Abstract
With many safety and technical limitations partly mitigated through chemical modifications, antisense oligonucleotides (ASOs) are gaining recognition as therapeutic entities. The increase in potency realized by ‘third generation chemistries’ may, however, simultaneously increase affinity to unintended targets with partial sequence complementarity. However, putative hybridization-dependent off-target effects (OTEs), a risk historically regarded as low, are not being adequately investigated. Here we show an unexpectedly high OTEs confirmation rate during screening of fully phosphorothioated (PS)-LNA gapmer ASOs designed against the BACH1 transcript. We demonstrate in vitro mRNA and protein knockdown of off-targets with a wide range of mismatch (MM) and gap patterns. Furthermore, with RNase H1 activity residing within the nucleus, hybridization predicted against intronic regions of pre-mRNAs was tested and confirmed. This dramatically increased ASO-binding landscape together with relatively high potency of such interactions translates into a considerable safety concern. We show here that with base pairing-driven target recognition it is possible to predict the putative off-targets and address the liability during lead design and optimization phases. Moreover, in silico analysis performed against both primary as well as spliced transcripts will be invaluable in elucidating the mechanism behind the hepatoxicity observed with some LNA-modified gapmers.
INTRODUCTION
The elegant and conceptually simple mechanism of RNase H1-mediated gene knockdown has long been viewed as having great therapeutic promise. By establishing an ‘antagonistic-like’ effect on a given target, antisense oligonucleotides (ASOs) have the potential to selectively target the entire transcriptome, with relatively short design and optimization cycle times. Therapeutic ASOs are not, however, without their shortfalls and challenges, some of which include safety concerns. Hybridization-dependent toxicities, i.e. those driven by Watson–Crick base-pairing, are perhaps unique to ASOs and short interfering RNAs (siRNAs) in terms of underlying mechanism. It has been well reported that siRNAs can be relatively non-selective, owing to their ability to exert ‘microRNA-like’ effects on the 3′ UTR of mRNAs with limited sequence complementarity (1). For ASOs, far less has been published on hybridization-dependent OTEs (2,3), although it has not generally been viewed as a major issue. The Oligonucleotide Safety Working Group (OSWG) ‘letter to the editor’ published in Nature Biotechnology in 2012 (4) however clearly recommends assessment of OTEs for ASOs during drug discovery and development, both computationally and experimentally.
The molecular characteristics and downstream effects of ASOs are, however, significantly altered by a wide range of chemical modifications to the backbone, heterocycle or sugar moieties of the oligonucleotide. The vast majority of antisense therapeutics in clinical development contain a phosphorothioate (PS) backbone modification (5), which by increasing systemic stability and protein binding (relative to the naturally occurring phosphodiester backbone), improves the pharmacokinetic and pharmacodynamic properties of ASOs. However, the so-called ‘first generation’ (i.e. containing fully-PS backbones) ASOs have a poor therapeutic index and a number of class toxicities were identified. These have been extensively reviewed by Henry et al. (6) and predominantly involve hybridization-independent mechanisms (e.g. accumulation-driven degenerative changes in the kidney proximal tubules, antagonism of receptors of the innate immune system and immunostimulatory effects, coagulopathy, complement activation in the non-human primate). Hybridization-dependent toxicities are also a risk for the class though they may arise through exaggerated primary pharmacology, in a similar manner to that observed with some small molecule drugs. Although theoretically possible, this has not been reported as a significant issue with first generation ASOs (6). Subsequent ‘second generation’ modifications such as 2′-O-methoxyethyl (2′ MOE) (7) have enhanced potency (due to greater binding strength) and in vivo stability relative to the first generation ASOs (8,9). Certain aspects of the PS ASO class toxicities were partly mitigated by the incorporation of these modifications, e.g. complement activation in non-human primate and immunostimulatory effects in rodents are generally reduced (10). These modifications were, however, not compatible with RNase H1 cleavage activity. This limitation was overcome by using the so-called ‘gapmer’ strategy (11), where a DNA/PS ‘core region’, acting as an RNase H1 substrate, is flanked by ‘wings’ of modified nucleotides that increase affinity and stability. More recently bicyclic nucleotide modifications have been developed and are generally referred to as ‘third generation chemistries’. The locked nucleic acid (LNA) and constrained ethyl (cEt) modifications, developed independently by the groups of Jesper Wengel/Takeshi Imanishi, and by Isis Pharmaceuticals respectively (12–14), fall within this category. These modifications markedly increase binding energy/potency for the intended target, enabling the design of shorter ASO sequences (15). This has the potential to enhance in vivo delivery and can further mitigate some of the typical class effects of PS-containing ASOs (e.g. shorter ASOs have a reduced potential for complement activation) (16,17). On the other hand, by decreasing the sequence length it becomes increasingly more challenging to identify a unique target site. Furthermore, increased binding energy has the capacity to not only increase potency against the intended target, but also to ‘off-targets’. As the binding affinity is increased and classical toxicities are eliminated through increasingly advanced oligonucleotide chemical modifications, hybridization-mediated off-target effects (OTEs) may become a more prevalent concern.
Following delivery into the cells, it was found that ASOs can shuttle between cytoplasm and nucleus in a process similar to active transport for oligonucleotide classes with a PS backbone, and by diffusion for unmodified oligonucleotides (18). The RNase H1 enzyme, initially reported to be present throughout the cell (3,19), was recently shown to be active mainly in the nucleus and to a lesser extent in mitochondria (20). The activity of ASOs in the nucleus is well documented with studies such as those showing intronic (21) and nuclear (22) RNA knockdown, or those based on alternative splicing by steric hindrance (23). We hypothesized that ASOs are as likely, if not more so, to interact with intronic sequences as with exonic regions. However, hybridization-dependent intronic OTEs, to our knowledge, have not been explored for RNase H1-utlizings ASOs. The sheer length of unspliced transcripts, and potentially less stable secondary structure, in theory increase the probability of finding a potent hit. This should be as applicable to unintended ‘off-target’ interactions as intended ones.
The phenomenon of off-target RNA effects should be viewed as analogous to secondary pharmacology for small molecule drugs (i.e. interacting with unintended proteins or receptors). Pharmaceutical companies often characterize this potential during early lead optimization by assessing binding against related targets, or those that are known to carry a significant safety liability, with a view to progressing leads with the greatest overall selectivity. One of the unique opportunities of ASO-based therapeutics is that putative off-target interactions can be predicted using sequence alignment algorithms. In this study, a number of 16 nt long, PS-LNA-containing ASO gapmers were designed and screened against the BACH1 transcript. A list of putative intronic and exonic OTEs was generated for each ASO with corresponding high level target annotation. A small panel of potential unintended interactions was selected and prioritized based on Gene Ontology terms and literature review. These were verified in vitro in cells known to express both the intended target and off-target mRNAs using standard quantitative polymerase chain reaction (qPCR). The effect of targeting intronic regions was examined at the mRNA, rather than the pre-mRNA, level to capture the downstream effect of the cleavage event. This was followed up by a more extensive evaluation of a single lead using a medium-throughput qPCR method (OpenArray). To rule out the potential of false positive effects being generated by ASO-mediated qPCR interference, a subset of OTEs were confirmed by branched DNA assay (bDNA) and immunoblotting. A correlation between binding strength of ASO–RNA target sequence and corresponding knockdown potency was also evaluated to examine the contribution of affinity to observed effects. This thorough approach was used to examine the occurrence, potency and patterns of both exon and intron OTEs when using ASOs with a ‘third generation’ bicyclic chemistry.
MATERIALS AND METHODS
Oligonucleotide sequence
The LNA-PS ASO gapmers were designed and supplied by Exiqon, Denmark A/S (Vedbaek, Denmark) as >85% pure. The sequence specificity consideration at the design phase followed the common practice of selecting leads with least number of potential interactions with mature, messenger RNA, at the perfect or near perfect level. Six distinct BACH1-targeting ASOs were selected (Table 1) based on in vitro pharmacology screening of 130 and 30 leads in Human Lung Adenocarcinoma Epithelial (A549) and Normal Human Bronchial Epithelial (NHBE) cells respectively. The ASOs were fully phosphorothioated, contained between five and six LNA nucleotides and had 5-methylcytosine incorporated into CpG motifs. Additionally, a BACH1-targeting siRNA was used to verify that the observed OTEs were not a consequence of downstream effects following BACH1 knockdown. Both ASO and siRNA knockdowns were normalized to corresponding Random Sequence Controls (RSC) which were designed with matching modification type.
