Abstract
Background: Rapidly improving protocols for the derivation of autologous cells from stem cell sources is a welcome development. However, there are many circumstances when off-the-shelf universally immunocompatible cells may be needed. Embryonic stem cells (ESCs) provide a unique opportunity to modify the original source of differentiated cells to minimize their rejection by nonautologous hosts.
Hypothesis: Immune rejection of nonautologous human embryonic stem cell (hESC) derivatives can be reduced by downregulating human leukocyte antigen (HLA) class I molecules, without affecting the ability of these cells to differentiate into specific lineages.
Methods and Results: Beta-2-microglobulin (B2M) expression was decreased by lentiviral transduction using human anti-HLA class I light-chain B2M short hairpin RNA. mRNA levels of B2M were decreased by 90% in a RUES2-modified hESC line, as determined by quantitative real time-polymerase chain reaction analysis. The transduced cells were selected under puromycin pressure and maintained in an undifferentiated state. The latter was confirmed by Oct4 and Nanog expression, and by the formation of characteristic round-shaped colonies. B2M downregulation led to diminished HLA-I expression on the cell surface, as determined by flow cytometry. When used as target cells in a mixed lymphocyte reaction assay, transduced hESCs and their differentiated derivatives did not stimulate allogeneic T-cell proliferation. Using a cardiac differentiation protocol, transduced hESCs formed a confluent layer of cardiac myocytes and maintained a low level of B2M expression. Transduced hESCs were also successfully differentiated into a hepatic lineage, validating their capacity to differentiate into multiple lineages.
Conclusions: HLA-I depletion does not preclude hESC differentiation into cardiac or hepatic lineages. This methodology can be used to engineer tissue from nonautologous hESC sources with improved immunocompatibility.
Introduction
Tissues engineered from autologous induced pluripotent stem cells (iPSCs) should, in theory, offer a great way to fully circumvent graft rejection. Yet, this concept has been recently challenged, as autologous iPSCs may be targets of immune rejection due to genomic alterations acquired during the reprogramming of somatic cells, as well as other poorly understood factors.1–3 Derivation of iPSCs from elderly patients can be also problematic.4 It will be difficult to restore organs using autologous cells that retain inherited or accumulated mutations and/or acquired malicious epigenetic changes.5 iPSCs from different donors also require different culturing and differentiation protocols, making the final products and therapies hard to compare.6–8 Finally, at least today, iPSC protocols involve high costs and prolonged periods to acquire and cultivate initial cell sources, isolate, scale-up, reprogram, and differentiate iPSCs.
If the cell sources used to engineer tissue grafts are not autologous, they are likely to be rejected. An immune response is triggered by recognition of a peptide antigen, which is presented by major histocompatibility complex (MHC) molecules.9 While present in all species, MHCs are highly polygenic and polymorphic, varying among organisms and within a species.10 In humans, MHC molecules are called human leukocyte antigens (HLA), and they are coded for by 21 core genes located on the short arm of chromosome six.11 Because multiple alleles exist for each polymorphic HLA gene, the task of matching a specific pair of HLA haplotypes to a prospective transplant recipient is a challenge. HLA class I genes are expressed on most nucleated cells, while HLA class II genes are expressed only in specialized or professional antigen-presenting cells. The success of solid organ and hematopoietic stem cell transplantation has been greatly improved by matching HLA class I and II antigens between donors and recipients, with immunosuppressive therapy still required in cases of HLA mismatch to prevent rejection. Unfortunately, prolonged use of immunosuppressive drugs can lead to a variety of undesirable side effects and immunosuppression is not always effective.12–14
One possible alternative to immunosuppression is to create donor cells with minimal surface expression of HLA class I molecules. Creation of an immune compatible human embryonic stem cell (hESC) line, which can be differentiated into any desired cell lineage, is an appealing goal with a wide range of clinical implications. In recent years, significant progress by a number of laboratories, including ours, has been made toward this goal.15–21
In this study, we hypothesized that the immunogenicity of hESCs and their derivatives can be decreased by constitutive expression of short hairpin RNA (shRNA) against beta-2-microglobulin (B2M), the invariable nontransmembrane 12 kDa conserved light-chain of HLA class I molecules. This approach should diminish the assembly and presence of functional HLA-I molecules on the cell surface, without impairing the cell's ability to differentiate into specific cell phenotypes, thus making them and their differentiated derivatives less susceptible to recognition by allogeneic T lymphocytes.
Due to our long-term interests in heart repair and recent successes in improving postinfarction cardiac performance by stem cell derivatives,22–24 the main goal of our studies was to document the ability of the modified hESCs to differentiate into fully functional cardiac cells. We also confirmed the ability of these cells to differentiate into a hepatic lineage. Hepatocytes generated from iPSCs of individual donors can mimic individual differences in drug metabolism causing unpredictable side effects.25,26 In contrast, cells derived from a B2M(-) hESC line can be a reliable and consistent hepatocyte source for cell-based therapies for liver diseases, including hepatitis, end-stage liver failure, cirrhosis, and hepatocellular carcinoma.
Materials and Methods
Cell sources
A federally approved hESC line from Rockefeller University (RUES2, NIH Stem Registry identification No. 0013) was modified to include a genetically encoded calcium indicator GCaMP, as recently described.24 In brief, a transgene encoding for the constitutive expression of GCaMP3 was inserted into the AAVS1 locus in hESCs, generated by ZFN-mediated targeting.27 Having an endogenous calcium sensor greatly facilitates the analysis of cardiomyocyte behavior—in both in vitro and in vivo settings.23,24 It also eases monitoring and functional comparison of the control and modified cells, as there is no need to take cells out of the media to add/wash the dye out or account for data variability related to differences in dye retention or loading conditions. Undifferentiated GCaMP+hESCs were then cultured under feeder-free conditions, using vitronectin-coated plates and fed daily with the mouse embryonic fibroblast-conditioned medium (MEF-CM) supplemented with human basic fibroblast growth factor (bFGF, 4 μg/L). Methods for the feeder-free growth of hESCs on coverslips28,29 or protocols for in vitro 3D cardiac constructs30,31 have been detailed elsewhere. GCaMP expression enabled direct visualization of calcium release in contracting hESC-derived cardiac cells, as described earlier.23
Generation of B2M depleted hESC
For efficient and stable delivery of shRNAi constructs against B2M, we used stable transfection with a vector that integrated into genomic DNA, followed by puromycin-based selection. MISSION® shRNA lentiviral transduction particles were purchased from Sigma-Aldrich (SHC008V, clone No. TRCN0000066424). The backbone vector (pLKO.1-Puro) used for the cloning of B2M shRNA contains a puromycin resistance gene. Cells were transduced with 3.4 × 105 TU/mL in the presence of hexadimethrine bromide (polybrene; Sigma-Aldrich) at a final concentration of 8 μg/mL (MOI 1–1.5). Media were replaced at 48 h post-transduction. Undifferentiated cells were placed under puromycin pressure at 10 μg/mL final concentration for at least 10 generations. Control (mock) samples were treated identically, but with lentiviral particles omitted.
Qualitative and quantitative polymerase chain reaction
Genomic DNA was obtained from parental and stably transduced cells using a Qiagen Purification Kit. DNA was quantitated using a NanoDrop 2000 Spectrophotometer (ThermoScientific), and equal amounts of genomic DNA were used as a template for each clone to validate transgene incorporation. Plasmid DNA encoding the transgene was used as positive control. For quantitative real-time PCR (qRT-PCR), total RNA was isolated using TRIzol (Invitrogen) as per the manufacturer's instructions. First-strand cDNA synthesis was generated using the AffinityScript qPCR cDNA Synthesis Kit (Stratagene). qRT-PCR was performed using the BioRad qPCR kit and CFX384 PCR detection system. To increase the confidence in measuring the level of B2M, we used two primers that map different regions of the gene (Fig. 5A). The qRT-PCR experiments shown in Figure 4 were performed in three independent biological triplicates per sample and condition. The qRT-PCR experiments shown in Figure 2D were acquired from three different differentiation experiments, following normalization to the housekeeping gene. They were then displayed as a fraction of the maximum expression level for each gene within the analyzed time frame. Primers were ordered from BioSynthesis, Inc. and from RealTimePrimers. The full list of primers is included in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/tea).
