Abstract
One of the major obstacles to the pharmaceutical success of oligonucleotide therapeutics (ONTs) is efficient delivery from the point of injection to the intracellular setting where functional gene silencing occurs. In particular, a significant fraction of internalized ONTs are nonproductively sequestered in endo-lysosomal compartments. Here, we describe a two-step, robust assay for high-throughput de novo detection of small bioactive molecules that enhance cellular uptake, endosomal escape, and efficacy of ONTs. Using this assay, we screened the LOPAC (Sigma–Aldrich) Library of Pharmacologically Active Compounds and discovered that Guanabenz acetate (Wytensin™), an FDA-approved drug formerly used as an antihypertensive agent, is capable of markedly increasing the cellular internalization and target mRNA silencing of hydrophobically modified siRNAs (hsiRNAs), yielding a ∼100-fold decrease in hsiRNA IC50 (from 132 nM to 2.4 nM). This is one of the first descriptions of a high-throughput small-molecule screen to identify novel chemistries that specifically enhance siRNA intracellular efficacy, and can be applied toward expansion of the chemical diversity of ONTs.
INTRODUCTION
Synthetic oligonucleotides, including antisense (ASOs), splice-switching (SSOs), N-acetylgalatosamine (GalNAc)-siRNAs, miRNA modulators, LNPs and hsiRNA, are abundantly used to manipulate gene expression in biomedical research and hold great promise for becoming a breakthrough treatment in a variety of human genetic diseases (1,2). A wide range of delivery issues limits the clinical utility of these classes of compounds. For instance, a relatively small fraction of intracellular ONTs are biologically active, with the vast majority being trapped unproductively in ‘oligonucleotide sinks’ (3–6). This defines the need for high doses, which in turn may lead to toxicity and off-target effects. Nonproductive accumulation in subcellular compartments is generally considered the rate-limiting step for productive mRNA silencing (7). With therapeutic IC50 values in the nano- to picomolar range, millions of copies of ONTs are delivered per cell while less than 500 RNA-loaded RNA-induced silencing complexes (RISC) are required for activity (8). Therefore, solving the intracellular bio-distribution challenge is a fundamental step in optimizing RNA therapeutics for clinical use.
A decade of research has focused on designing modifications, conjugates, and carriers for improving ONT delivery, yet no ideal configuration (with the exception of N-acetylgalatosamine (GalNAc)-conjugated siRNA for delivery to hepatocytes) has emerged (9). Here, we decided to explore whether co-delivery of small molecules could improve the uptake and activity of cholesterol-modified siRNA-antisense hybrids, termed hsiRNA (10). Large delivery constructs, including dendrimers (11), cell-penetrating peptides (12,13), polymers such as dynamic polyconjugates (14,15), and nanoparticles (7) have been used previously as part of siRNA formulations, but the use of small molecule drugs as modulators of ONT efficacy is relatively unexplored and limited to several recent studies. First, Gooding et al. have reported the synthesis of small molecule carriers (SMoC) of siRNA, which form non-covalent electrostatic interactions with unmodified siRNA to enhance plasma membrane translocation (16,17). Second, Juliano's group demonstrated that the small molecule Retro-1, which is involved in retrograde transport of bacterial toxins, appreciably improved the activity of nuclear-localized ASOs following simultaneous transfection. This year, a publication from the same group reported an extensive screen identifying compounds that enhance antisense ONT efficacy (18,19). Collectively, these studies highlight the potential for small molecules to act as powerful gene silencing effectors. Here, we developed an unbiased, systematic protocol to explore the ease and feasibility of using small molecule screening to identify novel modulators of oligonucleotide efficacy.
MATERIALS AND METHODS
Oligonucleotides and small molecule compounds
All oligonucleotide sequences and modification patterns synthesized in-house are summarized in Supplementary Table S1. Accell siRNA (Thermo Fisher) targeting HTT was purchased and used without further purification. Clonidine (Sigma–Aldrich), Retro-1 (Sigma–Aldrich) and Salubrinal (Sigma–Aldrich) were dissolved in DMSO and stored as 10 mM stock solutions at −20°C. Unless otherwise specified, all oligonucleotides and small molecules were diluted in OptiMEM (Gibco) prior to administration in vitro.
Cell culture
HeLa cells were maintained in DMEM (Invitrogen) supplemented with 100U/ml penicillin and streptomycin (Invitrogen) and 10% fetal bovine serum (Gibco) at 37°C and 5% CO2. Human primary hepatocytes (Gibco) were thawed and plated in Williams E Medium (Gibco) supplemented with 5% fetal bovine serum, 1 μM dexamethasone in DMSO, 100 U/ml penicillin and streptomycin, 4 μg/ml human recombinant insulin, 2 mM GlutaMAX™, and 15 mM HEPES pH 7.4 at 37°C and 5% CO2. Plated hepatocytes were allowed to recover for 6 h and subsequently maintained in Williams E Medium (Gibco) supplemented with 0.1 μM dexamethasone in DMSO, 50 U/ml penicillin and streptomycin, 6.25 μg/ml human recombinant insulin, 6.25 μg/ml human transferrin, 6.25 μg/ml selenous acid, 6.25 μg/ml bovine serum albumin, 6.25 μg/ml linoleic acid, 2 mM GlutaMAX™,15 mM HEPES pH 7.4 and 5 mM CaCl2 at 37°C and 5% CO2.