Table 1. Sequences of ASO and siRNA oligonucleotides used in this study.
| Oligonucleotide ID | Sequence (5′ -> 3′) | LNA content | Description |
|---|---|---|---|
| GSK2910546A | TCAGTTTAGCAGTGTA | 5 | Human BACH1 targeting (ASO) |
| GSK2910557A | AGTGTGATGAAAAGCA | 6 | Human BACH1 targeting (ASO) |
| GSK2910579A | GGTCATATGTGTGTAA | 5 | Human BACH1 targeting (ASO) |
| GSK2910594A | ATCGTTTCCAATTTCC | 5 | Human BACH1 targeting (ASO) |
| GSK2910613A | GAGTTATTACTAGAGT | 6 | Human BACH1 targeting (ASO) |
| GSK2910632A | TGAGAAGATCCATAGC | 6 | Human BACH1 targeting (ASO) |
| RSC ASO | TGGGCGTATAGACGTG | 4 | Random sequence control (ASO) |
| GSK2328969A | AUUUGAACCUUUAAUUCAG | Human BACH1 targeting (siRNA, antisense strand) | |
| RSC siRNA | AAUUAUCCAUACAAUAUAC | Random sequence control (siRNA, antisense strand) |
The ASOs were fully phosphorothioated and had 5-methylcytosine incorporated into CpG motifs.
Off-target effect prediction
To secure a high sensitivity of putative off-target detection, an implementation of rigorous Needleman–Wunsch algorithm (24) was selected and optimized. The alignment scoring system and sensitivity parameters were modified based on the length of the query ASO and database size. The genomic database was constructed using Ensembl's gene annotation repository (based on GRCh37 human assembly from the Genome Reference Consortium) (25). All non-functional (pseudo) genes were removed. The prediction results were further filtered, to discard hits that did not reach a set similarity threshold (at least 75% overlap with ASO sequences). The putative off-targets were annotated with reported cellular function (Ensembl, NCBI, WikiGenes), cell and tissue expression (eGenetics (26), Institute of the Novartis Research Foundation (GNF) (27) and in-house datasets) and Gene Ontology terms. Representative genes were selected following a manual, non-clinical safety profile review and assessment of the sequence identity relative to the ASOs (to ensure that a wide range of alignment patterns were represented). The sequence identity of representative predicted OTEs that were investigated in this study are listed in Tables 2–4. To facilitate the viewing of results, for OTE genes predicted to interact with ASOs at multiple sites, only the ‘top’ hit, with relatively highest sequence identity is shown. However, an additive or synergistic effect of more than one interaction site should not be discounted. In the case of OTEs such as with PHF6 or RAD51B, two potential interactions are listed as it was difficult to prioritize one over the other.
Table 2. Summary of representative OTEs identified by qPCR (single donor, six biological replicates) and verified with bDNA assay (two donors, four biological replicates each).
| ASO ID | Target region | OTE name | Top sequence identity | Top sequence alignment | Max. Kd.(qPCR, 5 μM) | t-test(qPCR, 5 μM) | Max. Kd.(bDNA, 50 μM) |
|---|---|---|---|---|---|---|---|
| GSK2910546A | Exon | RIF1 | 2MM | aTGTGaCGATTTGACT | 56% | 0.001 | NP |
| Exon | TPX2 | 2MM | AtGTGACGATTTGAcT | 35% | 0.0007 | 61% | |
| Intron | ADK | 1MM | ATGTGACGATTTgACT | 33% | 0.006 | 51% | |
| Intron | RUFY3 | 1G Oligo | ATGTGACGA-TTTGACT | 49% | 0.0003 | NP | |
| GSK2910557A | Exon | TMX2 | 2MM | AcGAAAaGTAGTGTGA | 16% | 0.01 | 47% |
| Intron | PHF6 | 3MM | ACGAAAAgTAGtGTgA | 64% | 9.8E-05 | 72% | |
| Exon | PHF6 | 4G Oligo | ACGAAA- - - -AGTAGTGTGA | ||||
| GSK2910632A | Exon | USP9X | 1G Oligo | CGAT-ACCTAGAAGAGT | 75% | 4.2E-06 | 83% |
| Exon | SRRM2 | 2MM | CgAtACCTAGAAGAGT | 38% | 0.002 | NP | |
| Intron | TLE3 | 1MM | CGAtACCTAGAAGAGT | 31% | 0.0001 | 48% | |
| Intron | RAD51B | 1G Oligo | CGATACCTA-GAAGAGT | 77% | 1.8E-05 | NP | |
| Intron | RAD51B | 2MM | CGaTACCTAgAAGAGT |
Dose response curves were generated from NHBE cells, gymnotically transfected with specified ASO and incubated for 48 h before lysis. Maximum knockdown was derived from the highest silencing efficiency observed in a dose response curve (0.1–5 μM for qPCR, 0.01–50 μM for bDNA). NP stands for ‘not performed’. A number of OTEs listed below were predicted to interact with the ASOs at more than one site—only the ‘top’ interaction is reported except in cases where it is challenging to select the best alignment (e.g. PHF6 and RAD51B). Within the sequence alignment column, ‘lower case’ represents a mismatch and a ‘hyphen’ represents a gap.
Table 4. Summary of knockdown results for 2MM intronic OTEs predicted for GSK2910546A.
| Target | Top sequence alignment | Max. Kd.(qPCR, 50 μM) | EC50 (μM) | Therapeutic index (fold) |
|---|---|---|---|---|
| BACH1 | Perfect Match - Exon | 95% | 0.6 | On-Target |
| FBXW11 | aTGTGACGATTTGACt | 97% | 0.3 | <1 |
| TIAM2 | aTGTGACGATTTGACt | 97% | 0.4 | <1 |
| POU2F1 | aTGTGACGATTTGACt | 95% | 0.5 | <1 |
| BBS9 | aTgTGACGATTTGACT | 95% | 0.4 | <1 |
| ZNF674 | aTGTGACGATTTGACt | 86% | 0.5 | <1 |
| AP3B1 | aTgTGACGATTTGACT | 84% | 4.4 | 7 |
| FGF5 | aTgTGACGATTTGACT | 89% | 5.3 | 9 |
| RIC8B | ATgtGACGATTTGACT | 76% | 5.3 | 9 |
| PRICKLE1 | aTGTgACGATTTGACT | 63% | 18 | 29 |
| TBCK | aTGtGACGATTTGACT | 63% | 21 | 36 |
| SPATS2L | AtGTGACGATTTGACt | 71% | 36 | 60 |
| SCAPER | aTGTGAcGATTTGACT | 53% | 38 | 63 |
| SCAPER | ATGTGACGATTTGAct | |||
| RBMS3 | aTGTGACGATTTGAcT | 52% | 49 | 81 |
| NRF1 | aTGTGACgATTTGACT | 48% | ∼50 | ∼94 |
| FUT8 | ATGTGaCgATTTGACT | 32% | >50 | >100 |
| VPS13C | ATgTgACGATTTGACT | 31% | >50 | >100 |
| USP45 | aTGTGACGATTTGAcT | 26% | >50 | >100 |
| FRS2 | ATgTGACgATTTGACT | 21% | >50 | >100 |
| MEIS2 | ATgTGACGATTtGACT | 18% | No Activity | |
| RBFOX2 | ATGtgACGATTTGACT | 18% | No Activity | |
| PTPRK | aTGTGACgATTTGACT | 17% | No Activity | |
| PTPRK | aTGTGACgATTTGACT | |||
| *XRRA1 | 1 Gap Target - Intron | 98% | 0.5 | <1 |
| *GNL3 | 2 Mismatches - Exon | 82% | 7.8 | 13 |
Mature mRNA knockdown was measured in NHBE cells transfected gymnotically for 48 h with a 10-point dose range (with six biological replicates). Therapeutic Index was calculated against the EC50 of on-target gene (BACH1, 0.6 μM) which was measured alongside the OTEs. To facilitate classification of low potency OTEs (i.e. maximal knockdown observed in dose response curve lower than 45%), we set a threshold of 20% below which the hit is no longer viewed as active and placed in the ‘No Activity’ category. Higher dose points are required to fully evaluate such hits and to accurately fit a dose response curve. Two genes (*XRRA1 and *GNL3) which were initially predicted as 2MM OTEs, were subsequently found to have different alignments in the newer release of genomic sequence and coordinate databases. ASO alignment against SCAPER and PTPRK genes showed two independent alignments against intronic regions at 2MM level. Within the sequence alignment column, ‘lower case’ represents a mismatch.