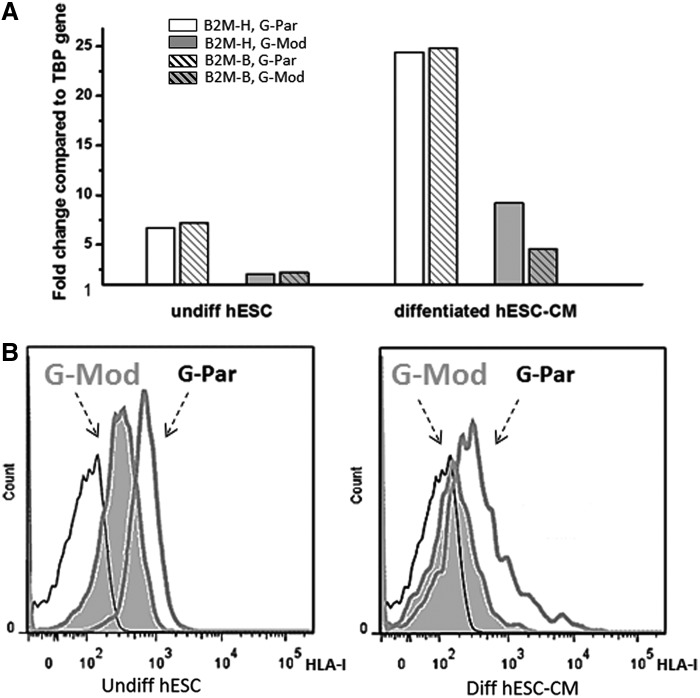
FIG. 5.
Expression levels of B2M versus the presence of functional human leukocyte antigen (HLA)-I on cell surface. (A) Data show decreased levels of B2M expression in a single round of parallel differentiation of G-Par and G-Mod cells to cardiac lineage. Two different primers (patterned vs. solid bars) mapping two different regions on the B2M gene: the 3′ end B2M-H (589–674) and the two exons spanning B2M-B (322–435) were used in this experiment. (B) Flow cytometry analysis confirmed a significant loss of HLA-I on the cell surface in both undifferentiated and differentiated G-Mod cells, compared to their G-Par counterparts. Thin line corresponds to unstained cell sample.
FIG. 4.
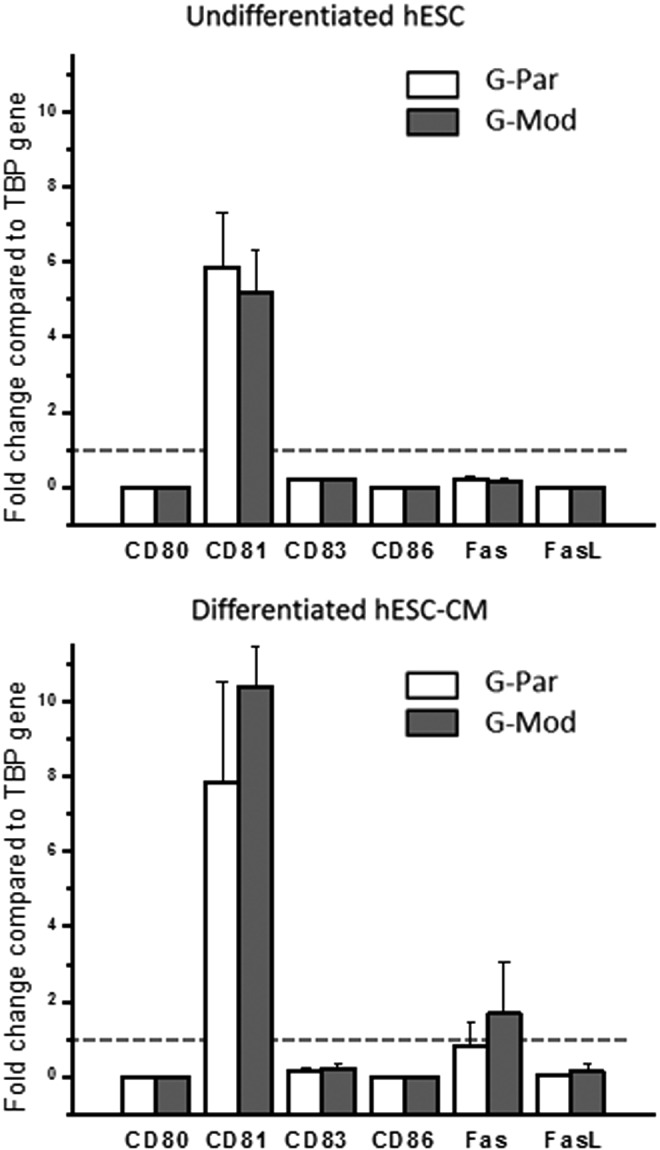
Levels of major costimulatory molecules remained unchanged in hESCs with diminished B2M expression in either undifferentiated or fully differentiated states. Gene expression levels of major costimulatory molecules, including CD80, CD81, CD83, CD86, FAS, and FAS-Ligand, after normalization to housekeeping gene TBP (human TATA box binding protein). No significant differences between G-Par and G-Mod cells were found in either undifferentiated (day 1) or fully differentiated (day 21) states.
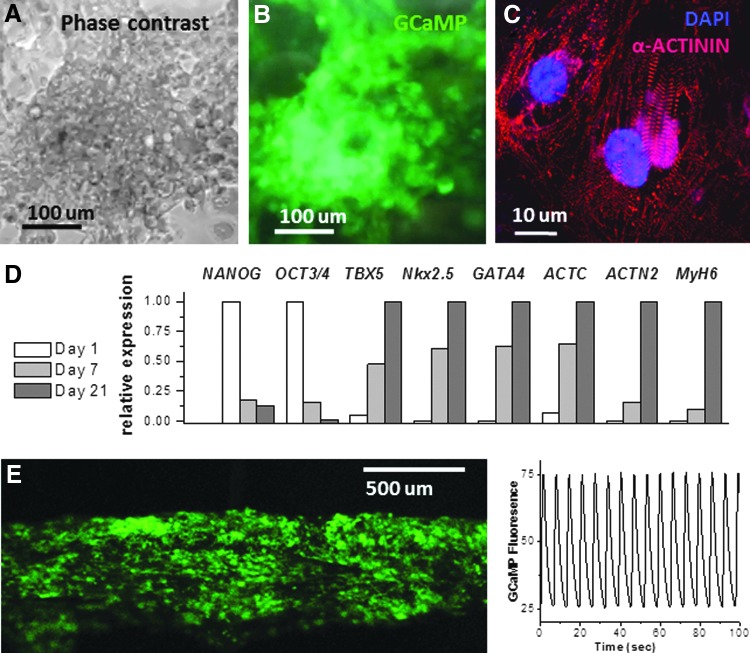
FIG. 2.
Properties of cardiac myocytes derived from hESCs with diminished B2M expression. The top row shows a typical appearance of a differentiated G-Mod cardiomyocyte layer in a phase-contrast (A) and a fluorescence mode (B). The presence of GCaMP sensor in G-Mod cells enables direct monitoring of rhythmical cell contractions upon cardiac lineage differentiation seen as transient increases in endogenous cell fluorescence. The panel (C) shows sarcomeric alpha-actinin immunostaining with regular striations indicative of proper structural development. The panel (D) illustrates changes in relative expression levels of pluripotency (NANOG, Oct3/4), mesodermal (TBX5), early and late (alpha-actin, GATA4, NkX2.5, alpha-2 actinin, myosin heavy-chain 6) markers in samples acquired on day 1, 7, and 21 of cardiac differentiation protocol (n = 3). The panel (E) shows 3D cardiac fiber composed of G-Mod cardiac myocytes with the trace of calcium transients acquired from it to illustrate regular spontaneous contractions.
