 from the Conjugate by Incubation with Bone Cells
The HPMA copolymer-PGE1 conjugate stock solution was prepared by dissolving the conjugate in RPMI 1640 (without phenol red and FBS) to the final concentration of 1 mg · ml−1. One day before PGE1 release test, the cells were placed in a 24-well cell culture dish at the density of 2 × 103/cm2 and cultured with corresponding culture media. Then the medium was carefully aspirated, washed twice with PBS. After that 0.2 ml of conjugate stock solution was added to the cells and incubated at 37 °C in a 5% CO2 humidified incubator. At chosen time intervals, the medium was collected, the cells were extracted twice with 0.2 ml of methanol containing 1% of acetic acid, and the solutions combined. The content of PGE1, PGA1, and PGB1 was measured using HPLC. The protein content was determined by Lowry assay.
Inhibition Study
Inhibitor stock solutions were freshly prepared by dissolving inhibitors in RPMI 1640 (without phenol red and FBS) (poorly soluble inhibitors were dissolved in a small amount of ethanol and then diluted with RPMI 1640) to the final concentration of 200 × 10−6 m (200 × 10−9 m for cathepsin K inhibitor III). HPMA copolymer-PGE1 conjugate stock solutions (2 mg · ml−1) were prepared by dissolving the conjugate in RPMI 1640 (without phenol red and FBS). In a typical experiment, 0.1 ml of inhibitor stock solution and 0.1 ml of RPMI 1640 were added to cells in a 24-well cell culture dish and incubated for 1 h in 37 °C, 5% CO2 humidified incubator. Then the inhibitor containing medium was carefully aspirated, 0.1 ml of inhibitor stock solution and 0.1 ml of conjugate stock solution were added to the cells and incubated for 24 h. The released PGE1 was measured by HPLC as described above.
Inhibition of the cleavage of the HPMA copolymer-PGE1 conjugate was also carried out using cell homogenates. RAW 264.7 and RAW-OC cells were harvested by centrifugation, re-suspended in buffer (200 × 10−3 m mannitol, 70 × 10−3 m sucrose, 1 × 10−3 m EGTA, and 10 × 10−3 m HEPES, pH 7.4) or acetate buffer (1 m sodium acetate, pH 5.5) and homogenized. The cell homogenates were equally separated into two tubes. One tube was pre-treated with cathepsin K inhibitor III (200 × 10−9 m) and other inhibitors (200 × 10−6 m) for 30 min individually. The other control tube was treated with the solvent that was used to dissolve the inhibitor. Subsequently, the HPMA copolymer-PGE1 conjugate was added into both control and sample tubes at the final concentration of 1 mg · ml−1 and incubated at 37 °C for 24 h. After incubation, cleavage products were extracted by methanol containing 1% of acetic acid and subjected to HPLC analysis.
Results and Discussion
We have previously designed a HPMA copolymer bone targeted drug conjugate containing d-aspartic acid octapeptide (D-Asp8) as the bone targeting moiety and PGE1 as the bone anabolic agent.[15] Biodistribution study in mice demonstrated the bone targeting capacity.[29] The HPMA copolymer-PGE1 conjugate contains a cathepsin K specific tetrapeptide spacer, Gly-Gly-Pro-Nle, linked to a 1,6-elimination 4-aminobenzyl alcohol group and attached to PGE1 via an ester bond (Figure 1).[15] The bond originating in Nle may be cleaved by cathepsin K, followed by a fast 1,6-elimination reaction to release (unmodified) PGE1.[15] Alternatively, PGE1 may be released by the cleavage of the ester bond by esterases.[30]
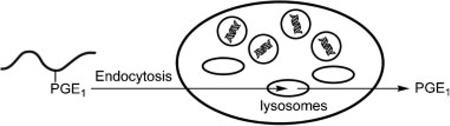
The release of PGE1 in vivo may occur both intracellularly and extracellularly. The conjugate may be internalized by endocytosis and PGE1 released in the lysosomal compartment of cathepsin K expressing cells. Alternatively, when osteoclasts migrate onto a particular bone area, the conjugate bound to the bone surface would be exposed directly to the secreted cathepsin K in the resorption lacuna resulting in PGE1 release. Then PGE1 could either diffuse out of the lacuna by crossing the ruffled border and basolateral membrane, or it could localize into the bone fluid as osteoclasts untie the sealing zone and migrate to another site to start a new cycle of bone resorption. In addition, bone resorption products from the resorption lacuna may be transcytosed to the secretory functional domain.[31] To mimic both scenarios, we have exposed the conjugate to bone (and control) cells and evaluated the internalization and subcellular trafficking of the conjugate as well as PGE1 release.