Cell culture and transfection
The human biological samples were sourced ethically and their research use was in accord with the terms of the informed consents. NHBE cells derived from two donors were purchased from Lonza and maintained in Bronchial Epithelial Cell Growth Medium (BEGM) (Lonza). Human lung carcinoma cells (A549) were purchased from ATCC and maintained in Dulbecco's Modified Eagle Medium (Gibco) supplemented with 10% fetal bovine serum, glutamine and penicillin/streptomycin. The ASO concentration used in each study was determined based on the cell type and experimental methodology. Cells were cultured in BD Biocoat Collagen I flasks (Becton Dickinson) and were not used beyond third passage. Once confluent, cells were seeded into collagen coated well plates (96 well—12 000 cells per well, plates from Greiner Bio-one; 12 well—200 000 cells per well, plates from Becton Dickinson; 6 well—300 000 cells per well, plates from Greiner Bio-one) and incubated for 24 h before transfection. For gymnotic (unassisted) transfections, the desired ASO concentration was prepared in antibiotic free medium (100 μl per well for 96-well plates and 2 ml per well for 6- and 12-well plates). The solution was added to cells which were then incubated for 48 h. ASO concentrations of up to 50 μM were used in gymnotic transfection experiments in NHBE cells. There was good tolerability based on an absence of a concentration-dependent reduced housekeeper signal and visual inspection of the cells during the study (data not shown). We have routinely assessed ASOs at 50 μM and above in a range of cells types using gymnosis and observed no notable effects on cell health based on a range of cytotoxicity endpoints (see ‘Results’ section for exemplar ASOs in Supplementary Figures S1 and S2). Lipofectamine 2000 (Invitrogen) was used for assisted transfections in 96-well plates. Oligonucleotides were diluted to the required concentration to a total volume of 10 μl per well and mixed with 10 μl of transfection complex (0.5 μl Lipofectamine 2000 and 9.5 μl of antibiotic and serum free media). After 20 min incubation, 80 μl of antibiotic free medium was added to the solution and mixed. After removing the medium from the wells, 100 μl oligonucleotide:transfection agent solution was added to the cells and incubated for 24 h. Experiments were repeated at least twice and dose-response curves were generated based on at least six ASO concentrations and plotted using quadratic polynomial function.
Transcript quantification
After the incubation period the medium was removed, cells were washed with phosphate-buffered saline (Sigma) and lysed using Promega SV RNA Lysis Buffer (Promega) or Buffer RLT (Qiagen) supplemented with β-mercaptoethanol (Promega). Total RNA was extracted using SV 96 Total RNA Isolation System (Promega) using Biomek 3000 automation workstation or RNeasy kit (Qiagen) using Qiacube robot (following the manufacturer's protocol). cDNA synthesis was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer's protocol. qPCR experiments were performed on an Applied Biosystems 7900HT System for standard qPCR (Taqman with FAM/TAMRA probes) and QuantStudio 12K Flex Real-Time PCR Platform for medium-throughput qPCR with OpenArrays. Primer and probe information are listed in Supplementary Tables S1 and S2 for standard qPCR and OpenArray respectively. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin (ACTB) were used as housekeeper genes for standard qPCR normalization and each data point was based on a measurement from six–eight biological replicates. For OpenArray experiments, four housekeeping genes (GAPDH, ACTB, RPLP0 and B2M) were used and eight data measurements taken, corresponding to two biological replicates for two donors, with two technical replicates per donor. Experiments were performed using protocols provided by Life Technologies. In short, each cDNA sample was combined with 2× TaqMan OpenArray Real-Time PCR Master Mix (Life Technologies) and loaded into custom designed OpenArray plates using a QuantStudio 12K flex AccuFill system. The OpenArrays were covered with immersion fluid (using reagents from QuantStudio 12K flex OpenArray accessories kit, Life Technologies) and loaded into the QuantStudio 12K flex instrument for RT-qPCR cycling. Samples were subjected to standard thermal cycling protocol provided by Life Technologies. Expression data from qPCR and OpenArray was normalized to corresponding housekeepers and logged before calculating the RNA level relative to control treatment. One way analysis of variance was used to calculate statistical significance of the treatment results. Statistical evaluation and plotting of data was performed in R software environment. Non-amplification based, branched DNA method was performed by Axolabs GmbH (Kulmbach, Germany) according to the manufacturer's protocol (Panomics).
Western blotting
Transfected cells were washed with cold Dulbecco's Phosphate-Buffered Saline (DPBS) (Life Technologies), treated with Radio-Immunoprecipitation Assay (RIPA) Buffer (Sigma) supplemented with Protease and Phosphatase Inhibitors (100X) (Sigma) and incubated for 30 min at 4°C. The cells were then centrifuged for 5 min at 3000 rpm. Bicinchoninic Acid (BCA) Protein Assay Kit (Pierce) was used to estimate total protein concentration in cell lysates—10 μg of protein was loaded in each well using Blue Loading Buffer (30X, New England Biolabs), DTT (30X) and XP Magic Marker (0.35 μl, Invitrogen). Samples were run on NuPAGE Novex 4–12% Bis-Tris Protein Gel (Life Technologies) at 130V (constant) using Bio-Rad Trans-Blot Semi-Dry (SD) Transfer Cell (0.8 mA/cm2, 44 mA for 90 min). Membranes were blocked with 5% milk powder for 1 h and incubated overnight at 4°C and 1 h at room temperature for primary and secondary antibodies respectively. Quantification was performed using Odyssey Imaging System (Li-COR). NuPAGE Tris-Acetate Gel with HiMark™ Pre-stained Protein Standard (Invitrogen) and 15 instead of 10 μg of sample was used for USP9X protein due to its size (290 kDa). The signal ratio between protein of interest and β-actin was calculated for each of the wells. Results were averaged across four biological replicates and expressed as a percentage change relative to control treatments with RSC ASO. Detail information in regards to antibodies used in the study is listed in Supplementary Table S3.
Binding affinity
ASO and target RNA oligonucleotides were diluted in a 100 mM NaCl/100 mM sodium phosphate/0.1 mM ethylenediaminetetraacetic acid (EDTA) buffer to a 10 ng/μl concentration, and mixed in a 1:1 ratio. EvaGreen dye (Biotium) was prepared in 1× Tris EDTA (TE) buffer (final concentration of 3 μM), mixed in 1:1 ratio with the duplex solution and transferred to a white 384-well plate (10 μl per well). The plate was sealed with adhesive tape, centrifuged briefly at 2500 rpm and placed into a Roche LightCycler 480 II. The melting temperature (Tm) analysis was carried out using the protocol described in Supplementary Table S4, which measures fluorescence at 465–510 nm with both ‘Max Peaks (two or less)’ and ‘SYBR Green I format’ options turned on. This produced melt peaks (based on the first negative derivative of the melt curves) and Tm values for each sample. The average Tm of an ASO/RNA duplex was determined from a minimum of four replicates per experiment, and the experiment was repeated a minimum of three times. The list of oligonucleotide sequences corresponding to predicted off-target binding sites is shown in Supplementary Table S5.
RESULTS
Modified ASOs efficiently silence both exonic and intronic off-targets with a wide range of mismatch/gap patterns
The prioritization and selection of putative OTEs was based on sequence alignment, mismatch pattern and review of high level information regarding the function of the encoded protein. A range of hits including near perfect matches against introns and reduced sequence similarity matches against exons were selected to initially explore the extent of unintended interactions for six BACH1-targeting ASOs. Predicted interaction sites together with their flanking regions (100 bp up- and downstream) were avoided when selecting primer/probe binding sites to mitigate potential qPCR interference. Primers were also designed to encompass all known splice variants. For each tested OTE a dose-response curve was generated to calculate EC50 value. To eliminate the potential for transfection agents complicating data interpretation, gymnotic transfection was used across the majority of experiments. Although gymnotic transfection is much less efficient than assisted transfection (we typically observe approximately three logs difference in EC50), it has been reported to be a ‘cleaner’ system and possibly more reflective of in vivo activity (28).