Cardiac differentiation protocol
hESC-CMs were generated using a previously described cardiac differentiation protocol.24,32 In brief, undifferentiated cells were expanded under standard feeder-free conditions using MEF-CM supplemented with human bFGF (4 μg/L). When the undifferentiated hESC colonies occupy approximately two-thirds of the surface area, they were dispersed using 0.2 mL/cm2 Versene, followed by a wash with MEF-CM containing 10 μM ROCK inhibitor Y-27632 dihydrochloride. Cells were then replated to form a dense monolayer. After a confluent monolayer was formed, hESCs were switched to the RPMI-B27 medium and serially pulsed with 100 μg/L activin A on day 1 and bone morphogenetic protein-4 (BMP4, 10 μg/L) from days 1 to 5. Thereafter, the differentiating cultures are grown in the RPMI-B27 medium supplemented with insulin and vitamin A. Spontaneous beating activity commenced on days 10–20.
Hepatic differentiation
hESCs were induced into definitive endoderm (DE) and subsequently hepatocytes using previously described protocols (Duan, Y., et al., 2010, Stem Cells). Briefly, hESCs were induced to DE in a serum-free condition with the RPMI medium (Invitrogen) containing activin A (100 ng/mL; Peprotech), Wnt3a (50 ng/mL; R&D Systems, Inc.), 2 mM L-glutamine (Gibco, Life Technologies), and 1% (vol/vol) penicillin/streptomycin (Gibco, Life Technologies) for 24 h. The medium was then changed to the RPMI medium with activin A (100 ng/mL), sodium butyrate (0.5 mM; Stemgent), and B27 supplement (Gibco, Life Technologies) for 6 days. DE cells were then passaged on collagen type I-coated 12-well plates in IMDM (Invitrogen) supplemented with 20% fetal bovine serum (FBS; Invitrogen), FGF4 (20 ng/mL), hepatocyte growth factor (HGF) (20 ng/mL), BMP2 (10 ng/mL), BMP4 (10 ng/mL), 0.3 mM L-thioglycerol (Sigma), 0.5% dimethyl sulfoxide (Sigma), 100 nM dexamethasone (Sigma), and 0.126 U/mL human insulin for 2 weeks. The growth factors were purchased from Invitrogen and the medium was changed every 2 days.
Expression of FASL and costimulatory molecules
The qRT-PCR assessment of major immune regulatory or costimulatory molecules, including CD80, CD81, CD83, CD86, as well as Fas and FasL, was conducted using undifferentiated cells (day 1, before activin A addition) and fully differentiated and actively beating cells (day 21). RNA was collected from three independent differentiation experiments with protocols running in parallel for control and modified cells. Three sets of RNA samples were collected and the RNA samples were prepared using the miRNeasy Mini Kit (Qiagen).
Immunocytochemistry and imaging protocols
For cardiac lineage-specific staining, samples were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked overnight in 1% bovine serum albumin (BSA). Samples were stained with mouse sarcomeric α-actinin for 2 h (1:500; Sigma-Aldrich), followed by goat anti-mouse Cy3 secondary antibodies (1:1000; Jackson ImmunoResearch Labs, Inc.). Nuclei were counterstained with DAPI (1:300; Molecular Probes). Calcium transient recordings were acquired using a Zeiss LSM 510 confocal imaging system using fluorescence from endogenous GCaMP sensor. For hepatic identification, cells were fixed in 4% paraformaldehyde+0.3% Triton X-100 in phosphate-buffered saline (PBS) for 15 min. The cells were then incubated in 1% BSA blocking solution for 1 h and exposed to goat anti-human Sox17 (R&D Systems, Inc.), goat anti-human Foxa2 (R&D Systems, Inc.), and goat anti-human HNF4α (Santa Cruz) antibodies for 90 min. The samples were washed thrice with PBS and incubated with a mixture of secondary antibodies and DAPI (Invitrogen) for 1 h. Donkey anti-goat IgG antibodies conjugated with Alexa-488 and Alexa-555 were used at 1:1000 dilutions. All incubations were performed at room temperature, unless otherwise mentioned. Fluorescent images were captured by a confocal microscope (Zeiss LSM Pascal).
Flow cytometric assessment of HLA-I presence on cell surface
A single-cell suspension of hESC or hESC-B2M(-) cells was first incubated with Fc block (BD Pharmingen Cat. No. 564219) to block nonspecific Ig-binding sites for 10 min at 4°C. Then, samples were stained with phycoerythrin fluorescent dye-conjugated anti-human HLA-A, B, C (clone DX17; BD Pharmingen, Cat. No. 560168). The DX17 monoclonal antibody reacts with a monomorphic epitope expressed on all HLA class I molecules examined. The cells were incubated with either DX17 or isotype control antibody in the FACS buffer (PBS+0.1% BSA+0.1% NaN3) at 1 μg/mL concentrations for 30 min, washed with FACS buffer thrice, resuspended, and examined using the BD FACSCalibur™ System.
Lymphocyte proliferation assay
Briefly, 10 mL of heparinized venous blood was drawn from a healthy donor and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density centrifugation. A total of 2 × 105 PBMCs/well were cultivated in 200 μL of complete culture medium (RPMI 1640 medium [Sigma Chemical Company], supplemented with penicillin [100 IU/mL], streptomycin [0.1 mg/mL], L-glutamine [0.29 m/L], and amphotericin B [5 mg/mL]), supplemented with 10% FBS. PBMC were cocultured with hESC or hESC-B2M(-) cells as targets. Cells were used in either undifferentiated stage or fully differentiated (day 21 of directed cardiac differentiation). PBMCs were also nonspecifically stimulated with phytohemagglutinin (PHA, 5 μg/mL) as a positive control. PBMCs cultured under similar conditions without any stimulation served as a negative control. The cultures were set up in triplicate and incubated for 4 days at 37°C in 5% CO2 atmosphere. For the thymidine proliferative assay, the transfected and control hESCs were incubated for 4 h with 1 μCi of tritiated [3H] thymidine. The cells were then transferred to a fiberglass membrane with MACH II harvester, and scintillation count was performed on a PerkinElmer Microbeta II scintillation counter. The proliferation was measured as uptake of [3H]-thymidine as count per minute units of the radioisotope compared between conditions.
Statistical analysis
Values are presented as mean ± SEM. Unless indicated otherwise, each quantitative or qualitative conclusion was derived from n ≥ 3 independent experiments. Representative images are shown. A Student's t-test was used to compare the differences between the means, with p < 0.05 considered significant.
Results
Derivation of beta-microglobulin depleted hESC line
The federally approved hESC-09-0013 cell line, deposited by the Rockefeller University (RUES2) was used in these studies. The RUES2 cell line was previously modified to include a genetically encoded calcium indicator, GCaMP, as described previously.24 This genetic modification allows live visualization of calcium transients in hESC-derived cardiac cells and in vivo grafts created from these cells.23 Expression of B2M in hESCs was permanently silenced by transduction with a lentiviral shRNA against human B2M. For simplicity, these cells are referred to as “G-Par” and “G-Mod” indicating parental RUES2-GCaMP and modified RUES2-GCaMP with downregulated B2M gene expression, respectively. B2M mRNA expression was decreased by 90% in G-Mod cells, compared with G-Par cells. Importantly, the expression levels of pluripotency genes were unchanged in G-Mod, as detected by qRT-PCR (Fig. 1A). Transduced undifferentiated G-Mod cells were indistinguishable from G-Par cells when visualized by phase-contrast microscopy (Fig. 1B). The transduced cells formed typical round-shaped colonies, characteristic of undifferentiated ESCs, and had to be passaged every 4–5 days, similarly to the parental cell line (Fig. 1C).
