Abstract
The germline-specific role of telomeres consists of chromosome end elongation and proper chromosome segregation during early developmental stages. Despite the crucial role of telomeres in germ cells, little is known about telomere biology in the germline. We analyzed telomere homeostasis in the Drosophila female germline and early embryos. A novel germline-specific function of deadenylase complex Ccr4-Not in the telomeric transcript surveillance mechanism is reported. Depletion of Ccr4-Not complex components causes strong derepression of the telomeric retroelement HeT-A in the germ cells, accompanied by elongation of the HeT-A poly(A) tail. Dysfunction of transcription factors Woc and Trf2, as well as RNA-binding protein Ars2, also results in the accumulation of excessively polyadenylated HeT-A transcripts in ovaries. Germline knockdowns of Ccr4-Not components, Woc, Trf2 and Ars2, lead to abnormal mitosis in early embryos, characterized by chromosome missegregation, centrosome dysfunction and spindle multipolarity. Moreover, the observed phenotype is accompanied by the accumulation of HeT-A transcripts around the centrosomes in early embryos, suggesting the putative relationship between overexpression of telomeric transcripts and mitotic defects. Our data demonstrate that Ccr4-Not, Woc, Trf2 and Ars2, components of different regulatory pathways, are required for telomere protection in the germline in order to guarantee normal development.
INTRODUCTION
Telomeres are nucleoprotein complexes at the ends of linear chromosomes that protect chromosome ends from fusion and degradation owing to the end under-replication process (1). Protection of chromosome ends is essential in all cell types to prevent chromosome end-to-end fusion. At the same time, loss of the terminal DNA is repaired only in germ and stem cells by the synthesis of tandem telomeric repeats (2,3). Elongation of chromosome ends in proliferating somatic cells is believed to lead to cancer (4). In germ cells, a special mechanism of telomere length maintenance is present, providing replicative potential for the shortened telomeres in normal terminally differentiated somatic cells (5). Telomere length control is an essential germline-specific function implying that telomere organization in the germ cells has distinctive characteristics. However, the basic aspects of telomere biology in the germline and in early development are far from being understood.
In most eukaryotres, telomeric DNA is maintained through telomerase action, which is responsible for the synthesis of 6–9 nucleotide repeats using its RNA component as a template (6). The telomeres of Drosophila are maintained as a result of the retrotransposition of specialized telomeric non-long terminal repeat retrotransposons (7–11). In all cases, telomere elongation utilizes reverse transcription, i.e. synthesis of DNA on the RNA template. Also, telomeric DNA serves as a platform for the binding of telomeric proteins, required for the protection of chromosome termini and the maintenance of specific chromatin structure. In particular, heterochromatic protein 1 (HP1) and the DNA repair complex provide protection against telomere fusions in both Drosophila and humans (12,13), demonstrating strong similarities in the telomere protection pathways in different organisms.
Recent data show that the transcription of telomeric DNA and the requirement for RNA-interference (RNAi) machinery in telomere function are also conserved among eukaryotes, irrespective of the mode of telomere elongation. Telomeres in organisms that use telomerase are transcribed into telomeric repeat-containing RNA (TERRA) (14,15), which serves as a precursor for small RNA production. Telomere-specific piRNA-like small RNAs have been detected in mouse embryonic stem cells (16) and in Arabidopsis (17). These small RNA species strongly resemble telomeric transposon-derived small RNA in Drosophila (18,19). In the Drosophila germline telomeric retrotransposons are silenced by the PIWI-interacting RNA (piRNA) pathway (18,19). Moreover, the increased telomeric transposon expression in flies with a disrupted piRNA pathway is correlated with an increased frequency of telomeric element transposition and, therefore, with telomere elongation (18). Recent studies have provided strong evidence that telomeric piRNAs impact the transcriptional status of telomeric retroelements (19,20) as well as formation of the telomere protection complex (21). Most likely, it is the piRNA pathway proteins that provide germline-specific peculiarities to telomeric complex formation. However, possible piRNA pathway partners in the germline telomeric protein complex are currently unknown.
HeT-A telomeric element is the main structural component of Drosophila telomeres present at all chromosome ends. In contrast, the other two telomere retrotransposons, TART and TAHRE, are represented by only a few copies (7,22). In order to discover new components of the HeT-A silencing complex, we performed a selective RNAi-based screen in the Drosophila female germline, using endogenous HeT-A expression as a read-out. We found that germline knockdown of the components of the Ccr4-Not deadenylase complex as well as the transcription factors Woc (without children) and Trf2 (TATA box-binding protein (TBP)-related factor 2) caused a strong derepression of HeT-A retrotransposon in ovaries. The Ccr4-Not complex is a conserved multi-subunit complex that regulates gene expression at different levels, including mRNA deadenylation, transcription, transport and degradation (23,24). In Drosophila, the Ccr4-Not deadenylase complex regulates the expression of specific germline mRNAs by controlling their poly(A)-tail length (25). Here, we provide evidence for the essential role of the Ccr4-Not complex in telomeric transposon HeT-A transcript adenylation status in the germ cells.
Germline knockdowns of factors affecting the HeT-A RNA level, such as Ccr4-Not components, the transcription factors Woc and Trf2 and RNA-binding protein Ars2 cause mitotic defects during the early stages of embryonic development. Our findings suggest that these factors act cooperatively to regulate telomeric repeat expression and telomere protection in the germline, providing genome integrity during early development.
MATERIALS AND METHODS
Drosophila strains
KD (from ‘knockdown’) flies were F1 of the cross of two strains bearing construct with short hairpin RNA (shRNA) (Supplementary Table S1) and, unless otherwise specified, strain #25751 (P{UAS-Dcr-2.D}1, w1118, P{GAL4-nos.NGT}40, Bloomington Stock Center) providing GAL4 expression under the control of the germline-specific promoter of the nanos (nos) gene. An additional Gal4 driver strain was #4937 (w1118; P{GAL4::VP16-nos.UTR}CG6325MVD1, Bloomington Stock Center). Control flies were F1 of the crossing of the white hairpin construct and #25751 (Bloomington Stock Center). The Trf2p1 allele was described in (26). Strains bearing spindle-E (spn-E) mutations were ru1 st1 spn-E1 e1 ca1/TM3, Sb1 es and ru1 st1 spn-Ehls3987 e1 ca1/TM3, Sb1 es. twin mutants were twinDG24102/Df(3R) crb87-4, twinS1/Df(3R) crb87-4 and twinS3/Df(3R) crb87-4. Not1 mutant was Not1MI07631/Not1G5743. Polymerase chain reaction (PCR) with genomic DNAs using Wolbachia-specific primers demonstrated that there was no contamination with Wolbachia for the strains used in the study.