The initial, small scale qPCR study in NHBE and A549 cells showed surprisingly high numbers of confirmed OTEs with potent, reproducible knockdown (representative results in Figure 1). The investigation revealed efficient silencing of hits predicted against both exonic and intronic regions, with a wide range of mismatch and/or gap patterns. Surprisingly, even a transcript with a low sequence identity (PHF6, 4 nt central gap against exon and three mismatches against intron), showed an EC50 of 2 μM (maximum 64% knockdown) as compared to the on-target knockdown of 0.3 μM (Figure 1B). A time course experiment revealed varying knockdown kinetics for GSK2910632A- and GSK2910557A-mediated OTEs. While USP9X and TLE3 showed similar dose response to the on-target BACH1 knockdown (with USP9X eventually showing greater activity than BACH1) (Figure 1C and E), PHF6 and AR showed delayed kinetics and required a 10 nM dose to match the on-target knockdown (Figure 1D and F). Several complementary approaches were undertaken to build confidence in the initial findings; multiple regions per gene were selected as binding sites for qPCR amplification, alternative cDNA amplification methods were compared and activity was verified in a second cell line (A549) (Figure 1C–F). In each assay, both reproducibility and potency of the observed OTE knockdown was further confirmed. To completely rule out any potential ASO-mediated qPCR artefact or bias, a branched DNA assay was performed for a selection of OTEs by Axolabs GmbH (Kulmbach, Germany). The transfection conditions mirrored those used in the qPCR study, though the knockdown was evaluated with higher maximum dose (5 versus 50 μM). The bDNA results (Table 2) confirmed all OTEs identified in the initial study.
Figure 1.
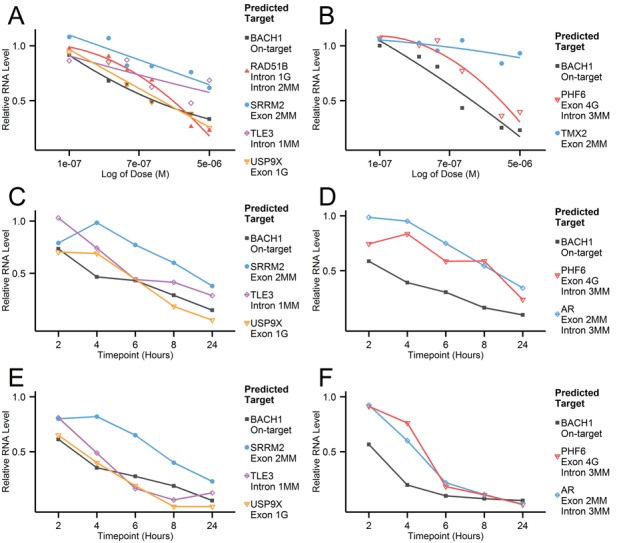
Representative qPCR results from NHBE cells gymnotically transfected with BACH1 targeting oligonucleotides; GSK2910632A (A) and GSK2910557A (B). Dose response curves were generated for on- (BACH1) and off-target genes predicted for a particular ASO. Oligonucleotide concentration ranged from 0.1 to 5 μM, each data point was derived from six biological replicates and ASO treatments were normalized to the non-targeting control (RSC ASO). Additional experiments were performed in an alternative cell line (A549) to examine knockdown kinetics at 2, 4, 6, 8 and 24 h after transfection. GSK2910632A [(C) 1 nM, (E) 10 nM] and GSK2910557A [(D) 1 nM, (F) 10 nM] were delivered to the cells using Lipofectamine 2000 and the knockdown of on-target and selected OTEs (relative to RSC ASO) was measured at the stated time intervals. Each data point was derived from two biological replicates and ASO treatments were normalized to the non-targeting control (RSC ASO). Sequence identity and target type information are included in the corresponding legend figures and are also listed in Table 2. ‘MM’ stands for mismatch, whereas ‘G’ stands for gap.
OTEs are sequence-specific, mediated through RNase H1 mechanism and reflected at the protein level
To differentiate between real OTEs and downstream effects caused by target knockdown or potential ASO class toxicity, we cross-checked the knockdown of off-targets in cells treated with a single siRNA and three LNA-ASOs molecules (GSK2910579A, GSK2910594A and GSK2910632A), all designed against BACH1 (Figure 2A). The tested oligonucleotides were not predicted to share any OTEs interactions. While all molecules showed potent knockdown of BACH1, off-target silencing was only visible with ASOs predicted to include such interactions (marked with an arrow). Similar validation was carried out in the vast majority of our assays and results always confirmed that OTEs are sequence specific and not caused by BACH1 knockdown or an ASO class effect. Two exploratory assays were set up to investigate the nature of the mechanism behind the OTE silencing. Firstly, transcript regions up (5′) and downstream (3′, Splice Junction) of the predicted cleavage site were chosen as binding sites for qPCR amplification (Figure 2B and C). With every OTE checked, there was good concordance between all the measurements, regardless of primer location. It can thus be assumed that the mechanism is based on full transcript degradation (exerted through RNase H1) rather than steric blocking of splicing (which could lead to aberrant, mis-spliced transcripts). Secondly, we examined if the observed off-target transcript knockdown resulted in a decrease in corresponding protein abundance. Western blotting was performed for PHF6 for GSK2910557A and USP9X for GSK2910632A (Figure 2D) as representative interactions. Protein levels were quantified and compared against non-targeting control treatments. With both OTEs, a substantial protein knockdown was observed just 48 h after gymnotic transfection. The confirmed off-target interactions were thus shown to be sequence-specific and leading to degradation of entire transcript and consequent protein knockdown.
Figure 2.
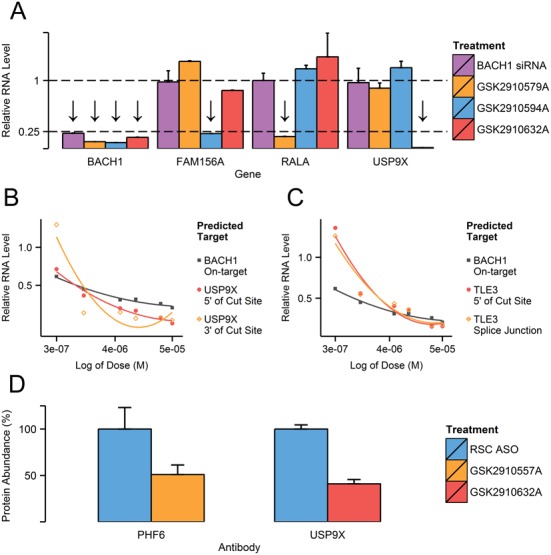
(A) On-target (BACH1) and selected exonic (FAM156A, USP9X) and intronic (RALA) OTE knockdown were tested with siRNA and ASO molecules designed against BACH1 but not predicted to share any OTE interactions. NHBE cells were transfected using Lipofectamine 2000 at 100 nM for siRNAs and 20 nM for ASOs, incubated for 24 h and lysed. Molecules predicted to interact with a particular gene are marked with an arrow above the corresponding column. Each column is derived from six biological replicates and normalized to corresponding RSC (siRNA or ASO), with error bars depicting standard error of the mean (SEM). Off-target knockdown was tested using qPCR primers designed against region upstream (5′) and downstream (3′, across Splice Junction) of the predicted ASO:OTE interaction site. Both exonic (USP9X) (B) and intronic (TLE3) (C) OTEs predicted for GSK2910632A were tested with two primer sets in gymnotically transfected NHBE cells (0.3–50 μM). All measurements are derived from six biological replicates and were normalized to RSC ASO controls of corresponding concentrations. Primer details are listed in Supplementary Table S1. (D) Abundance of PHF6 (left) and USP9X (right) proteins were quantified in samples treated with GSK2910557A and GSK2910632A respectively. NHBE cells were transfected gymnotically at 20 μM and incubated for 48 h before lysis. The results are shown as a percentage change relative to the non-targeting control (RSC ASO), with error bars representing SEM. Protein levels were measured in four biological replicates. Antibody details are listed in Supplementary Table S3. A representative Western blot with corresponding quantification is shown is Supplementary Figure S3.