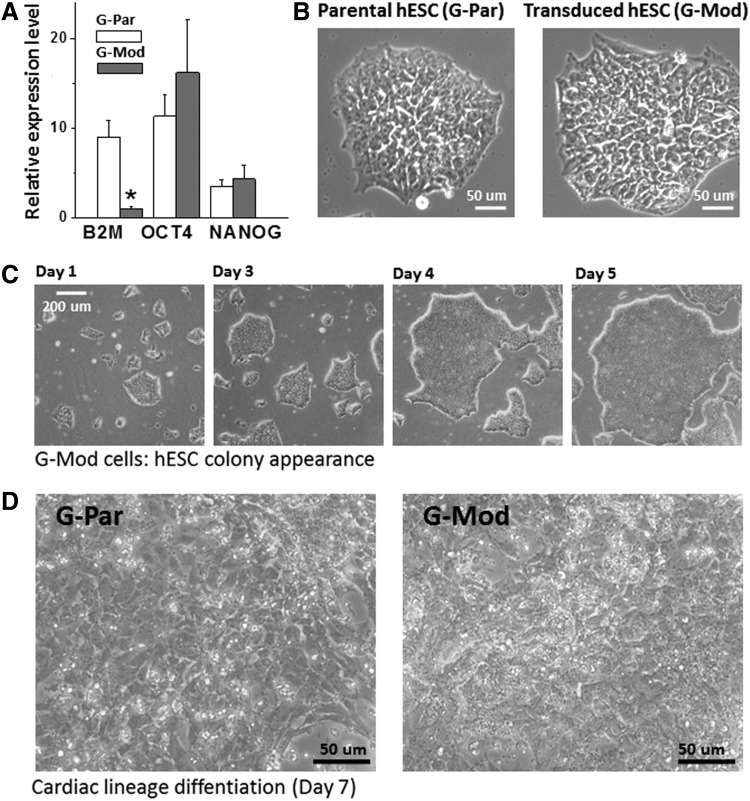
FIG. 1.
Transduced human embryonic stem cells (hESCs) (G-Mod) in an undifferentiated state are phenotypically similar to the parental (G-Par) cell line. (A) The expression levels of beta-2-microglobulin (B2M) are decreased by 80–90% in undifferentiated G-Mod cells compared to controls, while the expression levels of undifferentiated markers such as OCT4 or NANOG remained unchanged. G-Par and G-Mod samples were run in triplicate. Average fold-change in gene expression levels compared to two different housekeeping genes is shown. *p < 0.05. (B) Transduced cells formed round-shaped colonies undistinguishable from G-Par cells. (C) G-Mod cells grew at a similar rate compared to G-Par cells, with a similar number of days required to passage cell colonies (4–5 days). (D) When subjected to cardiac differentiation protocol, G-Mod underwent visible changes in cell morphology, similarly to those observed in G-Par cell layers.
Differentiation of beta-microglobulin depleted hESCs into cardiac muscle lineage
G-Par and G-Mod hESCs were subjected to a cardiac differentiation protocol, to confirm that the transduction procedure did not affect their differentiation potential or alter the expression of costimulatory molecules.24 Briefly, undifferentiated cells from dispersed colonies were cultured in media supplemented with activin A and BMP4. After 7 days, cells showed visible morphological changes that were suggestive of cardiac differentiation (Fig. 1D). On day 10–20 of the cardiac differentiation protocol, both G-Par and G-Mod cells started to visibly contract. The presence of GCaMP, an endogenous calcium sensor, made these cells brightly fluoresce in a 500–550 nm emission range with each contraction (Fig. 2A, B). When the cells were fixed and stained with classical cardiac muscle markers, G-Mod cells appeared similar to G-Par cells. This included a typical striated pattern of sarcomeric alpha-actinin (Fig. 2C) and an increase in expression of mesodermal (Tbx5), early (α-actin, Nkx2.5), and late cardiac markers (α-MYH6, α-actinin) (Fig. 2D). The expression of pluripotency genes (Oct3/4, Nanog) decreased with differentiation, which is consistent with the data by others.33
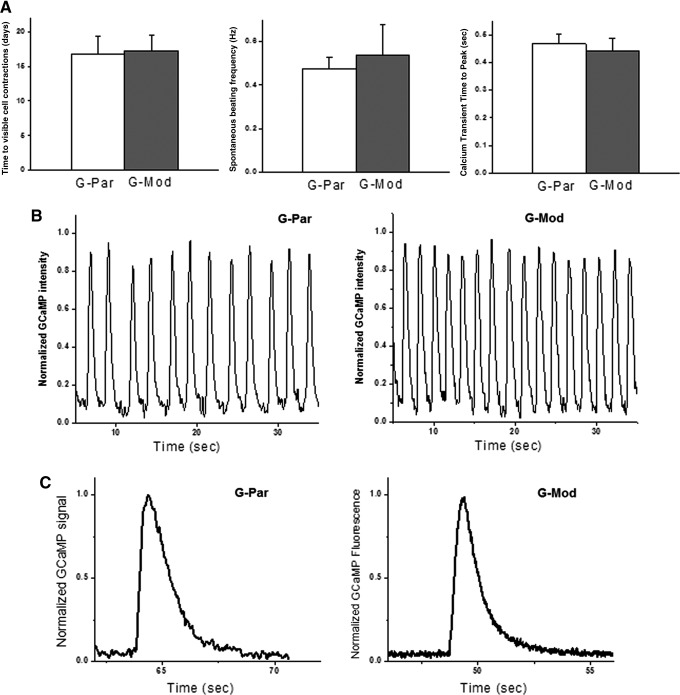
At optimal density, the cells formed robustly beating cardiac cell networks that continued to contract synchronously for several weeks in culture. When mixed with Matrigel or fibrin-based hydrogels,30 they formed regularly beating macroscopic 3D-cardiac fibers (Fig. 2E). Since these cells also expressed an endogenous calcium sensor (GCaMP), calcium transients were readily visualized as spikes in endogenous cell fluorescence that correlated with each episode of contraction.23,34 Analysis of calcium transients is commonly used as a sensitive cumulative index of excitation-coupling and cardiac contractions.35–37 It can reveal small changes in calcium handling machinery and, as such, serves as a useful indicator for cell maturation or physiologically adverse interventions.34,38–40 There were no detectable differences between G-Par-derived and G-Mod-derived cardiac myocytes as far as the number of days to develop calcium transients, their average frequency, or calcium transient time to peak (Fig. 3A). Sample recordings of calcium transients from confluent G-Par and G-Mod monolayers are shown in Figure 3B. When plated sparsely, individual cells often contracted less frequently with longer calcium transients (Fig. 3C), consistent with frequency-dependent acceleration of relaxation frequency,41 but again, no differences were noted between G-Par and G-Mod cells.
FIG. 3.
Functional assessment of cardiac myocytes derived from hESCs with diminished B2M expression. (A) There were no detectable functional differences between cardiac myocytes differentiated from G-Par and G-Mod cell lines. Neither the number of days to develop spontaneous contractions nor the average beating frequency or calcium transient time to peak was different between the two cell lines. (B) Typical recordings of calcium transients from spontaneously beating confluent cell monolayers. Frequency of spontaneous beating ranged from 0.1 to 1 Hz for individual coverslips with an average frequency of 0.45 Hz for both cell types. (C) Sparsely plated individual cells contracted less frequently with longer calcium transients, consistent with frequency-dependent acceleration of relaxation frequency.
Effects of beta-microglobulin depletion on other immune stimulatory molecules
To determine whether the transduction protocol had any unanticipated effects that could alter the immunogenicity of hESCs, we measured the mRNA expression levels of immune regulatory molecules (Fig. 4). CD80, CD83, and CD86 are members of the immunoglobulin supergene family and have been shown to have a costimulatory effect during T-cell activation.42,43 CD81 is a tetraspanin family member and has been implicated in regulating immune cell activation and cell–cell interaction.44 FAS and FAS-ligand, member of the tumor necrosis factor (TNF)-receptor and TNF superfamilies, are best described as regulators of cell death and in the transplant setting may promote downregulation of the immune response.45 As shown in Figure 4, no significant differences in the expression of these molecules in either undifferentiated or differentiated G-Par and G-Mod cells were found and the levels of CD80, CD86, and FASL were found to be negligible for both cell types.
B2M depleted hESCs have diminished HLA class I molecules on cell surface
B2M binds with major and minor gene subunits to produce functional HLA-I heterodimers on the cell surface. Decreased expression of B2M was confirmed using two primers that map two different regions on the B2M gene: the 3′ end B2M-H (589–674) and the two exons spanning B2M-B (322–435) (Fig. 5A).