LOPAC1280 small molecule primary screen
HeLa cells were purchased from the University of Massachusetts (Worcester, MA, USA) tissue culture facility and verified to be Mycoplasma negative. A hsiRNA-targeting sequence (from the MAP4K4 gene)(10) was cloned into the 3′UTR of Renilla luciferase within a standard psiCHECK2 vector. This plasmid also contains a Firefly luciferase reporter gene used for transfection normalization and cell variability. psiCHECK2 plasmid amplification was performed using the EndoFree Plasmid Maxi kit (Qiagen) following the manufacturer's recommended instructions. psiCHECK2 plasmid concentration was measured by Nanodrop and diluted to a final concentration of 1 μg/μl.
LOPAC1280 was purchased directly from Sigma-Aldrich in a 96-well plate format (16 plates total). Each small molecule was provided in a stock solution of 10 mM (250 μl) in DMSO. Each plate was diluted to 5 mM for screening.
For the primary screen, HeLa cells were seeded in T-175 flasks at 3 × 106 cells/ml in 10 ml and cultured for 20 h under standard conditions. The cells were then sub-cultured in 6% DMEM without antibiotics. After 6 h, psiCHECK2 plasmid was transfected using Lipofectamine2000 at a 1:2.5 ratio. After 7 h transfection, cell media was replaced with fresh DMEM and cells were incubated under standard conditions overnight. The following day, 50 μl of 0.02 μM MAP4K4-targeting hsiRNA dissolved in OptiMEM (Gibco) was added to 16 plates (Costar #3610, cell culture treated, bottom clear, white) in columns 2–11 and Wells E1-H1. This final concentration (10 nM) was selected as it typically induces minimal silencing (5–10%) after passive uptake over 24 h. In each plate, 50 μl of 1 μM hsiRNA (MAP4K4) was added in Wells A1-D1 as a positive control and 50 μl of 1 μM hsiRNA (NTC) was added in Wells A12-D12 as a negative control. Wells E12-H12 contained no hsiRNA (only 50 μl Opti-MEM minimal medium). Following addition of hsiRNA to each plate, 1 ul of 5 mM LOPAC compounds was added by a liquid handler (Tecan, TeMo).
psiCHECK2 transfected cells were collected by trypsinization and resuspended in DMEM containing 6% FBS without antibiotics. 15 000 cells/50 ul were added to each prepared well and incubated for 72 h under standard conditions. After 72 h, all 16 plates were assayed with the Dual-Glo kit (Promega) following the manufacturer's recommended protocol. Briefly, 50 μl of Dual-Glo luciferase substrate was added to each well. Firefly luminescence signal (fLUC) was read after 15 min using an EnVision plate reader (Perkin Elmer) with a 0.1 s integration time. Then, 50 μl of Stop-Glo substrate was added. Following a second 15 min incubation period, Renilla luciferase signal (rLUC) was recorded as described previously. The ratio of rLUC/fLUC was used for screening data analysis. The average calculated screening Z′ factor (Eq. 1) was 0.53, CV% = 10.7% and S/B = 13.9, representing a robotic performance.
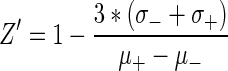 |
(1) |
Calculation of Z′ factor. σ+ and σ- represent standard deviation of positive and negative control. μ+ and μ- represent average of positive and negative controls, respectively. 65 small molecules that induced >50% enhancement in normalized luciferase knockdown were scored (5% hit rate). These primary hits were divided into three categories: (1) significant knockdown (15 hits), (2) knockdown that may be due to firefly signal increase (24 hits) and (3) much higher firefly signal (26 hits). We selected all hits from category 1, seven hits from category 2, and two hits from category 3 (24 total) for the secondary screen.
LOPAC1280 small molecule secondary screen
The QuantiGene 2.0 bDNA assay (Affymetrix, QS0011) was used to perform a secondary validation screen on the primary hits. 50 μl of 0.1 μM MAP4K4-targeting hsiRNA was added to a 96 well plate with 1 μM NTC and 1 μM MAP4K4-targeting hsiRNA as negative and positive controls, respectively. Then, 1 μL of 24 hits was added at three different concentrations (50 μM, 20 μM, or 5 μM). HeLa cells, previously resuspended in 6% FBS DMEM media without antibiotics, were added into each well with a density of 10 000 cells/50 μl per well. Mixture was incubated under standard conditions for 72 h. After 72 h, cells were lysed and processed according to the manufacturer's recommended protocol. Briefly, cells were lysed in 250 μl diluted lysis mixture (1:2 lysis mixture:H2O) with 0.167 ug/ul proteinase K (Affymetrix, QS0103) for 30 min at 55°C. Cell lysates were mixed thoroughly and 40 μl (∼8,000 cells) of lysate were added to a bDNA capture plate along with 40 μl additional diluted lysis mixture without proteinase K. Probe sets were diluted as specified in Affymetrix protocol and 20 μl of either human HTT or PPIB probes (Affymetrix: SA-50339, SA-10003) were added to each well of capture plate to a final volume of 100 μl. Luminescence was detected on either a Veritas Luminometer or a Tecan M 1000. Following analysis of MAP4K4 mRNA expression normalized to PPIB housekeeping mRNA expression, six small molecule hits out of 24 were confirmed to have constitutive activity.