Intronic regions are markedly more susceptible to ASO-silencing than exonic sequence
With the majority of the molecules being de-prioritized after the first round analysis, a more extensive follow-up assessment of larger numbers of putative off-targets was conducted using a reduced number of ASOs. The choice of the OTEs was based on an early safety-based literature review and more diverse sequence alignments with an exploratory aim in mind. Following the high confirmatory rate in the initial study and wide range of hits predicted against such regions, we focussed more heavily on putative intronic hits. Again, the sequence-specific nature of these interactions was demonstrated with the inclusion of both a RSC control and by evaluating the same off-targets in cells transfected with an alternative BACH1 ASO, not predicted to have the same interactions (data not shown). The percentage knockdown and therapeutic index, an approximate fold difference between the EC50 of intended therapeutic target (BACH1, 0.3 μM) and given OTE, are listed in Table 3.
Table 3. Results from medium-throughput qPCR study (OpenArray) performed on NHBE cells by unassisted transfection with GSK2910546A.
| Target region | OTE name | Top sequence identity | Top sequence alignment | Max. Kd.(qPCR, 50 μM) | EC50 (μM) | Therapeutic index (fold) |
|---|---|---|---|---|---|---|
| Exonic | RIF1 | 2MM | aTGTGaCGATTTGACT | 56% | 31 | >100 |
| EMB | 2MM | aTGTGACGATtTGACT | 36% | >50 | >100 | |
| TPX2 | 2MM | AtGTGACGATTTGAcT | 28% | >50 | >100 | |
| KCTD6 | 2MM | ATGTgACGATTTgACT | No Activity | |||
| TMEM185B | 2MM | ATGTGACGATTtGAcT | No Activity | |||
| CDH26 | 2MM | ATGTGACGATTtGaCT | No Activity | |||
| NBAS | 2MM | ATGTGACGATtTgACT | No Activity | |||
| UPF1 | 1MM + 1G Oligo | AtGTGACGATTT-GACT | 30% | >50 | >100 | |
| LAMA2 | 1MM + 1G Oligo | ATGT-GACGATTTGACt | No Activity | |||
| VTCN1 | 1MM + 1G Oligo | ATGT-GACGATTTGAcT | No Activity | |||
| IL36RN | 1MM + 1G Oligo | ATGT-GACGATTTgACT | No Activity | |||
| KBTBD3 | 1MM + 1G Oligo | ATGTgACGAT-TTGACT | No Activity | |||
| MOSPD2 | 1MM + 1G Oligo | ATGTgA-CGATTTGACT | No Activity | |||
| ZNF782 | 1MM + 1G Oligo | ATgTGACG-ATTTGACT | No Activity | |||
| Intronic | MAGI2 | 1MM | AtGTGACGATTTGACT | 90% | <0.3 | <1 |
| HDAC9 | 1MM | ATGTGACGAtTTGACT | 86% | 1.2 | 4 | |
| LARP1B | 1MM | ATGTGACgATTTGACT | 80% | 3.5 | 11 | |
| SLC12A2 | 1MM | ATGTGACGATTTgACT | 45% | >50 | >100 | |
| COG5 | 1MM | ATGTGACGATtTGACT | 40% | >50 | >100 | |
| ZNF385B | 1MM | ATGTGACGATTTGAcT | Possible Activity | ∼100 | ||
| VNN2 | 1MM | ATGTGACgATTTGACT | No Activity | |||
| GOT2 | 1G Target | ATGTGAcGATTTGACT | No Activity | |||
| FBXW11 | 2MM | aTGTGACGATTTGACt | 97% | 0.3 | Equipotent | |
| FGF5 | 2MM | aTgTGACGATTTGACT | 83% | 3.5 | 11 | |
| TGFA | 2MM | atGTGACGATTTGACT | 75% | 10 | 33 | |
| SCAPER | 2MM | ATGTGACGATTTGAct | 40% | >50 | >100 |
Dose response curve (0.3–50 μM) was generated for each off-target based on eight data measurements, corresponding to two biological replicates for two donors, with two technical replicates per donor. The data was normalized using four housekeepers (GAPDH, ACTB, PRLP0 and B2M) and calculated relative to RSC ASO. Primers and probes were pre-designed by Life Technologies, with corresponding assay IDs listed in Supplementary Table S2. Therapeutic Index shows approximate fold difference in EC50 between the intended (BACH1, 0.3 μM) and specified OTE (e.g. 10 μM/0.3 μM = TI of 33.3). Within the sequence alignment column, ‘lower case’ represents a mismatch or a gap in the target, a ‘hyphen’ represents a gap within the ASO sequence. Only the ‘top’ (i.e. of highest sequence identity) interaction is reported per OTE.
A range of exonic hits with 2MM or 1MM and 1 gap showed lower confirmatory rates and modest potency (TI > 100). For intronic hits however, we observed a much higher confirmatory rate, with some achieving very high potency (MAGI2, HDAC9, LARP1B, FBXW11 and FGF5). The measured difference in interaction suggests that intronic regions are more efficiently targeted and cleaved (summarized in Figure 3). This is especially visible with MAGI2 which showed 90% knockdown at all tested ASOs concentrations (0.3–50 μM), though the transcript showed low baseline expression. Moreover, our assay verified knockdown at a mature mRNA level ensuring that the ASO's interaction with the intron regions results in degradation of the whole transcript. Several of the high ranking intronic hits such as MAGI2 and HDAC9 were predicted to interact with the ASO at multiple sites—other than the reported ‘top’ alignment, the hits were of lower sequence identity (2MM + 1G or 3MM). Though the contribution of multiple sites to total knockdown cannot be overlooked, OTEs with a single predicted cut site (FBXW11, FGF5 and TGFA) also showed high silencing efficiency and potency. While among OTEs tested for GSK2910546A we did not identify very potent exonic hits, data from other leads (presented in Figures 1 and 2 and Table 4) confirmed that ASO:exon OTE interactions can also lead to highly efficient silencing. Based on the results presented here and our unpublished data, predicted exonic off-targets are however, relatively less likely to be confirmed.
Figure 3.
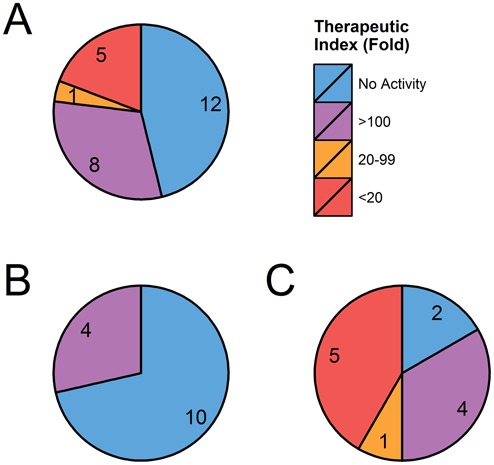
Graphical summary of 26 OTEs predicted and tested for GSK2910546A. Total (A), exonic (B) and intronic (C) OTEs are grouped by their potency relative to EC50 of intended target (BACH1, 0.3 μM). Data is derived from OpenArray results presented in Table 3. While among the exonic OTEs tested for GSK2910546A few showed high potency, results from other leads (Figures 1 and 2 and Table 4) confirmed that interactions with exonic regions can also lead to highly potent knockdown.