Reduced B2M gene expression in transduced hESCs does not definitively cause a reduction in assembled HLA-I molecules on the cell surface. Indeed, it was previously suggested that B2M present in serum can be used to reassemble functional HLA heterodimers.46 Since serum is a necessary component of most cell culture media and will also be present in vivo, it was important to verify that reduced B2M expression coincided with a decrease in assembled HLA-I on the cell surface. To do so, intact undifferentiated and differentiated G-Par and G-Mod cells (grown in standard, serum-containing media) were incubated with the HLA-I antibody, which recognizes a nonpolymorphic HLA-I epitope, and analyzed by flow cytometry. The results confirmed a significant loss of HLA-I on the cell surface of both undifferentiated and differentiated G-Mod cells, compared to their G-Par counterparts (Fig. 5B).
Lack of T-lymphocyte activation by beta-microglobulin depleted hESCs and their differentiated derivatives
A mixed lymphocyte reaction (MLR) assay was used to test whether G-Mod cells escape recognition by allogeneic T lymphocytes. In this assay, CD8 T cells recognize allogeneic HLA-I molecules on the surface of target cells and are stimulated to proliferate, as detected by the uptake and incorporation of the [3H]-thymidine in newly synthesized DNA molecules. Therefore, the extent of T-cell stimulation can be measured by accumulation of incorporated [3H]-thymidine after completion of the incubation period. The data showed a significant decrease in the stimulatory capacity of G-Mod cells to background levels, compared with their G-Par counterparts. This indicates that the decreased HLA class I expression is functionally significant and causes a lack of immune recognition of the genetically modified hESCs by T lymphocytes (Fig. 6). The same graph also shows [3H]-thymidine levels harvested from wells, in which media included PHA as a positive control, and from wells with no stimulants or target cells as negative controls. Notably, the presence of B2M depleted cells decreased incorporation levels of radioactive [3H]-thymidine in MLR below that of negative control. These findings are consistent with the existing literature on immunomodulatory effects of ESCs and their derivatives, as detailed in the Discussion section.
FIG. 6.
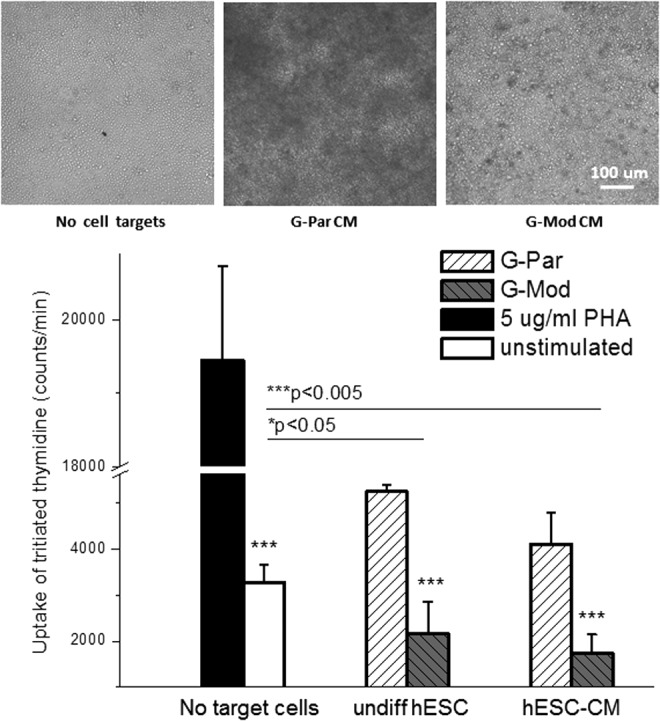
T-lymphocyte activation by parental versus transduced hESCs with diminished B2M expression and their differentiated derivatives. Mixed lymphocyte reaction assay was used to test whether G-Mod cells escaped recognition by allogeneic T lymphocytes. The graph also shows data from positive (phytohemagglutinin [PHA]) and negative (no target cells) controls that were run in parallel with G-Par and G-Mod samples. Images on the top illustrate the appearance of the wells with a large amount of proliferating allogeneic T lymphocytes (the latter are darker in appearance) that were present in G-Par cardiomyocyte samples, but not in the wells with G-Mod cells or negative control.
Differentiation of beta-microglobulin depleted hESCs into hepatic lineages
hESCs were differentiated into a hepatic lineage to further validate that HLA deficiency did not alter differentiation potential. First, transduced cells were differentiated into an endodermal fate through Activin-A treatment. G-Mod cells showed propensity to differentiate into DE, as determined by Foxa2 and Sox17 immunostaining (Fig. 7). The differentiation potential was found to be indistinguishable to that of the parental G-Par cell line. Next, we undertook a head-to-head comparison with hepatic differentiation. The DE cells were induced into hepatocyte progenitors using a combination of insulin, FGF4, HGF, BMP4, BMP2, and dexamethasone in serum-containing media. G-Mod and G-Par cells had comparable hepatic differentiation, as determined by induction of HNF4α expression shown in Figure 8.
FIG. 7.
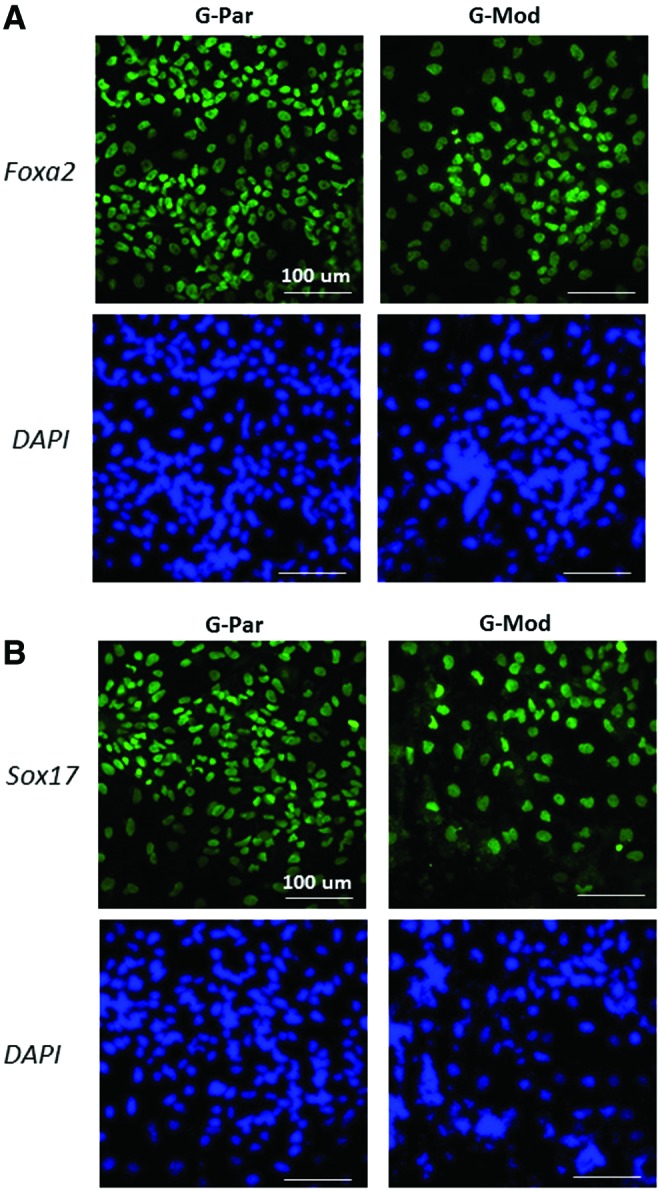
Induction of parental versus transduced hESCs with diminished B2M expression into definitive endoderm. Immunostaining images of differentiated G-Par and G-Mod cells for Foxa2 and Sox17 at 9 days in differentiation media. Nuclei were stained with DAPI (blue). Scale bar: 100 μm. Color images available online at www.liebertpub.com/tea
FIG. 8.