In situ RNA hybridization and immunochemistry
In situ RNA analysis was carried out according to the previously described procedure (27) using digoxigenin (DIG)-labeled strand-specific HeT-A, TART or TAHRE riboprobes and alkaline phosphatase (AP)-conjugated anti-DIG antibodies (Roche, diluted 1/2000). The HeT-A antisense probe contained a fragment of the ORF (nucleotides 4330–4690 of GenBank sequence DMU06920); the TART antisense probe contained a fragment corresponding to nucleotides 2377–2888 of GenBank sequence DMU02279. The TAHRE-specific probe contained a fragment corresponding to nucleotides 5717–6295 of GenBank sequence AJ542581.
Combined protein and RNA localization was done as described (28). Embryos were collected and fixed in methanol as described (29). The RNA fluorescence in situ hybridization (FISH) was done with DIG-labeled strand-specific riboprobes. For hybridization signal enhancement, we used incubation with anti-DIG- fluorescein (FITC) antibodies (Roche, diluted 1/500), followed by incubation with anti-FITC Alexa Fluor 488 antibodies (Life Technologies, diluted 1/500). Tyramide signal amplification was done as an alternative method of signal enhancement (30). Immunostaining was done using anti-γ-tubulin (Sigma), anti-α-tubulin (Sigma), anti-mouse-Alexa 546 (Life Technologies) and anti-rabbit-Alexa 546 (Life Technologies). Embryos were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in ProLong Gold antifade reagent (Life Technologies). Images were captured using a Zeiss LSM 510 Meta confocal microscope. Embryonic chromosome spreading was done as described (31).
RTqPCR analysis
cDNA was synthesized using random hexamer or oligo(dT) primers and SuperScriptII reverse transcriptase (Life Technologies). cDNA samples were analyzed by real-time quantitative PCR using SYTO-13 dye. qPCR was performed on DT-96, DNA Technology, Russian Federation. Eight serial three-fold dilutions of cDNA or DNA were amplified in duplicate with each primer pair to build standard curves. Values were averaged and normalized to the expression level of ribosomal protein gene rp49. Normalization of RT-qPCR data to the retrotransposon copy number in each strain was performed; however, no significant differences in copy number were detected. All experiments were performed with at least two independent RNA samples, and each sample was analyzed in triplicate. The primers used are listed in Supplementary Materials and Methods. Nuclear run-on (NRO) assay was performed as described previously (19).
Polyadenylation assay
Poly(A) tail length was analyzed using a poly(A) tail (PAT) assay as previously described (32) with an oligo (dT)-anchor primer and specific primers (Supplementary Materials and Methods). PCR was carried out in the presence of αdATP-P32, and the products were separated in a 6% non-denaturing polyacylamide gel. A polyadenylation assay based on RNA ligation-coupled RT-PCR was done as described (33) using the Illumina 3′linker RA3.
RESULTS
Trancription factors Woc and Trf2 suppress telomeric repeat HeT-A transcription in ovaries
In order to detect germline components contributing to telomeric homeostasis, we performed a selective RNAi screen for factors that downregulate HeT-A expression in Drosophila ovaries. This approach was based on the existing data that in somatic tissues certain factors of the telomeric complex are required for the silencing of telomeric repeats (34–36). As reported earlier, HP1, encoded by Su(var)205 gene, a component of the telomere protection complex (37,38), elicits transcriptional repression of telomeric retrotransposons both in soma (34) and in the germline (39,40). At the same time, transcription of telomeric and non-telomeric transposons in the germline is controlled by the piRNA system, supporting the idea that the telomeric complex in the germline consists of both germline-specific and ubiquitous components. To investigate the structure of telomeric chromatin in the germline, we examined HeT-A expression in the ovaries of flies depleted for known components of the somatic telomeric complex as well as factors involved in RNA surveillance. Here, we describe genes exerting the greatest effect on telomeric repeat expression in ovaries. Germline knockdown of several genes, previously implicated in telomere functioning in soma, did not noticeably affect HeT-A expression in our screen and were not included in further analysis (Supplementary Table S2).
In parallel to an independent study that describes Trf2 as a negative regulator of telomeric transposon expression in somatic tissues (41), we demonstrated that Trf2 is associated with the polytene chromosome telomeres of salivary glands (Supplementary Figure S1) and discovered a role of Trf2 in telomere functioning in the germline. RT-qPCR analysis of telomeric and non-telomeric retrotransposon expression demonstrated increased levels of HeT-A and TAHRE transcripts in ovaries upon germline knockdown of Trf2 (Trf2KDnos; nos designates the promoter of the germline-specific gene nanos that was used to express the Gal4 driver) (Figure 1A). Given that the HeT-A and TAHRE untranslated regions (UTR) are highly homologous (7), we suggest that Trf2 recognizes a similar sequence motif in the promoter regions of both elements.
Figure 1.