Considering the observed higher safety concern brought by unintended interaction between ASOs and intronic regions (Table 3), an additional study was performed to more comprehensively evaluate knockdown potential for intronic OTEs with two mismatches. A panel of 33 off-target genes predicted for GSK2910546A was chosen, encompassing a wide range of mismatch positions. The predictions were evaluated by unassisted transfection in NHBE cells based on a 10 point dose response curve. While the baseline expression for 10 genes was too low to obtain valid results, of the remaining 23 off-targets, several showed very potent knockdown (Table 4). Knockdown was also measured with alternative ASO that was not predicted to share any OTE interaction (data not shown). Whilst it is challenging to draw definitive patterns due to relatively small datasets and variable mechanistic aspects (accessibility, transcript abundance, exposure time in the nucleus), it is clear that for a 16 mer LNA-gapmer, OTEs with 2MM are still a significant concern. The alignment patterns of highest ranking off-targets (FBXW11, TIAM2, POU2F1 and ZNF674) suggest that mismatches at the terminal positions appear to be particularly well tolerated. Based on the results shown in Tables 2–4, LNA-PS ASO gapmers are able to tolerate mismatches across their entire length, regardless of whether they appear within the DNA core or in the ‘wings’.
Potency of OTEs is highly correlated with binding affinity between ASO and the target sequence
The presence of a mismatch or gap between ASO and the sequence of an off-target transcript reduces the affinity between the potential duplex, and is one of the key factors which determines the potency of knockdown. The affinity can be characterized through Tm, which is the temperature at which half of the oligonucleotides are in a single-stranded form, while the remaining half are forming a duplex. Several assays have been developed to experimentally measure the Tm of oligonucleotide duplexes. The original approach which used ultraviolet-visible (UV) spectroscopy is relatively low-throughput and requires significant quantities of oligonucleotides. Fluorescence-based assays are now more commonly used to measure nucleic acid hybridization, as these are more amenable to high-throughput analysis. The majority of such methodologies employ the use of a fluorophore dye and a quencher, which are attached to oligonucleotides. Denaturation of a nucleic acid duplex is coupled to the separation of the two probes, which results in a change of fluorescent signal (29–31). The approach does, however, require expensive labeled probes. As an alternative, a high-throughput method was developed which is based on an intercalating EvaGreen dye (Biotium), which fluoresces when bound to a double-stranded nucleic acids (e.g. ASO/RNA duplex). Although the mean Tm values were slightly lower when measured with the EvaGreen dye than by UV spectroscopy, results from both methods showed very high correlation (Pearson's r = 0.98, P = 0.0032) (Supplementary Figure S4, protocols for both methods are shown in Supplementary Table S4).
To explore the extent to which binding affinity alone may influence off-target activity, the binding strength between GSK2910546A and its on- (16 nt long perfectly complementary RNA sequence) and nine off-target sites (16 nt long RNA sequence corresponding to predicted target regions of 2MM OTEs) were analyzed using the EvaGreen fluorescence assay (sequences are listed in Supplementary Table S5). The decrease in Tm between fully complementary ASO–RNA duplex and ASO–2MM RNA duplexes varied between 4.2 and 11.6°C (Figure 4), although we have seen as little as 1.6 degree difference with RNA sequence which was initially predicted as a putative target site within XRRA1 gene (2MM, AUCACUGCUAAACUGA, data not shown). In general, a very high correlation was observed between measured Tm and percentage knockdown (r = 0.84, P-value = 0.0025). The most potent off-target activity observed was with those targets which retained the highest affinity for the ASO based on Tm. However there were outliers, which may indicate that some regions are more accessible to the ASO than others (context not evaluated in these analyses). For example, TBCK has similar affinity to ASO compared to FBXW11 but there is a 70-fold difference in their potency.
Figure 4.
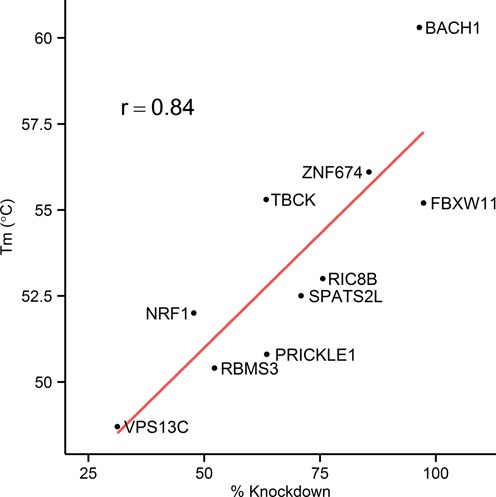
Correlation between Tm of ASO-target sequence duplexes and in vitro knockdown levels of corresponding genes (r = 0.84, P-value = 0.0025). The binding affinities between RNA sequences (listed in Supplementary Table S5) and GSK2910546A was measured using EvaGreen dye, following the protocol shown in Supplementary Table S4. The difference in Tm between fully complementary duplex (intended target, BACH1) and ASO-2MM RNA duplexes varied between 4.2 and 11.6°C, although it can be as low as 1.6 degrees (initially predicted 2MM target site within XRRA1 gene, data not shown).
DISCUSSION
The gapmer design of ASOs relies on the activity of RNase H1 to achieve gene knockdown following hybridization to the target transcript. RNase H1 activity occurs mainly within the nucleus (20) and there are numerous examples of interaction between ASOs and non-coding sequences (e.g. splice modulating therapeutics, intron or enhancer RNA targeting ASOs) (32,33). While ASOs interact with all RNA species, both spliced and unspliced, the concept does not appear to have been considered when assessing potential off-target interactions of RNase H1-utilizing ASOs. Specificity evaluations were historically performed exclusively against mature, mRNA sequence. Using a more thorough bioinformatic approach for our BACH1 targeting LNA-gapmers, a large number of potential off-target transcripts with partial homology to the ASOs were identified. These included between one and three mismatches in both exon and intron sequence and gaps in either the ASO or target sequences. While a relatively smaller number of non-coding RNA hits were also predicted, this study focussed on protein coding transcript as a proof of concept.
The extensive length of intronic regions translates into a substantial number of putative off-target predictions, though the efficiency of such hits was unknown. While identifying putative ASO interactions is relatively straightforward, no general method has been established to accurately predict the efficiency of on- or off-target knockdown, equivalent to those created for siRNAs (34). Our initial studies showed that interaction with both exonic and intronic regions (Figure 1) could lead to potent off-target hits. These results were confirmed using a number of techniques to rule out interference by the LNA-gapmer in the cDNA synthesis or qPCR assay (Table 2). Furthermore protein knockdown was demonstrated for a small number of confirmed off-targets (Figure 2D). After initially confirming the intron-based OTEs (Figure 1), a more thorough investigation was performed on higher numbers of predicted hits (with GSK2910546A, Table 3). While exon hits showed a relatively modest confirmatory rate (Figure 3B), active intron off-targets were abundant and in many cases highly potent (Figure 3C). Subsequent studies showed that of 21 putative intronic off-targets with two mismatches, 12 showed significant levels of mRNA knockdown following gymnotic delivery, with five displaying knockdown activity equivalent (or higher) to the on-target.
The potency, confirmation rate and breadth of mismatch and/or gap pattern of OTEs verified in our study is beyond anything previously reported in the literature. The main safety challenges have historically been attributed to hybridization-independent effects such as interaction with proteins (35) and Toll-like receptors of the innate immune system (36). While potent chemistries (such as, but not limited to, LNA) bring significant improvement to efficacy (32), there is a clear potential for these molecules to interact with transcripts that share complete or partial complementarity. Previous reports with earlier generations of ASO chemistry showed a decrease in activity against the target sequence with 1MM and essentially no activity with 2 (2,21) or 3 MM both in vitro and in vivo (3). These studies were, however, limited in scope, with very few targets and mismatches only being incorporated into a small number of the possible positions in the ASO/target duplex. The suggested improved mismatch discrimination brought by LNAs was mostly based on studies performed in the context of using LNA-oligos as hybridization probes (37,38) rather than as LNA-gapmers. The ASO-gapmer ability to distinguish between single nucleotide polymorphisms (39) appears to be limited to mismatches around the target cleavage site. Another study on the specificity of LNA-gapmers (40) reported reduced activity with 1MM and no activity with 2MM. As before, the conclusion was based on a very low sample size and small panel of mismatch position combinations. Furthermore, the mismatched ASOs have only been tested against a target region with the same accessibility. Our analysis (not shown) suggests that accessibility is one of the main determinants of ASO activity and can greatly affect the results. Moreover, position of the mismatch will affect both the affinity for the target RNA (Tm) and also interactions with RNase H1 enzyme. The introduction of mismatches into a MOE-gapmer has in some instances increased target RNA cleavage rate in a cell-free RNase H1 cleavage assay (41). Given the surprisingly high potency of 2MM hits (Table 4), it would be interesting to verify whether mismatches can affect the LNA-ASO:target:RNase H1 structure in a way that increases the cleavage efficiency.