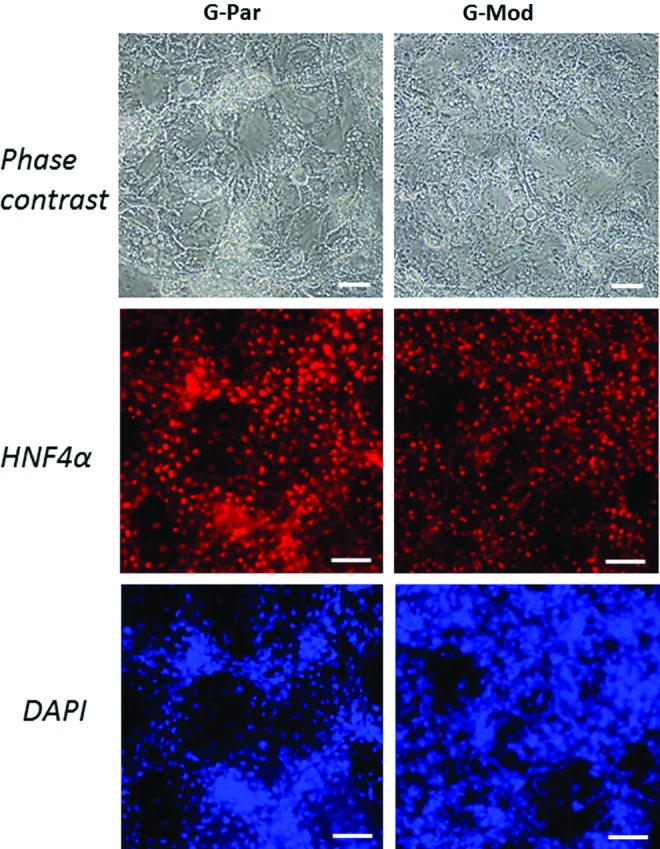
Induction of parental versus transduced hESCs with diminished B2M expression into hepatocytes. Brightfield and immunofluorescent staining images of differentiated G-Par and G-Mod for HNF4α (red) at 21 days. Nuclei were stained with DAPI (blue). Scale bar: 100 μm. Color images available online at www.liebertpub.com/tea
Discussion
The first approach to overcome immunological barriers for implantation of tissue-engineered organs is the creation of banks of human stem lines with different libraries of HLAs. As of January 2015, the HLA alleles nomenclature contained a total of 12542 alleles, of which 9437 are class I alleles (http://hla.alleles.org/nomenclature/stats.html). Yet, even large libraries only partially address this need, with an estimated 150 human embryonic cell lines matching with only 20% of the population in the United Kingdom.47 The estimated number of cell lines that would be required to ensure a match in a more ethnically diverse population, such as in the United States, is likely to be much higher.48 This approach also carries an additional burden of having line-specific maintenance and differentiation protocols for optimal results. Patients treated with these differently maintained stem cell lines will then likely have different therapeutic outcomes, thus complicating conclusions about treatment efficiency.
A second approach is the use of pluripotent stem cells from autologous cell sources.49 Yet, as of today, iPSC-based engineered tissues involve high costs and time commitment to acquire and cultivate initial cell sources, isolate, scale-up, reprogram, and differentiate iPSCs. Moreover, it will be difficult to repair organs using autologous cells that retain inherited or accumulated mutations and/or acquired epigenetic changes.50–54 There are also significant regulatory hurdles to utilizing autologous iPSC-based therapies. In the current system, each new batch of iPSCs would be a separate product requiring a new approval process.55
The third option is to create a universal donor cell with minimal surface expression of HLA class I molecules. The possibility of having a hypoimmunogenic or a universally immune compatible hESC line that can give rise to a specific phenotype is an appealing goal with a wide range of clinical implications.20 Creation of such a universal donor cell line may lead to off-the-shelf tissue-engineered products and the ability to treat patients for whom treatment with autologous cells is problematic due to genetic mutations. Since the ability to present peptide antigens is reduced, creation of such a cell line can also help to repair organs for patients with autoimmune disorders, including type-1 diabetes.56 Finally, tissue-engineered products made from a universal stem cell line will be much easier to standardize and regulate. Particularly, because different hESC lines have been shown to vary considerably in their differentiation potential, even when differentiation protocols are performed within the same laboratory.57
Depletion of B2M as a means to improve donor immune compatibility has been explored in the past at both the cellular and organ level. Specifically, it was shown that tissues from B2M knockout mice exhibited increased survival when transplanted into immunocompetent animals.58–60 This included a significant improvement in the function of renal allografts from MHC class I-deficient mice, compared with allografts from mice with normal MHC class I expression,58 or indefinite survival of pancreatic islet allografts from class I-deficient donors in fully allogeneic recipients.59 Similar effects were seen for cardiac allograft survival with some grafts accepted indefinitely.61 Interestingly, in contrast to the prolonged survival that has been observed for allografts deficient in MHC antigen expression, no significant extension of survival was observed in the case of xenografts, presumably due to a response through the innate immunity pathways.60
Recently, a number of laboratories have shown that downregulation of MHC class I molecules in ESCs can decrease their immunogenicity.15,17,62 Our work confirms these findings and shows, for the first time, that shRNA-based downregulation of MHC-I does not affect hESC ability to differentiate into specific lineages, including hepatic and cardiac. Furthermore, we show that the differentiated cells derived from MHC-I-deficient hESCs continue to be unrecognizable by allogeneic T lymphocytes. While in vivo implantation of these cells or tissue products made from them into allogeneic human being will be an ultimate proof of our hypothesis, such studies will require FDA approval for phase I clinical trials and are beyond the scope of this article. As an indirect in vivo evidence to support our main hypothesis, we can cite in our recently published findings that downregulation of B2M in mouse ESC-derived cardiac myocytes improves the survival in allogeneic mice.21
Our experiments confirmed the presence of classical cardiac and hepatic markers in cells differentiated from G-Mod cells with low B2M expression levels, which indicates that B2M and HLA class I expression is not essential for stem cell differentiation. This is supported by the observations that mice and humans with deficiencies in MHC class I expression, although immunodeficient, exhibit normal development of nonimmune cells and tissues. For example, B2M knockout mice have decreased MHC class I antigen on the cell surface, but these mice are fertile and healthy despite defects in T-cell-mediated immune responses.63 Individuals with a severe reduction in the cell surface expression of HLA class I molecule, due to rare genetic mutations, can suffer from increased susceptibility to infections, but do not display defects in nonimmune cells or tissues.64 The earlier studies indicate that the absence of B2M expression is not critical to normal cell or organ function.
The fact that shRNA transduction did not completely eliminate B2M expression in hESCs may have additional benefits. This is because a complete absence of functional HLA-I may cause natural killer (NK)-mediated cell killing. NK cell alloreactivity arises from the expression of inhibitory killer Ig-like receptors (KIRs) that do not recognize HLA.65 Inhibitory receptors such as KIR dampen NK cell activation upon interaction with their HLA class I ligands, including those of class C, -E, and -G.66,67 It has been previously shown that residual HLA expression in HLA-silenced cells is protective against NK-cell-mediated lysis.62,67 Specifically, it was previously shown that if at least 10% of initial B2M levels remain, the activation of NK is prevented. This can also be attributed to the fact that ESC derivatives consistently do not express NK stimulatory receptors such as NKG2D.68 We also felt that it was important to confirm that the diminished presence of B2M does not lead to any compensatory increases in the expression levels of other immune regulatory molecules (Fig. 4).
Interestingly, the data presented in Figure 6 suggest additional immunomodulatory effects of hESCs and their derivatives. Specifically, proliferation of allogeneic T cells was lower in wells with G-Mod cells, compared to wells with no target cells. Similar immunomodulatory effects were previously reported for mesenchymal stem cells and glial precursor cells.69–72 The exact mechanism of such immunomodulation remains to be established.