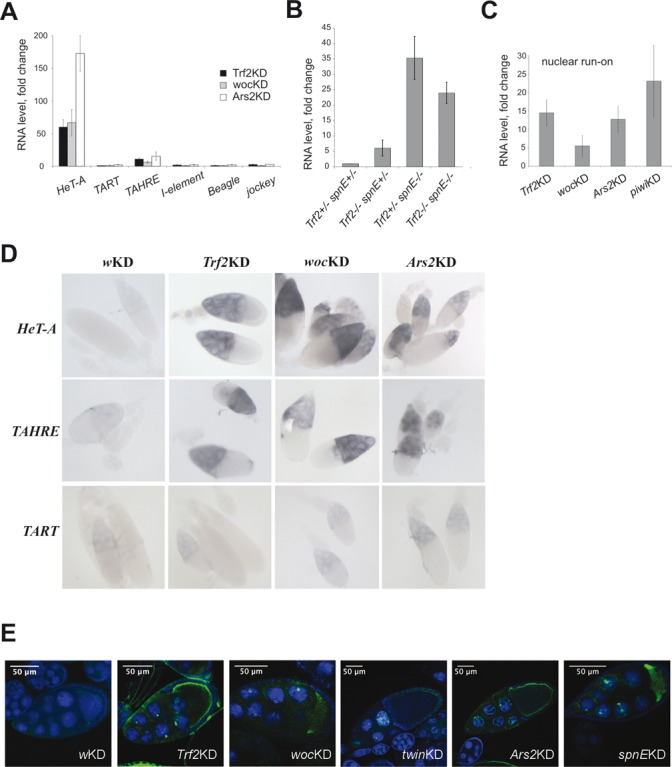
Telomeric element HeT-A expression is downregulated in ovaries by Trf2, Woc and Ars2. (A) RT-qPCR analysis of TE expression in ovaries of Trf2KDnos, wocKDnos and Ars2KDnos. (B) HeT-A transcript abundance in ovaries of Trf2/Trf2, spnE/spnE and Trf2,spnE double mutants. The fold change in the HeT-A RNA level is shown. (C) NRO analysis of HeT-A transcription in ovaries of Trf2KDnos, wocKDnos, Ars2KDnos and piwiKDnos flies. The RNA level was normalized to rp49. Mean values (±SD) for three biological samples normalized to control knockdown (white(w)KDnos) are shown. (D) In situ RNA hybridization of HeT-A, TART or TAHRE antisense probes with ovaries of the indicated genotypes (GAL4-nos driver was used). (E) RNA FISH of HeT-A antisense probe with ovaries of the indicated genotypes. The accumulation of HeT-A transcripts (green) in nuclei of nurse cells was observed. DNA was stained with DAPI (blue). GAL4-nos driver was used.
In order to determine if Trf2 functions in the piRNA pathway together with spindle-E (encoding DEAD box RNA helicase SpnE) to silence HeT-A, we applied a double mutant analysis. We used a viable Trf2p1 mutation (26) to generate Trf2/Trf2; spnE/spnE double mutants and compared HeT-A transcript abundance in their ovaries to that in ovaries of single Trf2/Trf2 or spnE/spnE mutants. HeT-A expression in the double mutants was higher than in Trf2 single mutants, but not in spnE single mutants (Figure 1B, Supplementary Figure S2). This could mean that in the absence of piRNAs, inhibitory complex containing Trf2 cannot be recruited to the HeT-A promoter. Northern blotting of short RNA did not reveal changes in the abundance of HeT-A-specific piRNA in ovaries of Trf2 mutants and Trf2KDnos flies (Supplementary Figure S3A). These data suggest that Trf2 may act downstream of piRNA processing to establish transcriptional silencing of HeT-A.
The zinc finger transcription factor Woc has been shown to bind Drosophila telomeres, exerting telomere-capping functions (42). wocKDnos causes a strong derepression of HeT-A and TAHRE but not TART and other TEs (Figure 1A), suggesting sequence-specific binding of this transcription factor to telomeric repeats in ovaries.
We extended the analysis of the telomeric protein complex by including an RNA binding protein, Ars2, involved in mi- and siRNA biogenesis (43), which was identified in an RNAi screen as a negative regulator of HeT-A expression (44). Particular interest in this protein was due to the fact that Ars2 downregulates the expression of human TERRA (45). RT-qPCR analysis revealed a strong derepression of HeT-A/TAHRE in ovaries of Ars2KDnos flies (Figure 1A). To address the question about the involvement of Trf2, Woc and Ars2 in the transcriptional regulation of HeT-A, we conducted an NRO assay, which allows estimating the level of nascent HeT-A transcripts (46). NRO on ovaries with Trf2, woc and Ars2 knockdowns demonstrates that all these factors provide transcriptional silencing of the telomeric retrotransposon HeT-A (Figure 1C).
In situ RNA hybridization analysis was performed to examine the effect of Trf2, Woc and Ars2 depletion on telomeric retroelement transcript distribution in ovaries (Figure 1D, Supplementary Figure S4). Knockdowns of Trf2, Woc and Ars2 exerted the most pronounced effect on HeT-A and TAHRE expression, resulting in the accumulation of their transcripts in nurse cells. The RNA FISH analysis on ovaries depleted for Trf2, Woc and Ars2 revealed HeT-A RNA signals not only in the cytoplasm, but also in the nuclei of nurse cells, where HeT-A transcripts were accumulated in distinct foci (Figure 1E). Similar nuclear foci were observed in spnEKD ovaries (Figure 1E). Previously, using combined DNA/RNA FISH analysis, it was shown that both HeT-A RNA and telomeric DNA were co-localized in nurse cell nuclei in spnE mutants (19). Most likely, we observed the accumulation of nascent transcripts at sites of transcription upon Trf2, Woc and Ars2 knockdown, in accordance with the result of NRO.
Taking into account the HeT-A/TAHRE-specific effects of Woc and Trf2, it is unlikely that these factors participate in piRNA biogenesis. However, it is possible that DNA-binding factors Woc and Trf2 are involved in the piRNA-mediated transcriptional silencing mechanism that is specific for telomeric repeats.
Ccr4-Not deadenylase complex provides telomeric element HeT-A silencing in the germline
mRNA turnover enzymes have been proposed to be involved in the piRNA-dependent degradation of transposon transcripts in cytoplasmic bodies (47). We observed a strong derepression of HeT-A upon germline knockdowns of twin (encoding Ccr4), Not1 and Pop2, which encode components of the Drosophila Ccr4-Not deadenylase complex (48) (Figure 2, Supplementary Figure S5). RT-qPCR analysis showed that changes in HeT-A expression level were considerably higher in comparison to other TEs (Figure 2A). We also examined additional mutant strains and shRNAs targeting Ccr4 and Not1 (Supplementary Figure S4, S5). Although the level of derepression varied, we always observed HeT-A RNA accumulation as a result of Ccr4-Not depletion. This indicates a specificity of the observed effects. piRNA pathway gene germline knockdowns exhibit a wide range of HeT-A upregulation comparable to the effects of Ccr4-Not knockdowns (Supplementary Figure S5). Increased HeT-A expression was detected in ovaries but not in the carcasses of twin mutants (Figure 2B), indicating that the Ccr4-Not complex represses HeT-A expression specifically in gonads. Northern blotting of short RNA did not show changes in HeT-A- and I-element-specific piRNA abundance in the ovaries of twinKDnos and Not1KDnos flies (Supplementary Figure S3B,C). Thus, HeT-A transcripts accumulated upon Ccr4-Not knockdown avoid piRNA-mediated degradation. In situ RNA hybridization on Ccr4, Pop2 and Not1 KD ovaries revealed HeT-A transcripts in nurse cells (Figure 2C). Importantly, RNA FISH revealed HeT-A RNA foci in the nuclei of twinKDnos nurse cells (Figure 1E) as well as upon Trf2, Woc, Ars2 and SpnE knockdown, suggesting that all these components regulate the telomeric transcript level at the site of transcription.