Our data shows that with a fully phosphorothioated 16 mer format (containing 5–6 LNAs) a wide range of patterns and mismatches can be tolerated. This is most likely due to compensation, as the remaining matched nucleotides exert high enough binding energy to maintain the interaction. This was confirmed in the binding assay (Figure 4), where experimentally measured Tm of ASO–OTE target sequence duplexes highly correlated with in vitro knockdown levels of a corresponding off-target genes (r = 0.84, P-value = 0.0025). We have also observed examples of OTEs with similar Tm but markedly different potency (TBCK and FBXW11), which underlines the likely importance of target site accessibility. In the context of shorter LNA probes, mismatches are more detrimental to the stability of the oligo:target duplex (38) though it has not been defined if this translates to decreased knockdown rates and potency versus off-targets in an LNA-gapmer context. Based on unpublished initial in-house data, we found that shorter gapmers have their own set of challenges. A number of 14 mer ASOs (containing 3–5 LNAs) covering a range of predicted Tm were designed against a second target and tested in vitro. Though some significant interactions do still occur, in general the confirmation rate of hits with 2MMs was reduced when compared to the 16 mer format (unpublished data). Unfortunately the confirmation rate and potency of OTEs with 1MM was considerable, with the EC50 values in some cases exceeding that of our intended target by several fold. While shorter ASOs can accommodate less mismatches and gaps, it is markedly more difficult to design a potent molecule while maintaining high specificity. Given the significant detrimental effect on the silencing efficiency brought by decreasing the ASO binding energy, it seems that with current technology certain potency:specificity trade-offs have to be accepted. Compounds with lower binding strength (e.g. with low modification content or less potent chemistry) would require a higher dose to establish a therapeutic effect, increasing the likelihood of encountering typical oligonucleotide class toxicities and possibly OTEs.
We observed that the scale of liability markedly differed from one ASO sequence to another, with a few designs showing relatively low confirmatory rates. It is possible that a certain combination of ASO length, gap size, sequence and positioning of chemically modified bases affect duplex stacking and increase tolerance for mismatches. Much more data is however needed to comprehensively explain the difference in OTEs liability and introduce a framework for design. Given the observed tolerance for mismatches and gaps, combined with confirmation of intron-OTEs and the sheer length of unspliced transcripts, it is very difficult to design a potent OTE-free lead, certainly for ASOs of ≤16 nt in length with currently available chemistries.
While both intronic and exonic off-targets can lead to potent knockdown, the clear differences in their confirmation rate (Figure 3) raises questions in regards to the cleavage event. As the RNase H1 enzyme is not capable of differentiating between coding and non-coding regions, the phenomenon must be driven by divergence in the molecular ‘environment’ between those sequence types. With the activity of cleavage occurring mainly in the nucleus (20), the length of time between the target being fully transcribed to the moment it is transported to the cytoplasm will affect ASO activity. The concept was particularly well demonstrated with a nuclear-retained transcript with expanded CUG repeats, which showed unusual sensitivity to RNase H1-mediated knockdown (42). Longer ‘exposure’ resulting from slower transcription, longer gene or target position (closer to the end of transcript or within 3′ UTR) would however, contribute equally to both regions. Additionally, as the introns are spliced out, exons would have the advantage of a longer ‘activity window’ in the nucleus. This would be expected to result in exonic hits exhibiting a higher overall confirmatory rate, which we have not observed in this study. With pre-mRNA structure shifting alongside transcription (43) and most likely being local (44), co-transcriptional accessibility could in theory encompass a stage of a more favorable structural variant for ASO binding. This concept was used to improve the in silico design of exon skipping oligonucleotides (43) but fails to explain the difference in observed confirmatory rate and potency between intronic and exonic targeting hits. We have observed examples where different accessibility most likely accounts for the differences in off-target activity, e.g. in the case of BBS9 and FGF5 which both have two mismatches in the same position but differ by almost 12-fold in EC50. The most likely explanation thus, lies in the different sets of RNA-binding proteins/other RNAs, ‘shielding’ those regions and affecting their secondary structure. With the co-transcriptional targeting of intronic regions and consequent depletion of mature transcript being confirmed, we can also safely assume that exonic regions can interact with ASO at both the unspliced as well as spliced RNA stage. This creates a question on the proportion of knockdown of ‘mature RNA’ targets such as BACH1 that is achieved by cleaving the transcript during and after transcription and splicing. The ratio is most likely determined by the time it takes for RNA to reach a mature stage and the time between reaching this stage and being exported from the nucleus.
Although the above mentioned characteristics of intron regions are a major hurdle to ASO specificity, they could also bring significant advantages to the therapeutic field. Several of the randomly selected intronic OTEs showed lower EC50s than our intended target (Tables 3 and 4), even though the six exon-targeting leads were among the most potent sequences in an in vitro pharmacology screening of 130 ASOs. The confirmation rate of the intron targets was also much higher relative to exonic hits (Figure 3). Designing ASOs specifically against intron regions might provide more potent molecules that would attain therapeutic effect with fewer chemical modifications (i.e. fewer LNAs) or at a lower dose. This in turn could reduce potential OTEs or class toxicities that are only observed at higher dose and/or binding strength. The efficiency of such ASOs is highly dependent on several intron characteristics (intron length, transcription time, size of gene, splicing pattern of entire transcript) and hence not all genes would be appropriate for such a strategy.
Our data demonstrate that a thorough OTEs assessment, utilizing both in silico predictions and in vitro confirmation in relevant cell lines, is a crucial aspect of lead optimization for RNase H1 recruiting ASOs. In terms of discharging and/or managing the risk around confirmed OTEs, there are a number of approaches that can be implemented. As expression of a given transcript varies greatly between different cells and tissues (45), it is tempting to de-prioritize OTEs based on their baseline expression levels. Whether this is a viable method depends on many factors such as route of administration, chemical modifications used or distribution to unintended organs/cells, especially those showing high levels of ASO accumulation (46). Based on sequence alignment the putative interactions should be examined and if verified, an EC50 established. Where there is a large separation in EC50s of the intended and unintended targets, this will build confidence that there is unlikely to be any safety-related consequence in vivo. Where significant activity versus an OTE is confirmed, a risk assessment may be necessary, which would include a literature-based review of the known (and theoretical) consequences to health of inhibiting the off-target gene. This paper-based exercise may discharge any risk or identify opportunities to do so experimentally. For certain OTEs with a theoretical safety liability, it may not always be possible to discharge the risk in vitro. For OTEs that have an ortholog sequence in animal models and show high sequence identity for the target region, any potential liability may be discharged by the lack of anticipated adverse effect observed in in vivo safety studies, or at least through demonstrating acceptable margins. It is also advantageous to design leads with perfect sequence complementarity against an ortholog (if any) of the intended target, to test the efficacy in an in vivo model. It also has to be noted that organism-specific toxicities are possible and OTEs predictions should be made separately for each animal model species.
Our study clearly demonstrates that hybridization-mediated OTEs are a significant concern for ASOs containing modifications that markedly increase binding energy (e.g. bicylic chemistries). Additional studies based on leads of varying chemical composition will help to determine whether the observed mismatch tolerance is a general property of ASO gapmers and to what extent the phenomenon is affected by structural changes brought by the various modifications. Critically, the confirmation rates and average potency of effect versus intronic regions were generally higher than for exonic hits, underscoring the importance of considering these off-targets in any such analyses. Importantly, these effects were measured at the mature mRNA level and also translated into protein knockdown when assessed. The overall selectivity does appear to vary from one ASO to another, making it possible to identify 16 mer LNA-PS ASO leads for progression with a relatively low potential for OTEs. A judicious computational and experimental analysis during lead development appears to be the best route to selecting molecules with lowest overall potential for hybridization-mediated OTEs and minimizing the likelihood of development failure and patient risk. A comprehensive computational pipeline (‘RNArcher’) geared toward development of ASO gapmers with minimized OTEs liability will be made available in the near future. Whether or not a given off-target interaction is acceptable or not should eventually be based on a risk:benefit analysis. It should also be remembered that hybridization-mediated OTEs for ASOs are analogous to secondary pharmacology targets for small molecules. With the former we are in the unique position of being able to predict those interactions, whereas for the latter in the main we are ignorant. The inclusion of intronic regions in OTE prediction can not only improve the selectivity of designed and progressed leads, it might also explain some of the LNA–ASO-mediated toxic phenotypes reported in the literature (47,48). Based on the comparison between exonic and intronic OTEs we also believe that designing RNase H1-utilizing ASOs specifically against introns, while highly influenced by intron characteristics and the splicing mechanism, has the potential to improve therapeutic index and possibly safety.