LOPAC1280 small molecule tertiary screen
Six secondary hits (0.5% hit rate) were analyzed as described above for the secondary screen using a PPIB-targeting hsiRNA to confirm that activity is not RNA sequence dependent. Based on reproducibility and toxicity profile (Supplementary Figures S6 and S7), Guanabenz acetate was selected as the best candidate for enhancing hsiRNA delivery and efficacy.
alamarBlue® cellular viability assay
HeLa cell viability following Guanabenz and hsiRNA administration was assessed by the alamarBlue® (Life Technologies) assay, a fluorescence-based protocol that measures metabolism in proliferating cells. HeLa cells were treated with 0–200 μM Guanabenz in the presence or absence of 0.5 μM hsiRNA for 24 h in a 96-well plate. Cells were then incubated for 0–2.5 h with fresh medium containing 10% alamarBlue. Absorbance at 570 and 600 nm was measured with a Tecan Infinite M200 microplate reader at various timepoints. Cellular viability was calculated according to the manufacturer's protocol and normalized to an untreated control (n = 3, mean ± SD).
Live cell imaging
For live cell uptake monitoring, cells were plated at a density of 2 × 105 cells per 35 mm glass-bottom dish and grown overnight. Prior to imaging, cell organelles were stained in HBSS (Gibco) using staining reagents purchased from Life Technologies unless specified: cell nuclei, endoplasmic reticulum and lysosomes were respectively stained using the NucBlue® Live ReadyProbe®, ER-Tracker™ Green (Bodipy FL Glibenclamide) and LysoTracker® Deep Red reagents as indicated by the manufacturer. Imaging was performed in non-supplemented DMEM without phenol red (Invitrogen). Cells were treated with 0.5 μM of Cy3-labeled hsiRNA, and live cell imaging was performed over time.
Confocal imaging
All confocal images were acquired with a CSU10B Spinning Disk Confocal System scan head (Solamere Technology Group) mounted on a TE-200E2 inverted microscope (Nikon) with a 60x Plan/APO oil lens and a Coolsnap HQ2 camera (Roper). Images were processed using ImageJ (1.47v) software. Number of neurons without or with hsiRNA was counted using ImageJ software.
Physicochemical properties of hsiRNA and Guanabenz
Dynamic light scattering measurements were acquired using a Nano-ZS (Malvern) and analyzed using the DTS (Nano) program. Two independent measurements, each with 24 runs, were performed.
Electron micrographs were collected on a transmission electron microscope (Tecnai Spirit 12, FEI) at the UMMS Electron Microscopy Core. Samples were prepared by depositing 10 μl of a hsiRNA and/or Guanabenz solution onto a carbon-coated copper grid and staining with 2% wt uranyl acetate.
IC50 calculation
Data analysis was done using GraphPad Prism 6 V6.04 software (GraphPad Software, Inc.).
Guanabenz derivative synthesis
All chemicals were purchased from commercial sources and were used without further purification. Deuterated solvents were purchased from Sigma-Aldrich. 1H NMR spectra were recorded on a Varian 400 MHz NMR spectrometer using the residual solvent signal as the reference. High-resolution mass spectra were obtained on a ThermoFinnigan LCQ ion trap mass spectrometer. Analytical thin-layer chromatography (TLC) was performed using silica gel 60 F254 using UV light as visualizing agent.
Two, four and eight were purchased from commercial sources. Three and seven were synthesized following previously reported procedures (20,21). Five and six are not reported in the literature and full synthesis is described in the Supplemental Methods.
RESULTS
Identification of small molecule enhancers of hsiRNA silencing in vitro
To directly assess whether co-delivery of small molecules with hsiRNA could enhance RNAi efficacy, we developed and validated a high-throughput two-step protocol (Z′ Factor = 0.74) (Supplementary Figures S1–S3) and performed a small molecule screen using LOPAC1280 (Library of Pharmacologically Active Compounds, Sigma–Aldrich) (Figure 1A). hsiRNA are small, asymmetric, ultra-stable, highly modified oligonucleotides which efficiently enter cells and induce silencing without the requirement for formulation (Figure 1B, modified sequences provided in Supplementary Table S1) (10). The difference in length between antisense and sense stands results in a fully phosphorothioated single-stranded tail, which is believed to work in conjunction with the sense strand 3′-conjugated cholesterol to promote cellular internalization (personal communication, Dr. Sylvia Corvera). HeLa cells were co-treated with a panel of 1,280 small molecules and a suboptimal concentration (10 nM) of MAP4K4-targeting hsiRNA (Figure 1C, Supplementary Table S2). A panel of 24 small molecules (1.8% hit rate) with >50% enhanced protein silencing and minimal toxicity scored as candidate hits (Dual-Glo®, Promega). These 24 compounds were subjected to a confirmatory assay (QuantiGene®, Affymetrix) to measure the impact of small molecules on mRNA expression levels using a MAP4K4- or distinct cyclophilin B (PPIB)-targeting hsiRNA sequence (Supplementary Figure S4), with six candidates confirmed to have activity (0.5% hit rate), including Phenamil, Caffeic acid, Doxepin, BF-170, Furegrelate, and Guanabenz acetate (Supplementary Figure S5). None of the six candidate hits shared apparent structural or functional homology, which is not surprising for such a diverse library as LOPAC. For all subsequent studies, we focused on the compound that induced the most reproducible shift in IC50, Guanabenz acetate (Wytensin™) (Figure 1C, Supplementary Figure S6).
Figure 1.