Our studies, similar to others, delivered short-hairpin RNAs through a lentiviral-based vector system.16,73–75 In cases when shRNA-based inhibition will be extended to clinical applications, there may be a valid concern regarding the carcinogenicity of introduced lentiviral genes. This problem can be solved by utilizing a self-inactivating lentiviral clone.76 Cells transduced with such clones cannot produce additional viral particles because the cells do not contain the genes for the viral capsid. Furthermore, upon integration into the target cell's genome, the 5′ LTR promoter is inactivated, which prevents replication of the viral sequences. Alternatively, one can use a combination of gene targeting and mitotic recombination to derive HLA-homozygous ESC subclones or use targeted disruption of both alleles of the B2M gene.15
For tissue engineering applications, we envision a general scenario whereby genetically engineered, hypoimmunogenic hESCs represent a universal donor cell, which may be differentiated into a desired tissue type and transplanted with a minimal immune response. Notably, cells differentiated from hESCs, including cardiac myocytes, retain their ability to withstand freeze–defrost cycles and therefore can be produced and stored in large quantities. These cells can then be defrosted upon request and seeded in premade tissue scaffolds, significantly speeding up the time needed to produce tissue-engineered products. In total, the availability of such cells represents an important step toward immunocompatible therapies that are based on cell and tissue-engineered constructs.
Conclusion
We confirmed that transduced hESCs retain a B2M shRNA insert through multiple passages in an undifferentiated state. We also have shown that the B2M(-) cells can form stable undifferentiated colonies and can be successfully differentiated into different phenotypes, such as cardiac muscle and hepatocytes. The differentiated cells maintain diminished levels of HLA-I on their surface and are not recognized by allogeneic T lymphocytes. Constitutive B2M shRNA expression did not cause any measureable changes in the expression levels of CD80, CD81, CD83, CD86, FAS, and FASL mRNAs. We conclude that stable shRNA B2M depletion is a promising approach to create a universal donor ESC that can be used to produce either cell grafts or engineered tissue constructs with improved survival and/or minimal need for coadministration of immunosuppressive drugs.
Supplementary Material
Acknowledgments
We thank Drs. Teresa Hawley, Larisa Dubrovsky, and Irene Riz for help with specific protocols. This work was supported by the American Heart Association (Z.K.), the National Science Foundation (N.S.), and the National Institutes of Health (N.G.P., N.S., M.A.L., G.S., A.R.) awards.
Authors' Contributions
Z.K. and N.S. conceived the study, participated in its experimental implementation, and wrote the manuscript. H.D. carried out shRNA transduction, PCR, and initial cardiac differentiation experiments. G.S., A.H., and A.R. conducted hepatic differentiation protocols. N.M. and I.I. maintained cell cultures in undifferentiated and differentiated states and conducted cardiac differentiation of parental and modified cells. N.G.P. helped with PCR, immunostaining, calcium transient protocols, and with the final editing of the manuscript. D.L. provided advice on FACS analysis, in designing MLR assay, and helped to edit the manuscript. I.T. participated in the design and discussion of MLR assay and qPCR of costimulatory molecules. M.A.L. provided GCaMP-expressing cell line, consulted on cardiac differentiation protocols, and provided valuable suggestions for the final version of the manuscript. All authors read and approved the final manuscript.
Disclosure Statement
The authors declare that they have no competing interests.
References
- 1.Pearl J.I., Kean L.S., Davis M.M., and Wu J.C. Pluripotent stem cells: immune to the immune system? Sci Transl Med 4, 164ps25, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao T., Zhang Z.-N., Rong Z., and Xu Y. Immunogenicity of induced pluripotent stem cells. Nature 474, 212, 2011 [DOI] [PubMed] [Google Scholar]
- 3.De Almeida P.E., Ransohoff J.D., Nahid A., and Wu J.C. Immunogenicity of pluripotent stem cells and their derivatives. Circ Res 112, 549, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batista L.F.Z., Pech M.F., Zhong F.L., et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 474, 399, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatima A., Kaifeng S., Dittmann S., et al. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS One 8, e83005, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwi-Dantsis L., and Gepstein L. Induced pluripotent stem cells for cardiac repair. Cell Mol Life Sci 69, 3285, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Wilson G.F., Soerens A.G., et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 104, e30, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Freire V., Lee A.S., Hu S., et al. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J Am Coll Cardiol 64, 436, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak T., Jett B., and Saunders M. Primer to the Immune Response, 1st edition. ISBN 9780080569659 [Internet] [cited 2015May14]. Available at http://store.elsevier.com/Primer-to-The-Immune-Response/Tak-Mak/isbn-9780080569659/
- 10.Kaufman J. Antigen processing and presentation: evolution from a bird's eye view. Mol Immunol 55, 159, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiina T., Hosomichi K., Inoko H., and Kulski J.K. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 54, 15, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Niethammer D. Side-effects of long-term immunosuppression versus morbidity in autologous stem cell rescue: striking the balance. Rheumatology 38, 747, 1999 [DOI] [PubMed] [Google Scholar]
- 13.Freytes D.O., Santambrogio L., and Vunjak-Novakovic G. Optimizing dynamic interactions between a cardiac patch and inflammatory host cells. Cells Tissues Organs 195, 171, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlando G. Finding the right time for weaning off immunosuppression in solid organ transplant recipients. Expert Rev Clin Immunol 6, 879, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Riolobos L., Hirata R.K., Turtle C.J., et al. HLA engineering of human pluripotent stem cells. Mol Ther 21, 1232, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueiredo C., Wedekind D., Müller T., et al. MHC universal cells survive in an allogeneic environment after incompatible transplantation. Biomed Res Int 2013, 796046, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu P., Chen J., He L., et al. Generating hypoimmunogenic human embryonic stem cells by the disruption of beta 2-microglobulin. Stem Cell Rev 9, 806, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Busch A., Marasco W.A., Doebis C., Volk H.-D., and Seifert M. MHC class I manipulation on cell surfaces by gene transfer of anti-MHC class I intrabodies—a tool for decreased immunogenicity of allogeneic tissue and cell transplants. Methods 34, 240, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Matsunaga Y., Fukuma D., Hirata S., et al. Activation of antigen-specific cytotoxic T lymphocytes by beta 2-microglobulin or TAP1 gene disruption and the introduction of recipient-matched MHC class I gene in allogeneic embryonic stem cell-derived dendritic cells. J Immunol 181, 6635, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Karabekian Z., Posnack N.G., and Sarvazyan N. Immunological barriers to stem-cell based cardiac repair. Stem Cell Rev 7, 315, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karabekian Z., Idress S., Jamshidi A., Posnack N.G., and Sarvazyan N. Downregulation of beta-microglobulin to diminish T-lymphocyte lysis of non-syngeneic cell sources of engineered heart tissue constructs. Biomed Mater 10, 034101, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laflamme M.A., Gold J., Xu C., et al. Formation of human myocardium in the rat heart from human embryonic stem cells. Am J Pathol 167, 663, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiba Y., Fernandes S., Zhu W.-Z., et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489, 332, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu W.-Z., Filice D., Palpant N.J., and Laflamme M.A. Methods for assessing the electromechanical integration of human pluripotent stem cell-derived cardiomyocyte grafts. Methods Mol Biol 1181, 229, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajiwara M., Aoi T., Okita K., et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A 109, 12538, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa T., Kobayashi M., Yanagi S., et al. Human induced hepatic lineage-oriented stem cells: autonomous specification of human iPS cells toward hepatocyte-like cells without any exogenous differentiation factors. PLoS One 10, e0123193, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockemeyer D., Soldner F., Beard C., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27, 851, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu W.-Z., Van Biber B., and Laflamme M.A. Methods for the derivation and use of cardiomyocytes from human pluripotent stem cells. Methods Mol Biol 767, 419, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C., Inokuma M.S., Denham J., et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol 19, 971, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Bakunts K., Gillum N., Karabekian Z., and Sarvazyan N. Formation of cardiac fibers in Matrigel matrix. Biotechniques 44, 341, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann W.-H., and Eschenhagen T. Engineered heart tissue. In: Vunjak-Novakovic G., and Freshney I., eds. Cult. Cells Tissue Eng. Hoboken, NJ: Wiley-Liss, 2006, pp. 259–292. [Google Scholar]