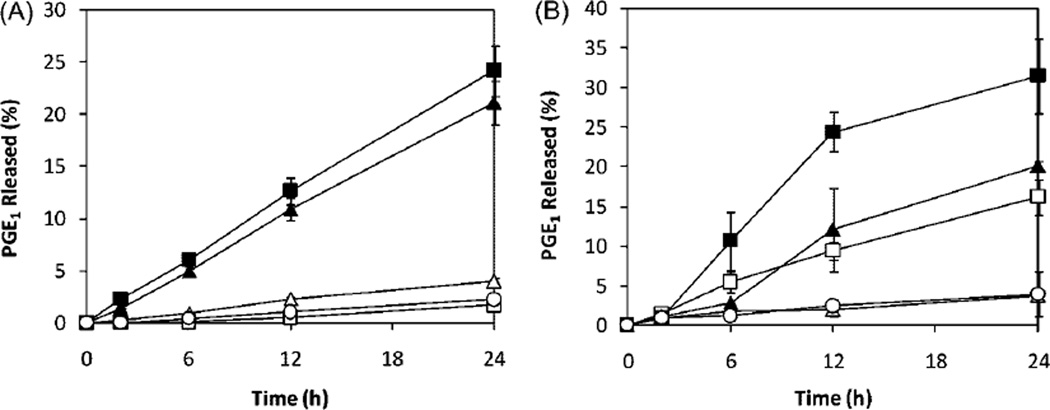
To evaluate the potential of different cell types to cleave the HPMA copolymer-PGE1 conjugate, we have incubated the conjugate with murine and human osteoclasts and osteoblasts, their precursors, and control (non-skeletal) cells (Figure 2). PGE1 was released from the conjugate during incubation with osteoclasts and osteoblasts. The release rate of PGE1 during incubation of the conjugate with osteoclast and osteoblast precursors and (non bone) control cells was lower than with (induced) osteoclasts and osteoblasts. For human cell lines, the release rate had an order of osteoclasts (FLG-OC) > osteoblasts (Saos-2-OB) ≫ osteoblast precursor (Saos-2), osteoclast precursor (FLG29.1), and control cells (HS-5). For murine cell lines, the order of release rates was osteoclasts (RAW-OC) > osteoblasts (MC3T3-E1-OB) > osteoclast precursor (RAW264.7) ≫ osteoblast precursor (MC3T3-E1) and control cells (C2C12).
Figure 2.
PGE1 release from HPMA copolymer-PGE1 conjugate (P-PGE1) during incubation with different cells (protein content was normalized to 0.6 ± 0.1 mg · ml−1). (A) Human cell lines. ■ FLG-OC; ▲ Saos-2-OB; △ Saos-2; ○ HS-5; □ FLG 29.1; (B) Murine cell lines. ■ RAW-OC; ▲ MC3T3-E1-OB; □ RAW 264.7; ○C2C12; △ MC3T3-E1.
These data validated the design of the cathepsin K sensitive spacer. The PGE1 release occurred fast in cells expressing cathepsin K.[5,32] The anabolic agent released in osteoclasts or osteoblasts can interact with its receptors on bone cell surfaces and initiate a cascade of bone modeling and remodeling that will result in net gain of bone mass. Indeed, our first in vivo experiments on the ovariectomized rat model demonstrated bone increases, sustained for 60 d after one administration of the HPMA copolymer-PGE1-D-Asp8 conjugate.[33]
However, other enzymes may contribute to the cleavage of the conjugate. There is an overlap in specificities of cathepsin K and cathepsins L and S,[4] so these enzymes may contribute to the cleavage of the tetrapeptide spacer. In addition, the ester bond connecting the HPMA copolymer with the C-1 position of PGE1 may be susceptible to cleavage by esterases.[30]
Based on published data,[30,34] we selected a series of potential enzyme inhibitors to identify additional enzymes that might be involved in the release of PGE1 from the HPMA copolymer conjugate. We used BW284c51 for acetylcholinesterase (AChE, EC 3.1.1.7), iso-OMPA for butyrylcholinesterase (BChE, EC 3.1.1.8), BNPP for carboxylesterase (CbE, EC 3.1.1.1), EDTA for paraoxonase (PON1, EC 3.1.8.1), orlistat for lysosomal acid lipase (LAL, EC 3.1.1.3), sPLA2in for secreted phospholipase A2 (sPLA, EC 3.1.1.4), cPLA2in for cytosolic phospholipase A2a (cPLA, EC 3.1.1.4), E-64 for cathepsin B (EC 3.4.22.1), and CatK III for cathepsin K (EC 3.4.22.38).