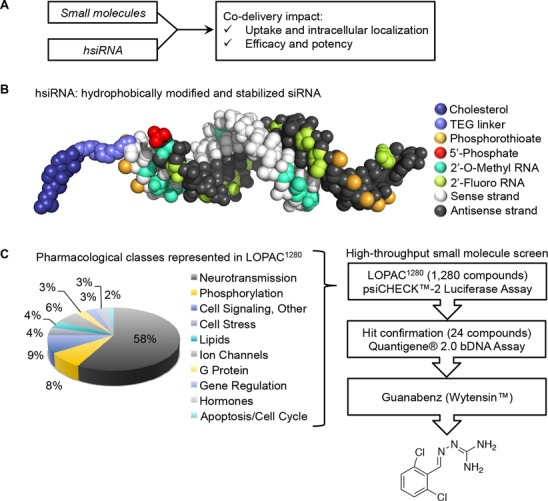
Screen for small molecules to enhance conjugated therapeutic oligonucleotide efficacy. (A) Small molecule screening strategy. (B) Schematic of the hydrophobically modified and stabilized siRNAs (hsiRNAs) used in study (see figure legend). (C) Distribution of the classes of bioactive small molecules in LOPAC library screened during high-throughput assay (Sigma–Aldrich). Workflow of high throughput small molecule screen and hit validation, yielding Guanabenz (Wytensin®) as a primary hit.
Guanabenz (Wytensin™) improves hsiRNA potency and efficacy in vitro
PPIB mRNA expression levels were monitored after treatment with 10 nM hsiRNA and increasing amounts of Guanabenz in HeLa cell culture (24 h, Figure 2A). A concentration-dependent enhancement of hsiRNA activity is observed, with target mRNA expression dropping from 90% to <10% following treatment with 0–80 μM Guanabenz (reported as percent of untreated control). Additionally, no effect is seen on mRNA expression following co-administration of Guanabenz and non-targeting (scrambled) hsiRNA (Supplementary Figure S7). Guanabenz-mediated protein silencing was evaluated using a luciferase reporter containing a hsiRNA-targeting sequence in the 3′-UTR (Dual-Glo®, Promega). In this experiment, treatment with 0–100 μM Guanabenz reduced the hsiRNA IC50 over 100-fold in a dose-dependent fashion from 132 to 2.4 μM (Figure 2B). Under these conditions, Guanabenz itself is non-toxic and has minimal effect on HeLa cell viability in the presence or absence of 0.5 μM hsiRNA (Figure 2C). The onset of Guanabenz toxicity occurs at concentrations of 125 μM and above for HeLa cells, while U87 and HepG2 cell lines are more sensitive (Supplementary Figure S8). These data suggest that Guanabenz is likely to work upstream of RISC-mediated silencing, as an improvement in efficacy was observed on both the RNA and protein level. This effect has been demonstrated with several hsiRNA compounds, targeting MAP4K4, PPIB, and HTT (Supplementary Figures S6, S10, S13). The use of direct mRNA quantification to assess hsiRNA activity against distinct targets ensured filtering against hits that were sequence, target or assay dependent.
Figure 2.
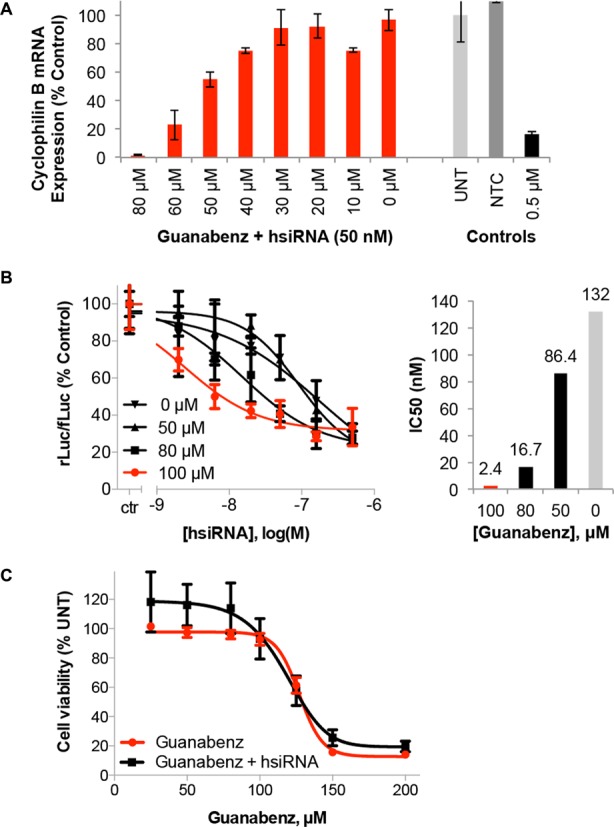
Guanabenz impacts hsiRNA potency and efficacy in a concentration-dependent manner. (A) Cyclophilin B (PPIB)-targeting hsiRNA was added to HeLa cells at 10 nM in the presence of increasing concentrations of Guanabenz. Level of PPIB mRNA was measured using QuantiGene® (Affymetrix) at 24 h normalized to housekeeping gene, MAP4K4 (mitogen-activated protein kinase 4), and presented as percent of untreated control (n = 3, mean ± SD). UNT – untreated cells, NTC – non-targeting control, 0.5 μM PPIB-targeting hsiRNA served as a positive control. (B) HeLa cells expressing psiCHECK™-2 Vector (Promega) containing PPIB target sequence were incubated with PPIB hsiRNA and Guanabenz solubilized in DMSO at concentrations shown for 24 h. Level of PPIB mRNA was measured using Dual-Glo® Luciferase Assay System (Promega) normalized to housekeeping gene, MAP4K4, and presented as percent of 1% DMSO treated cells (ctr) (n = 3, mean ± SD). IC50 values calculated using GraphPad Prism 6.04. (C) Cellular viability was measured by alamarBlue® (Life Technologies) following incubation of HeLa cells with 0–200 μM Guanabenz in the presence or absence of 0.5 μM hsiRNA. Data presented as percent of untreated control (n = 3, mean ± SD).