Figure 2.
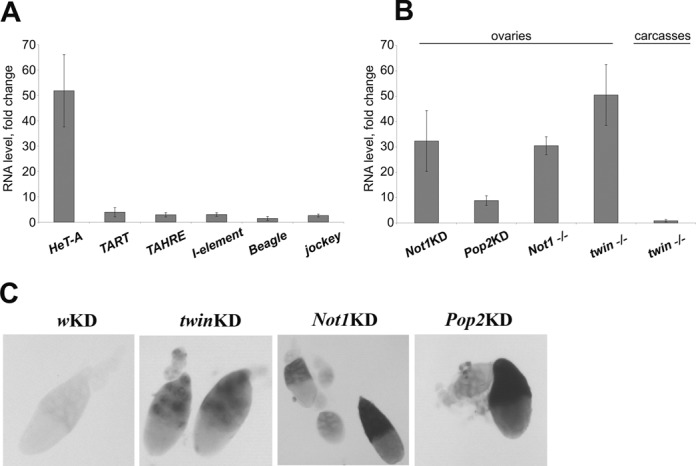
Ccr4-Not deadenylase complex suppresses telomeric element HeT-A expression in the germline. (A) RT-qPCR analysis of TE expression in ovaries of twinKDnos. (B) Changes in HeT-A steady state RNA levels in ovaries and carcasses of flies with indicated genotypes. twin mutant (twin-/-) is a trans-heterozygote twinDG24102/Df(3R) crb87-4. Error bars indicate SD. (C) In situ RNA hybridization of HeT-A antisense probes with ovaries of the indicated genotypes. GAL4-nos driver was used.
Polyadenylation state of HeT-A telomeric transposon transcripts
The Ccr4-Not complex is responsible for the cytoplasmic deadenylase activity in Drosophila ovaries and embryos (48–52). To verify whether stabilization of HeT-A transcripts upon germline knockdowns of Ccr4-Not components is due to increased polyadenylation, the poly(A) tail length of the HeT-A transcripts was measured in a PAT assay (32). Indeed, twin knockdown led to the appearance of additional PAT products with higher molecular weights (Figure 3A). To confirm that these products corresponded to HeT-A transcripts with longer poly(A) tails, but were not the result of alternative polyadenylation, the top and bottom PAT products were cloned and sequenced. These products corresponded to HeT-A transcripts with poly(A) tail lengths of 50–65 nt and 7–11 nt, respectively (Figure 3B). At the same time, the poly(A) tail length of TART was not affected by Ccr4 knockdown (Figure 3A). To confirm this result, we compared the data from the RT-qPCR analysis of TE expression obtained with oligo(dT) or random primers used in reverse transcription. Using oligo(dT) for reverse transcription priming allows for the estimation of the polyadenylation status of transcripts. We observed that the ratio of polyadenylated to total HeT-A transcripts increased upon Ccr4 knockdown. This ratio remained unaffected for TART transcripts (Supplementary Figure S6).
Figure 3.
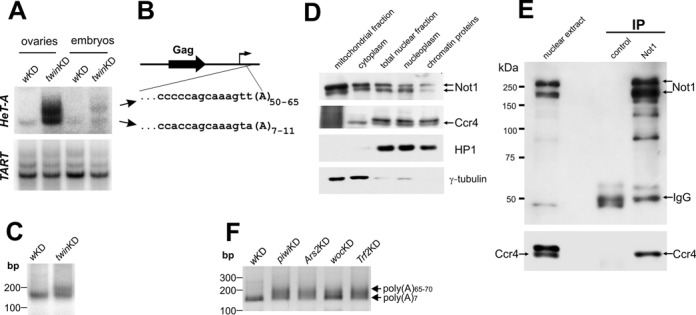
Polyadenylation state of HeT-A telomeric transposon transcripts. (A) PAT assay of HeT-A and TART RNA in ovaries and embryos of twinKDnos flies. (B) Sequencing of the top and bottom PCR products (three clones for each band) shows elongation of the poly(A) tail of HeT-A transcripts in ovaries of twinKDnos. The diagram on the top shows a scheme of HeT-A: open reading frame (black arrow), transcription start site (thin arrow) and the fragments of the sequenced regions. (C) PAT assay of HeT-A transcripts in the nuclear RNA fraction of control and twinKDnos flies. (D) Considerable amount of both Ccr4 and Not1 localize in ovarian nuclei. Western blot of w1118 ovary extract fractions probed with antibodies against Not1, Ccr4, HP1 and γ-tubulin. (E) Ccr4 was coimmunoprecipitated with Not1 in the nuclear extract from ovaries of w1118. Immunoprecipitation was performed with anti-Not1 antibodies or normal mouse serum (control IP). A Western blot was probed with anti-Not1 (top) or anti-Ccr4 (bottom). (F) Analysis of poly(A) tail length of HeT-A RNA using the 3′ ligation−PCR approach in ovaries upon white,Trf2, woc, Ars2 and piwi germline knockdown.
Given the accumulation of HeT-A transcripts in the nuclei upon Ccr4 depletion (Figure 1E), we investigated the poly(A) tail length of the HeT-A RNA in the nuclear fraction of ovarian RNA upon germline knockdown of Ccr4. The PAT assay demonstrated that in the nuclei of control ovaries, HeT-A transcripts were not adenylated, while Ccr4 KD led to an increase in HeT-A poly(A) length in the nucleus (Figure 3C). In yeast, the Ccr4-Not complex has been reported to have both cytoplasmic and nuclear localization and activity (24). Previously, it was shown by immunostaining that Pop2 (CAF1), Ccr4 and Not1 were localized mostly in the cytoplasm in Drosophila ovaries and embryos (48,53). To address the question about the presence of Ccr4-Not in the nuclei of Drosophila ovaries, we performed cellular fractionation and showed that both Ccr4 and Not1 are present not only in the cytoplasm but also in the nucleus (Figure 3D). Next, we co-immunoprecipitated Ccr4 from the ovarian nuclear extract using Not1 antibody (Figure 3E). Thus, our data show that the Ccr4-Not complex negatively regulates the poly(A) tail length of HeT-A transcripts. Taking into account the nuclear localization of the Ccr4-Not complex, we suggest that this process takes place in the nucleus; however, we cannot exclude the possibility that deadenylated HeT-A RNA could be imported back into the nucleus from the cytoplasm.