The results of cleavage of the conjugate (release of PGE1) after incubation of the conjugate with different types of cells in the presence of inhibitors are summarized in Table 1. It appears, that cathepsin K inhibitor III decreases the cleavage activity of RAW-OC and FLG-OC cells, but not remarkably. This might be related to the observation that procathepsin K is transformed to active cathepsin K during osteoclast migration to the bone.[35] E-64, cPLA2in, and sPLA2in exhibited also a modest effect; however, the evaluation of the efficacy of other inhibitors (Table 1) needs a more detailed study, such as the evaluation of the concentration dependence of inhibitor activity and inhibitors’ cytotoxicity.
Table 1.
Effect of inhibitors on the release of PGE1 from P-PGE1.a)
| Inhibitor | Cat K III | E-64 | Iso-OMPA | BW284c51 | BNPP | cPLA2in | sPLA2in | Orlistat |
|---|---|---|---|---|---|---|---|---|
| RAW 264.7 | 105.2 | 98.1 | 103.0 | 93.2 | 100.7 | 92.0 | 82.5 | 89.5 |
| RAW-OC | 63.8 | 86.2 | 99.4 | 93.2 | 100.0 | 119.7 | 101.6 | – |
| MC3T3-E1-OB | 87.5 | 102.2 | 95.1 | 100.2 | 106.4 | 124.7 | 118.8 | – |
| FLG 29.1 | 105.6 | 119.4 | 91.7 | 105.6 | 108.3 | 100.0 | 108.3 | – |
| FLG-OC | 69.1 | 75.5 | 82.8 | 88.1 | 81.6 | 75.1 | 82.4 | – |
| Saos-2 | 96.2 | 92.2 | 102.8 | 110.3 | 116.4 | 83.6 | 97.3 | – |
| Saos-2-OB | 84.3 | 86.0 | 96.3 | 108.2 | 106.4 | 95.5 | 105.0 | – |
| RAW 264.7(7.4)b) | 96.9 | 96.4 | 100.0 | 109.4 | 100.5 | 96.9 | 102.4 | 42.3 |
| RAW 264.7(5.5)b) | – | – | – | – | – | – | – | 37.5 |
| RAW-OC(7.4)b) | 74.5 | – | – | – | – | – | – | 33.1 |
| RAW-OC(5.5)b) | 84.5 | – | – | – | – | – | – | 38.4 |
The data were measured 3 times. PGE1 release in experiments without inhibitors was used as control (100). The standard deviations were ±10%;
Experiments with cell homogenates; pH was adjusted to 5.5 with acetate buffer and to 7.4 with PBS.
The inhibitors used possess different levels of hydrophobicity. Consequently, their subcellular location will be one of the factors influencing their activity. Therefore, inhibition experiments were also conducted with cell homogenates of RAW-OC and RAW264.7. Apparently, the activity of orlistat increased when compared to experiments with intact cells. These preliminary results imply that, in addition to cathepsin K, other enzymes contribute to the release of PGE1 from the conjugate. From the fact that E-64, cPLA2in, and orlistat demonstrated partial inhibitory activity in selected cells, focus of further studies should be on cathepsin L and lipases, in particular lysosomal acid lipase.