Guanabenz enhances cellular uptake of hsiRNA
We performed live cell imaging of Cy3-labeled hsiRNA using confocal microscopy to determine whether Guanabenz affected the rate or degree of passive hsiRNA cellular uptake. HeLa cells were treated with 0.5 μM Cy3-labeled hsiRNA in the presence or absence of 50 μM Guanabenz (Figure 3). Images were captured every 2 min over the course of 32 min at fixed laser intensity and acquisition time. Over this period, hsiRNA alone is capable of self-delivery and accumulates on plasma membranes (Figure 3A, left panel). However, Guanabenz markedly impacts the ability of hsiRNA to associate with the cell (Figure 3A, right panel, Supplementary Figure S9). Stable plasma membrane association could be a result of hsiRNA charge shielding via a non-covalent RNA-Guanabenz interaction. Indeed, the highly basic guanidinium moiety is analogous to an arginine side-chain, a feature commonly observed in cationic cell-penetrating peptides such as the HIV-derived TAT peptide or the non-viral penetratin (16,22). The observed impact on the level of hsiRNA cellular association is specific to early time points, as overall level of internalized oligonucleotides equilibrates by 24 h with no significant alteration in co-localization with lysosomal markers (Figure 3B). It appears that the major visible impact of Guanabenz is immediate enhancement of hsiRNA cellular association and internalization. To investigate if short-term Guanabenz exposure is sufficient to enhance hsiRNA activity, HeLa cells were treated with range of Guanabenz concentration for 1 h, with hsiRNA IC50 values collected at 24 h (Supplementary Figure S10). We did not observe any enhancement of PPIB mRNA silencing, suggesting that the perceived activity of Guanabenz entails continuous exposure. This might be due to a requirement for direct small molecule-RNA interactions and/or constitutive modulation of certain intracellular pathways.
Figure 3.
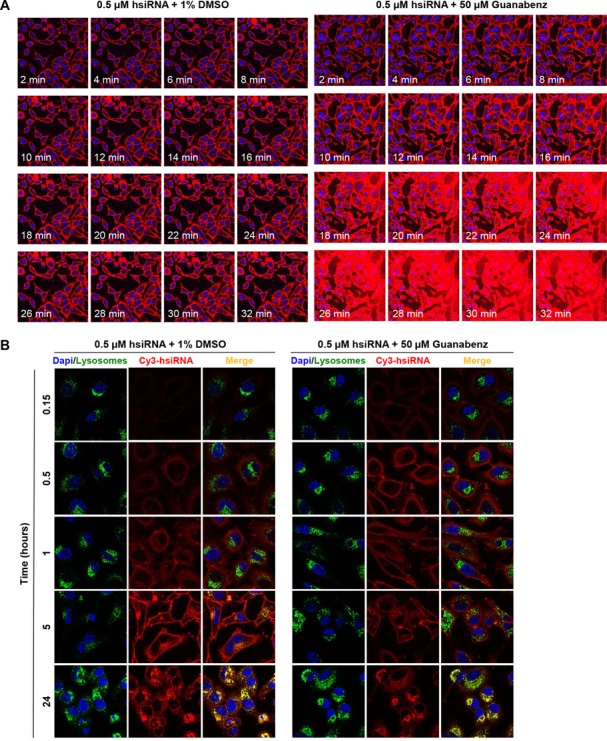
Guanabenz markedly impacts the rate and degree of intracellular uptake of hsiRNAs. (A) Uptake kinetics of hsiRNA. Cy3-PPIB hsiRNA (red), 0.5 μM, was added to HeLa cells in the presence of 1% DMSO or Guanabenz (50 μM). Imaged on a Leica confocal microscope, 63X, nuclei stained with Hoechst dye (blue). Laser intensity and acquisition time were fixed at the 2 min timepoint and kept constant throughout the experiment, resulting in saturation at later Guanabenz treatment timepoints. (B) Co-localization of hsiRNA with lysosomal markers. Cy3-HTT hsiRNA (red), 0.5 μM, was added in HeLa cells in presence of 1% DMSO or Guanabenz (50 μM). Nuclei stained with Hoechst dye (blue) and lysosomes stained with LysoTracker (green, Life Technologies). Cells were imaged on a Leica confocal microscope, with laser intensity and acquisition optimized for a 24 h experiment. Representative images taken at time points indicated are shown.