To explore a potential relationship of different factors affecting the HeT-A transcript level, we analyzed the poly(A) tail length of HeT-A transcripts in ovaries upon knockdown of Woc, Trf2, Ars2 and Piwi, factors that regulate HeT-A at a transcriptional level. We applied direct ligation of a 3′ adapter to RNA, followed by a PCR analysis of cDNA, since the oligo(dT)-based protocol of the PAT assay introduces a strong bias against transcripts with short poly(A) tails. Germline knockdowns of Woc, Trf2, Ars2 and Piwi caused the appearance of additional PCR products with higher molecular weights (Figure 3F) corresponding to HeT-A transcripts with longer poly(A) tails, as verified by sequencing. These results suggest that all these components act in a common pathway to regulate the life cycle of the telomeric transcripts, and that this process most likely occurs within the telomeric compartment in the nucleus.
Germline knockdowns of Ccr4, Not1, Woc and Trf2 cause abnormal mitosis in early stages of development.
In order to determine whether Trf2, Woc, Ars2 and Ccr4-Not depletion in the germline affects early stages of embryogenesis, we analyzed chromosome segregation in 0–2-h-old embryos during mitotic divisions. Maternally deposited RNA and proteins govern this stage until the onset of zygotic transcription, which occurs after two hours of development. Germline knockdowns of the Trf2, woc, twin and Ars2 genes led to asynchronous divisions during the syncytial stage of embryogenesis (Supplementary Figure S7), as well as a high lethality of embryos (Supplementary Table S3). Such embryos have extensive regions devoid of nuclei (Supplementary Figure S7) and exhibit various mitotic defects. Most frequently, we observed chromosome bridges, abnormal mitotic chromosome figures and multipolar spindles (Figure 4A,B, Supplementary Table S4). Surface optical sections of Trf2KDnos, wocKDnos, Ars2KDnos and twinKDnos embryos revealed free centrosomes that remained at the cortex while the nuclei sank into the interior of the embryo. Detached centrosomes are positive for γ-tubulin, the major centrosome component (Figure 4C, Table 1). Configurations of mitotic chromosomes in Trf2-/-, wocKDnos, Ars2KDnos and twinKDnos embryos were visualized using an embryonic squashing technique (31). We observed frequent chromosome end-to-end fusions, ring chromosomes and anaphase bridges, hallmarks of telomere dysfunction (Figure 4D, Supplementary Figure S8A). We performed DNA FISH on Trf2 mutant embryos with the HeT-A probe and showed that the HeT-A signal is present at the site of anaphase chromosome bridges (Supplementary Figure S8B). These data indicate that the mitotic bridges are caused by telomere fusions. Therefore, maternal depletion of Trf2, Woc, Ars2 and Ccr4 leads to HeT-A overexpression, which compromises the telomere protection complex (or vice versa), resulting in chromosome segregation defects. At later stages of syncytial development of such embryos, centrosomes frequently detach from the mitotic nuclei resulting in the accumulation of abundant γ-tubulin positive free centrosomes at the cortex while the defective nuclei are eliminated.
Figure 4.
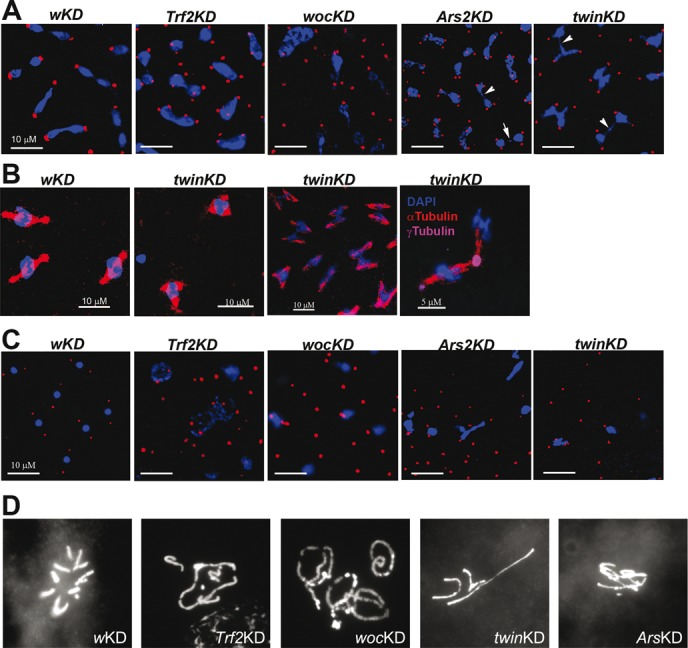
Mitotic defects observed in syncytial embryos from wocKDnos, Not1KDnos, twinKDnos, Ars2KDnos females. (A) Confocal images of embryos with the indicated genotypes. Abnormal chromatin figures, chromatin bridges (arrows) and free centrosomes were observed. Red: γ-tubulin; blue: DNA. Bar: 10 μm. (B) Multipolar spindles observed in syncytial embryos from twinKDnos. Red: α-tubulin; magenta: γ-tubulin; blue: DNA. (C) Cortical areas of embryos with the indicated genotypes. Abundant free centrosomes were observed. Nuclei fallen into the interior of the embryo are visible. Red: γ-tubulin; blue: DNA. Bar: 10 μm. (D) Mitotic chromosomes from embryos. Chromosome fusions were observed in Trf2KDnos, wocKDnos, Ars2KDnos and twinKDnos embryos.
Table 1. Quantification of free centrosomes at the late stages of syncytial development after migration of nuclei to the cortex.
| Genotype | Nuclei/centrosomesa | Free centrosomes associated with HeT-A RNAb |
|---|---|---|
| wKDnos | 46/93 | 0 |
| Trf2p1 | 69/246 | 26 |
| wocKDnos | 42/156 | 53 |
| Ars2KDnos | 78/232 | 36 |
| twinKDnos | 71/206 | 41 |
aThe number of nuclei/centrosomes was counted per optical field of samples immunostained with anti-γ-tubulin antibodies and hybridized with HeT-A antisense probe.
bOnly HeT-A signals that overlapped with γ-tubulin staining (or located at a distance less than one centrosome diameter away from it) were quantified.