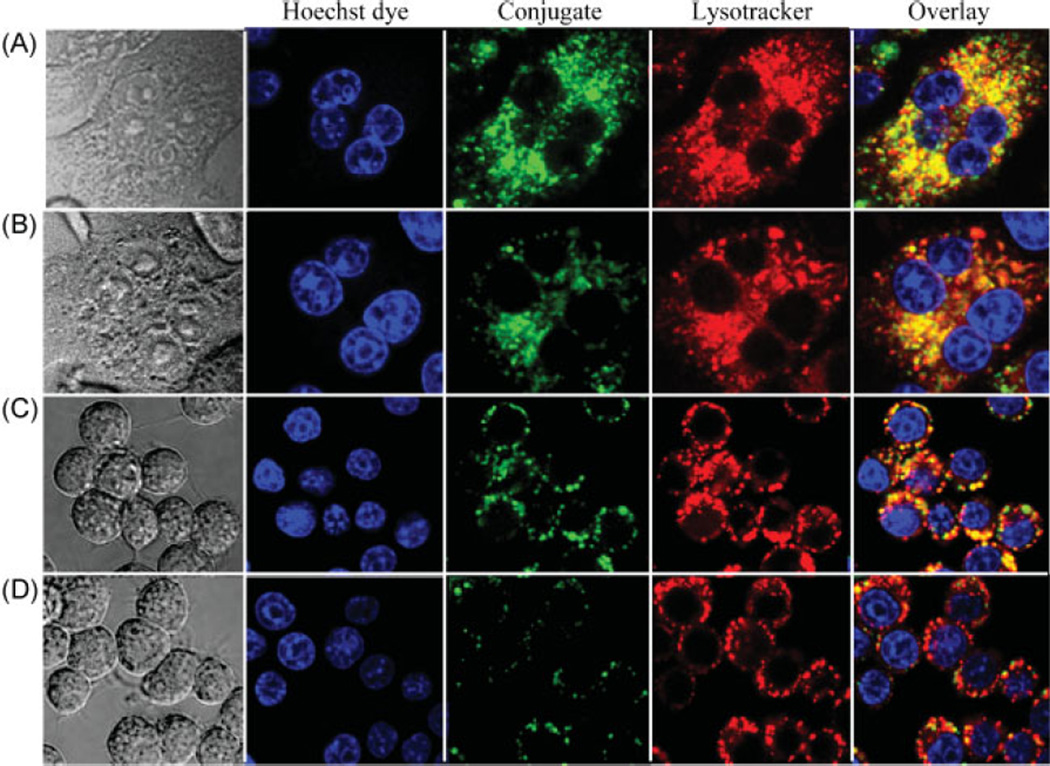
Confocal fluorescence microscopy was employed to evaluate the internalization and subcellular trafficking of HPMA copolymer-PGE1 conjugates, P-PGE1-FITC and P-PGE1-FITC-Asp8, in RAW264.7 and RAW-OC cells (Figure 3). RAW-OC giant cells displayed multiple nuclei, a morphologic characteristics of mature osteoclasts. In contrast, RAW264.7 cells were small, round, and single-nucleus cells. In all experiments, punctuate fluorescence in the perinuclear region was observed. Staining of nuclei with Hoechst 33342 and colocalization experiments with Lysotracker Red indicated clearly that the conjugates were internalized via endocytosis and localized in the late endosomal/lysosomal compartments. Introduction of negatively charged targeting moieties (D-Asp8) into the structure of conjugate decreases the rate of internalization as evidenced by the slower internalization of P-PGE1-FITC-Asp8 when compared to P-PGE1-FITC (Figure 3). This bodes well for the in vivo efficacy of the conjugates. The targeting moiety will change the biodistribution of the conjugate favoring the deposition to bone. Thus the participation of other enzymes, lysosomal acid lipase in particular, in the cleavage of the conjugate will be minimized. The internalization into the non-skeletal cells will be decreased due to a lower internalization rate and lower local concentration of the targeted conjugate when compared to a non-targeted conjugate. An alternative approach to be pursued is the design of HPMA copolymer- PGE1 conjugates which do not contain an ester bond. Synthesis of such conjugates is under way in our laboratory.
Figure 3.
Internalization and subcellular localization of the HPMA copolymer-PGE1 conjugates (green). Hoechst 33342 (50 × 10−9 m, blue) and Lysotracker Red DND-99 (50 × 10−9 m, red) were added for nuclear and lysosome staining. (A) RAW-OC cells incubated with P-PGE1-FITC; (B) RAW-OC cells incubated with P-PGE1-FITC-Asp8; (C) RAW 264.7 cells incubated with P-PGE1-FITC; (D) RAW 264.7 cells incubated with P-PGE1-FITC-Asp8.