We acquired dynamic light scattering (DLS) measurements and transmission electron microscopy (TEM) images to further investigate the physicochemical properties of hsiRNA and Guanabenz in solution (Figure 4A). By DLS, we attempted to detect particles following incubation of hsiRNA with varying concentrations of Guanabenz. No reliable measurements could be collected for 0.5 μM hsiRNA in the presence of 0–200 μM Guanabenz, or for 750 μM Guanabenz alone. However, we detected polydisperse aggregates following incubation of 0.5 μM hsiRNA with 500 or 750 μM Guanabenz. For 500 μM Guanabenz, the main species in solution had a calculated hydrodynamic radius (Z-average) of 63 nm. For 750 μM, the major particle had a hydrodynamic radius of 124 nm. These particles were highly disperse, with polydispersity indices (PdI) of 0.54 and 0.45, respectively. We confirmed these observations by TEM, where we observe crystalline hsiRNA particles of ∼100 nm in diameter in the presence of 500 or 750 μM Guanabenz. Furthermore, Cy3-labeled hsiRNA nanoparticles could be detected as a fluorescent pellet following ultra-centrifugation in the presence of 1 mM Guanabenz (Supplementary Figure S11). Mechanistically, a putative RNA-Guanabenz interaction may be mediated by the protonatable guanidinium moiety, as stable nucleotide-guanidinium complexes have been described previously (23). We directly observed weak Guanabenz-hsiRNA binding in vitro by isothermal calorimetry (ITC) (data not shown) and gel shift assay (Supplementary Figure S12), in which a large excess of Guanabenz is required for full complexation of siRNA. This is substantiated by the fact that over 100-fold molar excess of Guanabenz is required for enhanced siRNA biological activity in vitro. The discovery that Guanabenz could act as a small-molecule siRNA binder agrees well with reports from Selwood et. al. of guanidinium-based small molecule-carriers of siRNA, which are based on the alpha-helical structure of penetratin (16,17). In these studies, a weak non-covalent RNA-small molecule interaction is also observed. Taken together, these data imply that a direct, molecular-scale Guanabenz-hsiRNA interaction occurs and that Guanabenz-hsiRNA nanoaggregates form at high (non-pharmacologically relevant) concentrations of Guanabenz. We cannot rule out the possibility for smaller aggregates to form under our cell treatment conditions, but they likely represent a small percentage of the total sample and are undetectable by the methods used herein.
Figure 4.
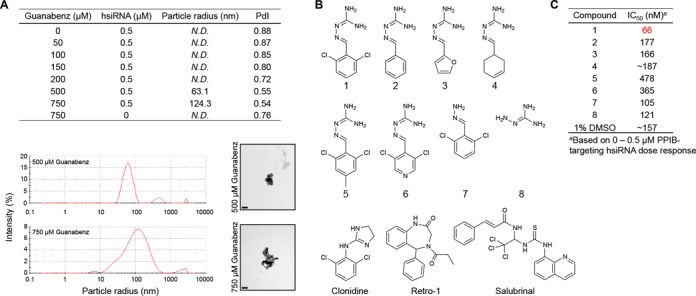
Physicochemical and biological properties of hsiRNA, Guanabenz, and Guanabenz derivatives (A) Characterization of nanoaggregates formed by hsiRNA and Guanabenz. (Top) Size distribution and intensity measured by dynamic light scattering (Nano-ZS, Malvern). N.D. = not detectable; PdI = polydispersity index. (Bottom right) hsiRNA nanoparticle size and morphology examined by transmission electron microscopy following incubation with 500 or 750 μM Guanabenz (Tecnai Spirit 12, FEI, 43,000X). Scale bar = 0.2 μM. (B) Chemical derivatives and/or functional analogs of Guanabenz (1) (C) IC50 values of PPIB-targeting hsiRNA following treatment with Guanabenz and derivatives [100 μM (1–4, 7,8) or 12.5 μM (5,6)] or 1% DMSO. Derivitization results in loss of effect of Guanabenz on hsiRNA IC50.
Guanabenz structure-activity relationship analysis
Based on this knowledge, a preliminary SAR analysis was performed to determine the basic chemical scaffold of Guanabenz required for improving hsiRNA efficacy (Figure 4B, Compound 1). Specifically, we were interested in determining whether structural perturbations at any positions were well tolerated and held potential for being modified into a linker for the covalent siRNA attachment. A panel of seven Guanabenz derivatives was prepared and their SAR activity was explored. We focused on making modifications on the benzene ring (Figure 4B, Compounds 2–6) and guanidinium group (Figure 4B, Compounds 7–8). Compounds 2, 3, 4, 7, and 8 were synthesized as described previously. Compounds 5 and 6 were prepared by the procedure described in the Supplemental Methods. To our surprise, none of the chemical derivatives showed dose-dependent enhancement of hsiRNA activity (Supplementary Figure S13). Elimination of chlorine, modification of the benzene ring, or loss of guanidinium group all resulted in near complete loss-of-function. Compounds 5 and 6 were also significantly more toxic. Calculated IC50 values for the derivatives are listed in Figure 4C.
This finding is supported by an in-depth structural analysis of the small molecule screening data, which revealed ∼30 compounds containing protonatable primary amines or guanidinium groups that were not identified as positive hits (Supplementary Table S3). This implies a mechanism more complex than a purely electrostatic-based interaction, in which a protonated Guanabenz masks the negative charge of siRNA and facilitates uptake. Collectively, this suggests that Guanabenz may be enhancing hsiRNA uptake through weak, non-covalent interactions, but also mediating enhanced gene silencing through independent modulation of cellular pathways.