To ask whether hyperexpression of HeT-A in ovaries leads to HeT-A transcript accumulation in early embryos, we examined HeT-A RNA abundance in 0−2-h-old Trf2-/-, wocKDnos, Ars2KDnos and twinKDnos embryos by RT-qPCR. We observed an accumulation of HeT-A transcripts in the embryos (Supplementary Figure S9), indicating that maternal HeT-A transcripts are effectively loaded in the embryo. Next, we studied the localization of HeT-A transcripts using fluorescence in situ RNA hybridization on 0–2-h-old embryos combined with immunostaining of γ-tubulin. HeT-A transcripts accumulated around the centrosomes at different stages of mitosis in the embryos upon germline knockdown of Trf2, Woc, Ccr4 and Ars2 (Figure 5AB, Supplementary Figure S10). Similar distribution of HeT-A RNA was observed in the embryos upon knockdown of the piRNA pathway genes spnE and rhino (rhi), which encodes HP1 family protein Rhino (Figure 5B). By contrast, transcripts of retrotransposons I-element, TART and jockey, which are overexpressed upon spnE and rhi depletion, were not detected around centrosomes (not shown). HeT-A RNA foci, associated with free centrosomes, were also visible in the cortex of wocKDnos, twinKDnos, Ars2KDnos, Trf2-/- and spnEKDnos embryos (Figure 5C, Supplementary Figure S11, Table 1). To detect RNA, we applied a tyramide signal amplification method (30) (Figure 5), as well as a traditional method using a DIG-labeled probe and anti-DIG antibodies (Supplementary Figures S10, S11). Similar patterns of HeT-A transcript localization revealed by both methods indicate that the observed signals were not artifacts of tyramide amplification. This was confirmed by the detection of oskar mRNA in the pole plasm using the tyramide amplification method (Supplementary Figure S12) (54). TART transcripts were not detected in the embryos upon germline knockdown of Trf2, Woc, Ccr4 and Ars2 (not shown).
Figure 5.
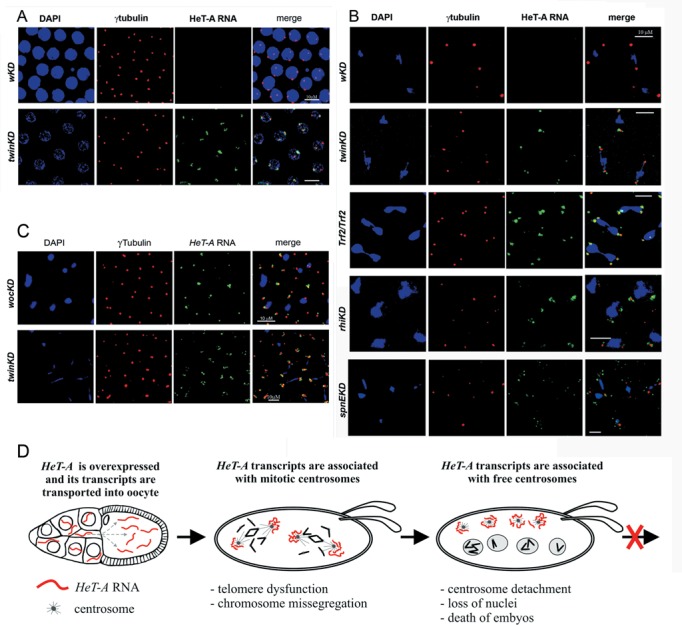
Telomeric transcripts are localized around centrosome in syncytial embryos. Confocal images of 0−2-h-old embryos with the indicated genotypes. RNA FISH of HeT-A antisense probe revealed HeT-A RNA (green) near centrosomes in the prophase of twinKDnos embryos (A), during metaphase-anaphase in twinKDnos, Trf2/Trf2, rhiKDnos and spnEKDnos embryos (B) and in the cortex of wocKDnos and twinKDnos embryos (C). Red: γ-tubulin; blue: DNA. Bar: 10 μm. (D) The scheme depicts mitotic defects in early developmental stages as a result of HeT-A derepression in the germline and the accumulation of abundant HeT-A transcripts around centrosomes in embryos.
DISCUSSION
In this study, we investigated the properties of Drosophila telomeric transcriptome in the germline, as well as the impact of the telomere repeat overexpression in the germline on the early development. We have shown that different components exert quantity and quality control of the telomeric transcriptome, resulting in the silencing of telomeric retroelements. The main and specific target of the silencing system is telomeric retrotransposon HeT-A, which is a major component of Drosophila telomeres. In addition to the piRNA system, transcriptional factors Woc and Trf2 are involved in the silencing of HeT-A in the germline. Expression of the major telomeric retrotransposon HeT-A and related telomeric element TAHRE, but not TART, is downregulated by Trf2 and Woc, indicating a specific interaction of these transcription factors with distinct sequences present in the HeT-A/TAHRE elements. This fact allows suggesting the HeT-A sequence as a possible platform for binding of specific telomeric proteins. In fission yeast, telomere-specific proteins Ccq1/Taz1 and the RNAi factors are involved in the recruitment of a higher-order chromatin complex at the telomeres (55). Assembly of the telomeric complex in the Drosophila germline, involving cooperation of DNA-binding proteins and piRNA pathway, resembles a mechanism operating at fission yeast telomeres.
Our data suggest an existence of an additional layer of control of the telomeric transcriptome. Deadenylase complex Ccr4-Not provides the deadenylated state of HeT-A transcripts in normal conditions. We do not know whether Ccr4-Not exerts HeT-A RNA deadenylation or prevents the polyadenylation of the nascent HeT-A transcripts. Moreover, depletion of different components affecting HeT-A silencing, such as Trf2, Woc and Ars2, leads to the accumulation of polyadenylated telomeric transcripts in ovaries. These data suggest that all of these components act cooperatively to regulate the life cycle of telomeric transcripts. Despite the fact that piRNA production was not affected by Trf2 and Ccr4-Not depletion, abundant HeT-A transcripts were accumulated upon knockdown of these factors.