Conclusion
Incubation of HPMA copolymer-PGE1 conjugate with induced murine and human osteoclasts and osteoblasts, precursor cells, and control non-skeletal cells revealed that the highest rate of cleavage occurred in osteoclasts, followed by osteoblasts. The high rate of cleavage in these cells is probably the result of cathepsin K catalyzed degradation of the Gly-Gly-Pro-Nle spacer followed by 1,6-elimination mediated by the 4-aminobenzylalcohol group. Other cell types were able to release PGE1 too, albeit with a substantially reduced rate. Cleavage experiments in the presence of selective enzyme inhibitors suggest that this cleavage might be mainly occurring via the ester bond and that lipases might be involved. Confocal fluorescence microscopy observation of osteoclasts and control cells incubated with HPMA copolymer-PGE1 conjugates divulged internalization and subcellular trafficking via endocytosis with ultimate location of the conjugates in the lysosomes.
Acknowledgements
We thank Drs. D. Wang and S. C. Miller for valuable discussions. The research was supported in part by NIH Grant GM069847.
Contributor Information
Huaizhong Pan, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA.
Jihua Liu, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA.
Yuanyi Dong, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA.
Monika Sima, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA.
Pavla Kopečková, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA; Department of Bioengineering, University of Utah, Salt Lake City, Utah 84112, USA.
Maria Luisa Brandi, Department of Clinical Physiopathology, University of Firenze, 50139 Firenze, Italy.
Jindřich Kopeček, Email: jindrich.kopecek@utah.edu, Department of Pharmaceutics and Pharmaceutical Chemistry/CCCD, University of Utah, Salt Lake City, Utah 84112, USA; Department of Bioengineering, University of Utah, Salt Lake City, Utah 84112, USA.
References
- 1.Wang D, Miller SC, Kopečková P, Kopeček J. Adv. Drug Delivery Rev. 2005;57:1049. doi: 10.1016/j.addr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Mostov K, Werb Z. Science. 1997;276:219. doi: 10.1126/science.276.5310.219. [DOI] [PubMed] [Google Scholar]
- 3.Teitelbaum SL. Science. 2000;289:1504. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 4.Brömme D, Okamoto K. Biol. Chem. Hoppe-Seyler. 1995;376:379. doi: 10.1515/bchm3.1995.376.6.379. [DOI] [PubMed] [Google Scholar]
- 5.Drake FH, Dodds RA, James IE, Connor JR, Debouck C, Richardson S, Lee-Rykaczewski E, Coleman L, Rieman D, Barthlow R, Hastings G, Gowen M. J. Biol. Chem. 1996;271:12511. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 6.Kafienah W, Brömme D, Buttle DJ, Croucher LJ, Hollander AP. Biochem. J. 1998;331:727. doi: 10.1042/bj3310727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay R. Endocrine. 2004;24:223. doi: 10.1385/ENDO:24:3:223. [DOI] [PubMed] [Google Scholar]
- 8.Cranney A, Adachi JD. Drug Saf. 2005;28:721. doi: 10.2165/00002018-200528080-00006. [DOI] [PubMed] [Google Scholar]
- 9.Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, Kotchen JM, Ockene J. J. Am. Med. Assoc. 2002;288:321. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 10.Espie M, Daures JP, Chevallier T, Mares P, Micheletti MC, De Reilhac P. Gynecol. Endocrinol. 2007;23:391. doi: 10.1080/09513590701382104. [DOI] [PubMed] [Google Scholar]
- 11.Cranney A, Tugwell P, Wells G, Guyatt G. Endocrinol. Rev. 2002;23:496. doi: 10.1210/er.2001-1002. [DOI] [PubMed] [Google Scholar]
- 12.Odvina CV, Zerwekh JE, Rao DS, Maalouf N, Gottschalk FA, Pak CY. J. Clin. Endocrinol. Metab. 2005;90:1294. doi: 10.1210/jc.2004-0952. [DOI] [PubMed] [Google Scholar]