Mechanistic analysis of observed guanabenz activity
Guanabenz acetate was FDA approved in 1982 as an alpha agonist of β-adrenoreceptor for the treatment of hypertension (24). Recently, it has been implicated as an inhibitor of several fundamental cellular pathways, including the protein-folding activity of the ribosome (PFAR) (25) and protein dephosphorylation by protein phosphatase 1 (PP1) (26–28). Both of these functionalities have been linked to neuroprotective anti-prion activity (29,30). To date, there has been no link between cellular pathways affected by Guanabenz and RNA trafficking, endocytosis, or therapeutic siRNA-dependent mRNA silencing. To verify that Guanabenz may act through a trans mechanism independent of RNA binding, we tested a panel of small molecule compounds that have alpha-2 adrenergic agonist or PP1-inhibitory activity. Clonidine is another FDA-approved therapeutic for hypertension and a chemical analog of Guanabenz (29). A dose response of a Huntingtin (HTT)-targeting hsiRNA was performed in the presence of 100 μM Clonidine or 60 μM Guanabenz (Supplementary Figure S14, structure Figure 4B). In this assay, Guanabenz enhanced HTT mRNA silencing by ∼30% over an untreated (1% DMSO) control. However, treatment with 100 μM Clonidine had no effect. A similar assay was done using Salubrinal, a specific inhibitor of GADD34, which is a non-enzymatic cofactor of PP1 (30). A dose response of HTT-targeting hsiRNA was performed in the presence of 25, 50, or 75 μM Salubrinal (Supplementary Figure S14, structure Figure 4B). Under these conditions, Salubrinal had no effect on hsiRNA efficacy. These data imply that the activity of Guanabenz towards hsiRNA is not related to alpha-2 adrenergic agonist or protein phosphatase-related functionality. Taken together, these data suggest that Guanabenz may be enhancing hsiRNA uptake and activity in a trans mechanism that has not been previously described.
Finally, we examined the impact of Guanabenz co-delivery on mRNA silencing efficacy of GalNAc-siRNA, LNA antisense, and sterol-conjugated siRNA (Accell siRNA, Thermo Fisher) in primary human hepatocytes (Supplementary Figure S15). We report that Guanabenz shows no activation of other classes of ONTs. This exemplifies the fact that small molecules can demonstrate specificity towards certain uptake and intracellular trafficking pathways, and points to the need for separate screening protocols for each class of compound. The only reported instance of small molecule-mediated ONT activation is Retro-1. Retro-1 was identified in a screen for compounds which block retrograde trafficking, and was shown to increase the activity of antisense and splice-switching oligonucleotides, but not siRNA (18). We evaluated the impact of Retro-1 on hsiRNA efficacy and observed that co-treatment with 100 μM Retro-1 had a slightly inhibitory effect, confirming that hsiRNA transport is not mediated through a Retro-1 dependent pathway (Supplementary Figure S14, structure Figure 4A). Taken together, these data indicate that compounds that are both cholesterol- and phosphorothioate-modified are internalized through a pathway selectively enhanced by Guanabenz.
DISCUSSION
In summary, in a proof-of-concept search for small molecule modulators of siRNA activity, we identified Guanabenz acetate (Wytensin™) from a high-throughput screen of 1280 pharmacologically active small molecules. We established that co-delivery of Guanabenz acetate with hsiRNA allows a 100-fold improvement in hsiRNA IC50, due in large part to a dramatic increase in hsiRNA cellular membrane association and internalization. This functionality is sequence independent, but ONT class specific. The identification of small molecules with a profound impact on hsiRNA efficacy was surprisingly straightforward, with several hits isolated from a highly diverse but relatively small sized library. A similar combinatorial approach previously revolutionized the field of lipidoid-mediated siRNA delivery, identifying novel chemical scaffolds with unprecedented efficacy (31,32). For gymnotic oligonucleotides (ASO, GalNAc-siRNAs, hsiRNA) our work illustrates the potential for small molecule screening to become the foundation for rational design of novel classes of conjugated ONTs or combinatorial small molecule-ONT formulations.
Supplementary Material
Acknowledgments
We acknowledge Dr Alexey Wolfson (Advirna) for providing the MAP4K4 psiCHECK-2 plasmid.