It is noteworthy that only a minor fraction of human and yeast TERRA molecules are polyadenylated (15,56). Human poly(A)-negative TERRA is chromatin-associated, suggesting its role in telomeric heterochromatin assembly (57). Ars2, which was previously identified as a regulator of human TERRA levels (45), is implicated in the regulation of HeT-A expression in Drosophila (present study), indicating a conservative mechanism of telomeric transcript turnover. In zebrafish and frog early embryos, mRNA poly(A) tail length correlated strongly with the translation efficiency (58). HeT-A-encoded Gag protein has been shown to localize at the telomeres in somatic tissues (36) and bind HeT-A transcripts (59). It is likely that polyadenylated HeT-A mRNA is effectively translated to produce abundant Gag protein that may be involved in telomere protein complex destabilization.
Our data show that the mechanisms of telomeric repeat silencing and assembly of the telomere protection complex are directly connected (Figure 5D). The germline knockdown of distinct factors affecting the HeT-A RNA level, such as Woc, Trf2, Ars2, Ccr4 and Not1, caused severe mitotic abnormalities during early development. Most likely, depletion of telomere silencing complex components in the germline causes a loss of telomere protection in the early embryos which leads to the occurrence of telomere fusions, chromosome-segregation failures and eventually to a ‘mitotic catastrophe’. Damaged nuclei sank to the interior of the embryo and free centrosomes were left in the cortex. DNA damage and replication stress have been shown to cause centrosome inactivation and elimination of the defective nuclei from the embryonic blastoderm (60,61). In those cases, detached centrosomes lacked γ-tubulin while the free centrosomes observed after Trf2, Woc, Ars2 and Ccr4 depletion were positive for γ-tubulin. Thus, the mechanisms of centrosome detachment caused by DNA damage and disruption of telomere silencing complex appear to be different. In latter case, an additional component, namely, HeT-A RNA, is accumulated as a result of loss of telomere protection.
We found that the Drosophila HeT-A RNA overexpressed in the germline was transported to the early embryos, where it was concentrated around the centrosomes. The association of telomeric transcripts with centrosomes is a novel observation. It is tempting to speculate that the localization of the HeT-A transcripts around the centrosome, per se, may be related to mitotic defects. Detached centrosomes present in the cortex of embryos with knockdowns of the telomeric silencing factors were frequently associated with HeT-A transcripts. HeT-A RNA and its encoded Gag protein form spherical ribonucleoprotein particles that are targeted to chromosome ends by a telomeric protein Verocchio (59). It remains to be tested whether HeT-A RNA in embryos can interact with certain proteins and how it may impact the observed mitotic defects. We do not exclude that HeT-A overexpression plays an important if not the critical role in the early developmental defects linked to disruption of the piRNA pathway. Indeed, biochemical data show that interplay between telomere and mitotic machinery does exist. In mammals, the telomere-associated proteins tankyrase 1 and telomere repeat binding factor 1 (TERF1) not only localize to telomeres but are also translocated to the spindle poles and have been proposed to mediate proper kinetochore-microtubule attachment during metaphase-anaphase progression (62–65). In Drosophila, BubR1, a component of kinetochore, was shown to bind unprotected telomeres, resulting in mitotic arrest (66). Although the functional significance of this dual distribution of telomere and mitotic control factors is not yet clear, these data suggest the existence of a new level of cell cycle control. Likely, it is the telomeric transcripts that provide the information link between telomeres and the mitotic spindle checkpoint components. Intriguingly, TERRA levels are elevated in cancer cells (67), which are characterized by centrosome amplification, aneuploidy and chromosome instability (68). We speculate that TERRA could mediate missegregation defects in cancer cells. Although speculative, this hypothesis opens up the possibility for better understanding the function of the telomeric transcriptome and the potential pathological significance of the telomeric repeat overexpression in cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for fly strains. The antibodies against Not1 and Ccr4 were kindly provided by Dr Wahle. We are very grateful to A.M. Olovnikov for fascinating discussion, A. Khodjakov for critical comments on the manuscript, A. Sergeeva for her contribution to several experiments.
Footnotes
Present addresses:
Sergey Shpiz. Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russian Federation.
Daria V. Kopytova and Sofia G. Georgieva. Institute of Gene Biology, Russian Academy of Sciences, Moscow, Russian Federation.
FUNDING
Russian Academy of Sciences program for Molecular and Cell Biology [to A.K.]; Russian Foundation for Basic Researches [15-04-02093 to A.K.]. Funding for open access charge: Russian Academy of Sciences program for Molecular and Cell Biology [to A.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Olovnikov A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 2.Lee H.W., Blasco M.A., Gottlieb G.J., Horner J.W. 2nd, Greider C.W., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 4.Shay J.W., Wright W.E. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 5.Bekaert S., Derradji H., Baatout S. Telomere biology in mammalian germ cells and during development. Dev. Biol. 2004;274:15–30. doi: 10.1016/j.ydbio.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 7.Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 8.Biessmann H., Champion L.E., O'Hair M., Ikenaga K., Kasravi B., Mason J.M. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levis R.W., Ganesan R., Houtchens K., Tolar L.A., Sheen F.M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 10.Biessmann H., Mason J.M., Ferry K., d'Hulst M., Valgeirsdottir K., Traverse K.L., Pardue M.L. Addition of telomere-associated HeT DNA sequences ‘heals’ broken chromosome ends in Drosophila. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 11.Shpiz S., Kwon D., Uneva A., Kim M., Klenov M., Rozovsky Y., Georgiev P., Savitsky M., Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- 12.Cenci G., Ciapponi L., Gatti M. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma. 2005;114:135–145. doi: 10.1007/s00412-005-0005-9. [DOI] [PubMed] [Google Scholar]
- 13.Raffa G.D., Ciapponi L., Cenci G., Gatti M. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus. 2011;2:383–391. doi: 10.4161/nucl.2.5.17873. [DOI] [PubMed] [Google Scholar]