- 13.Jee WS, Mori S, Li XJ, Chan S. Bone. 1990;11:253. doi: 10.1016/8756-3282(90)90078-d. [DOI] [PubMed] [Google Scholar]
- 14.Marks SC, Jr, Miller SC. J. Periodontal Res. 1994;29:103. doi: 10.1111/j.1600-0765.1994.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 15.Pan H, Kopečková P, Wang D, Yang J, Miller S, Kopeček J. J. Drug Targeting. 2006;14:425. doi: 10.1080/10611860600834219. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Miller SC, Shlyakhtenko LS, Portillo AM, Liu X-M, Papangkorn K, Kopečková P, Lyubchenko Y, Higuchi WI, Kopeček J. Bioconjugate Chem. 2007;18:1375. doi: 10.1021/bc7002132. [DOI] [PubMed] [Google Scholar]
- 17.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Proc. Natl. Acad. Sci. USA. 1999;96:3540. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam S, Hassan F, Tumurkhuu G, Dagvadorj J, Koide N, Naiki Y, Mori I, Yoshida T, Yokochi T. Biochem. Biophys. Res. Commun. 2007;360:346. doi: 10.1016/j.bbrc.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Gattei V, Bernabei PA, Pinto A, Bezzini R, Ringressi A, Formigli L, Tanini A, Attadia V, Brandi ML. J. Cell Biol. 1992;116:437. doi: 10.1083/jcb.116.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudo H, Kodama HA, Amagai Y, Yamamoto S, Kasai S. J. Cell Biol. 1983;96:191. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franceschi RT, Iyer BS. J. Bone Miner. Res. 1992;7:235. doi: 10.1002/jbmr.5650070216. [DOI] [PubMed] [Google Scholar]
- 22.Rodan SB, Imai Y, Thiede MA, Wesolowski G, Thompson D, Bar-Shavit Z, Shull S, Mann K, Rodan GA. Cancer Res. 1987;47:4961. [PubMed] [Google Scholar]
- 23.Jeschke M, Brandi ML, Susa M. J. Bone Miner. Res. 1998;13:1880. doi: 10.1359/jbmr.1998.13.12.1880. [DOI] [PubMed] [Google Scholar]
- 24.Quarles LD, Yohay DA, Lever LW, Caton R, Wenstrup RJ. J. Bone Miner. Res. 1992;7:683. doi: 10.1002/jbmr.5650070613. [DOI] [PubMed] [Google Scholar]
- 25.Nadiminty N, Lou W, Lee SO, Mehraein-Ghomi F, Kirk JS, Conroy JM, Zhang H, Gao AC. Clin. Cancer Res. 2006;12:1420. doi: 10.1158/1078-0432.CCR-05-1849. [DOI] [PubMed] [Google Scholar]
- 26.Kopeček J, Bažilová H. Eur. Polym. J. 1973;9:7. [Google Scholar]
- 27.Omelyanenko V, Kopečková P, Gentry C, Kopeček J. J. Controlled Release. 1998;53:25. doi: 10.1016/s0168-3659(97)00235-6. [DOI] [PubMed] [Google Scholar]
- 28.Collin-Osdoby P, Yu X, Zheng H, Osdoby P. Methods Mol. Med. 2003;80:153. doi: 10.1385/1-59259-366-6:153. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Sima M, Mosley RL, Davda JP, Tietze N, Miller SC, Gwilt PR, Kopečková P, Kopeček J. Mol. Pharm. 2006;3:717. doi: 10.1021/mp0600539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan H, Kopečková P, Liu J, Wang D, Miller SC, Kopeček J. Pharm. Res. 2007;24:2270. doi: 10.1007/s11095-007-9449-3. [DOI] [PubMed] [Google Scholar]
- 31.Nesbitt SA, Horton MA. Science. 1997;276:266. doi: 10.1126/science.276.5310.266. [DOI] [PubMed] [Google Scholar]
- 32.Mandelin J, Hukkanen M, Li TF, Korhonen M, Liljestrom M, Sillat T, Hanemaaijer R, Salo J, Santavirta S, Konttinen YT. Bone. 2006;38:769. doi: 10.1016/j.bone.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Miller S, Pan H, Wang D, Bowman BM, Kopečková P, Kopeček J. doi: 10.1007/s11095-008-9706-0. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheriff S, Du H, Grabowski GA. J. Biol. Chem. 1995;270:27766. doi: 10.1074/jbc.270.46.27766. [DOI] [PubMed] [Google Scholar]
- 35.Dodds RA, James IE, Rieman D, Ahern R, Hwang SM, Connor JR, Thompson SD, Veber DF, Drake FH, Holmes S, Lark MW, Gowen M. J. Bone Miner. Res. 2001;16:478. doi: 10.1359/jbmr.2001.16.3.478. [DOI] [PubMed] [Google Scholar]