Author Contribution: M.F.O, J.F.A., M.N., H.C., M.D., M.H., A.H.C., and A.K. performed experiments, analyzed data, and provided feedback on the manuscript. M.F.O. and A.K. prepared the figures and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Massachusetts Center for Clinical and Translational Science Pilot Project Program, NIH/NIGMS [GM1088030181]; CHDI Foundation (JSCAG367); Experiments performed at the UMMS Electron Microscopy Core were supported by the National Center For Research Resources [S10RR027897]. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Castanotto D., Rossi J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson B.L., McCray P.B. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J., Shum K.-T., Burnett J.C., Rossi J.J. Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals. 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M., et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 5.Gooding M., Browne L.P., Quinteiro F.M., Selwood D.L. siRNA delivery: from lipids to cell-penetrating peptides and their mimics. Chem. Biol. Drug Des. 2012;80:787–809. doi: 10.1111/cbdd.12052. [DOI] [PubMed] [Google Scholar]
- 6.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Dahlman J.E., Barnes C., Khan O.F., Thiriot A., Jhunjunwala S., Shaw T.E., Xing Y., Sager H.B., Sahay G., Speciner L., et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stalder L., Heusermann W., Sokol L., Trojer D., Wirz J., Hean J., Fritzsche A., Aeschimann F., Pfanzagl V., Basselet P., et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam R., Wang Q., Hoekstra M., Kandasamy P., Kel A.V., Milstein S., et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 10.Byrne M., Ford G., Tzekov R., Wang Y., Rodgers A., Cardia J., Holton K., Pandarinathan L., Lapierre J., Stanney W., et al. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 2013;29:855–864. doi: 10.1089/jop.2013.0148. [DOI] [PubMed] [Google Scholar]
- 11.Wu J., Huang W., He Z. Dendrimers as carriers for siRNA delivery and gene silencing: a review. ScientificWorldJournal. 2013;2013:630654. doi: 10.1155/2013/630654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S.W., Kim N.Y., Choi Y.B., Park S.H., Yang J.M., Shin S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. J. Control. Release. 2010;143:335–343. doi: 10.1016/j.jconrel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Crombez L., Aldrian-Herrada G., Konate K., Nguyen Q.N., McMaster G.K., Brasseur R., Heitz F., Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol. Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozema D.B., Lewis D.L., Wakefield D.H., Wong S.C., Klein J.J., Roesch P.L., Bertin S.L., Reppen T.W., Chu Q., Blokhin A.V., et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong S.C., Klein J.J., Hamilton H.L., Chu Q., Frey C.L., Trubetskoy V.S., Hegge J., Wakefield D., Rozema D.B., Lewis D.L. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic Acids Ther. 2012;22:380–390. doi: 10.1089/nat.2012.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gooding M., Adigbli D., Edith Chan A.W., Melander R.J., MacRobert A.J., Selwood D.L. A bifurcated proteoglycan binding small molecule carrier for siRNA delivery. Chem. Biol. Drug Des. 2014;84:24–35. doi: 10.1111/cbdd.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooding M., Tudzarova S., Worthington R.J., Kingsbury S.R., Rebstock A.-S., Dube H., Simone M.I., Visintin C., Lagos D., Quesada J.-M.F., et al. Exploring the interaction between siRNA and the SMoC biomolecule transporters: implications for small molecule-mediated delivery of siRNA. Chem. Biol. Drug Des. 2012;79:9–21. doi: 10.1111/j.1747-0285.2011.01249.x. [DOI] [PubMed] [Google Scholar]
- 18.Ming X., Carver K., Fisher M., Noel R., Cintrat J.-C., Gillet D., Barbier J., Cao C., Bauman J., Juliano R.L. The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Res. 2013;41:3673–3687. doi: 10.1093/nar/gkt066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B., Ming X., Cao C., Laing B., Yuan A., Porter M.A., Hull-Ryde E.A, Maddry J., Suto M., Janzen W.P., et al. High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic Acids Res. 2015;43:1987–1996. doi: 10.1093/nar/gkv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baum T., Bruce W. (2,6-disubstituted benzylidene)amino guanidines and related compounds. 1974 US 3,816,531 A. [Google Scholar]
- 21.Kodama J.K., Haynes G.R., Albert J.R. Therapeutic Agents. 1976 US 3,975,533 A. [Google Scholar]
- 22.Moschos S.A., Jones S.W., Perry M.M., Williams A.E., Erjefalt J.S., Turner J.J., Barnes P.J., Sproat B.S., Gait M.J., Lindsay M.A. Lung delivery studies using siRNA conjugated to TAT (48 - 60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug. Chem. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohara K., Smietana M., Vasseur J.-J. Characterization of specific noncovalent complexes between guanidinium derivatives and single-stranded DNA by MALDI. J. Am. Soc. Mass Spectrom. 2006;17:283–291. doi: 10.1016/j.jasms.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Holmes B., Brogden R., Heel R., Speight T., Avery R. Guanabenz. A review of its pharmacodynamic properties and therapeutic efficacy in hypertension. Drugs. 1983;26:212–229. doi: 10.2165/00003495-198326030-00003. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen P., Hammoud H., Halliez S., Pang Y., Evrard J., Blondel M., Strasbourg U., De F. Structure−activity relationship study around guanabenz identifies two derivatives retaining antiprion activity but having lost α2-adrenergic receptor agonistic activity. ACS Chem. Neurosci. 2014;5:1075–1082. doi: 10.1021/cn5001588. [DOI] [PubMed] [Google Scholar]
- 26.Tsaytler P., Harding H.P., Ron D., Bertolotti A. Selective Inhibition of a Regulatory Subunit of a Protein Phosphatase 1 Restores Proteostasis. Science. 2002;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H.-Q., Ren M., Jiang H.-Z., Wang J., Zhang J., Yin X., Wang S.-Y., Qi Y., Wang X.-D., Feng H.-L. Guanabenz delays the onset of disease symptoms, extends lifespan, improves motor performance and attenuates motor neuron loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neuroscience. 2015;277:132–138. doi: 10.1016/j.neuroscience.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Popko B., Tixier E., Roos R.P. Guanabenz, which enhances the unfolded protein response, ameliorates mutant SOD1-induced amyotrophic lateral sclerosis. Neurobiol. Dis. 2014;71:317–324. doi: 10.1016/j.nbd.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tribouillard-Tanvier D., Béringue V., Desban N., Gug F., Bach S., Voisset C., Galons H., Laude H., Vilette D., Blondel M. Antihypertensive drug guanabenz is active in vivo against both yeast and mammalian prions. PLoS One. 2008;3:e1981. doi: 10.1371/journal.pone.0001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A., et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahon K.P., Love K.T., Whitehead K.A., Qin J., Akinc A., Leshchiner E., Leshchiner I., Langer R., Anderson D.G. Combinatorial approach to determine functional group effects on lipidoid-mediated siRNA delivery. Bioconjug. Chem. 2010;21:1448–1454. doi: 10.1021/bc100041r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