- 14.Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 15.Bah A., Wischnewski H., Shchepachev V., Azzalin C.M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao F., Li X., Hiew S., Brady H., Liu Y., Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15:1274–1281. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savitsky M., Kwon D., Georgiev P., Kalmykova A., Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shpiz S., Olovnikov I., Sergeeva A., Lavrov S., Abramov Y., Savitsky M., Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozhkov N.V., Hammell M., Hannon G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurana J.S., Xu J., Weng Z., Theurkauf W.E. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 2010;6:e1001246. doi: 10.1371/journal.pgen.1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- 23.Collart M.A., Panasenko O.O. The Ccr4–not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Reese J.C. The control of elongation by the yeast Ccr4-not complex. Biochim. Biophys. Acta. 2013;1829:127–133. doi: 10.1016/j.bbagrm.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasko P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a012294. doi: 10.1101/cshperspect.a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopytova D.V., Krasnov A.N., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., et al. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol. Cell. Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogan G.L., Tulin A.V., Aravin A.A., Abramov Y.A., Kalmykova A.I., Maisonhaute C., Gvozdev V.A. The GATE retrotransposon in Drosophila melanogaster: mobility in heterochromatin and aspects of its expression in germline tissues. Mol. Genet. Genomics. 2003;269:234–242. doi: 10.1007/s00438-003-0827-1. [DOI] [PubMed] [Google Scholar]
- 28.Shpiz S., Kwon D., Rozovsky Y., Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothwell W.F., Sullivan W. In: Drosophilia protocols. Sullivan W, Ashburner M, Hawley RS, editors. Cold Spring Harbor, NY: CSHL Press; 2000. pp. 141–157. [Google Scholar]
- 30.Shpiz S., Lavrov S., Kalmykova A. Combined RNA/DNA fluorescence in situ hybridization on whole-mount Drosophila ovaries. Methods Mol. Biol. 2014;1093:161–169. doi: 10.1007/978-1-62703-694-8_13. [DOI] [PubMed] [Google Scholar]
- 31.Gao G., Bi X., Chen J., Srikanta D., Rong Y.S. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10728–10733. doi: 10.1073/pnas.0902707106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salles F.J., Strickland S. Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol. 1999;118:441–448. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
- 33.Charlesworth A., Cox L.L., MacNicol A.M. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savitsky M., Kravchuk O., Melnikova L., Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell. Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torok T., Benitez C., Takacs S., Biessmann H. The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster. Chromosoma. 2007;116:185–195. doi: 10.1007/s00412-006-0090-4. [DOI] [PubMed] [Google Scholar]
- 36.Silva-Sousa R., Lopez-Panades E., Pineyro D., Casacuberta E. The chromosomal proteins JIL-1 and Z4/Putzig regulate the telomeric chromatin in Drosophila melanogaster. PLoS Genet. 2012;8:e1003153. doi: 10.1371/journal.pgen.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanti L., Giovinazzo G., Berloco M., Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 38.Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., Berloco M., Turano C., Ferraro A., Pimpinelli S. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 39.Klenov M.S., Lavrov S.A., Stolyarenko A.D., Ryazansky S.S., Aravin A.A., Tuschl T., Gvozdev V.A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S.H., Elgin S.C. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva-Sousa R., Varela M.D., Casacuberta E. The Putzig partners DREF, TRF2 and KEN are involved in the regulation of the Drosophila telomere retrotransposons, HeT-A and TART. Mob. DNA. 2013;4:18. doi: 10.1186/1759-8753-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raffa G.D., Cenci G., Siriaco G., Goldberg M.L., Gatti M. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol. Cell. 2005;20:821–831. doi: 10.1016/j.molcel.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Sabin L.R., Zhou R., Gruber J.J., Lukinova N., Bambina S., Berman A., Lau C.K., Thompson C.B., Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czech B., Preall J.B., McGinn J., Hannon G.J. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 2013;50:749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheibe M., Arnoult N., Kappei D., Buchholz F., Decottignies A., Butter F., Mann M. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 2013;23:2149–2157. doi: 10.1101/gr.151878.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shpiz S., Kalmykova A. Analyses of piRNA-mediated transcriptional transposon silencing in Drosophila: nuclear run-on assay on ovaries. Methods Mol. Biol. 2014;1093:149–159. doi: 10.1007/978-1-62703-694-8_12. [DOI] [PubMed] [Google Scholar]
- 47.Lim A.K., Tao L., Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semotok J.L., Cooperstock R.L., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 50.Chicoine J., Benoit P., Gamberi C., Paliouras M., Simonelig M., Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Zaessinger S., Busseau I., Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 52.Morris J.Z., Hong A., Lilly M.A., Lehmann R. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005;132:1165–1174. doi: 10.1242/dev.01672. [DOI] [PubMed] [Google Scholar]
- 53.Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ephrussi A., Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 55.Sugiyama T., Cam H.P., Sugiyama R., Noma K., Zofall M., Kobayashi R., Grewal S.I. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 56.Azzalin C.M., Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 57.Porro A., Feuerhahn S., Reichenbach P., Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 2010;30:4808–4817. doi: 10.1128/MCB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L., Beaucher M., Cheng Y., Rong Y.S. Coordination of transposon expression with DNA replication in the targeting of telomeric retrotransposons in Drosophila. EMBO J. 2014;33:1148–1158. doi: 10.1002/embj.201386940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sibon O.C., Kelkar A., Lemstra W., Theurkauf W.E. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 61.Takada S., Kelkar A., Theurkauf W.E. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura M., Zhou X.Z., Kishi S., Kosugi I., Tsutsui Y., Lu K.P. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr. Biol. 2001;11:1512–1516. doi: 10.1016/s0960-9822(01)00456-0. [DOI] [PubMed] [Google Scholar]
- 63.Munoz P., Blanco R., de Carcer G., Schoeftner S., Benetti R., Flores J.M., Malumbres M., Blasco M.A. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 2009;29:1608–1625. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lan J., Zhu Y., Xu L., Yu H., Yu J., Liu X., Fu C., Wang X., Ke Y., Huang H., et al. The 68-kDa telomeric repeat binding factor 1 (TRF1)-associated protein (TAP68) interacts with and recruits TRF1 to the spindle pole during mitosis. J. Biol. Chem. 2014;289:14145–14156. doi: 10.1074/jbc.M113.526244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith S., de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 1999;112:3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 66.Musaro M., Ciapponi L., Fasulo B., Gatti M., Cenci G. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat. Genet. 2008;40:362–366. doi: 10.1038/ng.2007.64. [DOI] [PubMed] [Google Scholar]
- 67.Deng Z., Wang Z., Xiang C., Molczan A., Baubet V., Conejo-Garcia J., Xu X., Lieberman P.M., Dahmane N. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J. Cell Sci. 2012;125:4383–4394. doi: 10.1242/jcs.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pihan G.A. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front. Oncol. 2013;3:277. doi: 10.3389/fonc.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