Supplementary Material
Acknowledgments
We would like to thank and acknowledge Axolabs GmbH (Kulmbach, Germany) for performing the branched DNA assay and associated cell culture, Ben Andrews and Sabine Fenner for synthesizing oligonucleotides used in the binding affinity assay, Helen Wood for assistance in measuring Tm using UV spectroscopy and Erika Cule for support in statistical analysis.
Footnotes
Present addresses:
Gemma Turner, College of Medical, Veterinary & Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK
Sofie Eriksson, The Francis Crick Institute, London WC2A 3LY, UK
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EPSRC Industrial CASE Award (Imperial College London, GlaxoSmithKline and Public Health England) (EP/J502017/1 to P.J. Kamola). Funding for open access charge: GlaxoSmithKline.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., Cook J., Nickel J., Yu R., Stecker K., Myers K., Dean N.M. Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat. Biotechnol. 2000;18:862–867. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- 3.Crooke S.T. Antisense Drug Technology: Principles, Strategies, and Applications. 2nd edn. NY: CRC Press; 2007. [Google Scholar]
- 4.Lindow M., Vornlocher H.-P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H., et al. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
- 5.Eckstein F. A dinucleoside phosphorothioate. Tetrahedron Lett. 1967;8:1157–1160. [Google Scholar]
- 6.Henry S.P., Bolte H., Auletta C., Kornbrust D.J. Evaluation of the toxicity of ISIS 2302, a phosphorothioate oligonucleotide, in a four-week study in cynomolgus monkeys. Toxicology. 1997;120:145–155. doi: 10.1016/s0300-483x(97)03661-5. [DOI] [PubMed] [Google Scholar]
- 7.Altmann K.-H., Dean N.M., Fabbro D., Freier S.M., Geiger T., Hanera R., Hiisken D.A., Martina P., Monia B.P., Miiller M., et al. Second generation of antisense oligonucleotides: from nuclease resistance to biological efficacy in animals. Chim. Int. J. Chem. 1996;50:168–176. [Google Scholar]
- 8.Zellweger T., Miyake H., Cooper S., Chi K., Conklin B.S., Monia B.P., Gleave M.E. Antitumor activity of antisense clusterin oligonucleotides is improved in vitro and in vivo by incorporation of 2′-O-(2-methoxy)ethyl chemistry. J. Pharmacol. Exp. Ther. 2001;298:934–940. [PubMed] [Google Scholar]
- 9.Geary R.S., Yu R.Z., Watanabe T., Henry S.P., Hardee G.E., Chappell A., Matson J., Sasmor H., Cummins L., Levin A.A. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab. Dispos. Biol. Fate Chem. 2003;31:1419–1428. doi: 10.1124/dmd.31.11.1419. [DOI] [PubMed] [Google Scholar]
- 10.Henry S.P., Monteith D., Bennett F., Levin A.A. Toxicological and pharmacokinetic properties of chemically modified antisense oligonucleotide inhibitors of PKC-alpha and C-raf kinase. Anticancer. Drug Des. 1997;12:409–420. [PubMed] [Google Scholar]
- 11.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 12.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Kinberger G., Migawa M.T., Gaus H., Bhat B., et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp. Ser. 2008;52:553–554. doi: 10.1093/nass/nrn280. [DOI] [PubMed] [Google Scholar]
- 13.Koshkin A.A., Singh S.K., Nielsen P., Rajwanshi V.K., Kumar R., Meldgaard M., Olsen C.E., Wengel J. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 14.Obika S., Nanbu D., Hari Y., Morio K., In Y., Ishida T., Imanishi T. Synthesis of 2′-O, 4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 15.Straarup E.M., Fisker N., Hedtjärn M., Lindholm M.W., Rosenbohm C., Aarup V., Hansen H.F., Ørum H., Hansen J.B.R., Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iversen P.L., Cornish K.G., Iversen L.J., Mata J.E., Bylund D.B. Bolus intravenous injection of phosphorothioate oligonucleotides causes hypotension by acting as alpha(1)-adrenergic receptor antagonists. Toxicol. Appl. Pharmacol. 1999;160:289–296. doi: 10.1006/taap.1999.8771. [DOI] [PubMed] [Google Scholar]
- 17.Geary R.S., Watanabe T.A., Truong L., Freier S., Lesnik E.A., Sioufi N.B., Sasmor H., Manoharan M., Levin A.A. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001;296:890–897. [PubMed] [Google Scholar]
- 18.Lorenz P., Misteli T., Baker B.F., Bennett C.F., Spector D.L. Nucleocytoplasmic shuttling: a novel in vivo property of antisense phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 2000;28:582–592. doi: 10.1093/nar/28.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., Crouch R.J. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 22.Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 25.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien S., Smedley D., Otgaar D., Greyling G., Jongeneel C.V., McCarthy M.I., et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein C.A., Hansen J.B., Lai J., Wu S., Voskresenskiy A., Høg A., Worm J., Hedtjärn M., Souleimanian N., Miller P., et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You Y., Tataurov A.V., Owczarzy R. Measuring thermodynamic details of DNA hybridization using fluorescence. Biopolymers. 2011;95:472–486. doi: 10.1002/bip.21615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnet G., Tyagi S., Libchaber A., Kramer F.R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardullo R.A., Agrawal S., Flores C., Zamecnik P.C., Wolf D.E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaukowitch K., Joo J.-Y., Liu X., Watts J.K., Martinez C., Kim T.-K. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol. Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ui-Tei K. Optimal choice of functional and off-target effect-reduced siRNAs for RNAi therapeutics. Front. Genet. 2013;4:107. doi: 10.3389/fgene.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 36.Krieg A.M. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 37.Koch T. Locked nucleic acids: a family of high affinity nucleic acid probes. J. Phys. Condens. Matter. 2003;15:1861–1871. [Google Scholar]
- 38.You Y., Moreira B.G., Behlke M.A., Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Østergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F., et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laxton C., Brady K., Moschos S., Turnpenny P., Rawal J., Pryde D.C., Sidders B., Corbau R., Pickford C., Murray E.J. Selection, Optimization, and Pharmacokinetic Properties of a Novel, Potent Antiviral Locked Nucleic Acid-Based Antisense Oligomer Targeting Hepatitis C Virus Internal Ribosome Entry Site. Antimicrob. Agents Chemother. 2011;55:3105–3114. doi: 10.1128/AAC.00222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Vasquez G., Swayze E.E., Crooke S.T. The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol. Pharmacol. 2007;71:73–82. doi: 10.1124/mol.106.025429. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wee K.B., Pramono Z.A.D., Wang J.L., MacDorman K.F., Lai P.S., Yee W.C. Dynamics of co-transcriptional pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. PLoS One. 2008;3:e1844. doi: 10.1371/journal.pone.0001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiller M., Zhang Z., Backofen R., Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapushesky M., Adamusiak T., Burdett T., Culhane A., Farne A., Filippov A., Holloway E., Klebanov A., Kryvych N., Kurbatova N., et al. Gene Expression Atlas update–a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;40:D1077–D1081. doi: 10.1093/nar/gkr913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lendvai G., Velikyan I., Bergström M., Estrada S., Laryea D., Välilä M., Salomäki S., Långström B., Roivainen A. Biodistribution of 68Ga-labelled phosphodiester, phosphorothioate, and 2′-O-methyl phosphodiester oligonucleotides in normal rats. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2005;26:26–38. doi: 10.1016/j.ejps.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 47.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., Kreeger J., DiMattia D., Chen T., et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22:344–359. doi: 10.1089/nat.2012.0366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