- 32.Zhu W.-Z., Xie Y., Moyes K.W., Gold J.D., Askari B., and Laflamme M.A. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res 107, 776, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romito A., Lonardo E., Roma G., Minchiotti G., Ballabio A., and Cobellis G. Lack of sik1 in mouse embryonic stem cells impairs cardiomyogenesis by down-regulating the cyclin-dependent kinase inhibitor p57kip2. PLoS One 5, e9029, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posnack N.G., Idrees R., Ding H., et al. Exposure to phthalates affects calcium handling and intercellular connectivity of human stem cell-derived cardiomyocytes. PLoS One 10, e0121927, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biktashev V.N., Arutunyan A., and Sarvazyan N.A. Generation and escape of local waves from the boundary of uncoupled cardiac tissue. Biophys J 94, 3726, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillum N., Karabekian Z., Swift L.M., Brown R.P., Kay M.W., and Sarvazyan N. Clinically relevant concentrations of di (2-ethylhexyl) phthalate (DEHP) uncouple cardiac syncytium. Toxicol Appl Pharmacol 236, 25, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viatchenko-Karpinski S., and Györke S. Modulation of the Ca(2+)-induced Ca(2+) release cascade by beta-adrenergic stimulation in rat ventricular myocytes. J Physiol 533(Pt 3), 837, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arutunyan A., Pumir A., Krinsky V., Swift L., and Sarvazyan N. Behavior of ectopic surface: effects of beta-adrenergic stimulation and uncoupling. Am J Physiol Hear Circ Physiol 285, H2531, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Lieu D.K., Siu C.W., Fu J.-D., Tse H.-F., and Li R.A. Facilitated maturation of Ca2+ handling properties of human embryonic stem cell-derived cardiomyocytes by calsequestrin expression. Am J Physiol Cell Physiol 297, C152, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satin J., Itzhaki I., Rapoport S., et al. Calcium handling in human embryonic stem cell-derived cardiomyocytes. Stem Cells 26, 1961, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Endoh M. Force-frequency relationship in intact mammalian ventricular myocardium: physiological and pathophysiological relevance. Eur J Pharmacol 500, 73, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Breloer M., and Fleischer B. CD83 regulates lymphocyte maturation, activation and homeostasis. Trends Immunol 29, 186, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Chen L., and Flies D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 13, 227, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy S., and Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol 5, 136, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Green D.R., and Ferguson T.A. The role of Fas ligand in immune privilege. Nat Rev Mol Cell Biol 2, 917, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Li X., and Faustman D. Use of donor beta 2-microglobulin-deficient transgenic mouse liver cells for isografts, allografts, and xenografts. Transplantation 55, 940, 1993 [DOI] [PubMed] [Google Scholar]
- 47.Taylor C.J., Bolton E.M., Pocock S., Sharples L.D., Pedersen R.A., and Bradley J.A. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet 366, 2019, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Solomon S., Pitossi F., and Rao M.S. Banking on iPSC—is it doable and is it worthwhile. Stem Cell Rev 11, 1, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi K., Tanabe K., Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Mayshar Y., Ben-David U., Lavon N., et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7, 521, 2010 [DOI] [PubMed] [Google Scholar]
- 51.Gore A., Li Z., Fung H.-L., et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lister R., Pelizzola M., Kida Y.S., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pera M.F. Stem cells: the dark side of induced pluripotency. Nature 471, 46, 2011 [DOI] [PubMed] [Google Scholar]
- 54.Hussein S.M., Batada N.N., Vuoristo S., et al. Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58, 2011 [DOI] [PubMed] [Google Scholar]
- 55.Carpenter M.K., and Rao M.S. Concise review: making and using clinically compliant pluripotent stem cell lines. Stem Cells Transl Med 4, 381, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prange S., Zucker P., Jevnikar A.M., and Singh B. Transplanted MHC class I-deficient nonobese diabetic mouse islets are protected from autoimmune injury in diabetic nonobese recipients. Transplantation 71, 982, 2001 [DOI] [PubMed] [Google Scholar]
- 57.Pekkanen-Mattila M., Kerkelä E., Tanskanen J.M.A., et al. Substantial variation in the cardiac differentiation of human embryonic stem cell lines derived and propagated under the same conditions—a comparison of multiple cell lines. Ann Med 41, 360, 2009 [DOI] [PubMed] [Google Scholar]
- 58.Coffman T., Geier S., Ibrahim S., et al. Improved renal function in mouse kidney allografts lacking MHC class I antigens. J Immunol 151, 425, 1993 [PubMed] [Google Scholar]
- 59.Markmann J.F., Bassiri H., Desai N.M., et al. Indefinite survival of MHC class I-deficient murine pancreatic islet allografts. Transplantation 54, 1085, 1992 [DOI] [PubMed] [Google Scholar]
- 60.Markmann J.F., Campos L., Bhandoola A., et al. Genetically engineered grafts to study xenoimmunity: a role for indirect antigen presentation in the destruction of major histocompatibility complex antigen deficient xenografts. Surgery 116, 242, 1994 [PubMed] [Google Scholar]
- 61.Campos L., Naji A., Deli B.C., et al. Survival of MHC-deficient mouse heterotopic cardiac allografts. Transplantation 59, 187, 1995 [PubMed] [Google Scholar]
- 62.Figueiredo C., Seltsam A., and Blasczyk R. Class-, gene-, and group-specific HLA silencing by lentiviral shRNA delivery. J Mol Med (Berl) 84, 425, 2006 [DOI] [PubMed] [Google Scholar]
- 63.Zijlstra M., Bix M., Simister N.E., Loring J.M., Raulet D.H., and Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature 344, 742, 1990 [DOI] [PubMed] [Google Scholar]
- 64.Zimmer J., Andrès E., Donato L., Hanau D., Hentges F., and de la Salle H. Clinical and immunological aspects of HLA class I deficiency. QJM 98, 719, 2005 [DOI] [PubMed] [Google Scholar]
- 65.Thielens A., Vivier E., and Romagné F. NK cell MHC class I specific receptors (KIR): from biology to clinical intervention. Curr Opin Immunol 24, 239, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Anfossi N., André P., Guia S., et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Borrego F., Ulbrecht M., Weiss E.H., Coligan J.E., and Brooks A.G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med 187, 813, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frenzel L.P., Abdullah Z., Kriegeskorte A.K., et al. Role of natural-killer group 2 member D ligands and intercellular adhesion molecule 1 in natural killer cell-mediated lysis of murine embryonic stem cells and embryonic stem cell-derived cardiomyocytes. Stem Cells 27, 307, 2009 [DOI] [PubMed] [Google Scholar]
- 69.Liu J., Götherström C., Forsberg M., et al. Human neural stem/progenitor cells derived from embryonic stem cells and fetal nervous system present differences in immunogenicity and immunomodulatory potentials in vitro. Stem Cell Res 10, 325, 2013 [DOI] [PubMed] [Google Scholar]
- 70.Kimbrel E.A., Kouris N.A., Yavanian G.J., et al. Mesenchymal stem cell population derived from human pluripotent stem cells displays potent immunomodulatory and therapeutic properties. Stem Cells Dev 23, 1611, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gebler A., Zabel O., and Seliger B. The immunomodulatory capacity of mesenchymal stem cells. Trends Mol Med 18, 128, 2012 [DOI] [PubMed] [Google Scholar]
- 72.Kim H., Walczak P., Muja N., Campanelli J.T., and Bulte J.W.M. ICV-transplanted human glial precursor cells are short-lived yet exert immunomodulatory effects in mice with EAE. Glia 60, 1117, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Figueiredo C., Horn P.A., Blasczyk R., and Seltsam A. Regulating MHC expression for cellular therapeutics. Transfusion 47, 18, 2007 [DOI] [PubMed] [Google Scholar]
- 74.Figueiredo C., Goudeva L., Horn P.A., Eiz-Vesper B., Blasczyk R., and Seltsam A. Generation of HLA-deficient platelets from hematopoietic progenitor cells. Transfusion 50, 1690, 2010 [DOI] [PubMed] [Google Scholar]
- 75.Haga K., Lemp N.A., Logg C.R., et al. Permanent, lowered HLA class I expression using lentivirus vectors with shRNA constructs: averting cytotoxicity by alloreactive T lymphocytes. Transplant Proc 38, 3184, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zufferey R., Dull T., Mandel R.J., et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72, 9873, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.